

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
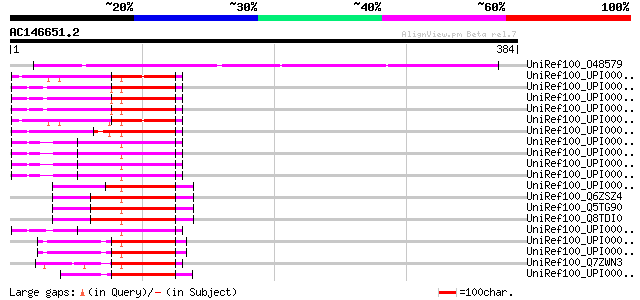
Query= AC146651.2 - phase: 0
(384 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O48579 Mi-2 autoantigen-like protein [Arabidopsis thal... 191 3e-47
UniRef100_UPI000036393E UPI000036393E UniRef100 entry 92 3e-17
UniRef100_UPI000024A9C6 UPI000024A9C6 UniRef100 entry 87 8e-16
UniRef100_UPI0000249F39 UPI0000249F39 UniRef100 entry 87 8e-16
UniRef100_UPI000024756B UPI000024756B UniRef100 entry 87 8e-16
UniRef100_UPI000036393F UPI000036393F UniRef100 entry 86 1e-15
UniRef100_UPI00001D044A UPI00001D044A UniRef100 entry 84 5e-15
UniRef100_UPI0000436C5C UPI0000436C5C UniRef100 entry 83 1e-14
UniRef100_UPI0000436C5A UPI0000436C5A UniRef100 entry 83 1e-14
UniRef100_UPI0000436C59 UPI0000436C59 UniRef100 entry 83 1e-14
UniRef100_UPI0000436C58 UPI0000436C58 UniRef100 entry 83 1e-14
UniRef100_UPI0000367DE6 UPI0000367DE6 UniRef100 entry 82 2e-14
UniRef100_Q6ZSZ4 Hypothetical protein FLJ45103 [Homo sapiens] 82 2e-14
UniRef100_Q5TG90 Chromodomain helicase DNA binding protein 5 [Ho... 82 2e-14
UniRef100_Q8TDI0 Chromodomain helicase-DNA-binding protein 5 [Ho... 82 2e-14
UniRef100_UPI0000436C5D UPI0000436C5D UniRef100 entry 82 3e-14
UniRef100_UPI0000428AD3 UPI0000428AD3 UniRef100 entry 81 4e-14
UniRef100_UPI00001CF8B3 UPI00001CF8B3 UniRef100 entry 81 6e-14
UniRef100_Q7ZWN3 B230399n07-prov protein [Xenopus laevis] 80 1e-13
UniRef100_UPI0000337430 UPI0000337430 UniRef100 entry 80 1e-13
>UniRef100_O48579 Mi-2 autoantigen-like protein [Arabidopsis thaliana]
Length = 2228
Score = 191 bits (485), Expect = 3e-47
Identities = 128/354 (36%), Positives = 189/354 (53%), Gaps = 9/354 (2%)
Query: 19 LKRKRRKLPIGPDQSSGKEQSNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHD 78
+K+KRRKLP D K S+ D+ + SS+ + K+ LKT+ + SSK+KG+D
Sbjct: 1 MKQKRRKLPSILDILDQKVDSSMAFDSPEYTSSSKPS--KQRLKTDSTPERNSSKRKGND 58
Query: 79 GYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFEENDQLKPLNNL 138
G ++ECVICDLGG+LLCCDSCPRTYH CL+PPLKRIP GKW CP C ++ LKP+N L
Sbjct: 59 GNYFECVICDLGGDLLCCDSCPRTYHTACLNPPLKRIPNGKWICPKCSPNSEALKPVNRL 118
Query: 139 DSISRRARTKTVPVKSKAGVNPVNLEKVSGIFGNKHISKKRST-KAKSISTMGGKFFGMK 197
D+I++RARTKT SKAG E+ S I+ + IS ++S+ K KSIS K G +
Sbjct: 119 DAIAKRARTKTKKRNSKAG---PKCERASQIYCSSIISGEQSSEKGKSISAEESKSTGKE 175
Query: 198 PVLSPVDATCSDKPMDPSLESCMEGTSCADADEKNLNLSPTV-APMDRMSVSPDKEVLSP 256
SP+D CS + + +S D+ + PT P D E LS
Sbjct: 176 VYSSPMDG-CSTAELGHASADDRPDSSSHGEDDLGKPVIPTADLPSDAGLTLLSCEDLSE 234
Query: 257 SKITNLDANDDLLEEKPDLSCDKIPFRKTLVLAITVGGEEMGKRKHKVIGDNANQKKRRT 316
SK+++ + + EK + + +I KT V + G + KRK ++ + ++ +
Sbjct: 235 SKLSDTEKTHEAPVEKLEHASSEIVENKT-VAEMETGKGKRKKRKRELNDGESLERCKTD 293
Query: 317 EKGKKVVITPIKSKSGNNKVQTKQKSKTHKISISASKGDVGKKKSDAQQKDKVL 370
