

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146650.9 - phase: 0 /pseudo
(314 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
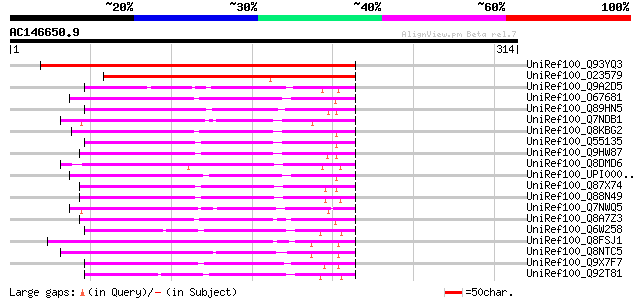
UniRef100_Q93YQ3 Formyltetrahydrofolate deformylase-like [Arabid... 294 2e-78
UniRef100_O23579 Formyltransferase purU homolog [Arabidopsis tha... 241 1e-62
UniRef100_Q9A2D5 Formyltetrahydrofolate deformylase [Caulobacter... 109 1e-22
UniRef100_O67681 Formyltetrahydrofolate deformylase [Aquifex aeo... 108 2e-22
UniRef100_Q89HN5 Formyltetrahydrofolate deformylase [Bradyrhizob... 99 1e-19
UniRef100_Q7NDB1 Formyltetrahydrofolate deformylase [Gloeobacter... 96 1e-18
UniRef100_Q8KBG2 Formyltetrahydrofolate deformylase [Chlorobium ... 96 2e-18
UniRef100_Q55135 Formyltetrahydrofolate deformylase (EC 3.5.1.10... 95 3e-18
UniRef100_Q9HW87 Formyltetrahydrofolate deformylase [Pseudomonas... 93 1e-17
UniRef100_Q8DMD6 Formyltetrahydrofolate deformylase [Synechococc... 92 2e-17
UniRef100_UPI0000270C13 UPI0000270C13 UniRef100 entry 91 3e-17
UniRef100_Q87X74 Formyltetrahydrofolate deformylase [Pseudomonas... 91 4e-17
UniRef100_Q88N49 Formyltetrahydrofolate deformylase [Pseudomonas... 90 9e-17
UniRef100_Q7NWQ5 Formyltetrahydrofolate deformylase [Chromobacte... 90 9e-17
UniRef100_Q8A7Z3 Formyltetrahydrofolate deformylase [Bacteroides... 89 2e-16
UniRef100_Q6W258 Formyltetrahydrofolate deformylase [Rhizobium sp.] 87 4e-16
UniRef100_Q8FSJ1 Putative formyltetrahydrofolate deformylase [Co... 86 1e-15
UniRef100_Q8NTC5 Formyltetrahydrofolate hydrolase [Corynebacteri... 85 3e-15
UniRef100_Q9X7F7 PurU protein [Methylobacterium chloromethanicum] 84 4e-15
UniRef100_Q92T81 PROBABLE FORMYLTETRAHYDROFOLATE DEFORMYLASE PRO... 83 8e-15
>UniRef100_Q93YQ3 Formyltetrahydrofolate deformylase-like [Arabidopsis thaliana]
Length = 323
Score = 294 bits (752), Expect = 2e-78
Identities = 138/195 (70%), Positives = 164/195 (83%)
Query: 20 IRNFSFKSLDLPPLPSPSLSHGIHVFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNK 79
+++ F L SP L G+HVFHC DAVGIVAKLS+CIA++GGNIL DVFVP+N
Sbjct: 19 LKSSRFHGESLDSSVSPVLIPGVHVFHCQDAVGIVAKLSDCIAAKGGNILGYDVFVPENN 78
Query: 80 HVFYSRSDFVFDPVKWPRKQMEEDFLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHC 139
+VFYSRS+F+FDPVKWPR Q++EDF ++Q + A SVVRVP++DPKYKIA+L SKQDHC
Sbjct: 79 NVFYSRSEFIFDPVKWPRSQVDEDFQTIAQRYGALNSVVRVPSIDPKYKIALLLSKQDHC 138
Query: 140 LVDLLHGWQDGKLPVDITCVISNHHRDSNTHVIRFLERHGIPYHCLSTTNENKREGEILE 199
LV++LH WQDGKLPVDITCVISNH R SNTHV+RFLERHGIPYH +STT ENKRE +ILE
Sbjct: 139 LVEMLHKWQDGKLPVDITCVISNHERASNTHVMRFLERHGIPYHYVSTTKENKREDDILE 198
