

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146632.4 - phase: 0
(1038 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
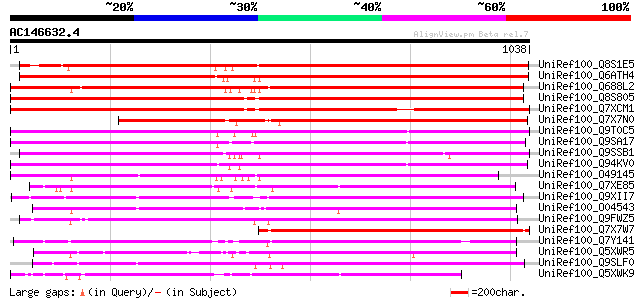
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8S1E5 Putative gag/pol polyprotein [Oryza sativa] 1092 0.0
UniRef100_Q6ATH4 Putative polyprotein [Oryza sativa] 1084 0.0
UniRef100_Q688L2 Putative polyprotein [Oryza sativa] 1073 0.0
UniRef100_Q8S805 Putative copia-type polyprotein [Oryza sativa] 1050 0.0
UniRef100_Q7XCM1 Putative gag-pol polyprotein [Oryza sativa] 1012 0.0
UniRef100_Q7X7N0 OSJNBa0035I04.5 protein [Oryza sativa] 832 0.0
UniRef100_Q9T0C5 Retrotransposon like protein [Arabidopsis thali... 784 0.0
UniRef100_Q9SA17 F28K20.17 protein [Arabidopsis thaliana] 783 0.0
UniRef100_Q9SSB1 T18A20.5 protein [Arabidopsis thaliana] 768 0.0
UniRef100_Q94KV0 Polyprotein [Arabidopsis thaliana] 754 0.0
UniRef100_O49145 Polyprotein [Arabidopsis arenosa] 736 0.0
UniRef100_Q7XE85 Putative pol polyprotein [Oryza sativa] 676 0.0
UniRef100_Q9XII7 Putative retroelement pol polyprotein [Arabidop... 652 0.0
UniRef100_O04543 F20P5.25 protein [Arabidopsis thaliana] 642 0.0
UniRef100_Q9FWZ5 Putative retroelement polyprotein [Arabidopsis ... 642 0.0
UniRef100_Q7X7W7 OSJNBa0061G20.2 protein [Oryza sativa] 637 0.0
UniRef100_Q7Y141 Putative polyprotein [Oryza sativa] 620 e-176
UniRef100_Q5XWR5 Putative retroelement pol polyprotein-like [Sol... 608 e-172
UniRef100_Q9SLF0 Putative retroelement pol polyprotein [Arabidop... 608 e-172
UniRef100_Q5XWK9 Gag-pol polyprotein-like [Solanum tuberosum] 599 e-169
>UniRef100_Q8S1E5 Putative gag/pol polyprotein [Oryza sativa]
Length = 1090
Score = 1092 bits (2825), Expect = 0.0
Identities = 562/1064 (52%), Positives = 709/1064 (65%), Gaps = 68/1064 (6%)
Query: 19 GIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNLVSVRKFTTDNSVSVEFDP 78
G G G ++P G + ++ P ++NL+SVR+FT DN S+EFD
Sbjct: 35 GFSTGGGGALPGGGGGYQGVANP---------------VRNLLSVRQFTRDNKCSIEFDE 79
Query: 79 FGFSVKDFQTGMRLMRCESRGDLYPITTSQAISPSTFA----ALAPSLWHARLGHPGAPV 134
FGFSVKD QT ++RC SRG+LY T A +PS+ A A + +LWH RLGHPG
Sbjct: 80 FGFSVKDLQTRRVILRCNSRGELY---TLPAATPSSAAHGLLATSSTLWHCRLGHPGPAA 136
Query: 135 VDSLRKNKFIECNRASGSHICHSCSLGKHIKLPFVSSNSCTIMPFDIIHSDIWTSPVLSS 194
+ LR I CN+ S +CH+C LGKH +LPF +S+S T +PF+++H D+WTSPV+S+
Sbjct: 137 IHGLRNIASISCNKIDTS-LCHACQLGKHTRLPFHNSSSRTSVPFELVHCDVWTSPVMST 195
Query: 195 SGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQFEREVKNIQCDNGKEFDN 254
SG +YY++ +DD+S+F WTF L KS V + F ++ TQF +K+ Q DNG+EF N
Sbjct: 196 SGFKYYLVVLDDFSHFCWTFLLRLKSDVHRHIVEFVEYVSTQFGLPLKSFQADNGREFVN 255
Query: 255 RHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLLVHASLPPSFWHHALQMA 314
F G RLSCP+TS QNGKAER +RTINN IRTLL+ AS+PPS+W AL A
Sbjct: 256 TAITTFLASRGTQLRLSCPYTSPQNGKAERMLRTINNSIRTLLIQASMPPSYWAEALATA 315
Query: 315 TYLINILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCYPLFPSTTINKLQARSTPCVFL 374
TYL+N P+ + P ++L++ P +SHLRVFGCLCYP +TT +KL RST CVFL
Sbjct: 316 TYLLNRRPSSSIHQSLPFQLLHRTIPDFSHLRVFGCLCYPNLSATTPHKLSPRSTACVFL 375
Query: 375 GYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFA------------------------K 410
GYP++H+ Y+C DLS+ +IIISRHV+FDE+QFPFA +
Sbjct: 376 GYPTSHKGYRCLDLSTHRIIISRHVVFDESQFPFAATPPAASSFDFLLQGLSPADAPSLE 435
Query: 411 LHNPQPYTYGFMDDGPSPYV-----------IHHLTSQPSLGQP------AQHDLPNTQP 453
+ P+P T + PY+ + + PS G P A P +
Sbjct: 436 VEQPRPLTVAPSTEVEQPYLPLPSRRLSAGTVTVASEAPSAGAPLVGTSSADATPPGSAT 495
Query: 454 TTQPTTPEEQHAHSPPSSSPNPSPSTTATPSPPYQPTPISVPKPVTRSQHGIFKPKRQLN 513
+H ++ + P S+TA + P P S+ VTRSQ G +P +L
Sbjct: 496 RASTIVSPFRHVYTRRPVTTVPPSSSTAVTNAVAAPQPHSM---VTRSQSGSLRPVDRLT 552
Query: 514 LN-TSVPRSPLPRNPVSALRDPNWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFT 572
T SP+P N SAL DPNW+ AM DE+ L+ N TW LV RPP N+ WIF
Sbjct: 553 YTATQAAASPVPANYHSALADPNWRAAMADEYKELVDNGTWRLVSRPPRANIATGKWIFK 612
Query: 573 HKEKSDGVFERYKARLVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLD 632
HK SDG RYKAR V G +QQ G+D ETFSPVVK ATIR VLS+A S+AW IHQLD
Sbjct: 613 HKFHSDGSLARYKARWVVRGYSQQHGIDYDETFSPVVKLATIRVVLSIAASRAWPIHQLD 672
Query: 633 VKNAFLHGELKETVYMHQPMGFRDPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIG 692
VKNAFLHG LKETVY QP GF DP P+ VCLL+KSLYGLKQAPRAWY+RFA Y+ +G
Sbjct: 673 VKNAFLHGHLKETVYCQQPSGFVDPTAPDAVCLLQKSLYGLKQAPRAWYQRFATYIRQMG 732
Query: 693 FSHSTSDHSLFIYRKGTAMAYILLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHY 752
F S SD SLF+Y+ G +AY+LLYVDDIILTAS+ L + + L +EFAM DLG LH+
Sbjct: 733 FMPSASDTSLFVYKDGDRIAYLLLYVDDIILTASTTTLLQQLTARLHSEFAMTDLGDLHF 792
Query: 753 FLGIAVTHHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHY 812
FLGI+V GLFLSQR+YA ++++RAGMA C S+STPVDT KLSA P ADPS Y
Sbjct: 793 FLGISVKRSPDGLFLSQRQYAVDLLQRAGMAECHSTSTPVDTHAKLSATDGLPVADPSAY 852
Query: 813 RSLAGALQYLTFTRPDIAYAVQQVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSS 872
RS+AGALQYLT TRPD+AYAVQQVCLFMHDPRE H+ +KRI+RY++G+L GLH+
Sbjct: 853 RSIAGALQYLTLTRPDLAYAVQQVCLFMHDPREPHLALVKRILRYVKGSLSIGLHIGSGP 912
Query: 873 TSTLISYTDADWGGCPDTRRSTSGYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVV 932
+L +Y+DADW GCP++RRSTSGYCV+LGDNL+SWS+KRQ T+SRSSAEAEYR VA+ V
Sbjct: 913 IQSLTAYSDADWAGCPNSRRSTSGYCVYLGDNLVSWSSKRQTTVSRSSAEAEYRAVAHAV 972
Query: 933 SESCWLRNLLLELHCPIRKASLVYCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARG 992
+E CWLR LL ELH PI A++VYCDNVSA+Y++ NPV H+RTKHIE+DIH VREKVA G
Sbjct: 973 AECCWLRQLLQELHVPIASATIVYCDNVSAVYMTANPVHHRRTKHIEIDIHFVREKVALG 1032
Query: 993 EVRVLHVPSRYQIADIFTKGLPLVLFEDFRNNLSVRQPPVSTAG 1036
+VRVL+VPS +Q ADI TKGLP+ LF DFR++L VR PP +TAG
Sbjct: 1033 QVRVLYVPSSHQFADIMTKGLPVQLFTDFRSSLCVRAPPAATAG 1076
>UniRef100_Q6ATH4 Putative polyprotein [Oryza sativa]
Length = 1480
Score = 1084 bits (2803), Expect = 0.0
Identities = 561/1096 (51%), Positives = 716/1096 (65%), Gaps = 82/1096 (7%)
Query: 20 IIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNLVSVRKFTTDNSVSVEFDPF 79
I VGNGH+IP+ + L LKN+L +P L++NL+ VR+FT DN S EFD F
Sbjct: 386 ITVGNGHTIPVICRGTSFLPIGTTRFALKNILVAPSLVRNLLFVRQFTRDNKCSFEFDEF 445
Query: 80 GFSVKDFQTGMRLMRCESRGDLYPI-TTSQAISPSTFAALAPSLWHARLGHPGAPVVDSL 138
GFSVKD T ++RC SRGDLY + TT AI+ +F A + +LWH RLGHP V +L
Sbjct: 446 GFSVKDLPTRRVILRCNSRGDLYTLPTTVPAITAHSFLAKSSTLWHHRLGHPSPAAVQTL 505
Query: 139 RKNKFIECNRASGSHICHSCSLGKHIKLPFVSSNSCTIMPFDIIHSDIWTSPVLSSSGHR 198
K + C R S + +CH+C LGKH +L F S+S T PF+++H D+WTSPVLS SG +
Sbjct: 506 HKLAILSCTR-SNNKLCHACHLGKHTRLSFSKSSSSTSSPFELVHCDVWTSPVLSLSGFK 564
Query: 199 YYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQFEREVKNIQCDNGKEFDNRHFW 258
YY++ +DD+++F WTFPL KS V L F +++TQF ++ Q DNG +F N
Sbjct: 565 YYLVVLDDFTHFCWTFPLRHKSDVHQHLLEFVAYVKTQFSLPIRCFQADNGTKFVNHATT 624
Query: 259 EFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLLVHASLPPSFWHHALQMATYLI 318
F G+ RLSCP+TS QNGKAER +RTIN IRTLL+ AS+PPS+W AL ATYL+
Sbjct: 625 SFFASRGIVLRLSCPYTSPQNGKAERVLRTINKSIRTLLIQASMPPSYWAEALATATYLL 684
Query: 319 NILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCYPLFPSTTINKLQARSTPCVFLGYPS 378
N P+ + P ++L+ K P YS+L+VFGCLCYP + T +KL RS P VFLGY +
Sbjct: 685 NRRPSTSVRNSIPYQLLHNKLPDYSNLQVFGCLCYPNLSAMTSHKLSPRSAPYVFLGYSA 744
Query: 379 NHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLHNPQ-PYTYGFMDDG------------ 425
+H+ ++C D+S+R++ ISRHV+FDE FPFA + PQ ++ F+ G
Sbjct: 745 SHKGFRCLDISTRRLYISRHVVFDEKTFPFAAI--PQDASSFDFLLQGFSIAVAPSSEVE 802
Query: 426 --------PSPYVIHHL---------------------TSQPSLGQPAQHDLPN---TQP 453
PSP V + +P G P LP
Sbjct: 803 RPRFSSMTPSPEVEQPIPDDDTSGTELFQLLPGLRSSAAGRPLAGSPVDARLPGGCANDA 862
Query: 454 TTQPTTPEEQHAHSPPSSS---PNPSPSTTATPSPPY--------QPTPISVPKP----- 497
++ PP++S P PS T + PY QP P ++ +P
Sbjct: 863 ANGSSSSNLSPVMDPPAASVVRPAPSEGPTTSLISPYRHTYLRRSQPAPTAIHRPIRASR 922
Query: 498 ----------------VTRSQHGIFKPKRQLNLN-TSVPRSPLPRNPVSALRDPNWKMAM 540
VTRSQ G +P ++ T SP+P N SAL DPNW+ A
Sbjct: 923 AFHSATDQQQQTGHTMVTRSQTGHLRPIQRFTYTATHDVVSPVPSNYRSALADPNWRAAT 982
Query: 541 DDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKEKSDGVFERYKARLVGDGKTQQVGVD 600
+E+ AL+ N TW LVPRPP NV+ WIF HK SDG R+KAR V G +QQ G+D
Sbjct: 983 ANEYKALVDNNTWRLVPRPPGANVVTGKWIFKHKFHSDGTLARHKARWVVRGYSQQHGID 1042
Query: 601 CGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKNAFLHGELKETVYMHQPMGFRDPNLP 660
ETFSPVVKPATI VLS+A S++W IHQLDVKNAFLHG L+ETVY QP GF DP+ P
Sbjct: 1043 YDETFSPVVKPATIHVVLSIAASRSWPIHQLDVKNAFLHGNLEETVYYQQPSGFVDPSAP 1102
Query: 661 NHVCLLKKSLYGLKQAPRAWYKRFADYVSTIGFSHSTSDHSLFIYRKGTAMAYILLYVDD 720