+K K ++ + S S K K K K ++ + K++ +K K L
Sbjct: 294 KKRAKKSLSKVGSSSQTTKSPESSKKKKKKNRVTLKSLSKPQSKTETPEKVKKL 347
>UniRef100_UPI000036393E UPI000036393E UniRef100 entry
Length = 1076
Score = 91.7 bits (226), Expect = 3e-17
Identities = 54/147 (36%), Positives = 75/147 (50%), Gaps = 19/147 (12%)
Query: 2 KKESKSSAPTMLNKNWVLKRKRRKLPI----GPDQSSGK---------EQSNGKEDNSVA 48
KK SKS P + +K + ++ +PI P + K E + +ED+SV
Sbjct: 182 KKRSKS--PRVSDKKKAAAKIKKMVPIRIKLSPISAKRKKSCSSDDLDEDESEQEDSSVH 239
Query: 49 SESSRSASAKRMLKTEEGTAQFSSKKKGHDGYFYE----CVICDLGGNLLCCDSCPRTYH 104
S S RS S+ R+ K + G KK DGY + C +C GG ++ CD+CPR YH
Sbjct: 240 SSSVRSDSSGRVKKNKRGRPAKKKKKSKGDGYETDHQDYCEVCQQGGEIILCDTCPRAYH 299
Query: 105 LQCLDPPLKRIPMGKWQCPSCFEENDQ 131
L CL+P L + P GKW CP C +E Q
Sbjct: 300 LVCLEPELDKAPEGKWSCPHCEKEGIQ 326
Score = 63.9 bits (154), Expect = 7e-09
Identities = 25/48 (52%), Positives = 32/48 (66%), Gaps = 1/48 (2%)
Query: 78 DGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
D + C +C GG LLCCD+C +YH+ CL+PPL IP G+W CP C
Sbjct: 362 DDHMEFCRVCKDGGELLCCDTCT-SYHIHCLNPPLPEIPNGEWLCPRC 408
>UniRef100_UPI000024A9C6 UPI000024A9C6 UniRef100 entry
Length = 1837
Score = 87.0 bits (214), Expect = 8e-16
Identities = 51/138 (36%), Positives = 70/138 (49%), Gaps = 12/138 (8%)
Query: 2 KKESKSSAPTMLNKNWVLKRKRRKLPIGPDQSSGKEQSNGKEDNSVASESSRSASAKRML 61
K + K AP + K + KR+K E + +E++SV S S RS S+ R+
Sbjct: 191 KPKPKKMAPLRI-KLGAIGNKRKK---SSSSEEVDEDDSEQENSSVHSSSVRSDSSGRVK 246
Query: 62 KTEEGTAQFSSKKK--GHDGYFYE------CVICDLGGNLLCCDSCPRTYHLQCLDPPLK 113
K + G KK DG YE C +C GG ++ CD+CPR YHL CL+P L+
Sbjct: 247 KNKRGRPAKKKKKALGDEDGDGYETDHQDYCEVCQQGGEIILCDTCPRAYHLVCLEPELE 306
Query: 114 RIPMGKWQCPSCFEENDQ 131
+ P GKW CP C +E Q
Sbjct: 307 KAPEGKWSCPHCEKEGIQ 324
Score = 72.4 bits (176), Expect = 2e-11
Identities = 27/48 (56%), Positives = 33/48 (68%)
Query: 78 DGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
D + C +C GG LLCCDSCP +YH+ CL+PPL IP G+W CP C
Sbjct: 356 DDHMEFCRVCKDGGELLCCDSCPSSYHIHCLNPPLPEIPNGEWLCPRC 403
>UniRef100_UPI0000249F39 UPI0000249F39 UniRef100 entry
Length = 1754
Score = 87.0 bits (214), Expect = 8e-16
Identities = 51/138 (36%), Positives = 70/138 (49%), Gaps = 12/138 (8%)
Query: 2 KKESKSSAPTMLNKNWVLKRKRRKLPIGPDQSSGKEQSNGKEDNSVASESSRSASAKRML 61
K + K AP + K + KR+K E + +E++SV S S RS S+ R+
Sbjct: 184 KPKPKKMAPLRI-KLGAIGNKRKK---SSSSEEVDEDDSEQENSSVHSSSVRSDSSGRVK 239
Query: 62 KTEEGTAQFSSKKK--GHDGYFYE------CVICDLGGNLLCCDSCPRTYHLQCLDPPLK 113
K + G KK DG YE C +C GG ++ CD+CPR YHL CL+P L+
Sbjct: 240 KNKRGRPAKKKKKALGDEDGDGYETDHQDYCEVCQQGGEIILCDTCPRAYHLVCLEPELE 299
Query: 114 RIPMGKWQCPSCFEENDQ 131
+ P GKW CP C +E Q
Sbjct: 300 KAPEGKWSCPHCEKEGIQ 317
Score = 72.4 bits (176), Expect = 2e-11