Query: 200 LVQNTDFLVLARYMQ 214
LV++TDFLVLARYMQ
Sbjct: 199 LVKDTDFLVLARYMQ 213
>UniRef100_O23579 Formyltransferase purU homolog [Arabidopsis thaliana]
Length = 295
Score = 241 bits (616), Expect = 1e-62
Identities = 116/165 (70%), Positives = 137/165 (82%), Gaps = 9/165 (5%)
Query: 59 ECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFLKLSQAFNATRSVV 118
+CIA++GGNIL DV VP+NK+VFYSRS+F+FDPVKWPR+QM+EDF ++Q F+A SVV
Sbjct: 21 DCIAAKGGNILGYDVLVPENKNVFYSRSEFIFDPVKWPRRQMDEDFQTIAQKFSALSSVV 80
Query: 119 RVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVI---------SNHHRDSNT 169
RVP+LDPKYKIA+L SKQDHCLV++LH WQDGKLPVDITCVI SNH R NT
Sbjct: 81 RVPSLDPKYKIALLLSKQDHCLVEMLHKWQDGKLPVDITCVISDSGIFGVFSNHERAPNT 140
Query: 170 HVIRFLERHGIPYHCLSTTNENKREGEILELVQNTDFLVLARYMQ 214
HV+RFL+RHGI YH L TT++NK E EILELV+ TDFLVLARYMQ
Sbjct: 141 HVMRFLQRHGISYHYLPTTDQNKIEEEILELVKGTDFLVLARYMQ 185
>UniRef100_Q9A2D5 Formyltetrahydrofolate deformylase [Caulobacter crescentus]
Length = 280
Score = 109 bits (272), Expect = 1e-22
Identities = 72/172 (41%), Positives = 99/172 (56%), Gaps = 13/172 (7%)
Query: 47 CPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFLK 106
CPD GIVAK+S + RG NIL A F Q F+ R VFD R+ + DF
Sbjct: 7 CPDQRGIVAKVSAFLFERGCNILDAQQFDDQETGQFFMR--VVFDADGADREALRGDFGA 64
Query: 107 LSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNHHRD 166
L+ F + +R A +Y++ +LASK DHCL DL++ W+ G+LP+DIT V+SNH
Sbjct: 65 LADGFKM-KWTLRNRA--DRYRVLLLASKFDHCLADLVYRWRIGELPMDITGVVSNHPAQ 121
Query: 167 SNTHVIRFLERHGIPYHCLSTTNENK--REGEILELVQ--NTDFLVLARYMQ 214
+ HV + G+ +H L T E K +E E+ +L+Q TD +VLARYMQ
Sbjct: 122 TYAHV----DLSGLDFHHLPVTKETKFEQEAELWKLIQETKTDIVVLARYMQ 169
>UniRef100_O67681 Formyltetrahydrofolate deformylase [Aquifex aeolicus]
Length = 283
Score = 108 bits (270), Expect = 2e-22
Identities = 61/181 (33%), Positives = 99/181 (53%), Gaps = 12/181 (6%)
Query: 38 LSHGIHVFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPR 97
+ + I + CPD G+V ++S IA GGNI++ D + + F +R ++ + K PR
Sbjct: 1 MQNAILLVSCPDRKGLVKEISSFIADNGGNIVSFDQHIDEQTKTFLARVEWSLEDFKIPR 60
Query: 98 KQMEEDFLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDIT 157
+++E +F K++Q F+ ++ D K+A+ SKQ+HC DL+H + G+L ++
Sbjct: 61 EKIENEFKKVAQKFSMN---FQISFSDYVKKVAIFVSKQEHCFYDLMHRFYSGELKGEVK 117
Query: 158 CVISNHHRDSNTHVIRFLERHGIPYHCLSTTNENKREGEILEL----VQNTDFLVLARYM 213
VISNH + T E G+P++ + T ENK E E EL + +VLARYM
Sbjct: 118 LVISNHEKARKT-----AEFFGVPFYHIPKTKENKLEAEKRELELLKEYGVELVVLARYM 172
Query: 214 Q 214
Q
Sbjct: 173 Q 173