N VCLL+KSLYGLKQAPRAWY+RFA Y+ +GF+ S S+ SLF+Y+ G +AY+LLYVDD
Sbjct: 1103 NAVCLLQKSLYGLKQAPRAWYQRFATYIRQLGFTSSASNTSLFVYKDGDNIAYLLLYVDD 1162
Query: 721 IILTASSDALRVTIISLLSTEFAMKDLGSLHYFLGIAVTHHTGGLFLSQRKYAAEIIERA 780
IILTASS L I + L +EFAM DLG LH+FLGI+VT + GLFLSQR+Y ++++RA
Sbjct: 1163 IILTASSATLLHHITARLHSEFAMTDLGDLHFFLGISVTRSSDGLFLSQRQYVVDLLKRA 1222
Query: 781 GMAACKSSSTPVDTKPKLSANSSAPHADPSHYRSLAGALQYLTFTRPDIAYAVQQVCLFM 840
GM+ C S++TPVD + KLSA AP +DP+ YRSL GALQYLT TRP++AYAVQQVCLFM
Sbjct: 1223 GMSECHSTATPVDARSKLSATDGAPVSDPTEYRSLVGALQYLTLTRPELAYAVQQVCLFM 1282
Query: 841 HDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTSTLISYTDADWGGCPDTRRSTSGYCVF 900
HDPRE H+ +KRI+RYI+GTL GLHL + +L +Y+DADW GCPD+ RSTSGYCVF
Sbjct: 1283 HDPREPHLALIKRILRYIKGTLHIGLHLGTTPVDSLTAYSDADWAGCPDSHRSTSGYCVF 1342
Query: 901 LGDNLISWSAKRQATLSRSSAEAEYRGVANVVSESCWLRNLLLELHCPIRKASLVYCDNV 960
LGDNL+SWS+KRQ T+SRSSAEAEYR VA+ ++E CWLR LL ELH + A++VYCDNV
Sbjct: 1343 LGDNLVSWSSKRQTTVSRSSAEAEYRAVAHAIAECCWLRQLLSELHISLTSATVVYCDNV 1402
Query: 961 SAIYLSGNPVQHQRTKHIEMDIHSVREKVARGEVRVLHVPSRYQIADIFTKGLPLVLFED 1020
SA+Y++ NPV H+RTKHIE+DIH VREKVA G+VRVLH+PS +Q ADI TKGLP+ LF +
Sbjct: 1403 SAVYMAANPVHHRRTKHIEIDIHFVREKVALGQVRVLHLPSSHQFADIMTKGLPVQLFTE 1462
Query: 1021 FRNNLSVRQPPVSTAG 1036
FR++L VR P STAG
Sbjct: 1463 FRSSLCVRDNPASTAG 1478
>UniRef100_Q688L2 Putative polyprotein [Oryza sativa]
Length = 1679
Score = 1073 bits (2775), Expect = 0.0
Identities = 575/1112 (51%), Positives = 727/1112 (64%), Gaps = 88/1112 (7%)
Query: 1 MTSEQG--NLSSYFNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIK 58
MTS+ G +LS N ++ I+VGNG +IP+ H+ + PN TL+++L SP +IK
Sbjct: 570 MTSDTGILSLSHPPNPNSPSHIVVGNGSTIPVTSIGHSKICHPNCSFTLRDILCSPAIIK 629
Query: 59 NLVSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLYPITTSQAISPSTFAAL 118
NL+SVR+F DN SVEFDPFGFSVKD +T + R S G LY + + P+ A L
Sbjct: 630 NLISVRRFVIDNWCSVEFDPFGFSVKDLRTRTVIARFNSSGPLYSLHHALPPPPAATALL 689
Query: 119 APS----LWHARLGHPGAPVVDSLRKNKFIECNRASGSHICHSCSLGKHIKLPFVSSNSC 174
A S LWH RLGH G ++ L + R + +CH+C LG+H++LPF SS S
Sbjct: 690 ANSSTDLLWHRRLGHLGHDALNRLAA--VVPMTRGDLTGVCHACQLGRHVRLPFASSTSR 747
Query: 175 TIMPFDIIHSDIWTSPVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIR 234
F++ H D+WTSPV+S+SG +YY++ +DD S+++WTFPL KS F+ F +++
Sbjct: 748 ASTNFELFHCDLWTSPVVSASGFKYYLVILDDCSHYVWTFPLRFKSDTFTTLSHFFAYVK 807
Query: 235 TQFEREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIR 294
TQF +++IQCDNG+EFDN F NGV R+SCPHTS QNGKAER +R++NNI+R
Sbjct: 808 TQFATNIRSIQCDNGREFDNSAARTFFLTNGVHLRMSCPHTSPQNGKAERILRSLNNIVR 867
Query: 295 TLLVHASLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCYP 354
++L A LP SFW AL AT+LIN P K L +P LY PSYSHLRVFGC CYP
Sbjct: 868 SMLFQAKLPGSFWVEALHTATHLINRHPTKTLDRHTPHFALYGTHPSYSHLRVFGCKCYP 927
Query: 355 LFPSTTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLHN- 413
+TT +KL RST CVFLGYP H+ Y+C+D S ++IISRHV+FDE FPF +L N
Sbjct: 928 NLSATTPHKLAPRSTMCVFLGYPLYHKGYRCFDPLSNRVIISRHVVFDEHSFPFTELTNG 987
Query: 414 -PQPYTYGFMDDGPSPY----------------------VIHHLTSQPSL---------G 441
F++D +P ++H L P G
Sbjct: 988 VSNATDLDFLEDFTAPAQAPIGATRRPAVAPTTQTASSPMVHGLERPPPCSPTRPVSTPG 1047
Query: 442 QPAQHDLPNTQPTTQP-------TTPEEQHAHSPPSSSPNPSPSTTAT----PSPPY--- 487
P+ D P+ P T+P A PS+S P+ ST T P+PP
Sbjct: 1048 GPSSPDSRLGPPSPTPALIGPASTSPGPPSAGPAPSASTCPASSTWETVARPPTPPGLPR 1107
Query: 488 -----------------------QPTPISVPK--PV-------TRSQHGIFKPKRQLNLN 515
PTP+S + PV TR++ G KP +LNL+
Sbjct: 1108 LDGPHLPPAPHVPRRLRSVRATGAPTPLSGLEISPVVNDHVMTTRAKSGHHKPVHRLNLH 1167
Query: 516 TSVPRSPLPRNPVSALRDPNWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKE 575
+ P S +P+ +AL DP W+ AM++E+NAL+ N+TW+LVPRP VNV+ WIF HK
Sbjct: 1168 AA-PLSLVPKTYRAALADPLWRAAMEEEYNALLANRTWDLVPRPAGVNVVTGKWIFKHKF 1226
Query: 576 KSDGVFERYKARLVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKN 635
+DG +RYKAR V G TQ+ GVD ETFSPVVKPAT+RTVLSLA+S+ W +HQLDVKN
Sbjct: 1227 HADGSLDRYKARWVLRGFTQRPGVDFDETFSPVVKPATVRTVLSLAVSRDWPVHQLDVKN 1286
Query: 636 AFLHGELKETVYMHQPMGFRDPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIGFSH 695
AFLHG L+ETVY QP GF D P+ VC L KSLYGLKQAPRAWY RF ++ +IGF
Sbjct: 1287 AFLHGTLQETVYCTQPPGFVDSAKPDMVCCLNKSLYGLKQAPRAWYSRFTTFLQSIGFVE 1346
Query: 696 STSDHSLFIYRKGTAMAYILLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFLG 755
+ SD SLFI +G Y+LLYVDDI+LTASS L IS L EF+MKDLG+LH+FLG
Sbjct: 1347 AKSDTSLFILHRGNDTVYLLLYVDDIVLTASSRTLLHWTISALQGEFSMKDLGALHHFLG 1406
Query: 756 IAVTHHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHYRSL 815
++VT ++ GL LSQR+Y +I+ERAGMA CK +TPVDT KLS++ P ADP+ +RSL
Sbjct: 1407 VSVTRNSAGLVLSQRQYCIDILERAGMADCKPCNTPVDTTAKLSSSDGPPVADPTDFRSL 1466
Query: 816 AGALQYLTFTRPDIAYAVQQVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTST 875
AGALQYLTFTRPDI+YAVQQVCL MHDPRE H+ ALKRI+ YI+G++D GLH+ SS
Sbjct: 1467 AGALQYLTFTRPDISYAVQQVCLHMHDPREPHLAALKRILHYIRGSVDLGLHIQRSSACD 1526
Query: 876 LISYTDADWGGCPDTRRSTSGYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVSES 935
L Y+DADW GCPDTRRSTSGY VFLGDNL+SWS+KRQ T+SRSSAEAEYR VAN V+E
Sbjct: 1527 LAVYSDADWAGCPDTRRSTSGYAVFLGDNLVSWSSKRQHTVSRSSAEAEYRAVANAVAEV 1586
Query: 936 CWLRNLLLELHCPIRKASLVYCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARGEVR 995
WLR LL ELH P +A+LVYCDNVSA+YLS NPVQHQRTKH+E+D+H VRE+VA G VR
Sbjct: 1587 TWLRQLLQELHSPPSRATLVYCDNVSAVYLSSNPVQHQRTKHVEIDLHFVRERVAVGAVR 1646
Query: 996 VLHVPSRYQIADIFTKGLPLVLFEDFRNNLSV 1027
VLHVP+ Q ADIFTKGLP +F +FR++L+V
Sbjct: 1647 VLHVPTTSQYADIFTKGLPTPVFTEFRSSLNV 1678
>UniRef100_Q8S805 Putative copia-type polyprotein [Oryza sativa]
Length = 1803
Score = 1050 bits (2716), Expect = 0.0
Identities = 531/1032 (51%), Positives = 698/1032 (67%), Gaps = 16/1032 (1%)
Query: 1 MTSEQGNLSSYFNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNL 60
M+S G L+ L + I VGNG +P+ + T++ + L L NVL SP LIKNL
Sbjct: 378 MSSTPGILAHPRPLPFSSCITVGNGAKLPVTHTASTHIPTSSTDLHLHNVLVSPPLIKNL 437
Query: 61 VSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLYPITTSQAISPSTFAALAP 120
+SV+K T DN+VS+EFDP GFS+KD QT + +RC+S GDLYP+ + S ++ +
Sbjct: 438 ISVKKLTRDNNVSIEFDPTGFSIKDLQTQVVKLRCDSPGDLYPLRLPSPHALSATSSPSV 497
Query: 121 SLWHARLGHPGAPVVDSLRKNKFIECNRASGSHICHSCSLGKHIKLPFVSSNSCTIMPFD 180
WH RLGHPG+ + + + +CN+ S H C +C +G +++LPF SS+S T+ PF
Sbjct: 498 EHWHLRLGHPGSASLSKVLGSFDFQCNK-SAPHHCSACHVGTNVRLPFHSSSSQTLFPFQ 556
Query: 181 IIHSDIWTSPVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQFERE 240
++H+D+WTSP+ S+SG++YYV+F+DD+++++WTFP+ KS+VF SF + TQF
Sbjct: 557 LVHTDVWTSPIYSNSGYKYYVVFLDDFTHYIWTFPVRNKSEVFHTVRSFFAYAHTQFGLP 616
Query: 241 VKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLLVHA 300
V +Q DNGKE+D+ +G RLSCP++S QNGKAER +RTIN+ +RT+LVH+
Sbjct: 617 VLALQTDNGKEYDSYALRSLLSLHGAVLRLSCPYSSQQNGKAERILRTINDCVRTMLVHS 676
Query: 301 SLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCYPLFPSTT 360
+ P SFW ALQ A +LIN P + P ++L P+Y HLRVFGCLCYP +T
Sbjct: 677 AAPLSFWAEALQTAMHLINRRPCRATGSLKPYQLLLGAPPTYDHLRVFGCLCYPNTIATA 736
Query: 361 INKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLHNPQPYTYG 420
+KL RS CVF+GYP++HR Y+CYD+ SR++ SRHV F E FPF +P+P
Sbjct: 737 PHKLSPRSLACVFIGYPADHRGYRCYDMVSRRVFTSRHVTFVEDVFPFRDAPSPRPSAPP 796
Query: 421 FMDDGPSPYVIHHLTSQ---PSLGQPAQHDL--PNTQPTTQPTTPEEQHAHSPPSSSPNP 475
D G V+ +Q +G HD P + ++ P++ H +PP P+P
Sbjct: 797 PPDHGDDTIVLLPAPAQHVVTPVGTAPAHDAASPPSPASSTPSSAAPAHDVAPP---PSP 853
Query: 476 SPSTTATPSPPYQPTPISVPKPVTRSQHGIFKPKRQLNLNTSVPRSPLPRNPVSALRDPN 535
S+ A+ SPP TR++ GI KP + + + SP P + ALRDPN
Sbjct: 854 ETSSPASASPPRHAM-------TTRARAGISKPNPRYAMTATSTLSPTPSSVRVALRDPN 906
Query: 536 WKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKEKSDGVFERYKARLVGDGKTQ 595
W+ AM EF+AL+ N+TW LVPRPP +I W+F K +DG ++YKAR V G Q
Sbjct: 907 WRAAMQAEFDALLANRTWTLVPRPPGARIITGKWVFKTKLHADGSLDKYKARWVVRGFNQ 966
Query: 596 QVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKNAFLHGELKETVYMHQPMGFR 655
+ GVD GETFSPVVKPATIRTVL+L SK W HQLDV NAFLHG L+E V QP GF
Sbjct: 967 RPGVDFGETFSPVVKPATIRTVLTLISSKQWPAHQLDVSNAFLHGHLQERVLCQQPTGFE 1026
Query: 656 DPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIGFSHSTSDHSLFIYRKGTAMAYIL 715
D P VCLL +SLYGL+QAPRAW+KRFAD+ +++GF S +D SLF+ R+G+ AY+L
Sbjct: 1027 DAARPADVCLLSRSLYGLRQAPRAWFKRFADHATSLGFVQSRADPSLFVLRRGSDTAYLL 1086
Query: 716 LYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFLGIAVTHHTGGLFLSQRKYAAE 775
LYVDD+IL+ASS +L II L EF +KD+G L YFLGI V G LSQ KYA +
Sbjct: 1087 LYVDDMILSASSSSLLQRIIDRLQAEFKVKDMGPLKYFLGIEVQRTADGFVLSQSKYATD 1146