Identities = 27/48 (56%), Positives = 33/48 (68%)
Query: 78 DGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
D + C +C GG LLCCDSCP +YH+ CL+PPL IP G+W CP C
Sbjct: 349 DDHMEFCRVCKDGGELLCCDSCPSSYHIHCLNPPLPEIPNGEWLCPRC 396
>UniRef100_UPI000024756B UPI000024756B UniRef100 entry
Length = 1904
Score = 87.0 bits (214), Expect = 8e-16
Identities = 51/138 (36%), Positives = 70/138 (49%), Gaps = 12/138 (8%)
Query: 2 KKESKSSAPTMLNKNWVLKRKRRKLPIGPDQSSGKEQSNGKEDNSVASESSRSASAKRML 61
K + K AP + K + KR+K E + +E++SV S S RS S+ R+
Sbjct: 198 KPKPKKMAPLRI-KLGAIGNKRKK---SSSSEEVDEDDSEQENSSVHSSSVRSDSSGRVK 253
Query: 62 KTEEGTAQFSSKKK--GHDGYFYE------CVICDLGGNLLCCDSCPRTYHLQCLDPPLK 113
K + G KK DG YE C +C GG ++ CD+CPR YHL CL+P L+
Sbjct: 254 KNKRGRPAKKKKKALGDEDGDGYETDHQDYCEVCQQGGEIILCDTCPRAYHLVCLEPELE 313
Query: 114 RIPMGKWQCPSCFEENDQ 131
+ P GKW CP C +E Q
Sbjct: 314 KAPEGKWSCPHCEKEGIQ 331
Score = 72.4 bits (176), Expect = 2e-11
Identities = 27/48 (56%), Positives = 33/48 (68%)
Query: 78 DGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
D + C +C GG LLCCDSCP +YH+ CL+PPL IP G+W CP C
Sbjct: 363 DDHMEFCRVCKDGGELLCCDSCPSSYHIHCLNPPLPEIPNGEWLCPRC 410
>UniRef100_UPI000036393F UPI000036393F UniRef100 entry
Length = 1052
Score = 86.3 bits (212), Expect = 1e-15
Identities = 54/151 (35%), Positives = 76/151 (49%), Gaps = 23/151 (15%)
Query: 2 KKESKSSAPTMLNKNWVLKRKRRKLPI----GPDQSSGK---------EQSNGKEDNSVA 48
KK SKS P + +K + ++ +PI P + K E + +ED+SV
Sbjct: 16 KKRSKS--PRVSDKKKAAAKIKKMVPIRIKLSPISAKRKKSCSSDDLDEDESEQEDSSVH 73
Query: 49 SESSRSASAKRMLKTEEGTAQFSSKK----KGHDGYFYE----CVICDLGGNLLCCDSCP 100
S S RS S+ R+ K + G KK + DGY + C +C GG ++ CD+CP
Sbjct: 74 SSSVRSDSSGRVKKNKRGRPAKKKKKIPGDEEGDGYETDHQDYCEVCQQGGEIILCDTCP 133
Query: 101 RTYHLQCLDPPLKRIPMGKWQCPSCFEENDQ 131
R YHL CL+P L + P GKW CP C +E Q
Sbjct: 134 RAYHLVCLEPELDKAPEGKWSCPHCEKEGIQ 164
Score = 63.9 bits (154), Expect = 7e-09
Identities = 25/48 (52%), Positives = 32/48 (66%), Gaps = 1/48 (2%)
Query: 78 DGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
D + C +C GG LLCCD+C +YH+ CL+PPL IP G+W CP C
Sbjct: 200 DDHMEFCRVCKDGGELLCCDTCT-SYHIHCLNPPLPEIPNGEWLCPRC 246
>UniRef100_UPI00001D044A UPI00001D044A UniRef100 entry
Length = 2007
Score = 84.3 bits (207), Expect = 5e-15
Identities = 50/138 (36%), Positives = 66/138 (47%), Gaps = 9/138 (6%)
Query: 2 KKESKSSAPTMLNKNWVLKRKRRKLPIGPDQSSGKEQSNGKEDNSVASESSRSASAKRML 61
K K AP + K +L +RRK G + + + + + SV S S R R
Sbjct: 298 KLRGKKMAPLKI-KLGLLGGRRRKAGSGDEGPEPEAEESDLDSGSVHSASGRPDGPVRAK 356
Query: 62 KTEEGTAQFSSKK----KGHDGYFYE----CVICDLGGNLLCCDSCPRTYHLQCLDPPLK 113
K + G KK + DGY + C +C GG ++ CD+CPR YHL CLDP L
Sbjct: 357 KLKRGRPGRKKKKVTGEEEVDGYETDHQDYCEVCQQGGEIILCDTCPRAYHLVCLDPELD 416
Query: 114 RIPMGKWQCPSCFEENDQ 131
R P GKW CP C +E Q