>UniRef100_Q89HN5 Formyltetrahydrofolate deformylase [Bradyrhizobium japonicum]
Length = 287
Score = 99.4 bits (246), Expect = 1e-19
Identities = 66/172 (38%), Positives = 92/172 (53%), Gaps = 11/172 (6%)
Query: 47 CPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFLK 106
CPD GIV+ +S +A G NIL A F F+ R F + ++ F
Sbjct: 12 CPDRPGIVSAVSTFLAHNGQNILDAQQFDDVETKKFFMRVVFTAADLAVELTALQTGFAA 71
Query: 107 LSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNHHRD 166
+++ F + A K+ +L SK DHCLVD+L+ W+ G+LP+ T ++SNH R+
Sbjct: 72 IAERFGMEWQMRDRAA---HRKVMLLVSKSDHCLVDILYRWRTGELPMVPTAIVSNHPRE 128
Query: 167 SNTHVIRFLERHGIPYHCLSTTNENKREGE--ILELV--QNTDFLVLARYMQ 214
V L+ GIP+H L T E+KRE E IL+LV TD +VLARYMQ
Sbjct: 129 ----VYAGLDFGGIPFHHLPVTKESKREQEAQILDLVAKTGTDLVVLARYMQ 176
>UniRef100_Q7NDB1 Formyltetrahydrofolate deformylase [Gloeobacter violaceus]
Length = 300
Score = 95.9 bits (237), Expect = 1e-18
Identities = 65/189 (34%), Positives = 100/189 (52%), Gaps = 14/189 (7%)
Query: 32 PLPSPSLSHGIH--VFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKH-VFYSRSDF 88
P P ++S + CPD GIVA +S+C+ GGNIL +D +F+ R +F
Sbjct: 10 PAPDTAMSPATRTLLISCPDRRGIVAAVSQCVLEAGGNILRSDQHTTDPLGGIFFMRLEF 69
Query: 89 VFDPVKWPRKQMEEDFLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQ 148
++ F +++ F +VR +PK ++A+ S+ DHC VDLL Q
Sbjct: 70 LYANAPADDDGFARCFAPVAERFAMDWRLVRTG--EPK-RMALFVSRLDHCFVDLLWRRQ 126
Query: 149 DGKLPVDITCVISNHHRDSNTHVIRFLERHGIPYHCLS--TTNENKREGEILELVQ-NTD 205
G+LPV I V+SNH + ++G+PYH L+ TN+ RE ++L L++ D
Sbjct: 127 SGELPVKIPLVVSNH-----PDLEPVAAQYGLPYHYLAIDKTNQPAREAQMLNLLEGEVD 181
Query: 206 FLVLARYMQ 214
F+VLARYM+
Sbjct: 182 FIVLARYMR 190
>UniRef100_Q8KBG2 Formyltetrahydrofolate deformylase [Chlorobium tepidum]
Length = 289
Score = 95.5 bits (236), Expect = 2e-18
Identities = 60/180 (33%), Positives = 90/180 (49%), Gaps = 12/180 (6%)
Query: 39 SHGIHVFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRK 98
S I + CPD VG+VA+++ I RGGNIL + V ++ F+ R + D P +
Sbjct: 8 SKAILLLSCPDRVGLVARIANFIYERGGNILDLNEHVDVDERQFFLRVSWSLDHFSIPAE 67
Query: 99 QMEEDFLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITC 158
+E F L + F A ++ + ++AV SK DHCL ++L G+ +D+
Sbjct: 68 DLESAFAPLGREFRAN---WQIRLSGKRSRMAVFVSKYDHCLREILWRHSLGEFDIDLPL 124
Query: 159 VISNHHRDSNTHVIRFLERHGIPYHCLSTTNENKREGEILELV----QNTDFLVLARYMQ 214
VISNH + +E HGIP+H + T E K E ++ D +VLARYMQ
Sbjct: 125 VISNH-----PDLAPLVEAHGIPFHVIPVTPEAKAAAEQRQMALCDEHGIDTIVLARYMQ 179
>UniRef100_Q55135 Formyltetrahydrofolate deformylase (EC 3.5.1.10) (Formyl-FH(4)
hydrolase) [Synechocystis sp.]