Query: 776 IIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHYRSLAGALQYLTFTRPDIAYAVQQ 835
++ERAGMA CK+ +TP D KPKLS++ D S YRS+AGALQYLT TRPDIAYAVQQ
Sbjct: 1147 VLERAGMANCKAVATPADAKPKLSSDEGPLFQDSSWYRSIAGALQYLTLTRPDIAYAVQQ 1206
Query: 836 VCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTSTLISYTDADWGGCPDTRRSTS 895
VCL MH PRE H+ LKRI+RYI+GT GLHL S++ TL +++DADW GCPDTRRSTS
Sbjct: 1207 VCLHMHAPREAHVTLLKRILRYIKGTAAFGLHLRASTSPTLTAFSDADWAGCPDTRRSTS 1266
Query: 896 GYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVSESCWLRNLLLELHCPIRKASLV 955
G+C+FLGD+LISWS+KRQ T+SRSSAEAEYRGVAN V+E WLR LL ELHC + +A++
Sbjct: 1267 GFCIFLGDSLISWSSKRQTTVSRSSAEAEYRGVANAVAECTWLRQLLGELHCRVPQATIA 1326
Query: 956 YCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARGEVRVLHVPSRYQIADIFTKGLPL 1015
YCDN+S++Y+S NPV H+RTKHIE+DIH VREKVA GE+RVL +PS +Q AD+FTKGLP
Sbjct: 1327 YCDNISSVYMSKNPVHHKRTKHIELDIHFVREKVALGELRVLPIPSAHQFADVFTKGLPS 1386
Query: 1016 VLFEDFRNNLSV 1027
+F +FR +L V
Sbjct: 1387 SMFNEFRASLCV 1398
>UniRef100_Q7XCM1 Putative gag-pol polyprotein [Oryza sativa]
Length = 1417
Score = 1012 bits (2616), Expect = 0.0
Identities = 520/1043 (49%), Positives = 684/1043 (64%), Gaps = 48/1043 (4%)
Query: 1 MTSEQGNLSSYFNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNL 60
M+S G L+ L + I VGNG +P+ + T++ + L L NVL SP LIKNL
Sbjct: 378 MSSTPGILAHPRPLPFSSCITVGNGAKLPVTHTASTHIPTSSTDLHLHNVLVSPPLIKNL 437
Query: 61 VSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLYPITTSQAISPSTFAALAP 120
+SV+K T DN+VS+EFDP GFS+KD QT + +RC+S GDLYP+ + S ++ +
Sbjct: 438 ISVKKLTRDNNVSIEFDPTGFSIKDLQTQVVKLRCDSPGDLYPLRLPSPHALSATSSPSV 497
Query: 121 SLWHARLGHPGAPVVDSLRKNKFIECNRASGSHICHSCSLGKHIKLPFVSSNSCTIMPFD 180
WH RLGHPG+ + + + +CN+ S H C +C +G +++LPF SS+S T+ PF
Sbjct: 498 EHWHLRLGHPGSASLSKVLGSFDFQCNK-SAPHHCSACHVGTNVRLPFHSSSSQTLFPFQ 556
Query: 181 IIHSDIWTSPVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQFERE 240
++H+D+WTSP+ S+SG++YYV+F+DD+++++WTFP+ KS+VF SF + TQF
Sbjct: 557 LVHTDVWTSPIYSNSGYKYYVVFLDDFTHYIWTFPVRNKSEVFHTVRSFFAYAHTQFGLP 616
Query: 241 VKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLLVHA 300
V +Q DNGKE+D+ +G RLSCP++S QNGKAER +RTIN+ +RT+LVH+
Sbjct: 617 VLALQTDNGKEYDSYALRSLLSLHGAVLRLSCPYSSQQNGKAERILRTINDYVRTMLVHS 676
Query: 301 SLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCYPLFPSTT 360
+ P SFW ALQ AT+LIN P + +P ++L P+Y HLRVFGCLCYP +T
Sbjct: 677 AAPLSFWAEALQTATHLINRRPCRATGSLTPYQLLLGAPPTYDHLRVFGCLCYPNTIATA 736
Query: 361 INKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLHNPQPYTYG 420
+KL RS CVF+GYP++HR Y+CYD+ SR++ SRHV F E FPF +P+P
Sbjct: 737 PHKLSPRSLACVFIGYPADHRGYRCYDMVSRRVFTSRHVTFVEDVFPFRDAPSPRPSAPP 796
Query: 421 FMDDGPSPYVIHHLTSQ---PSLGQPAQHDL--PNTQPTTQPTTPEEQHAHSPPSSSPNP 475
D G V+ +Q +G HD P + ++ P++ H +PP P+P
Sbjct: 797 PPDHGDDTIVLLPAPAQHVVTPVGTAPAHDAASPPSPASSTPSSAAPAHDVAPP---PSP 853
Query: 476 SPSTTATPSPPYQPTPISVPKPVTRSQHGIFKPKRQLNLNTSVPRSPLPRNPVSALRDPN 535
S+ A+ SPP TR++ GI KP + + + SP P + +ALRDPN
Sbjct: 854 ETSSPASASPPRHAM-------TTRARAGISKPNPRYAMTATSTLSPTPSSVRAALRDPN 906
Query: 536 WKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKEKSDGVFERYKARLVGDGKTQ 595
W+ AM EF+AL+ N+TW LVPRPP +I W+F K +DG ++YKAR V G Q
Sbjct: 907 WRAAMQAEFDALLANRTWTLVPRPPGARIITGKWVFKTKLHADGSLDKYKARWVVRGFNQ 966
Query: 596 QVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKNAFLHGELKETVYMHQPMGFR 655
+ GVD GETFSPVVKPATIRTVL+L SK W HQLDV NAFLHG L+E V QP GF
Sbjct: 967 RPGVDFGETFSPVVKPATIRTVLTLISSKQWPAHQLDVSNAFLHGHLQERVLCQQPTGFE 1026
Query: 656 DPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIGFSHSTSDHSLFIYRKGTAMAYIL 715
D P VCLL +SLYGL+QAPRAW+KRFAD+ +++GF S +D SLF+ R+G+ AY+L
Sbjct: 1027 DAARPADVCLLSRSLYGLRQAPRAWFKRFADHATSLGFVQSRADPSLFVLRRGSDTAYLL 1086
Query: 716 LYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFLGIAVTHHTGGLFLSQRKYAAE 775
LYVDD+IL+ASS +L II L EF +KD+G L YFLGI V G LSQ KYA +
Sbjct: 1087 LYVDDMILSASSSSLLQRIIDRLQAEFKVKDMGPLKYFLGIEVQRTADGFVLSQSKYATD 1146
Query: 776 IIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHYRSLAGALQYLTFTRPDIAYAVQQ 835
D S YRS+AGALQYLT TRPDIAYAVQQ
Sbjct: 1147 --------------------------------DSSWYRSIAGALQYLTLTRPDIAYAVQQ 1174
Query: 836 VCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTSTLISYTDADWGGCPDTRRSTS 895
VCL MH PRE H+ LKRI+RYI+GT GLHL S++ TL +++DADW GCPDTRRSTS
Sbjct: 1175 VCLHMHAPREAHVTLLKRILRYIKGTAAFGLHLRASTSPTLTAFSDADWAGCPDTRRSTS 1234
Query: 896 GYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVSESCWLRNLLLELHCPIRKASLV 955
G+C+FLGD+LISWS+KRQ T+SRSSAEAEYRGVAN V+E WLR LL ELHC + +A++
Sbjct: 1235 GFCIFLGDSLISWSSKRQTTVSRSSAEAEYRGVANAVAECTWLRQLLGELHCRVPQATIA 1294
Query: 956 YCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARGEVRVLHVPSRYQIADIFTKGLPL 1015
YCDN+S++Y+S NPV H+RTKHIE+DIH VREKVA GE+RVL +PS +Q AD+FTKGLP
Sbjct: 1295 YCDNISSVYMSKNPVHHKRTKHIELDIHFVREKVALGELRVLPIPSAHQFADVFTKGLPS 1354
Query: 1016 VLFEDFRNNLSVRQPPVSTAGVC 1038
+F +FR +L V +TAG C
Sbjct: 1355 SMFNEFRASLCVGDTTAATAGGC 1377
>UniRef100_Q7X7N0 OSJNBa0035I04.5 protein [Oryza sativa]
Length = 1246
Score = 832 bits (2150), Expect = 0.0
Identities = 445/838 (53%), Positives = 552/838 (65%), Gaps = 28/838 (3%)
Query: 217 SKKSQVFSIFLSFRTFIRTQFEREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTS 276
S KS V + F + TQF +K +Q DNG EF N + F G RLSCP+TS
Sbjct: 415 SLKSDVHRHIVEFVEYAHTQFGHRLKCLQADNGTEFVNHNTTSFLAGRGSLLRLSCPYTS 474
Query: 277 SQNGKAERKIRTINNIIRTLLVHASLPPSFWHHALQMATYLINILPNKQLAYQSPLKILY 336
QNGKAER IRT+NN IRTLL+ AS+PPS+W L ATYL+N P+ + P ++L+
Sbjct: 475 PQNGKAERMIRTLNNSIRTLLLQASMPPSYWAEGLATATYLLNRRPSSSVNNSIPFQLLH 534
Query: 337 QKEPSYSHLRVFGCLCYPLFPSTTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIIS 396
+K P+YS LRVFGCLCYP +T +KL S CVFLGYPS+H+ Y C ++S+R+IIIS
Sbjct: 535 RKIPNYSMLRVFGCLCYPNLSATAAHKLAPYSAACVFLGYPSSHKGYCCLNISTRRIIIS 594
Query: 397 RHVIFDETQFPFAKLHNPQPYTYGFMDDGPSPYVIHHLTSQPSLGQPAQHDLPNTQ---- 452
HVIFDETQFPF+ + D P+P VI PSL Q LP+
Sbjct: 595 CHVIFDETQFPFSGDPVDASSLDFLLQDAPAPSVI-----APSLAGVEQPHLPHAPFPVN 649
Query: 453 -----PTTQPTTPEEQHAHSPPSSSPNPSPSTTATPSPPYQPTPISVPKPVTR--SQHGI 505
PT P+T +E + ++ + + + VT + H
Sbjct: 650 VEQRLPTGAPSTKDEHLPYYVQPAAHCGQDDGKFHTAGCHVSIFLKQGTIVTSYPAVHVF 709
Query: 506 FKPKRQLNLNTSVPRSPLPRNPVSALRDPNWKM--------AMDDEFNALIKNKTWELVP 557
F R ++P PLP + P+ + AM +EF ALI N TW LVP
Sbjct: 710 FTTSRH---GGAIP-VPLPTTTGADDSAPSHRCGCTHTSPAAMAEEFKALIDNGTWRLVP 765
Query: 558 RPPDVNVIRSMWIFTHKEKSDGVFERYKARLVGDGKTQQVGVDCGETFSPVVKPATIRTV 617
RPP NV+ WIF HK SDG R+KAR V G +QQ G+D ETFSPVVKP+TIR V
Sbjct: 766 RPPGANVVTGKWIFKHKFHSDGSLARHKARWVVHGYSQQHGIDYDETFSPVVKPSTIRVV 825
Query: 618 LSLALSKAWSIHQLDVKNAFLHGELKETVYMHQPMGFRDPNLPNHVCLLKKSLYGLKQAP 677
LS+A S++W IHQLDVKNAFLHG L ETVY Q GF DP P+ VCLL++SLYGLKQAP
Sbjct: 826 LSIATSRSWPIHQLDVKNAFLHGTLDETVYCQQSSGFIDPAAPDAVCLLQRSLYGLKQAP 885
Query: 678 RAWYKRFADYVSTIGFSHSTSDHSLFIYRKGTAMAYILLYVDDIILTASSDALRVTIISL 737
RAWY+RFA Y+ +GF+ S SD SLFI R G +AY+LLYVDDIIL ASS L I +
Sbjct: 886 RAWYQRFATYIRQLGFTPSASDTSLFILRDGDRLAYLLLYVDDIILMASSAELLCHITAR 945
Query: 738 LSTEFAMKDLGSLHYFLGIAVTHHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVDTKPK 797
L TEFAM DLG LH+FLGI+V LFLSQR+YA ++++RAGM+ ++TPVD + K
Sbjct: 946 LHTEFAMTDLGDLHFFLGISVRRTPDDLFLSQRQYAVDLLQRAGMSEYHPTATPVDARAK 1005
Query: 798 LSANSSAPHADPSHYRSLAGALQYLTFTRPDIAYAVQQVCLFMHDPREEHMHALKRIVRY 857
LSA+ AP ADP+ YRSLA ALQYLT TRP+IAYAVQQVC FMHDP E H+ +KRI+RY
Sbjct: 1006 LSASEGAPVADPTEYRSLADALQYLTLTRPEIAYAVQQVCRFMHDPHEPHLALVKRIMRY 1065
Query: 858 IQGTLDHGLHLYPSSTSTLISYTDADWGGCPDTRRSTSGYCVFLGDNLISWSAKRQATLS 917
I+G+L GLH+ +L +Y+DADW GCPD+RRSTSGYC+FLGDNL+SWS+KRQ T+S
Sbjct: 1066 IKGSLSAGLHIGTGPVGSLTAYSDADWAGCPDSRRSTSGYCIFLGDNLVSWSSKRQTTVS 1125
Query: 918 RSSAEAEYRGVANVVSESCWLRNLLLELHCPIRKASLVYCDNVSAIYLSGNPVQHQRTKH 977
RSSAEAEYR VA+ ++E C LR LL ELH PI ++V+CDNVSA Y++ NPV H+ TKH
Sbjct: 1126 RSSAEAEYRAVAHAIAECCCLRQLLQELHAPISTTTVVFCDNVSAAYMTANPVHHRCTKH 1185
Query: 978 IEMDIHSVREKVARGEVRVLHVPSRYQIADIFTKGLPLVLFEDFRNNLSVRQPPVSTA 1035
IE+DIH VREKVA G+VRVLHVPS +Q D TKGLP+ LF DFR++L VR PP +TA
Sbjct: 1186 IEIDIHFVREKVALGQVRVLHVPSTHQFPDFMTKGLPVQLFTDFRSSLCVRDPPAATA 1243
>UniRef100_Q9T0C5 Retrotransposon like protein [Arabidopsis thaliana]
Length = 1515
Score = 784 bits (2025), Expect = 0.0
Identities = 436/1104 (39%), Positives = 624/1104 (56%), Gaps = 71/1104 (6%)
Query: 1 MTSEQGNLSSYFNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNL 60
+T+ L + S + +IVGNG +PI L+ L L++VL P + K+L
Sbjct: 334 ITNTTDGLQNSQTYSGDDSVIVGNGDFLPITHIGTIPLNISQGTLPLEDVLVCPGITKSL 393