Sbjct: 417 RAPEGKWSCPHCEKEGVQ 434
Score = 69.3 bits (168), Expect = 2e-10
Identities = 29/62 (46%), Positives = 38/62 (60%), Gaps = 3/62 (4%)
Query: 64 EEGTAQFSSKKKGHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCP 123
EEG +K+ D + C +C GG LLCCD+C +YH+ CL+PPL IP G+W CP
Sbjct: 446 EEGE---EGEKEEEDDHMEYCRVCKDGGELLCCDACISSYHIHCLNPPLPDIPNGEWLCP 502
Query: 124 SC 125
C
Sbjct: 503 RC 504
>UniRef100_UPI0000436C5C UPI0000436C5C UniRef100 entry
Length = 1908
Score = 83.2 bits (204), Expect = 1e-14
Identities = 48/136 (35%), Positives = 65/136 (47%), Gaps = 16/136 (11%)
Query: 2 KKESKSSAPTMLNKNWVLKRKRRKLPIGPDQSSGKEQSNGKEDNSVASESSRSASAKRML 61
K ++K AP + K KRKR SS E+ +G+ D S S S S
Sbjct: 284 KSKAKKVAPLKI-KLGNFKRKR---------SSSGEEDDGESDFDSFSVSDGSNSRSSRS 333
Query: 62 KTEEGTAQFSSKKKGHDGYFYE------CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRI 115
K ++ + KK D YE C +C GG ++ CD+CPR YH+ CLDP ++R
Sbjct: 334 KNKKAKSSKKKKKMDEDADGYETDHQDYCEVCQQGGEIILCDTCPRAYHMVCLDPDMERA 393
Query: 116 PMGKWQCPSCFEENDQ 131
P G W CP C +E Q
Sbjct: 394 PEGTWSCPHCEKEGIQ 409
Score = 73.2 bits (178), Expect = 1e-11
Identities = 30/74 (40%), Positives = 44/74 (58%)
Query: 52 SRSASAKRMLKTEEGTAQFSSKKKGHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPP 111
+R S++ + ++G ++ D + C +C GG LLCCD+CP +YHL CL+PP
Sbjct: 412 AREESSEGEEENDDGRRDDGDVEEEDDHHMEFCRVCKDGGELLCCDTCPSSYHLHCLNPP 471
Query: 112 LKRIPMGKWQCPSC 125
L IP G+W CP C
Sbjct: 472 LPDIPNGEWICPRC 485
>UniRef100_UPI0000436C5A UPI0000436C5A UniRef100 entry
Length = 1865
Score = 83.2 bits (204), Expect = 1e-14
Identities = 48/136 (35%), Positives = 65/136 (47%), Gaps = 16/136 (11%)
Query: 2 KKESKSSAPTMLNKNWVLKRKRRKLPIGPDQSSGKEQSNGKEDNSVASESSRSASAKRML 61
K ++K AP + K KRKR SS E+ +G+ D S S S S
Sbjct: 271 KSKAKKVAPLKI-KLGNFKRKR---------SSSGEEDDGESDFDSFSVSDGSNSRSSRS 320
Query: 62 KTEEGTAQFSSKKKGHDGYFYE------CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRI 115
K ++ + KK D YE C +C GG ++ CD+CPR YH+ CLDP ++R
Sbjct: 321 KNKKAKSSKKKKKMDEDADGYETDHQDYCEVCQQGGEIILCDTCPRAYHMVCLDPDMERA 380
Query: 116 PMGKWQCPSCFEENDQ 131
P G W CP C +E Q
Sbjct: 381 PEGTWSCPHCEKEGIQ 396
Score = 73.2 bits (178), Expect = 1e-11
Identities = 30/74 (40%), Positives = 44/74 (58%)
Query: 52 SRSASAKRMLKTEEGTAQFSSKKKGHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPP 111
+R S++ + ++G ++ D + C +C GG LLCCD+CP +YHL CL+PP
Sbjct: 399 AREESSEGEEENDDGRRDDGDVEEEDDHHMEFCRVCKDGGELLCCDTCPSSYHLHCLNPP 458
Query: 112 LKRIPMGKWQCPSC 125
L IP G+W CP C
Sbjct: 459 LPDIPNGEWICPRC 472
>UniRef100_UPI0000436C59 UPI0000436C59 UniRef100 entry
Length = 1909
Score = 83.2 bits (204), Expect = 1e-14
Identities = 48/136 (35%), Positives = 65/136 (47%), Gaps = 16/136 (11%)
Query: 2 KKESKSSAPTMLNKNWVLKRKRRKLPIGPDQSSGKEQSNGKEDNSVASESSRSASAKRML 61