Length = 284
Score = 94.7 bits (234), Expect = 3e-18
Identities = 57/172 (33%), Positives = 92/172 (53%), Gaps = 12/172 (6%)
Query: 47 CPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFLK 106
CPD GIVA++++ I GNI+ AD + +F +R ++ D + R ++ + +
Sbjct: 12 CPDQPGIVAQIAQFIYQNQGNIIHADQHTDFSSGLFLNRVEWQLDNFRLSRPELLSAWSQ 71
Query: 107 LSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNHHRD 166
L++ AT ++ D ++A+ SKQDHCL+D+L W+ G+L +I +ISNH
Sbjct: 72 LAEQLQATW---QIHFSDQLPRLALWVSKQDHCLLDILWRWRSGELRCEIPLIISNH--- 125
Query: 167 SNTHVIRFLERHGIPYHCLSTTNENKREGEILELV----QNTDFLVLARYMQ 214
+ ++ GI +HCL T ENK E EL D +VLA+Y+Q
Sbjct: 126 --PDLKSIADQFGIDFHCLPITKENKLAQETAELALLKQYQIDLVVLAKYLQ 175
>UniRef100_Q9HW87 Formyltetrahydrofolate deformylase [Pseudomonas aeruginosa]
Length = 283
Score = 92.8 bits (229), Expect = 1e-17
Identities = 58/175 (33%), Positives = 90/175 (51%), Gaps = 12/175 (6%)
Query: 44 VFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEED 103
V CPD VGIVAK+S +A+ G I A + F+ R + D + + +
Sbjct: 7 VIACPDGVGIVAKVSNFLATYNGWITEASHHSDNDNGWFFMRHEIRADSLPFDLDGFRQA 66
Query: 104 FLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNH 163
F +++ F+ R+ + K ++ ++ASK+ HCL DLLH W G+L +I CVI+NH
Sbjct: 67 FAPIAREFSMEW---RITDSEVKKRVVLMASKESHCLADLLHRWHSGELDCEIPCVIANH 123
Query: 164 HRDSNTHVIRFLERHGIPYHCLSTTNENKREG--EILELV--QNTDFLVLARYMQ 214
+ +E HGIPY + ++K+ E+ L+ D +VLARYMQ
Sbjct: 124 -----DDLRSMVEWHGIPYFHVPVDPQDKQPAFDEVSRLIDEHGADCIVLARYMQ 173
>UniRef100_Q8DMD6 Formyltetrahydrofolate deformylase [Synechococcus elongatus]
Length = 291
Score = 92.0 bits (227), Expect = 2e-17
Identities = 65/191 (34%), Positives = 103/191 (53%), Gaps = 18/191 (9%)
Query: 32 PLPSPSLSHGIHVFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFD 91
P+P+ +LS CPD G+VAKL++ + GNI+ AD +F SR ++ +
Sbjct: 2 PMPTMTLS-----ISCPDQRGLVAKLAQFVYRYNGNIVHADHHTDAVAGIFLSRLEWELE 56
Query: 92 PVKWPRKQMEEDFLKLSQ---AFNATRSV-VRVPALDPKYKIAVLASKQDHCLVDLLHGW 147
+ PR Q+ F+ +Q F + + V ++ A D Y++A+ S+QDHCL DLL
Sbjct: 57 GFEIPRDQIATTFINYAQREKIFTSWQGVRWQLRASDIPYRLAIWVSRQDHCLWDLLLRQ 116
Query: 148 QDGKLPVDITCVISNHHRDSNTHVIRFLERHGIPYHCLSTTNENK--REGEILELVQN-- 203
+ G L +I +ISNH H+ E+ GI +H + T E K E + L+L+++
Sbjct: 117 RAGDLFAEIPLIISNHE-----HLRPIAEQFGIDFHYIPVTPETKPLAEAKQLQLLKDYR 171
Query: 204 TDFLVLARYMQ 214
D +VLA+YMQ
Sbjct: 172 IDLVVLAKYMQ 182
>UniRef100_UPI0000270C13 UPI0000270C13 UniRef100 entry
Length = 272
Score = 91.3 bits (225), Expect = 3e-17
Identities = 60/182 (32%), Positives = 94/182 (50%), Gaps = 13/182 (7%)
Query: 38 LSHGIHVFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPR 97
+++ + + C D GIV+ ++E I S GNI+ D + + F+ R ++ D R
Sbjct: 1 MANAVLLIECEDQQGIVSSITEFIYSNQGNIIDLDQYTDHDNTHFFMRLEWSLDSFTIQR 60
Query: 98 KQMEEDFLKL-SQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDI 156