Query: 61 VSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLYPI--TTSQAISPSTFAAL 118
+SV K T D S FD +KD +T L + LY + Q + +
Sbjct: 394 LSVSKLTDDYPCSFTFDSDSVVIKDKRTQQLLTQGNKHKGLYVLKDVPFQTYYSTRQQSS 453
Query: 119 APSLWHARLGHPGAPVVDSLRKNKFIECNRASGSHICHSCSLGKHIKLPFVSSNSCTIMP 178
+WH RLGHP V+ L K K I N+ S S++C +C +GK +LPFV+S + P
Sbjct: 454 DDEVWHQRLGHPNKEVLQHLIKTKAIVVNKTS-SNMCEACQMGKVCRLPFVASEFVSSRP 512
Query: 179 FDIIHSDIW-TSPVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQF 237
+ IH D+W +PV S+ G +YYV+F+D+YS F W +PL KS FS+F+ F+ + Q+
Sbjct: 513 LERIHCDLWGPAPVTSAQGFQYYVIFIDNYSRFTWFYPLKLKSDFFSVFVLFQQLVENQY 572
Query: 238 EREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLL 297
+ ++ QCD G EF + F G+ +SCPHT QNG AER+ R + + +L+
Sbjct: 573 QHKIAMFQCDGGGEFVSYKFVAHLASCGIKQLISCPHTPQQNGIAERRHRYLTELGLSLM 632
Query: 298 VHASLPPSFWHHALQMATYLINILPNKQLA-YQSPLKILYQKEPSYSHLRVFGCLCYPLF 356
H+ +P W A + +L N+LP+ L+ +SP ++L+ P Y+ LRVFG CYP
Sbjct: 633 FHSKVPHKLWVEAFFTSNFLSNLLPSSTLSDNKSPYEMLHGTPPVYTALRVFGSACYPYL 692
Query: 357 PSTTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLHN--- 413
NK +S CVFLGY + ++ Y+C + K+ I RHV+FDE +FP++ +++
Sbjct: 693 RPYAKNKFDPKSLLCVFLGYNNKYKGYRCLHPPTGKVYICRHVLFDERKFPYSDIYSQFQ 752
Query: 414 ----------------------------------PQPYTYGFMDDGPSPYVIHHLTSQPS 439
P + G +P + T+ P
Sbjct: 753 TISGSPLFTAWQKGFSSTALSRETPSTNVEDIIFPSATVSSSVPTGCAPNIAETATA-PD 811
Query: 440 LGQPAQHDL-----PNTQPTTQPTTPEEQHAHSPPSSSPNPSPSTTA-TPS--------- 484
+ A HD+ P T T+ PT PEE + S+ + + ++A TP
Sbjct: 812 VDVAAAHDMVVPPSPITS-TSLPTQPEESTSDQNHYSTDSETAISSAMTPQSINVSLFED 870
Query: 485 ---PPYQPT-------PISVPKPVTRSQHGIFKPKRQLNLNTSVPRSPLPRNPVSALRDP 534
PP Q P + +TR++ GI KP + L + P P++ AL+D
Sbjct: 871 SDFPPLQSVISSTTAAPETSHPMITRAKSGITKPNPKYALFSVKSNYPEPKSVKEALKDE 930
Query: 535 NWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKEKSDGVFERYKARLVGDGKT 594
W AM +E + + TW+LVP ++ W+F K SDG +R KARLV G
Sbjct: 931 GWTNAMGEEMGTMHETDTWDLVPPEMVDRLLGCKWVFKTKLNSDGSLDRLKARLVARGYE 990
Query: 595 QQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKNAFLHGELKETVYMHQPMGF 654
Q+ GVD ET+SPVV+ AT+R++L +A WS+ QLDVKNAFLH ELKETV+M QP GF
Sbjct: 991 QEEGVDYVETYSPVVRSATVRSILHVATINKWSLKQLDVKNAFLHDELKETVFMTQPPGF 1050
Query: 655 RDPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIGFSHSTSDHSLFIYRKGTAMAYI 714
DP+ P++VC LKK++Y LKQAPRAW+ +F+ Y+ GF S SD SLF+Y KG + ++
Sbjct: 1051 EDPSRPDYVCKLKKAIYDLKQAPRAWFDKFSSYLLKYGFICSFSDPSLFVYLKGRDVMFL 1110
Query: 715 LLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFLGIAVTHHTGGLFLSQRKYAA 774
LLYVDD+ILT ++D L ++++LSTEF MKD+G+LHYFLGI +H GLFLSQ KY +
Sbjct: 1111 LLYVDDMILTGNNDVLLQQLLNILSTEFRMKDMGALHYFLGIQAHYHNDGLFLSQEKYTS 1170
Query: 775 EIIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHYRSLAGALQYLTFTRPDIAYAVQ 834
+++ AGM+ C S TP+ L ++ P +P+++R LAG LQYLT TRPDI +AV
Sbjct: 1171 DLLVNAGMSDCSSMPTPLQL--DLLQGNNKPFPEPTYFRRLAGKLQYLTLTRPDIQFAVN 1228
Query: 835 QVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTSTLISYTDADWGGCPDTRRST 894
VC MH P H LKRI+ Y++GT+ G++L ++ S L Y+D+DW GC DTRRST
Sbjct: 1229 FVCQKMHAPTMSDFHLLKRILHYLKGTMTMGINLSSNTDSVLRCYSDSDWAGCKDTRRST 1288
Query: 895 SGYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVSESCWLRNLLLELHCPIRKASL 954
G+C FLG N+ISWSAKR T+S+SS EAEYR ++ SE W+ LL E+ P ++
Sbjct: 1289 GGFCTFLGYNIISWSAKRHPTVSKSSTEAEYRTLSFAASEVSWIGFLLQEIGLPQQQIPE 1348
Query: 955 VYCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARGEVRVLHVPSRYQIADIFTKGLP 1014
+YCDN+SA+YLS NP H R+KH ++D + VRE+VA G + V H+P+ Q+ADIFTK LP
Sbjct: 1349 MYCDNLSAVYLSANPALHSRSKHFQVDYYYVRERVALGALTVKHIPASQQLADIFTKSLP 1408
Query: 1015 LVLFEDFRNNLSVRQPPVSTAGVC 1038
F D R L V PP ++ C
Sbjct: 1409 QAPFCDLRFKLGVVLPPDTSLRGC 1432
>UniRef100_Q9SA17 F28K20.17 protein [Arabidopsis thaliana]
Length = 1415
Score = 783 bits (2021), Expect = 0.0
Identities = 426/1044 (40%), Positives = 593/1044 (55%), Gaps = 23/1044 (2%)
Query: 1 MTSEQGNLSSYFNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNL 60
+TS L S + ++VG+G +PI T + N + L VL P + K+L
Sbjct: 332 VTSSTNGLQSATEYEGDDAVLVGDGTYLPITHTGSTTIKSSNGKIPLNEVLVVPNIQKSL 391
Query: 61 VSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLYPITTSQ--AISPSTFAAL 118
+SV K D V FD + D QT + R LY + + A+ + A
Sbjct: 392 LSVSKLCDDYPCGVYFDANKVCIIDLQTQKVVTTGPRRNGLYVLENQEFVALYSNRQCAA 451
Query: 119 APSLWHARLGHPGAPVVDSLRKNKFIECNRASGSHICHSCSLGKHIKLPFVSSNSCTIMP 178
+WH RLGH + + L+ +K I+ N++ S +C C +GK +LPF+ S+S + P
Sbjct: 452 TEEVWHHRLGHANSKALQHLQNSKAIQINKSRTSPVCEPCQMGKSSRLPFLISDSRVLHP 511
Query: 179 FDIIHSDIW-TSPVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQF 237
D IH D+W SPV+S+ G +YY +FVDDYS + W +PL KS+ S+F+SF+ + Q
Sbjct: 512 LDRIHCDLWGPSPVVSNQGLKYYAIFVDDYSRYSWFYPLHNKSEFLSVFISFQKLVENQL 571
Query: 238 EREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLL 297
++K Q D G EF + E+G+ R+SCP+T QNG AERK R + + ++L
Sbjct: 572 NTKIKVFQSDGGGEFVSNKLKTHLSEHGIHHRISCPYTPQQNGLAERKHRHLVELGLSML 631
Query: 298 VHASLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCYPLFP 357
H+ P FW + A Y+IN LP+ L SP + L+ ++P YS LRVFG CYP
Sbjct: 632 FHSHTPQKFWVESFFTANYIINRLPSSVLKNLSPYEALFGEKPDYSSLRVFGSACYPCLR 691
Query: 358 STTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLHNP--- 414
NK RS CVFLGY S ++ Y+C+ + K+ ISR+VIF+E++ PF + +
Sbjct: 692 PLAQNKFDPRSLQCVFLGYNSQYKGYRCFYPPTGKVYISRNVIFNESELPFKEKYQSLVP 751
Query: 415 -------QPYTYGFMDDGPSPYVIHHLTSQPSLGQPAQHDLPNTQPTTQPTTPEEQHAHS 467
Q + + + + P L S+P + +Q T Q T PE +
Sbjct: 752 QYSTPLLQAWQHNKISEISVPAAPVQLFSKPI----DLNTYAGSQVTEQLTDPEPTSNNE 807
Query: 468 PPSSSPNPSPSTTATPSPPYQPTPISVPKPVTRSQHGIFKPKRQLNLNTSVPRSPLPRNP 527
NP A Q I+ TRS+ GI KP + L TS + P+
Sbjct: 808 GSDEEVNPVAEEIAAN----QEQVINSHAMTTRSKAGIQKPNTRYALITSRMNTAEPKTL 863
Query: 528 VSALRDPNWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKEKSDGVFERYKAR 587
SA++ P W A+ +E N + TW LVP D+N++ S W+F K DG ++ KAR
Sbjct: 864 ASAMKHPGWNEAVHEEINRVHMLHTWSLVPPTDDMNILSSKWVFKTKLHPDGSIDKLKAR 923
Query: 588 LVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKNAFLHGELKETVY 647
LV G Q+ GVD ETFSPVV+ ATIR VL ++ SK W I QLDV NAFLHGEL+E V+
Sbjct: 924 LVAKGFDQEEGVDYLETFSPVVRTATIRLVLDVSTSKGWPIKQLDVSNAFLHGELQEPVF 983
Query: 648 MHQPMGFRDPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIGFSHSTSDHSLFIYRK 707
M+QP GF DP P HVC L K++YGLKQAPRAW+ F++++ GF S SD SLF+ +
Sbjct: 984 MYQPSGFIDPQKPTHVCRLTKAIYGLKQAPRAWFDTFSNFLLDYGFVCSKSDPSLFVCHQ 1043
Query: 708 GTAMAYILLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFLGIAVTHHTGGLFL 767
+ Y+LLYVDDI+LT S +L ++ L F+MKDLG YFLGI + + GLFL
Sbjct: 1044 DGKILYLLLYVDDILLTGSDQSLLEDLLQALKNRFSMKDLGPPRYFLGIQIEDYANGLFL 1103
Query: 768 SQRKYAAEIIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHYRSLAGALQYLTFTRP 827
Q YA +I+++AGM+ C TP+ +L +S A+P+++RSLAG LQYLT TRP
Sbjct: 1104 HQTAYATDILQQAGMSDCNPMPTPL--PQQLDNLNSELFAEPTYFRSLAGKLQYLTITRP 1161
Query: 828 DIAYAVQQVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTSTLISYTDADWGGC 887
DI +AV +C MH P LKRI+RYI+GT+ GL + +ST TL +Y+D+D GC
Sbjct: 1162 DIQFAVNFICQRMHSPTTSDFGLLKRILRYIKGTIGMGLPIKRNSTLTLSAYSDSDHAGC 1221
Query: 888 PDTRRSTSGYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVSESCWLRNLLLELHC 947
+TRRST+G+C+ LG NLISWSAKRQ T+S SS EAEYR + E W+ LL +L
Sbjct: 1222 KNTRRSTTGFCILLGSNLISWSAKRQPTVSNSSTEAEYRALTYAAREITWISFLLRDLGI 1281
Query: 948 PIRKASLVYCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARGEVRVLHVPSRYQIAD 1007
P + VYCDN+SA+YLS NP H R+KH + D H +RE+VA G + H+ + +Q+AD
Sbjct: 1282 PQYLPTQVYCDNLSAVYLSANPALHNRSKHFDTDYHYIREQVALGLIETQHISATFQLAD 1341
Query: 1008 IFTKGLPLVLFEDFRNNLSVRQPP 1031
+FTK LP F D R+ L V P
Sbjct: 1342 VFTKSLPRRAFVDLRSKLGVSGSP 1365
>UniRef100_Q9SSB1 T18A20.5 protein [Arabidopsis thaliana]
Length = 1522
Score = 768 bits (1983), Expect = 0.0
Identities = 428/1090 (39%), Positives = 608/1090 (55%), Gaps = 76/1090 (6%)
Query: 20 IIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNLVSVRKFTTDNSVSVEFDPF 79
I+V +G+ +PI +++ + + LK VL P ++K+L+SV K T+D SVEFD
Sbjct: 355 IMVADGNFLPITHTGSGSIASSSGKIPLKEVLVCPDIVKSLLSVSKLTSDYPCSVEFDAD 414
Query: 80 GFSVKDFQTGMRLMRCESRGDLYPITTS--QAISPSTFAALAPSLWHARLGHPGAPVVDS 137
+ D T L+ +R LY + Q + + + + +WH RLGH A V+
Sbjct: 415 SVRINDKATKKLLVMGRNRDGLYSLEEPKLQVLYSTRQNSASSEVWHRRLGHANAEVLHQ 474
Query: 138 LRKNKFIECNRASGSHICHSCSLGKHIKLPFVSSNSCTIMPFDIIHSDIW-TSPVLSSSG 196
L +K I +C +C LGK +LPF+ S P + IH D+W SP S G
Sbjct: 475 LASSKSIIIINKVVKTVCEACHLGKSTRLPFMLSTFNASRPLERIHCDLWGPSPTSSVQG 534
Query: 197 HRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQFEREVKNIQCDNGKEFDNRH 256