K ++K AP + K KRKR SS E+ +G+ D S S S S
Sbjct: 300 KSKAKKVAPLKI-KLGNFKRKR---------SSSGEEDDGESDFDSFSVSDGSNSRSSRS 349
Query: 62 KTEEGTAQFSSKKKGHDGYFYE------CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRI 115
K ++ + KK D YE C +C GG ++ CD+CPR YH+ CLDP ++R
Sbjct: 350 KNKKAKSSKKKKKMDEDADGYETDHQDYCEVCQQGGEIILCDTCPRAYHMVCLDPDMERA 409
Query: 116 PMGKWQCPSCFEENDQ 131
P G W CP C +E Q
Sbjct: 410 PEGTWSCPHCEKEGIQ 425
Score = 73.2 bits (178), Expect = 1e-11
Identities = 30/74 (40%), Positives = 44/74 (58%)
Query: 52 SRSASAKRMLKTEEGTAQFSSKKKGHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPP 111
+R S++ + ++G ++ D + C +C GG LLCCD+CP +YHL CL+PP
Sbjct: 428 AREESSEGEEENDDGRRDDGDVEEEDDHHMEFCRVCKDGGELLCCDTCPSSYHLHCLNPP 487
Query: 112 LKRIPMGKWQCPSC 125
L IP G+W CP C
Sbjct: 488 LPDIPNGEWICPRC 501
>UniRef100_UPI0000436C58 UPI0000436C58 UniRef100 entry
Length = 1855
Score = 83.2 bits (204), Expect = 1e-14
Identities = 48/136 (35%), Positives = 65/136 (47%), Gaps = 16/136 (11%)
Query: 2 KKESKSSAPTMLNKNWVLKRKRRKLPIGPDQSSGKEQSNGKEDNSVASESSRSASAKRML 61
K ++K AP + K KRKR SS E+ +G+ D S S S S
Sbjct: 211 KSKAKKVAPLKI-KLGNFKRKR---------SSSGEEDDGESDFDSFSVSDGSNSRSSRS 260
Query: 62 KTEEGTAQFSSKKKGHDGYFYE------CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRI 115
K ++ + KK D YE C +C GG ++ CD+CPR YH+ CLDP ++R
Sbjct: 261 KNKKAKSSKKKKKMDEDADGYETDHQDYCEVCQQGGEIILCDTCPRAYHMVCLDPDMERA 320
Query: 116 PMGKWQCPSCFEENDQ 131
P G W CP C +E Q
Sbjct: 321 PEGTWSCPHCEKEGIQ 336
Score = 73.2 bits (178), Expect = 1e-11
Identities = 30/74 (40%), Positives = 44/74 (58%)
Query: 52 SRSASAKRMLKTEEGTAQFSSKKKGHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPP 111
+R S++ + ++G ++ D + C +C GG LLCCD+CP +YHL CL+PP
Sbjct: 339 AREESSEGEEENDDGRRDDGDVEEEDDHHMEFCRVCKDGGELLCCDTCPSSYHLHCLNPP 398
Query: 112 LKRIPMGKWQCPSC 125
L IP G+W CP C
Sbjct: 399 LPDIPNGEWICPRC 412
>UniRef100_UPI0000367DE6 UPI0000367DE6 UniRef100 entry
Length = 1952
Score = 82.4 bits (202), Expect = 2e-14
Identities = 42/113 (37%), Positives = 60/113 (52%), Gaps = 6/113 (5%)
Query: 33 SSGKEQSNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHDGYFYE------CVI 86
SS +E + D AS S S ++ + + + KK+ DG YE C +
Sbjct: 289 SSSEEDEREESDFDSASIHSASVRSECSAALGKKSKRRRKKKRIDDGDGYETDHQDYCEV 348
Query: 87 CDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFEENDQLKPLNNLD 139
C GG ++ CD+CPR YHL CLDP L++ P GKW CP C +E Q +P ++ D
Sbjct: 349 CQQGGEIILCDTCPRAYHLVCLDPELEKAPEGKWSCPHCEKEGIQWEPKDDDD 401
Score = 72.4 bits (176), Expect = 2e-11
Identities = 27/53 (50%), Positives = 36/53 (66%)
Query: 73 KKKGHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
+++ D + C +C GG LLCCD+CP +YHL CL+PPL IP G+W CP C
Sbjct: 409 EEEEEDDHMEFCRVCKDGGELLCCDACPSSYHLHCLNPPLPEIPNGEWLCPRC 461