++ E F L Q F+ ++ D ++ + +K HCL D+L WQ G+L ++I
Sbjct: 61 GKIGEYFETLIGQRFSIRFNLYFT---DEHPRMGLFVTKLSHCLFDILGRWQSGELEIEI 117
Query: 157 TCVISNHHRDSNTHVIRFLERHGIPYHCLSTTNENKREGEILELV----QNTDFLVLARY 212
VISNH +T +ER IP+ + TNENK E E +L + DF+VLARY
Sbjct: 118 PVVISNHENLRST-----VERFDIPFEYIPITNENKAEQEQKQLKVLSDYDVDFVVLARY 172
Query: 213 MQ 214
MQ
Sbjct: 173 MQ 174
>UniRef100_Q87X74 Formyltetrahydrofolate deformylase [Pseudomonas syringae]
Length = 283
Score = 90.9 bits (224), Expect = 4e-17
Identities = 60/175 (34%), Positives = 86/175 (48%), Gaps = 12/175 (6%)
Query: 44 VFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEED 103
V CPD VGIVAK+S +AS G I A F+ R + D + + E
Sbjct: 7 VIACPDRVGIVAKVSNFLASHNGWITEASHHSDNLSGWFFMRHEIRADTLPFDLDGFREA 66
Query: 104 FLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNH 163
F +++ F+ R+ K ++ ++AS++ HCL DLLH W +L DI CVISNH
Sbjct: 67 FTPIAEEFSMDW---RITDSAQKKRVVLMASRESHCLADLLHRWHSDELDCDIACVISNH 123
Query: 164 HRDSNTHVIRFLERHGIPYHCLSTTNENKRE--GEILELV--QNTDFLVLARYMQ 214
+ +E H IPY+ + ++K E+ LV D +VLARYMQ
Sbjct: 124 Q-----DLRSMVEWHDIPYYHVPVDPKDKEPAFAEVSRLVGHHQADVVVLARYMQ 173
>UniRef100_Q88N49 Formyltetrahydrofolate deformylase [Pseudomonas putida]
Length = 283
Score = 89.7 bits (221), Expect = 9e-17
Identities = 59/175 (33%), Positives = 90/175 (50%), Gaps = 12/175 (6%)
Query: 44 VFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEED 103
V CPD VGIVAK+S +A G I A + F+ R + + + + + E
Sbjct: 7 VIACPDRVGIVAKVSNFLALYNGWINEASHHSDEQSGWFFMRHEIRAESLPFGIEAFREA 66
Query: 104 FLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNH 163
F +++ F+ T R+ K ++ ++AS++ HCL DLLH W +L +I CVISNH
Sbjct: 67 FAPIAEEFSMTW---RITDSAQKKRVVLMASRESHCLADLLHRWHTDELDCEIPCVISNH 123
Query: 164 HRDSNTHVIRFLERHGIPYHCLSTTNENKRE--GEILELVQN--TDFLVLARYMQ 214
+ + +E HGIP+ + ++K E+ LVQ D +VLARYMQ
Sbjct: 124 N-----DLRSMVEWHGIPFFHVPVDPKDKAPAFAEVSRLVQEHAADVVVLARYMQ 173
>UniRef100_Q7NWQ5 Formyltetrahydrofolate deformylase [Chromobacterium violaceum]
Length = 289
Score = 89.7 bits (221), Expect = 9e-17
Identities = 61/186 (32%), Positives = 92/186 (48%), Gaps = 18/186 (9%)
Query: 38 LSHGIH----VFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPV 93
+SH H + PD G+VA ++ + + NI+ AD ++++F R + D
Sbjct: 3 MSHDKHSATLLISAPDKKGLVAAIANFLMTYNANIMHADQHQDTSENLFLMRVQWDLDGF 62
Query: 94 KWPRKQMEEDFLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLP 153
P F ++ V + A P+ +A+ S+ +HCLVDL+H W+ G+L
Sbjct: 63 TLPMDSFAAAFQPIADEHGMNWKV-SLSARKPR--MAIFVSQYEHCLVDLMHRWRIGELD 119
Query: 154 VDITCVISNHHRDSNTHVIRFLERHGIPYHCLSTTNENKREGE-----ILELVQNTDFLV 208
DI VISNH R +E +GIP+H + T +NK E E +LE DF+V
Sbjct: 120 CDIPLVISNHET-----CRRLVEFNGIPFHVIKVTKDNKAEAEAEQFRLLE-EAGVDFIV 173
Query: 209 LARYMQ 214
LARYMQ
Sbjct: 174 LARYMQ 179
>UniRef100_Q8A7Z3 Formyltetrahydrofolate deformylase [Bacteroides thetaiotaomicron]