RYYV+F+D YS F W +PL KS FS F+ F+ + Q ++K QCD G EF +
Sbjct: 535 FRYYVVFIDHYSRFTWFYPLKLKSDFFSTFVMFQKLVENQLGHKIKIFQCDGGGEFISSQ 594
Query: 257 FWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLLVHASLPPSFWHHALQMATY 316
F + +++G+ +SCP+T QNG AERK R I + +++ + LP +W + A +
Sbjct: 595 FLKHLQDHGIQQNMSCPYTPQQNGMAERKHRHIVELGLSMIFQSKLPLKYWLESFFTANF 654
Query: 317 LINILPNKQL-AYQSPLKILYQKEPSYSHLRVFGCLCYPLFPSTTINKLQARSTPCVFLG 375
+IN+LP L +SP + LY K P YS LRVFGC CYP K RS CVFLG
Sbjct: 655 VINLLPTSSLDNNESPYQKLYGKAPEYSALRVFGCACYPTLRDYASTKFDPRSLKCVFLG 714
Query: 376 YPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLH---NPQPYTYGFMDDGPSPYVIH 432
Y ++ Y+C + +I ISRHV+FDE PF ++ +PQ T S H
Sbjct: 715 YNEKYKGYRCLYPPTGRIYISRHVVFDENTHPFESIYSHLHPQDKTPLLEAWFKS---FH 771
Query: 433 HLT-SQP--------SLGQPAQHDL-------------PNTQPTTQPT---------TPE 461
H+T +QP S+ QP DL PN T +PE
Sbjct: 772 HVTPTQPDQSRYPVSSIPQPETTDLSAAPASVAAETAGPNASDDTSQDNETISVVSGSPE 831
Query: 462 E----------QHAHSPPSSSPNPSPSTTATPSPPYQPTPISVPKP-------------V 498
HSP + S +PSP+ ++ S P Q +PI + V
Sbjct: 832 RTTGLDSASIGDSYHSPTADSSHPSPARSSPASSP-QGSPIQMAPAQQVQAPVTNEHAMV 890
Query: 499 TRSQHGIFKPKRQLNLNTSVPRSPLPRNPVSALRDPNWKMAMDDEFNALIKNKTWELVPR 558
TR + GI KP ++ L T P P+ AL+ P W AM +E + +TW LVP
Sbjct: 891 TRGKEGISKPNKRYVLLTHKVSIPEPKTVTEALKHPGWNNAMQEEMGNCKETETWTLVPY 950
Query: 559 PPDVNVIRSMWIFTHKEKSDGVFERYKARLVGDGKTQQVGVDCGETFSPVVKPATIRTVL 618
P++NV+ SMW+F K +DG ++ KARLV G Q+ G+D ET+SPVV+ T+R +L
Sbjct: 951 SPNMNVLGSMWVFRTKLHADGSLDKLKARLVAKGFKQEEGIDYLETYSPVVRTPTVRLIL 1010
Query: 619 SLALSKAWSIHQLDVKNAFLHGELKETVYMHQPMGFRDPNLPNHVCLLKKSLYGLKQAPR 678
+A W + Q+DVKNAFLHG+L ETVYM QP GF D + P+HVCLL KSLYGLKQ+PR
Sbjct: 1011 HVATVLKWELKQMDVKNAFLHGDLTETVYMRQPAGFVDKSKPDHVCLLHKSLYGLKQSPR 1070
Query: 679 AWYKRFADYVSTIGFSHSTSDHSLFIYRKGTAMAYILLYVDDIILTASSDALRVTIISLL 738
AW+ RF++++ GF S D SLF+Y + +LLYVDD+++T ++ +++ L
Sbjct: 1071 AWFDRFSNFLLEFGFICSLFDPSLFVYSSNNDVILLLLYVDDMVITGNNSQSLTHLLAAL 1130
Query: 739 STEFAMKDLGSLHYFLGIAVTHHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVDTKPKL 798
+ EF MKD+G +HYFLGI + + GGLF+SQ+KYA +++ A MA C TP+ +
Sbjct: 1131 NKEFRMKDMGQVHYFLGIQIQTYDGGLFMSQQKYAEDLLITASMANCSPMPTPLPLQLDR 1190
Query: 799 SANSSAPHADPSHYRSLAGALQYLTFTRPDIAYAVQQVCLFMHDPREEHMHALKRIVRYI 858
+N +DP+++RSLAG LQYLT TRPDI +AV VC MH P + LKRI+RYI
Sbjct: 1191 VSNQDEVFSDPTYFRSLAGKLQYLTLTRPDIQFAVNFVCQKMHQPSVSDFNLLKRILRYI 1250
Query: 859 QGTLDHGLHLYPSSTSTLIS----------YTDADWGGCPDTRRSTSGYCVFLGDNLISW 908
+GT+ G+ Y S++S+++S Y+D+D+ C +TRRS GYC F+G N+ISW
Sbjct: 1251 KGTVSMGIQ-YNSNSSSVVSAYESDYDLSAYSDSDYANCKETRRSVGGYCTFMGQNIISW 1309
Query: 909 SAKRQATLSRSSAEAEYRGVANVVSESCWLRNLLLELHCPIRKASLVYCDNVSAIYLSGN 968
S+K+Q T+SRSS EAEYR ++ SE W+ ++L E+ + ++CDN+SA+YL+ N
Sbjct: 1310 SSKKQPTVSRSSTEAEYRSLSETASEIKWMSSILREIGVSLPDTPELFCDNLSAVYLTAN 1369
Query: 969 PVQHQRTKHIEMDIHSVREKVARGEVRVLHVPSRYQIADIFTKGLPLVLFEDFRNNLSVR 1028
P H RTKH ++D H +RE+VA + V H+P Q+ADIFTK LP F R L V
Sbjct: 1370 PAFHARTKHFDVDHHYIRERVALKTLVVKHIPGHLQLADIFTKSLPFEAFTRLRFKLGVD 1429
Query: 1029 QPPVSTAGVC 1038
PP + C
Sbjct: 1430 FPPTPSLRGC 1439
>UniRef100_Q94KV0 Polyprotein [Arabidopsis thaliana]
Length = 1453
Score = 754 bits (1947), Expect = 0.0
Identities = 429/1064 (40%), Positives = 593/1064 (55%), Gaps = 34/1064 (3%)
Query: 1 MTSEQGNLSSYFNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNL 60
+TS NL + + + ++VG+G +PI T +S + L L VL P + K+L
Sbjct: 335 VTSSTNNLQAASPYNGSDTVLVGDGAYLPITHVGSTTISSDSGTLPLNEVLVCPDIQKSL 394
Query: 61 VSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLYPITTSQAIS--PSTFAAL 118
+SV K D V FD + D T + + LY + + ++ + A
Sbjct: 395 LSVSKLCDDYPCGVYFDANKVCIIDINTQKVVSKGPRSNGLYVLENQEFVAFYSNRQCAA 454
Query: 119 APSLWHARLGHPGAPVVDSLRKNKFIECNRASGSHICHSCSLGKHIKLPFVSSNSCTIMP 178
+ +WH RLGH + ++ L+ +K I N++ S +C C +GK KL F SSNS +
Sbjct: 455 SEEIWHHRLGHSNSRILQQLKSSKEISFNKSRMSPVCEPCQMGKSSKLQFFSSNSRELDL 514
Query: 179 FDIIHSDIW-TSPVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQF 237
IH D+W SPV+S G +YYV+FVDDYS + W +PL KS F++F++F+ + QF
Sbjct: 515 LGRIHCDLWGPSPVVSKQGFKYYVVFVDDYSRYSWFYPLKAKSDFFAVFVAFQNLVENQF 574
Query: 238 EREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLL 297
++K Q D G EF + + + G+ R+SCP+T QNG AERK R + +++
Sbjct: 575 NTKIKVFQSDGGGEFTSNLMKKHLTDCGIQHRISCPYTPQQNGIAERKHRHFVELGLSMM 634
Query: 298 VHASLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCYPLFP 357
H+ P FW A A++L N+LP+ L SPL+ L +++P+Y+ LRVFG CYP
Sbjct: 635 FHSHTPLQFWVEAFFTASFLSNMLPSPSLGNVSPLEALLKQKPNYAMLRVFGTACYPCLR 694
Query: 358 STTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLHN---P 414
+K + RS CVFLGY S ++ Y+C + ++ ISRHVIFDE FPF + + P
Sbjct: 695 PLGEHKFEPRSLQCVFLGYNSQYKGYRCLYPPTGRVYISRHVIFDEETFPFKQKYQFLVP 754
Query: 415 QPYTYGFMDDGPS--PYVIHHLTSQ------PSLGQPAQHDLPNTQ-PTTQPT------- 458
Q Y + S P L Q SL +P Q TTQP
Sbjct: 755 Q-YESSLLSAWQSSIPQADQSLIPQAEEGKIESLAKPPSIQKNTIQDTTTQPAILTEGVL 813
Query: 459 -------TPEEQHAHSPPSSSPNPSPSTTATPSPPYQPTPISVPKPVTRSQHGIFKPKRQ 511
+ EE S + + T Q P + TRS+ GI K +
Sbjct: 814 NEEEEEDSFEETETESLNEETHTQNDEAEVTVEEEVQQEPENTHPMTTRSKAGIHKSNTR 873
Query: 512 LNLNTSVPRSPLPRNPVSALRDPNWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIF 571
L TS P++ AL P W A++DE + TW LV D+N++ W+F
Sbjct: 874 YALLTSKFSVEEPKSIDEALNHPGWNNAVNDEMRTIHMLHTWSLVQPTEDMNILGCRWVF 933
Query: 572 THKEKSDGVFERYKARLVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQL 631
K K DG ++ KARLV G Q+ G+D ETFSPVV+ ATIR VL +A +K W+I QL
Sbjct: 934 KTKLKPDGSVDKLKARLVAKGFHQEEGLDYLETFSPVVRTATIRLVLDVATAKGWNIKQL 993
Query: 632 DVKNAFLHGELKETVYMHQPMGFRDPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTI 691
DV NAFLHGELKE VYM QP GF D P++VC L K+LYGLKQAPRAW+ ++Y+
Sbjct: 994 DVSNAFLHGELKEPVYMLQPPGFVDQEKPSYVCRLTKALYGLKQAPRAWFDTISNYLLDF 1053
Query: 692 GFSHSTSDHSLFIYRKGTAMAYILLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLH 751
GFS S SD SLF Y K +LLYVDDI+LT S L ++ L+ F+MKDLG+
Sbjct: 1054 GFSCSKSDPSLFTYHKNGKTLVLLLYVDDILLTGSDHNLLQELLMSLNKRFSMKDLGAPS 1113
Query: 752 YFLGIAVTHHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVDTKPKLSAN-SSAPHADPS 810
YFLG+ + GLFL Q YA +I+ +A M+ C S TP+ P+ N +S +P+
Sbjct: 1114 YFLGVEIESSPEGLFLHQTAYAKDILHQAAMSNCNSMPTPL---PQHIENLNSDLFPEPT 1170
Query: 811 HYRSLAGALQYLTFTRPDIAYAVQQVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYP 870
++RSLAG LQYLT TRPDI +AV +C MH P LKRI+RY++GT+ GLH+
Sbjct: 1171 YFRSLAGKLQYLTITRPDIQFAVNFICQRMHSPTTADFGLLKRILRYVKGTIHLGLHIKK 1230
Query: 871 SSTSTLISYTDADWGGCPDTRRSTSGYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVAN 930
+ +L++Y+D+DW GC +TRRST+G+C LG NLISWSAKRQ T+S+SS EAEYR +
Sbjct: 1231 NQNLSLVAYSDSDWAGCKETRRSTTGFCTLLGCNLISWSAKRQETVSKSSTEAEYRALTA 1290
Query: 931 VVSESCWLRNLLLELHCPIRKASLVYCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVA 990
V E WL LL ++ +LV CDN+SA+YLS NP H R+KH + D H +RE+VA
Sbjct: 1291 VAQELTWLSFLLRDIGVTQTHPTLVKCDNLSAVYLSANPALHNRSKHFDTDYHYIREQVA 1350
Query: 991 RGEVRVLHVPSRYQIADIFTKGLPLVLFEDFRNNLSVRQPPVST 1034
G V H+ + Q+ADIFTK LP F D R L V +PP ++
Sbjct: 1351 LGLVETKHISATLQLADIFTKPLPRRAFIDLRIKLGVAEPPTTS 1394
>UniRef100_O49145 Polyprotein [Arabidopsis arenosa]
Length = 1390
Score = 736 bits (1900), Expect = 0.0
Identities = 415/1051 (39%), Positives = 584/1051 (55%), Gaps = 78/1051 (7%)
Query: 1 MTSEQGNLSSYFNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNL 60
+TS+ NL+ + + + + +G +PI L P L LK+VL+ P + KNL
Sbjct: 343 LTSDLNNLALHQPYNGGEEVTIADGSGLPISHSGSALLPTPTRSLDLKDVLYVPDIQKNL 402
Query: 61 VSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLYPITTSQAISPSTFAALAP 120
+SV + N VSVEF P F VKD TG RL++ +++ +LY + +I+ S FA+ P
Sbjct: 403 ISVYRMCNTNGVSVEFFPAHFQVKDLSTGARLLQGKTKNELYEWPVNSSIATSMFASPTP 462
Query: 121 SL----WHARLGHPGAPVVDSLRKNKFIECNRASGSHI-CHSCSLGKHIKLPFVSSNSCT 175
WHARLGHP P++ +L + + + + + C CS+ K KLPF S+ +
Sbjct: 463 KTDLPSWHARLGHPSLPILKTLISKFSLPISHSLQNQLLCSDCSINKSHKLPFYSNTIAS 522
Query: 176 IMPFDIIHSDIWTSPVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRT 235
P + +++D+WTSP+ S ++YY++ VD Y+ + W +PL +KSQV F++F +
Sbjct: 523 SHPLEYLYTDVWTSPITSIDNYKYYLVIVDHYTRYTWLYPLRQKSQVRETFITFTALVEN 582
Query: 236 QFEREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRT 295
+F+ ++ + DNG EF F +G++ + PHT NG +ERK R I T
Sbjct: 583 KFKSKIGTLYSDNGGEFIALR--SFLASHGISHMTTPPHTPELNGISERKHRHIVETGLT 640
Query: 296 LLVHASLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCYPL 355
LL AS+ +W +A A YLIN + L +SP L+ + P+Y LRVFGCLC+P
Sbjct: 641 LLSTASMSKEYWSYAFTTAVYLINRMLTPVLGNESPYMKLFGQPPNYLKLRVFGCLCFPW 700