>UniRef100_Q6ZSZ4 Hypothetical protein FLJ45103 [Homo sapiens]
Length = 1225
Score = 82.4 bits (202), Expect = 2e-14
Identities = 42/113 (37%), Positives = 60/113 (52%), Gaps = 6/113 (5%)
Query: 33 SSGKEQSNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHDGYFYE------CVI 86
SS +E + D AS S S ++ + + + KK+ DG YE C +
Sbjct: 289 SSSEEDEREESDFDSASIHSASVRSECSAALGKKSKRRRKKKRIDDGDGYETDHQDYCEV 348
Query: 87 CDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFEENDQLKPLNNLD 139
C GG ++ CD+CPR YHL CLDP L++ P GKW CP C +E Q +P ++ D
Sbjct: 349 CQQGGEIILCDTCPRAYHLVCLDPELEKAPEGKWSCPHCEKEGIQWEPKDDDD 401
Score = 72.4 bits (176), Expect = 2e-11
Identities = 28/64 (43%), Positives = 39/64 (60%)
Query: 62 KTEEGTAQFSSKKKGHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQ 121
K ++ + ++ D + C +C GG LLCCD+CP +YHL CL+PPL IP G+W
Sbjct: 397 KDDDDEEEEGGCEEEEDDHMEFCRVCKDGGELLCCDACPSSYHLHCLNPPLPEIPNGEWL 456
Query: 122 CPSC 125
CP C
Sbjct: 457 CPRC 460
>UniRef100_Q5TG90 Chromodomain helicase DNA binding protein 5 [Homo sapiens]
Length = 1225
Score = 82.4 bits (202), Expect = 2e-14
Identities = 42/113 (37%), Positives = 60/113 (52%), Gaps = 6/113 (5%)
Query: 33 SSGKEQSNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHDGYFYE------CVI 86
SS +E + D AS S S ++ + + + KK+ DG YE C +
Sbjct: 289 SSSEEDEREESDFDSASIHSASVRSECSAALGKKSKRRRKKKRIDDGDGYETDHQDYCEV 348
Query: 87 CDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFEENDQLKPLNNLD 139
C GG ++ CD+CPR YHL CLDP L++ P GKW CP C +E Q +P ++ D
Sbjct: 349 CQQGGEIILCDTCPRAYHLVCLDPELEKAPEGKWSCPHCEKEGIQWEPKDDDD 401
Score = 72.4 bits (176), Expect = 2e-11
Identities = 28/64 (43%), Positives = 39/64 (60%)
Query: 62 KTEEGTAQFSSKKKGHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQ 121
K ++ + ++ D + C +C GG LLCCD+CP +YHL CL+PPL IP G+W
Sbjct: 397 KDDDDEEEEGGCEEEEDDHMEFCRVCKDGGELLCCDACPSSYHLHCLNPPLPEIPNGEWL 456
Query: 122 CPSC 125
CP C
Sbjct: 457 CPRC 460
>UniRef100_Q8TDI0 Chromodomain helicase-DNA-binding protein 5 [Homo sapiens]
Length = 1954
Score = 82.4 bits (202), Expect = 2e-14
Identities = 42/113 (37%), Positives = 60/113 (52%), Gaps = 6/113 (5%)
Query: 33 SSGKEQSNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHDGYFYE------CVI 86
SS +E + D AS S S ++ + + + KK+ DG YE C +
Sbjct: 289 SSSEEDEREESDFDSASIHSASVRSECSAALGKKSKRRRKKKRIDDGDGYETDHQDYCEV 348
Query: 87 CDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFEENDQLKPLNNLD 139
C GG ++ CD+CPR YHL CLDP L++ P GKW CP C +E Q +P ++ D
Sbjct: 349 CQQGGEIILCDTCPRAYHLVCLDPELEKAPEGKWSCPHCEKEGIQWEPKDDDD 401
Score = 72.4 bits (176), Expect = 2e-11
Identities = 28/64 (43%), Positives = 39/64 (60%)
Query: 62 KTEEGTAQFSSKKKGHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQ 121