Length = 284
Score = 89.0 bits (219), Expect = 2e-16
Identities = 59/176 (33%), Positives = 92/176 (51%), Gaps = 13/176 (7%)
Query: 44 VFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEED 103
+ HCPD GI+A++++ I GNI+ D +V +++F+ R ++ P++++E+
Sbjct: 7 LLHCPDKPGILAEVTDFITVNKGNIIYLDQYVDHVENIFFMRIEWELKDFLVPQEKIEDY 66
Query: 104 FLKL-SQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISN 162
F L Q + R+ D K ++A+ SK HCL D+L + G+ V+I +ISN
Sbjct: 67 FRTLYGQKYEMD---FRLYFSDVKPRMAIFVSKMSHCLFDMLARYTAGEWNVEIPLIISN 123
Query: 163 HHRDSNTHVIRFLERHGIPYHCLSTTNENKREGEILEL----VQNTDFLVLARYMQ 214
H HV ER GIP++ T E K E E E+ F+VLARYMQ
Sbjct: 124 H--PDLQHV---AERFGIPFYLFPITKETKEEQERKEMELLAKHKITFIVLARYMQ 174
>UniRef100_Q6W258 Formyltetrahydrofolate deformylase [Rhizobium sp.]
Length = 295
Score = 87.4 bits (215), Expect = 4e-16
Identities = 58/172 (33%), Positives = 87/172 (49%), Gaps = 13/172 (7%)
Query: 47 CPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFLK 106
C GIVA LS +A +G NI+ + F +F+ R F+ + R +EE
Sbjct: 11 CKSTRGIVAALSGYLAEQGCNIIDSSQFDDLQTGLFFMRISFISEQGVG-RAALEEGLKP 69
Query: 107 LSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNHHRD 166
++ F ++ + K ++ S+ HCL DLL+ W+ G LP+DI V+SNH
Sbjct: 70 IAATFAMETAL---HDQSERTKALLMVSRFGHCLNDLLYRWKIGALPIDIVGVVSNHFDY 126
Query: 167 SNTHVIRFLERHGIPYHCLSTTNEN--KREGEILELVQNT--DFLVLARYMQ 214
V H IP+HC+ T EN K E ++L+ V+ T + +VLARYMQ
Sbjct: 127 QKVVV-----NHDIPFHCIKVTKENKPKAEAQLLDFVEQTGAELIVLARYMQ 173
>UniRef100_Q8FSJ1 Putative formyltetrahydrofolate deformylase [Corynebacterium
efficiens]
Length = 305
Score = 85.9 bits (211), Expect = 1e-15
Identities = 56/197 (28%), Positives = 95/197 (47%), Gaps = 11/197 (5%)
Query: 24 SFKSLDLPPLPSPSLSHGIHVFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFY 83
SF + P +P + F CPD+ GIVAKLS +A RGG I A F + + F+
Sbjct: 4 SFTEIRNRPGTAPEERQYVLTFGCPDSTGIVAKLSSFLAERGGWITEAGYFTDPDSNWFF 63
Query: 84 SRSDFVFDPVKWPRKQMEEDFLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDL 143
+R + + +++ E+F ++ R+ + K +L SK+ HCL DL
Sbjct: 64 TRQAVRAESIDMEIEELREEFAAFAEEEFGPRARWQFTDTAQVKKAVILVSKEGHCLHDL 123
Query: 144 LHGWQDGKLPVDITCVISNHHRDSNTHVIRFLERHGIPYHCL----STTNENKREGEILE 199
L + P+++ VI NH D+ ++ + HG+P+H + + + + E
Sbjct: 124 LGRVAENDYPMEVVAVIGNH--DNLEYIAK---NHGVPFHHIPFPKDAVGKRRAFDAVTE 178
Query: 200 LVQ--NTDFLVLARYMQ 214
+V N D +V+AR+MQ
Sbjct: 179 IVNELNPDAIVMARFMQ 195
>UniRef100_Q8NTC5 Formyltetrahydrofolate hydrolase [Corynebacterium glutamicum]
Length = 304
Score = 84.7 bits (208), Expect = 3e-15
Identities = 56/190 (29%), Positives = 91/190 (47%), Gaps = 14/190 (7%)
Query: 32 PLPSPSLSHGIHVFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFD 91
P +P + F CPD+ GIVAKLS +A RGG I A F + + F++R +
Sbjct: 12 PSAAPEERQFVLTFGCPDSTGIVAKLSSFLAERGGWITEAGYFTDPDSNWFFTRQAIRAE 71
Query: 92 PVKWPRKQMEEDFLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGK 151
+ +Q+ E+F L++ F K K +L SK+ HCL DLL +
Sbjct: 72 SIDTTIEQLREEFAPLAEEFGPRAKWSFTDTAQVK-KAVLLVSKEGHCLHDLLGRVAEND 130
Query: 152 LPVDITCVISNHHRDSNTHVIRFL-ERHGIPYHCL----STTNENKREGEILELVQ--NT 204
P+++ V+ NH +R++ E H +P+ + + K ++ E+V +
Sbjct: 131 YPMEVVAVVGNHEN------LRYIAENHNVPFFHVPFPKDAVGKRKAFDQVAEIVNGYDP 184
Query: 205 DFLVLARYMQ 214
D +VLAR+MQ
Sbjct: 185 DAIVLARFMQ 194
>UniRef100_Q9X7F7 PurU protein [Methylobacterium chloromethanicum]
Length = 287
Score = 84.3 bits (207), Expect = 4e-15
Identities = 59/172 (34%), Positives = 92/172 (53%), Gaps = 11/172 (6%)
Query: 47 CPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFLK 106
C D GIVA +S + R NI+ A F + F+ R F + ++ +E
Sbjct: 12 CADRPGIVAAVSGALLERDCNIVEAKQFEDVIEKRFFMRVVFSGINGQNSLREHQEAMTP 71
Query: 107 LSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNHHRD 166
L+Q F S+ + K ++ +L S+ DHCLVD+L+ + G+LP+D+T V++NH +
Sbjct: 72 LAQRFEMDWSI---RDCETKRRVMILVSRFDHCLVDILYRKRIGELPMDLTAVVTNHAAE 128
Query: 167 SNTHVIRFLERHGIPYHCLSTTNENKR--EGEILELVQ--NTDFLVLARYMQ 214
+ H L+ G P L T E KR E ++LEL++ T+ +VLARYMQ
Sbjct: 129 NYAH----LDLCGAPLISLPVTAETKRAQEDKLLELIERTGTEVVVLARYMQ 176
>UniRef100_Q92T81 PROBABLE FORMYLTETRAHYDROFOLATE DEFORMYLASE PROTEIN [Rhizobium
meliloti]
Length = 296
Score = 83.2 bits (204), Expect = 8e-15
Identities = 54/172 (31%), Positives = 88/172 (50%), Gaps = 13/172 (7%)
Query: 47 CPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFLK 106
C G+VA LS +A +G NI + F + F+ R+ F+ + + +EE
Sbjct: 12 CKSTRGVVAALSGYLAEQGCNIADSSQFDDLDTGKFFMRTSFISEE-RVGLAALEEGLKP 70
Query: 107 LSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNHHRD 166
++ F ++ + K+ ++ S+ HCL DLL+ W+ G LP+DI V+SNH
Sbjct: 71 IASKFEMETALHEQ---SERMKVLLMVSRFGHCLNDLLYRWKIGALPIDIVGVVSNHFDY 127
Query: 167 SNTHVIRFLERHGIPYHCLSTTNEN--KREGEILELVQNT--DFLVLARYMQ 214
V H IP+H + T EN K E +++++V+ T + +VLARYMQ
Sbjct: 128 QKVVV-----NHDIPFHHIKVTKENKPKAEAQLMDVVEQTGAELIVLARYMQ 174
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.333 0.144 0.459
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 484,009,422
Number of Sequences: 2790947
Number of extensions: 18496367
Number of successful extensions: 73317
Number of sequences better than 10.0: 194
Number of HSP's better than 10.0 without gapping: 153
Number of HSP's successfully gapped in prelim test: 41
Number of HSP's that attempted gapping in prelim test: 72922
Number of HSP's gapped (non-prelim): 201
length of query: 314
length of database: 848,049,833
effective HSP length: 127
effective length of query: 187
effective length of database: 493,599,564
effective search space: 92303118468
effective search space used: 92303118468
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 74 (33.1 bits)
Medicago: description of AC146650.9