Query: 356 FPSTTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKL---- 411
T +KL RS PCV LGY + +Y C D ++ ++ SRHV F E+ FPF+
Sbjct: 701 LRPYTAHKLDNRSMPCVLLGYSLSQSAYLCLDRATGRVYTSRHVQFAESIFPFSTTSPSV 760
Query: 412 ---------HNPQPYTYGFM--------DDGPSPYVIHHLTSQPSLGQPAQHDLPN---- 450
+ +P + + PS H SQP + P+ P+
Sbjct: 761 TPPSDPPLSQDTRPISVPILARPLTTAPPSSPSCSAPHRSPSQPGILSPSAPFQPSPPSS 820
Query: 451 -TQPTTQPTTPEEQH------------------------AHSPPSSSPNP---------- 475
T P T P+ EE H + SP S+SP P
Sbjct: 821 PTSPITSPSLSEESHVGHNQETGPTGSSPPVSPQPQSEQSTSPRSTSPQPNSPHTQHSPR 880
Query: 476 --SPSTTATPSPPYQPTPISVPKPV-----TRSQHGIFKPKRQL-NLNTS-VPRSP-LPR 525
+P+ T +PSP P P + P P+ TRS++ I KP + NL T P P +P+
Sbjct: 881 SITPALTPSPSPSPPPNP-NPPPPIQHTMRTRSKNNIVKPNPKFANLATKPTPLKPIIPK 939
Query: 526 NPVSALRDPNWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKEKSDGVFERYK 585
AL DPNW+ AM DE NA +N T++LVP P+ NVI W+FT K +GV +RYK
Sbjct: 940 TVAEALLDPNWRQAMCDEINAQTRNGTFDLVPPAPNQNVIGCKWVFTLKYLPNGVLDRYK 999
Query: 586 ARLVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKNAFLHGELKET 645
ARLV G QQ G D ETFSPV+K T+R+VL +A+SK WSI Q+DV NAFL G L +
Sbjct: 1000 ARLVAKGFHQQYGHDFKETFSPVIKSTTVRSVLHVAVSKGWSIRQIDVNNAFLQGTLSDE 1059
Query: 646 VYMHQPMGFRDPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIGFSHSTSDHSLFIY 705
VY+ QP GF D + P+HVC L K+LYGLKQAPRAWY+ Y+ T GF +S +D SLF
Sbjct: 1060 VYVMQPPGFVDKDNPHHVCRLHKALYGLKQAPRAWYQELRSYLLTQGFVNSIADTSLFTL 1119
Query: 706 RKGTAMAYILLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFLGIAVTHHTGGL 765
R + Y+L+YVDD+++T S + I+ L+ F++KDLG + YFLGI T + GL
Sbjct: 1120 RHKRTILYVLVYVDDMLITGSDTNIITRFIANLAARFSLKDLGEMSYFLGIEATRTSKGL 1179
Query: 766 FLSQRKYAAEIIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHYRSLAGALQYLTFT 825
L Q++Y +++E+ M A TP+ PKLS S P PS YR++ G+LQYL+FT
Sbjct: 1180 HLMQKRYVLDLLEKTNMLAAHPVLTPMSPTPKLSLTSGTPLDKPSEYRAVLGSLQYLSFT 1239
Query: 826 RPDIAYAVQQVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTSTLISYTDADWG 885
RPDIAYAV ++ +MH P + H A KRI+RY+ GT HG+ + + L +Y+DADW
Sbjct: 1240 RPDIAYAVNRLSQYMHCPTDLHWQAAKRILRYLAGTPSHGIFIRADTPLKLHAYSDADWA 1299
Query: 886 GCPDTRRSTSGYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVSESCWLRNLLLEL 945
G D ST+ Y ++LG ISWS+K+Q ++RSS EAEYR VAN SE W+ +LL EL
Sbjct: 1300 GDTDNYNSTNAYILYLGSTPISWSSKKQNGVARSSTEAEYRAVANATSEIRWVCSLLTEL 1359
Query: 946 HCPIRKASLVYCDNVSAIYLSGNPVQHQRTK 976
+ +VYCDNV A YLS NPV R K
Sbjct: 1360 GITLSSPPVVYCDNVGATYLSANPVFDSRMK 1390
>UniRef100_Q7XE85 Putative pol polyprotein [Oryza sativa]
Length = 1688
Score = 676 bits (1743), Expect = 0.0
Identities = 391/1022 (38%), Positives = 552/1022 (53%), Gaps = 54/1022 (5%)
Query: 39 SFPNPPLTLKNVLHSPQLIKNLVSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRL----MR 94
S +P T+ NV P+L NL+SV + T D + V FD V+D TG + +
Sbjct: 228 SISSPQFTVPNVSLVPKLSMNLISVGQLT-DTNCFVGFDDTSCFVQDRHTGAVIGTGHRQ 286
Query: 95 CESRG----DLYPITTSQAISPSTFAALAPSL------WHARLGHPGAPVVDSLRKNKFI 144
S G D + +S +PS ++ + + WH RLGH + +L +
Sbjct: 287 KRSCGLYILDSLSLPSSSTNTPSVYSPMCSTACKSFPQWHHRLGHLCGSRLATLINQGVL 346
Query: 145 ECNRASGSHICHSCSLGKHIKLPFVSSNSCTIMPFDIIHSDIW-TSPVLSSSGHRYYVLF 203
+ +C C LGK ++LP+ SS S + PFD++HSD+W SP S GH YYV+F
Sbjct: 347 GSVPVDTTFVCKGCKLGKQVQLPYPSSTSRSSRPFDLVHSDVWGKSPFPSKGGHNYYVIF 406
Query: 204 VDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQFEREVKNIQCDNGKEFDNRHFWEFCKE 263
VDDYS + W + + +SQ+ SI+ SF I TQF ++ + D+G E+ + F EF
Sbjct: 407 VDDYSRYTWIYFMKHRSQLISIYQSFAQMIHTQFSSAIRIFRSDSGGEYMSNAFREFLVS 466
Query: 264 NGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLLVHASLPPSFWHHALQMATYLINILPN 323
G +LSCP +QNG AERK R I RTLL+ + +P FW A+ A YLIN+ P+
Sbjct: 467 QGTLPQLSCPGAHAQNGVAERKHRHIIETARTLLIASFVPAHFWAEAISTAVYLINMQPS 526
Query: 324 KQLAYQSPLKILYQKEPSYSHLRVFGCLCYPLFPSTTINKLQARSTPCVFLGYPSNHRSY 383
L +SP ++L+ P Y HLRVFGC CY L KL A+S CVFLGY H+ Y
Sbjct: 527 SSLQGRSPGEVLFGSPPRYDHLRVFGCTCYVLLAPRERTKLTAQSVECVFLGYSLEHKGY 586
Query: 384 KCYDLSSRKIIISRHVIFDETQFPFAKLHNPQP------YTYGFMDDGPSPYVIHHLTSQ 437
+CYD S+R+I ISR V FDE + PF QP ++ ++ PSP +
Sbjct: 587 RCYDPSARRIRISRDVTFDENK-PFFYSSTNQPSSPENSISFLYLPPIPSPESLPSSPIT 645
Query: 438 PSLGQ-----PAQHDLPNTQPTTQPTTPEEQHAHSPPSSSPNPSPSTTATPSPP--YQPT 490
PS P+ +P P+ P+ +H P SSSP PST + P Y
Sbjct: 646 PSPSPIPPSVPSPTYVPPPPPSPSPSPVSPPPSHIPASSSPPHVPSTITLDTFPFHYSRR 705
Query: 491 PISVPKPVTRSQHGIFKPKRQLNLNTSVPRSPL---------------------PRNPVS 529
P +P SQ + P ++ ++ PR L P
Sbjct: 706 P-KIPNESQPSQPTLEDPTCSVDDSSPAPRYNLRARDALRAPNRDDFVVGVVFEPSTYQE 764
Query: 530 ALRDPNWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKEKSDGVFERYKARLV 589
A+ P+WK+AM +E AL + TW++VP P I W++ K KSDG ERYKARLV
Sbjct: 765 AIVLPHWKLAMSEELAALERTNTWDVVPLPSHAVPITCKWVYKVKTKSDGQVERYKARLV 824
Query: 590 GDGKTQQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKNAFLHGELKETVYMH 649
G Q G D ETF+PV T+RT++++A +++W+I Q+DVKNAFLHG+L E VYMH
Sbjct: 825 ARGFQQAHGRDYDETFAPVAHMTTVRTLIAVAATRSWTISQMDVKNAFLHGDLHEEVYMH 884
Query: 650 QPMGFRDPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIGFSHSTSDHSLFIYRKGT 709
P G P P HV L+++LYGLKQAPRAW+ RF+ V GFS S D +LFI+
Sbjct: 885 PPPGVEAP--PGHVFRLRRALYGLKQAPRAWFARFSSVVLAAGFSPSDHDPALFIHTSSR 942
Query: 710 AMAYILLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFLGIAVTHHTGGLFLSQ 769
+LLYVDD+++T + LS +F M DLG L YFLGI VT G +LSQ
Sbjct: 943 GRTLLLLYVDDMLITGDDLEYIAFVKGKLSEQFMMSDLGPLSYFLGIEVTSTVDGYYLSQ 1002
Query: 770 RKYAAEIIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHYRSLAGALQYLTFTRPDI 829
+Y +++ ++G+ ++++TP++ +L + P DPS YR L G+L YLT TRPDI
Sbjct: 1003 HRYIEDLLAQSGLTDSRTTTTPMELHVRLRSTDGTPLDDPSRYRHLVGSLVYLTVTRPDI 1062
Query: 830 AYAVQQVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTSTLISYTDADWGGCPD 889
AYAV + F+ P H L R++RY++GT L SS L +++D+ W P
Sbjct: 1063 AYAVHILSQFVSAPISVHYGHLLRVLRYLRGTTTQCLFYAASSPLQLRAFSDSTWASDPI 1122
Query: 890 TRRSTSGYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVSESCWLRNLLLELHCPI 949
RRS +GYC+FLG +L++W +K+Q +SRSS EAE R +A SE WLR LL +
Sbjct: 1123 DRRSVTGYCIFLGTSLLTWKSKKQTAVSRSSTEAELRALATTTSEIVWLRWLLADFGVSC 1182
Query: 950 RKASLVYCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARGEVRVLHVPSRYQIADIF 1009
+ + CDN AI ++ +P++H+ TKHI +D R + + + +VPS Q+AD F
Sbjct: 1183 DVPTPLLCDNTGAIQIANDPIKHELTKHIGVDASFTRSHCQQSTIALHYVPSELQVADFF 1242
Query: 1010 TK 1011
TK
Sbjct: 1243 TK 1244
>UniRef100_Q9XII7 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1454
Score = 652 bits (1683), Expect = 0.0
Identities = 383/1031 (37%), Positives = 554/1031 (53%), Gaps = 32/1031 (3%)
Query: 3 SEQGNLSSYFNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNLVS 62
S +L S + S + + G ++ I G L N + LKNVL P+ NL+S
Sbjct: 442 SHDRSLFSSLDTSVLSAVNLPTGPTVKISGVGTLKL---NDDILLKNVLFIPEFRLNLIS 498
Query: 63 VRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLYPITTSQAISPSTFAALAPSL 122
+ T D V FD ++D G L + +LY + S S A + S+
Sbjct: 499 ISSLTDDIGSRVIFDKNSCEIQDLIKGRMLGQGRRVANLYLLDVGDQ-SISVNAVVDISM 557
Query: 123 WHARLGHPGAPVVDSLRKNKFIECNRASGSHICHSCSLGKHIKLPFVSSNSCTIMPFDII 182
WH RLGH +D++ + ++ GS CH C L K KL F +SN FD++
Sbjct: 558 WHRRLGHASLQRLDAISDSLGTTRHKNKGSDFCHVCHLAKQRKLSFPTSNKVCKEIFDLL 617
Query: 183 HSDIWTS-PVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQFEREV 241
H D+W V + G++Y++ VDD+S W + L KS+V ++F +F + Q++ +V
Sbjct: 618 HIDVWGPFSVETVEGYKYFLTIVDDHSRATWMYLLKTKSEVLTVFPAFIQQVENQYKVKV 677
Query: 242 KNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLLVHAS 301
K ++ DN E F F E G+ SCP T QN ERK + I N+ R L+ +
Sbjct: 678 KAVRSDNAPELK---FTSFYAEKGIVSFHSCPETPEQNSVVERKHQHILNVARALMFQSQ 734
Query: 302 LPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCYPLFPSTTI 361
+P S W + A +LIN P++ L ++P +IL P Y LR FGCLCY
Sbjct: 735 VPLSLWGDCVLTAVFLINRTPSQLLMNKTPYEILTGTAPVYEQLRTFGCLCYSSTSPKQR 794
Query: 362 NKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLHNPQPYTYGF 421
+K Q RS C+FLGYPS ++ YK DL S + ISR+V F E FP AK + F
Sbjct: 795 HKFQPRSRACLFLGYPSGYKGYKLMDLESNTVFISRNVQFHEEVFPLAKNPGSESSLKLF 854
Query: 422 MDDGP-SPYVIHHLTSQPSLGQPAQHDLPNTQPTTQPTTPEEQHAHSPPSSSPNPSPSTT 480
P S +I T PS LP+ Q + P Q PP+ N T
Sbjct: 855 TPMVPVSSGIISDTTHSPS-------SLPS-QISDLPPQISSQRVRKPPAHL-NDYHCNT 905
Query: 481 ATPSPPYQPTPISVPKPVTRSQHGIFKPKRQLNLNTSVPRSPLPRNPVSALRDPNWKMAM 540
Y PIS + + P +N ++ + P+P N A W A+
Sbjct: 906 MQSDHKY---PIS-----STISYSKISPSHMCYIN-NITKIPIPTNYAEAQDTKEWCEAV 956
Query: 541 DDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKEKSDGVFERYKARLVGDGKTQQVGVD 600
D E A+ K TWE+ P + W+FT K +DG ERYKARLV G TQ+ G+D