K ++ + ++ D + C +C GG LLCCD+CP +YHL CL+PPL IP G+W
Sbjct: 397 KDDDDEEEEGGCEEEEDDHMEFCRVCKDGGELLCCDACPSSYHLHCLNPPLPEIPNGEWL 456
Query: 122 CPSC 125
CP C
Sbjct: 457 CPRC 460
>UniRef100_UPI0000436C5D UPI0000436C5D UniRef100 entry
Length = 1532
Score = 81.6 bits (200), Expect = 3e-14
Identities = 48/136 (35%), Positives = 66/136 (48%), Gaps = 11/136 (8%)
Query: 2 KKESKSSAPTMLNKNWVLKRKRRKLPIGPDQSSGKEQSNGKEDNSVASESSRSASAKRML 61
K ++K AP + K KRKR + I S E+ +G+ D S S S S
Sbjct: 161 KSKAKKVAPLKI-KLGNFKRKRSSVCI----FSLGEEDDGESDFDSFSVSDGSNSRSSRS 215
Query: 62 KTEEGTAQFSSKKKGHDGYFYE------CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRI 115
K ++ + KK D YE C +C GG ++ CD+CPR YH+ CLDP ++R
Sbjct: 216 KNKKAKSSKKKKKMDEDADGYETDHQDYCEVCQQGGEIILCDTCPRAYHMVCLDPDMERA 275
Query: 116 PMGKWQCPSCFEENDQ 131
P G W CP C +E Q
Sbjct: 276 PEGTWSCPHCEKEGIQ 291
Score = 73.2 bits (178), Expect = 1e-11
Identities = 30/74 (40%), Positives = 44/74 (58%)
Query: 52 SRSASAKRMLKTEEGTAQFSSKKKGHDGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPP 111
+R S++ + ++G ++ D + C +C GG LLCCD+CP +YHL CL+PP
Sbjct: 294 AREESSEGEEENDDGRRDDGDVEEEDDHHMEFCRVCKDGGELLCCDTCPSSYHLHCLNPP 353
Query: 112 LKRIPMGKWQCPSC 125
L IP G+W CP C
Sbjct: 354 LPDIPNGEWICPRC 367
>UniRef100_UPI0000428AD3 UPI0000428AD3 UniRef100 entry
Length = 2095
Score = 81.3 bits (199), Expect = 4e-14
Identities = 43/119 (36%), Positives = 63/119 (52%), Gaps = 12/119 (10%)
Query: 22 KRRKLPIGPDQSSGKEQSNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHDGYF 81
KR+K G + + + ++ S+ S S RS + + K + + KK+ DG
Sbjct: 269 KRKK---GSSSEEDEREDSDLDNASIHSSSVRSECSAALGKKNKRRRK---KKRIDDGDG 322
Query: 82 YE------CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFEENDQLKP 134
YE C +C GG ++ CD+CPR YHL CLDP L++ P GKW CP C +E Q +P
Sbjct: 323 YETDHQDYCEVCQQGGEIILCDTCPRAYHLVCLDPELEKAPEGKWSCPHCEKEGIQWEP 381
Score = 72.0 bits (175), Expect = 3e-11
Identities = 27/48 (56%), Positives = 33/48 (68%)
Query: 78 DGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
D + C +C GG LLCCD+CP +YHL CL+PPL IP G+W CP C
Sbjct: 398 DDHMEFCRVCKDGGELLCCDACPSSYHLHCLNPPLPEIPNGEWLCPRC 445
>UniRef100_UPI00001CF8B3 UPI00001CF8B3 UniRef100 entry
Length = 2141
Score = 80.9 bits (198), Expect = 6e-14
Identities = 43/119 (36%), Positives = 63/119 (52%), Gaps = 12/119 (10%)
Query: 22 KRRKLPIGPDQSSGKEQSNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHDGYF 81
KR+K G + + + ++ S+ S S RS + + K + + KK+ DG
Sbjct: 323 KRKK---GSSSEEDEPEDSDLDNASIHSSSVRSECSAALGKKNKRRRK---KKRIDDGDG 376
Query: 82 YE------CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCFEENDQLKP 134
YE C +C GG ++ CD+CPR YHL CLDP L++ P GKW CP C +E Q +P
Sbjct: 377 YETDHQDYCEVCQQGGEIILCDTCPRAYHLVCLDPELEKAPEGKWSCPHCEKEGIQWEP 435
Score = 72.0 bits (175), Expect = 3e-11