Sbjct: 957 DAEIGAMEKTNTWEITTLPKGKKAVGCKWVFTLKFLADGNLERYKARLVAKGYTQKEGLD 1016
Query: 601 CGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKNAFLHGELKETVYMHQPMGFRDPN-- 658
+TFSPV K TI+ +L ++ SK W + QLDV NAFL+GEL+E ++M P G+ +
Sbjct: 1017 YTDTFSPVAKMTTIKLLLKVSASKKWFLKQLDVSNAFLNGELEEEIFMKIPEGYAERKGI 1076
Query: 659 -LPNHVCL-LKKSLYGLKQAPRAWYKRFADYVSTIGFSHSTSDHSLFIYRKGTAMAYILL 716
LP++V L LK+S+YGLKQA R W+K+F+ + ++GF + DH+LF+ +L+
Sbjct: 1077 VLPSNVVLRLKRSIYGLKQASRQWFKKFSSSLLSLGFKKTHGDHTLFLKMYDGEFVIVLV 1136
Query: 717 YVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFLGIAVTHHTGGLFLSQRKYAAEI 776
YVDDI++ ++S+A + L F ++DLG L YFLG+ V T G+ + QRKYA E+
Sbjct: 1137 YVDDIVIASTSEAAAAQLTEELDQRFKLRDLGDLKYFLGLEVARTTAGISICQRKYALEL 1196
Query: 777 IERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHYRSLAGALQYLTFTRPDIAYAVQQV 836
++ GM ACK S P+ K+ + D YR + G L YLT TRPDI +AV ++
Sbjct: 1197 LQSTGMLACKPVSVPMIPNLKMRKDDGDLIEDIEQYRRIVGKLMYLTITRPDITFAVNKL 1256
Query: 837 CLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTSTLISYTDADWGGCPDTRRSTSG 896
C F PR H+ A R+++YI+GT+ GL SS TL + D+DW C D+RRST+
Sbjct: 1257 CQFSSAPRTTHLTAAYRVLQYIKGTVGQGLFYSASSDLTLKGFADSDWASCQDSRRSTTS 1316
Query: 897 YCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVSESCWLRNLLLELHCPIRKASLVY 956
+ +F+GD+LISW +K+Q T+SRSSAEAEYR +A E WL LL+ L ++Y
Sbjct: 1317 FTMFVGDSLISWRSKKQHTVSRSSAEAEYRALALATCEMVWLFTLLVSLQAS-PPVPILY 1375
Query: 957 CDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARGEVRVLHVPSRYQIADIFTKGLPLV 1016
D+ +AIY++ NPV H+RTKHI++D H+VRE++ GE+++LHV + Q+ADI TK L
Sbjct: 1376 SDSTAAIYIATNPVFHERTKHIKLDCHTVRERLDNGELKLLHVRTEDQVADILTKPLFPY 1435
Query: 1017 LFEDFRNNLSV 1027
FE ++ +S+
Sbjct: 1436 QFEHLKSKMSI 1446
>UniRef100_O04543 F20P5.25 protein [Arabidopsis thaliana]
Length = 1315
Score = 642 bits (1656), Expect = 0.0
Identities = 365/982 (37%), Positives = 522/982 (52%), Gaps = 23/982 (2%)
Query: 45 LTLKNVLHSPQLIKNLVSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLYPI 104
L L +VL PQ NL+SV T + FD ++D + + + +LY +
Sbjct: 323 LILNDVLFIPQFKFNLLSVSSLTKSMGCRIWFDETSCVLQDATRELMVGMGKQVANLYIV 382
Query: 105 TTSQAISPSTFAALAPS------LWHARLGHPGAPVVDSLRKNKFIECNRASGSHICHSC 158
P T +++ + LWH RLGHP + + + + C C
Sbjct: 383 DLDSLSHPGTDSSITVASVTSHDLWHKRLGHPSVQKLQPMSSLLSFPKQKNNTDFHCRVC 442
Query: 159 SLGKHIKLPFVSSNSCTIMPFDIIHSDIWTS-PVLSSSGHRYYVLFVDDYSNFLWTFPLS 217
+ K LPFVS N+ + PFD+IH D W V + G+RY++ VDDYS W + L
Sbjct: 443 HISKQKHLPFVSHNNKSSRPFDLIHIDTWGPFSVQTHDGYRYFLTIVDDYSRATWVYLLR 502
Query: 218 KKSQVFSIFLSFRTFIRTQFEREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSS 277
KS V ++ +F T + QFE +K ++ DN E + F +F G+ SCP T
Sbjct: 503 NKSDVLTVIPTFVTMVENQFETTIKGVRSDNAPELN---FTQFYHSKGIVPYHSCPETPQ 559
Query: 278 QNGKAERKIRTINNIIRTLLVHASLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQ 337
QN ERK + I N+ R+L + +P S+W + A YLIN LP L + P ++L +
Sbjct: 560 QNSVVERKHQHILNVARSLFFQSHIPISYWGDCILTAVYLINRLPAPILEDKCPFEVLTK 619
Query: 338 KEPSYSHLRVFGCLCYPLFPSTTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISR 397
P+Y H++VFGCLCY +K R+ C F+GYPS + YK DL + II+SR
Sbjct: 620 TVPTYDHIKVFGCLCYASTSPKDRHKFSPRAKACAFIGYPSGFKGYKLLDLETHSIIVSR 679
Query: 398 HVIFDETQFPFAKLHNPQPYTYGFMDDGPSPYVIHHLTS--QPSLGQPAQHDLPNTQPTT 455
HV+F E FPF Q F D P+P + + PS + LP+ PT
Sbjct: 680 HVVFHEELFPFLGSDLSQEEQNFFPDLNPTPPMQRQSSDHVNPSDSSSSVEILPSANPTN 739
Query: 456 QPTTPEEQHAHSPPSSSPNPSPSTTATPSPPYQPTPISVPKPVTRSQHGIFKPKRQLNLN 515
P Q +H + P+ TP + K + S I P L
Sbjct: 740 NVPEPSVQTSH---RKAKKPAYLQDYYCHSVVSSTPHEIRKFL--SYDRINDP--YLTFL 792
Query: 516 TSVPRSPLPRNPVSALRDPNWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKE 575
+ ++ P N A + W+ AM EF+ L TWE+ P D I WIF K
Sbjct: 793 ACLDKTKEPSNYTEAEKLQVWRDAMGAEFDFLEGTHTWEVCSLPADKRCIGCRWIFKIKY 852
Query: 576 KSDGVFERYKARLVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKN 635
SDG ERYKARLV G TQ+ G+D ETFSPV K +++ +L +A S+ QLD+ N
Sbjct: 853 NSDGSVERYKARLVAQGYTQKEGIDYNETFSPVAKLNSVKLLLGVAARFKLSLTQLDISN 912
Query: 636 AFLHGELKETVYMHQPMGFR----DPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTI 691
AFL+G+L E +YM P G+ D PN VC LKKSLYGLKQA R WY +F+ + +
Sbjct: 913 AFLNGDLDEEIYMRLPQGYASRQGDSLPPNAVCRLKKSLYGLKQASRQWYLKFSSTLLGL 972
Query: 692 GFSHSTSDHSLFIYRKGTAMAYILLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLH 751
GF S DH+ F+ +L+Y+DDII+ +++DA + S + + F ++DLG L
Sbjct: 973 GFIQSYCDHTCFLKISDGIFLCVLVYIDDIIIASNNDAAVDILKSQMKSFFKLRDLGELK 1032
Query: 752 YFLGIAVTHHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSH 811
YFLG+ + G+ +SQRKYA ++++ G CK SS P+D + +S +
Sbjct: 1033 YFLGLEIVRSDKGIHISQRKYALDLLDETGQLGCKPSSIPMDPSMVFAHDSGGDFVEVGP 1092
Query: 812 YRSLAGALQYLTFTRPDIAYAVQQVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPS 871
YR L G L YL TRPDI +AV ++ F PR+ H+ A+ +I++YI+GT+ GL +
Sbjct: 1093 YRRLIGRLMYLNITRPDITFAVNKLAQFSMAPRKAHLQAVYKILQYIKGTIGQGLFYSAT 1152
Query: 872 STSTLISYTDADWGGCPDTRRSTSGYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANV 931
S L Y +AD+ C D+RRSTSGYC+FLGD+LI W +++Q +S+SSAEAEYR ++
Sbjct: 1153 SELQLKVYANADYNSCRDSRRSTSGYCMFLGDSLICWKSRKQDVVSKSSAEAEYRSLSVA 1212
Query: 932 VSESCWLRNLLLELHCPIRKASLVYCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVAR 991
E WL N L EL P+ K +L++CDN +AI+++ N V H+RTKHIE D HSVRE++ +
Sbjct: 1213 TDELVWLTNFLKELQVPLSKPTLLFCDNEAAIHIANNHVFHERTKHIESDCHSVRERLLK 1272
Query: 992 GEVRVLHVPSRYQIADIFTKGL 1013
G + H+ + QIAD FTK L
Sbjct: 1273 GLFELYHINTELQIADPFTKPL 1294
>UniRef100_Q9FWZ5 Putative retroelement polyprotein [Arabidopsis thaliana]
Length = 1404
Score = 642 bits (1655), Expect = 0.0
Identities = 371/1042 (35%), Positives = 543/1042 (51%), Gaps = 57/1042 (5%)
Query: 20 IIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNLVSVRKFTTDNSVSVEFDPF 79
+I+ NG +PI G + L + P+ NL+SV++ T D + F P
Sbjct: 356 VIIANGDKVPIEGIGNLKLFNKD-----SKAFFMPKFTSNLLSVKRTTRDLNCYAIFGPN 410
Query: 80 GFSVKDFQTGMRLMRCESRGDLYPITTSQAISPSTFAALAP------SLWHARLGHPGAP 133
+D +TG + S+G+LY + S S F++ + +LWHARLGHP
Sbjct: 411 DVYFQDIETGKVIGEGGSKGELYVLEDLSPNSSSCFSSKSHLGISFNTLWHARLGHPHTR 470
Query: 134 VVDSLRKNKFIECNRASGSHICHSCSLGKHIKLPFVSSNSCTIMPFDIIHSDIWTSPVLS 193
+ + N I + S C +C LGKH K F S + FD++HSD+WTSP +S
Sbjct: 471 ALKLMLPN--ISFDHTS----CEACILGKHCKSVFPKSLTIYEKCFDLVHSDVWTSPCVS 524
Query: 194 SSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQFEREVKNIQCDNGKEFD 253
++Y+V F+++ S + W L K +VF F +F T++ QF ++K + DNG E+
Sbjct: 525 RDNNKYFVTFINEKSKYTWITLLPSKDRVFEAFTNFETYVTNQFNAKIKVFRTDNGGEYT 584
Query: 254 NRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLLVHASLPPSFWHHALQM 313
++ F + + G+ + SCP+T QNG AERK R + + R+++ H S+P FW A+
Sbjct: 585 SQKFRDHLAKRGIIHQTSCPYTPQQNGVAERKNRHLMEVARSMMFHTSVPKRFWGDAVLT 644
Query: 314 ATYLINILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCYPLFPSTTINKLQARSTPCVF 373
A YLIN P K L+ SP ++L +P HLRVFGC+C+ L P +KL A+ST C+F
Sbjct: 645 ACYLINRTPTKVLSDLSPFEVLNNTKPFIDHLRVFGCVCFVLIPGEQRSKLDAKSTKCMF 704
Query: 374 LGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAK--LHNPQPYTYGFMDDGPS-PYV 430
LGY + + YKC+D + + ISR V F E Q K N + T+ D + ++
Sbjct: 705 LGYSTTQKGYKCFDPTKNRTFISRDVKFLENQDYNNKKDWENLKDLTHSTSDRVETLKFL 764
Query: 431 IHHLTSQPSLGQPAQHDLPNTQPTTQPTTPEEQHAHSPPSSSPNPSPSTTATPSPPYQ-- 488
+ HL + + Q ++ Q E H + P T S Q
Sbjct: 765 LDHLGNDSTSTTQHQPEMTQDQEDLNQENEEVSLQHQENLTHVQEDPPNTQEHSEHVQEI 824
Query: 489 ---------PTPISVPKPVTRSQHGIFKPKRQLNLN-----------------------T 516
PT + P P R I + K N N +
Sbjct: 825 QDDSSEDEEPTQVLPPPPPLRRSTRIRRKKEFFNSNAVAHPFQATCSLALVPLDHQAFLS 884
Query: 517 SVPRSPLPRNPVSALRDPNWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKEK 576
+ +P+ A+ W+ A+ DE NA+ +N TW+ P + S W+FT K K
Sbjct: 885 KISEHWIPQTYEEAMEVKEWRDAIADEINAMKRNHTWDEDDLPKGKKTVSSRWVFTIKYK 944
Query: 577 SDGVFERYKARLVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKNA 636
S+G ERYK RLV G TQ G D ETF+PV K T+R VL+LA + +W + Q+DVKNA
Sbjct: 945 SNGDIERYKTRLVARGFTQTYGSDYMETFAPVAKLHTVRVVLALATNLSWGLWQMDVKNA 1004
Query: 637 FLHGELKETVYMHQPMGFRDPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIGFSHS 696
FL GEL++ VYM P G D + V L+K++YGLKQ+PRAWY + + + GF S
Sbjct: 1005 FLQGELEDDVYMTPPPGLEDTIPCDKVLRLRKAIYGLKQSPRAWYHKLSRTLKDHGFKKS 1064
Query: 697 TSDHSLFIYRKGTAMAYILLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFLGI 756
SDH+LF + + +L+YVDD+I+T + + + L + F +KDLG L YFLGI
Sbjct: 1065 ESDHTLFTLQSPQGIVVVLIYVDDLIITGDNKDGIDSTKTFLKSCFDIKDLGELKYFLGI 1124
Query: 757 AVTHHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVDTKPKLS---ANSSAPHADPSHYR 813