Identities = 27/48 (56%), Positives = 33/48 (68%)
Query: 78 DGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
D + C +C GG LLCCD+CP +YHL CL+PPL IP G+W CP C
Sbjct: 452 DDHMEFCRVCKDGGELLCCDACPSSYHLHCLNPPLPEIPNGEWLCPRC 499
>UniRef100_Q7ZWN3 B230399n07-prov protein [Xenopus laevis]
Length = 1893
Score = 80.1 bits (196), Expect = 1e-13
Identities = 48/125 (38%), Positives = 61/125 (48%), Gaps = 20/125 (16%)
Query: 20 KRKRRK-----LPIGPDQSSGKEQSNGKEDNSVASESSRSA----SAKRMLKTEEGTAQF 70
KRKR + D G S D S S+SSRS + K+ K EEG A
Sbjct: 294 KRKRSSSEEEDIDADSDYDEGSMNSLSVSDGST-SKSSRSRKKLKATKKKKKEEEGAAA- 351
Query: 71 SSKKKGHDGYFYE----CVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSCF 126
G DGY + C +C GG ++ CD+CPR YH+ CLDP + + P GKW CP C
Sbjct: 352 -----GADGYETDHQDYCEVCQQGGEIILCDTCPRAYHMVCLDPDMDKAPEGKWSCPHCE 406
Query: 127 EENDQ 131
+E Q
Sbjct: 407 KEGVQ 411
Score = 69.7 bits (169), Expect = 1e-10
Identities = 26/48 (54%), Positives = 32/48 (66%)
Query: 78 DGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
D + C +C GG LLCCD CP +YH+ CL+PPL IP G+W CP C
Sbjct: 436 DHHMEFCRVCKDGGELLCCDVCPSSYHIHCLNPPLPEIPNGEWLCPRC 483
>UniRef100_UPI0000337430 UPI0000337430 UniRef100 entry
Length = 1989
Score = 79.7 bits (195), Expect = 1e-13
Identities = 35/100 (35%), Positives = 57/100 (57%), Gaps = 7/100 (7%)
Query: 39 SNGKEDNSVASESSRSASAKRMLKTEEGTAQFSSKKKGHDGYFYECVICDLGGNLLCCDS 98
S+G +S + +S+ K+ ++TE+G + H Y C +C GG ++ CD+
Sbjct: 269 SDGSSRSSRPKKKPKSSKKKKKVETEDGDGY----ETDHQDY---CEVCQQGGEIILCDT 321
Query: 99 CPRTYHLQCLDPPLKRIPMGKWQCPSCFEENDQLKPLNNL 138
CPR YH+ CLDP +++ P GKW CP C +E Q + ++L
Sbjct: 322 CPRAYHMVCLDPDMEKAPEGKWSCPHCEKEGIQWEARDDL 361
Score = 70.5 bits (171), Expect = 8e-11
Identities = 26/48 (54%), Positives = 33/48 (68%)
Query: 78 DGYFYECVICDLGGNLLCCDSCPRTYHLQCLDPPLKRIPMGKWQCPSC 125
D + C +C GG LLCCD+CP +YH+ CL+PPL IP G+W CP C
Sbjct: 382 DHHIEFCRVCKDGGELLCCDTCPSSYHIHCLNPPLPEIPNGEWICPRC 429
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.311 0.129 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 652,198,656
Number of Sequences: 2790947
Number of extensions: 28468252
Number of successful extensions: 78632
Number of sequences better than 10.0: 1257
Number of HSP's better than 10.0 without gapping: 844
Number of HSP's successfully gapped in prelim test: 414
Number of HSP's that attempted gapping in prelim test: 76141
Number of HSP's gapped (non-prelim): 2448
length of query: 384
length of database: 848,049,833
effective HSP length: 129
effective length of query: 255
effective length of database: 488,017,670
effective search space: 124444505850
effective search space used: 124444505850
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 76 (33.9 bits)
Medicago: description of AC146651.2