V GLFLSQRKY +++ G K + TP++ K++ D YR
Sbjct: 1125 EVCRSNAGLFLSQRKYTLDLLNETGFMDAKPARTPLEDGYKVNRKGEKEDEKFGDAPLYR 1184
Query: 814 SLAGALQYLTFTRPDIAYAVQQVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSST 873
L G L YLT TRPDI +AV QV M P H + ++RI+RY++G+ G+ + +S+
Sbjct: 1185 KLVGKLIYLTNTRPDICFAVNQVSQHMKVPMVYHWNMVERILRYLKGSSGQGIWMGKNSS 1244
Query: 874 STLISYTDADWGGCPDTRRSTSGYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVS 933
+ ++ Y DAD+ G RRS +GYC F+G NL +W K+Q +S SSAE+EYR + + +
Sbjct: 1245 TEIVGYCDADYAGDRGDRRSKTGYCTFIGGNLATWKTKKQKVVSCSSAESEYRAMRKLTN 1304
Query: 934 ESCWLRNLLLELHCPIRKASLVYCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARGE 993
E WL+ LL +L ++CDN +AIY++ N V H+RTKHIE+D H VREK+ G
Sbjct: 1305 ELTWLKALLKDLGIEQHMPITMHCDNKAAIYIASNSVFHERTKHIEVDCHKVREKIIEGV 1364
Query: 994 VRVLHVPSRYQIADIFTKGLPL 1015
+ S Q+ADIFTK L
Sbjct: 1365 TLPCYTRSEDQLADIFTKAASL 1386
>UniRef100_Q7X7W7 OSJNBa0061G20.2 protein [Oryza sativa]
Length = 542
Score = 637 bits (1642), Expect = 0.0
Identities = 316/542 (58%), Positives = 397/542 (72%), Gaps = 5/542 (0%)
Query: 498 VTRSQHGIFKPK-RQLNLNTSVPRSPLPRNPVSALRDPNWKMAMDDEFNALIKNKTWELV 556
VTR + GI +P R +L T+ +P + LRDP W AM DE+ AL N TW LV
Sbjct: 2 VTRWKAGIVQPNPRYAHLATA---DDVPTTVRAVLRDPAWFAAMQDEYRALQDNGTWALV 58
Query: 557 PRPPDVNVIRSMWIFTHKEKSDGVFERYKARLVGDGKTQQVGVDCGETFSPVVKPATIRT 616
PRP +VI WIF +K +DG ER KAR V G TQ+ G+D +TFSPVVKPATIRT
Sbjct: 59 PRPRGAHVITGKWIFKNKFHADGTLERRKARWVARGFTQRPGLDFDKTFSPVVKPATIRT 118
Query: 617 VLSLALSKAWSIHQLDVKNAFLHGELKETVYMHQPMGFRDPNLPNHVCLLKKSLYGLKQA 676
VL LA + W +HQLDVKNAFLHG+L E Y HQP GF DP+ P+ VCLL+KSLYGLKQA
Sbjct: 119 VLHLAAMRDWPVHQLDVKNAFLHGDLTEHFYCHQPAGFVDPSQPDAVCLLRKSLYGLKQA 178
Query: 677 PRAWYKRFADYVSTIGFSHSTSDHSLFIYRKGTAMAYILLYVDDIILTASSDALRVTIIS 736
PRAW++RF ++ +GF S SD+SLF+ R+GT A++LLYVDDI+L ASS L II+
Sbjct: 179 PRAWFQRFGTHLHHLGFVSSKSDNSLFVLRRGTDEAHLLLYVDDIVLAASSQRLLQHIIN 238
Query: 737 LLSTEFAMKDLGSLHYFLGIAVTHHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVDTKP 796
L EFAMKDLG +H+FLGI V G FLSQ +YA ++++RAG+A CK + TP+DTK
Sbjct: 239 QLRVEFAMKDLGPVHFFLGIQVRRTADGFFLSQEQYAGDVLDRAGLADCKPAPTPIDTKA 298
Query: 797 KLSANSSAPHADPSHYRSLAGALQYLTFTRPDIAYAVQQVCLFMHDPREEHMHALKRIVR 856
K+S+ + P++DP+ YRS+ GALQYLT TRPD++YAVQQVCL MH PR+ H +KRI+R
Sbjct: 299 KVSSTTGQPYSDPTFYRSIVGALQYLTLTRPDLSYAVQQVCLHMHSPRDVHWTLVKRILR 358
Query: 857 YIQGTLDHGLHLYPSSTSTLISYTDADWGGCPDTRRSTSGYCVFLGDNLISWSAKRQATL 916
Y++GT GL L SST +L +Y+D DW GCPDTRRSTSG+CVF GD+ SWS+KRQ+ +
Sbjct: 359 YVRGTTHKGLQLRRSSTPSLTAYSDVDWAGCPDTRRSTSGFCVFFGDSSESWSSKRQSVV 418
Query: 917 SRSSAEAEYRGVANVVSESCWLRNLLLELHCPIRKASLVYCDNVSAIYLSGNPVQHQRTK 976
SRSSAEAEYRGVAN +E CWLR+LL ELH + KA+LVYCDN+SA+YLS NP+ H R K
Sbjct: 419 SRSSAEAEYRGVANAAAECCWLRHLLGELHVKLDKATLVYCDNISAVYLSKNPLHHGRAK 478
Query: 977 HIEMDIHSVREKVARGEVRVLHVPSRYQIADIFTKGLPLVLFEDFRNNLSVRQPPVSTAG 1036
H+E+D+H VREKVA G++RV H+P+R Q+ADI TK LP LFEDFR++L + TAG
Sbjct: 479 HVELDVHFVREKVAVGDIRVAHIPTRQQLADIMTKRLPTALFEDFRSSLCI-VDDAPTAG 537
Query: 1037 VC 1038
C
Sbjct: 538 GC 539
>UniRef100_Q7Y141 Putative polyprotein [Oryza sativa]
Length = 1335
Score = 620 bits (1599), Expect = e-176
Identities = 372/1026 (36%), Positives = 548/1026 (53%), Gaps = 67/1026 (6%)
Query: 7 NLSSYFNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNLVSVRKF 66
NL + S + I +GNG G + + P +K+VL P L +NL+S+ +
Sbjct: 341 NLFREMDSSYHAKIHMGNGSIAQSEGKGTVAVQTADGPKFIKDVLLVPDLKQNLLSIGQL 400
Query: 67 TTDNSVSVEFDPFGFSVKDFQTGMRLMRC---ESRGDLYPI--TTSQAISPSTFAALAPS 121
++ +V F+ F + D + + + ++R L + TT A+ +
Sbjct: 401 L-EHGYAVYFEDFSCKILDRKNNRLVAKINMEKNRNFLLRMNHTTQMALRSEVDIS---D 456
Query: 122 LWHARLGHPGAPVVDSLRKNKFIECNR--ASGSHICHSCSLGKHIKLPFVSSNSCTIM-P 178
LWH R+GH + LR ++ S C C GK I+ F S + P
Sbjct: 457 LWHKRMGHLNYRALKLLRTKGMVQGLPFITLKSDPCEGCVFGKQIRASFPHSGAWRASAP 516
Query: 179 FDIIHSDIWTS-PVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFLSFRTFIRTQF 237
+++H+DI P +S G+ Y++ F+DDY+ +W + L +KS IF F+ + Q
Sbjct: 517 LELVHADIVGKVPTISEGGNWYFITFIDDYTRMIWVYFLKEKSAALEIFKKFKAMVENQS 576
Query: 238 EREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIRTINNIIRTLL 297
R++K ++ D G+E+ ++ F ++C+ G+ +L+ +++ QNG AERK RTIN++ ++L
Sbjct: 577 NRKIKVLRSDQGREYISKEFEKYCENAGIRRQLTAGYSAQQNGVAERKNRTINDMANSML 636
Query: 298 VHASLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQKEPSYSHLRVFGCLCYPLFP 357
+P SFW A+ A Y++N P K + ++P + Y K+P H+RVFGC+CY P
Sbjct: 637 QDKGMPKSFWAEAVNTAVYILNRSPTKAVTNRTPFEAWYGKKPVIGHMRVFGCICYAQVP 696
Query: 358 STTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFPFAKLHNPQPY 417
+ K +S C+F+GY + Y+ Y+L +KIIISR IFDE+ +
Sbjct: 697 AQKRVKFDNKSDRCIFVGYADGIKGYRLYNLEKKKIIISRDAIFDESA----------TW 746
Query: 418 TYGFMDDGPSPYVIHHLTSQPSLGQPAQHDLPNTQPTTQPTTPEEQHAHSPPSSSPNPSP 477
+ + +P + T+ +LGQP H T E H SP SSP S
Sbjct: 747 NWKSPEASSTPLLP---TTTITLGQPHMHG----------THEVEDHTPSPQPSSPMSSS 793
Query: 478 STTATPSPPYQP---TPISVPKPVTRSQHGIFKPKRQLN-------LNTSVPRSPLPRNP 527
S ++ SP + TP S P+ V RS + + Q N SV P++
Sbjct: 794 SASSDSSPSSEEQISTPESAPRRV-RSMVELLESTSQQRGSEQHEFCNYSVVE---PQSF 849
Query: 528 VSALRDPNWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKEKSDGVFERYKAR 587
A + NW AM+DE + + KN TWELV RP D VI W++ K DG ++YKAR
Sbjct: 850 QEAEKHDNWIKAMEDEIHMIEKNNTWELVDRPRDREVIGVKWVYKTKLNPDGSVQKYKAR 909
Query: 588 LVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKNAFLHGELKETVY 647
LV G Q+ G+D ET++PV + TIRT+++LA K W I+QLDVK+AFL+G L E +Y
Sbjct: 910 LVAKGFKQKPGIDYYETYAPVARLETIRTIIALAAQKRWKIYQLDVKSAFLNGYLDEEIY 969
Query: 648 MHQPMGFRDPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIGFSHSTSDHSLFIYRK 707
+ QP GF N V LKK+LYGLKQAPRAWY + Y GF+ S S+ +L++ +
Sbjct: 970 VEQPEGFSVQGGENKVFRLKKALYGLKQAPRAWYSQIDKYFIQKGFAKSISEPTLYVNKT 1029
Query: 708 GTAMAYILLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFLGIAVTHHTGGLFL 767
GT + + LYVDD+I T +S+ + + + M DLG LHYFLG+ V G+F+
Sbjct: 1030 GTDILIVSLYVDDLIYTGNSEKMMQDFKKDMMHTYEMSDLGLLHYFLGMEVHQSDEGIFI 1089
Query: 768 SQRKYAAEIIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHYRSLAGALQYLTFTRP 827
SQRKYA I+++ M CKS +TP+ K A A ADP+ YRSL G+L YLT TRP
Sbjct: 1090 SQRKYAENILKKFKMDNCKSVTTPLLPNEKQKARDGADKADPTIYRSLVGSLLYLTATRP 1149
Query: 828 DIAYAVQQVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTSTLISYTDADWGGC 887
DI +A + +M P + + A KR++RYI+GT D+G+ P S LI YTD+DW GC
Sbjct: 1150 DIMFAASLLSRYMSSPSQLNFTAAKRVLRYIKGTADYGIWYKPVKESKLIGYTDSDWAGC 1209
Query: 888 PDTRRSTSGYCVFLGDNLISWSAKRQATLSRSSAEAEYRGVANVVSESCWLRNLLLELHC 947
D +STSGY LG SAEAEY + VS+ WLR ++ +L
Sbjct: 1210 LDDMKSTSGYAFSLG-----------------SAEAEYVAASKAVSQVVWLRRIMEDLGE 1252
Query: 948 PIRKASLVYCDNVSAIYLSGNPVQHQRTKHIEMDIHSVREKVARGEVRVLHVPSRYQIAD 1007
+ + +YCD+ SAI +S NPV H RTKHI + H +RE V R EV++ + Q+AD
Sbjct: 1253 KQYQPTTIYCDSKSAIAISENPVSHDRTKHIAIKYHYIREAVDRQEVKLEFCRTDEQLAD 1312
Query: 1008 IFTKGL 1013
IFTK L
Sbjct: 1313 IFTKAL 1318
>UniRef100_Q5XWR5 Putative retroelement pol polyprotein-like [Solanum tuberosum]
Length = 1476
Score = 608 bits (1569), Expect = e-172
Identities = 375/1001 (37%), Positives = 525/1001 (51%), Gaps = 68/1001 (6%)
Query: 47 LKNVLHSPQLIKNLVSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLYPITT 106
+KNVL P NL+SV K T + + V F P F ++D TG E LY
Sbjct: 471 VKNVLCVPTFQFNLLSVSKLTKELNCCVIFFPDFFIIQDLFTGKVKEIGEEINGLYITRP 530
Query: 107 SQA--ISPSTFAALA----PSLWHARLGHPGAPVVDSLRKNKFIECNRASGSHICHSCSL 160
Q S T AA+ +WH RLGH V LRK K + + C C L
Sbjct: 531 HQHHDTSKKTLAAIKGCEEAEMWHKRLGHIPMSV---LRKIKMFDSPQKLVLPSCDVCPL 587
Query: 161 GKHIKLPFVSSNSCTIMPFDIIHSDIWTSPVLSSSGH--RYYVLFVDDYSNFLWTFPLSK 218
+ ++LPF S S + FD+IH D+W P +++ + RY++ VDD+S + W F +
Sbjct: 588 ARQVRLPFPISQSRSENCFDLIHLDVW-GPYKAATHNKMRYFLTVVDDHSRWTWIFLMHL 646
Query: 219 KSQVFSIFLSFRTFIRTQFEREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQ 278
KS V ++ +F I TQF +++K + DNG EF N K +G+ + SCPHT Q
Sbjct: 647 KSDVSTVLQNFILMIDTQFGQKIKIFRSDNGTEFFNAQCDGLFKSHGIVHQSSCPHTPQQ 706
Query: 279 NGKAERKIRTINNIIRTLLVHASLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQK 338
NG ER+ + I R L LP FW + A ++IN +P+ L +SP +++Y++
Sbjct: 707 NGVVERRHKHILETARALRFQGHLPIRFWGECVLSAVHIINRIPSSVLHNKSPFELMYKR 766
Query: 339 EPSYSHLRVFGCLCYPLFPSTTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRH 398
P S++RV GCLC+ + +N S + YK YDL + +SR
Sbjct: 767 SPDLSYMRVIGCLCHA---TNLVNT--------------STQKGYKLYDLEHQHFFVSRD 809
Query: 399 VIFDETQFPFAKLHNPQPY-TYGFMDDGPSPYVIHHLTSQPSLGQPA----QHDLPNTQP 453
++F+E FPF P+ T F+ SP H T QPA + +P P
Sbjct: 810 MVFNEAVFPFQSPALADPHDTPVFL---ASPPCSSH-TEDADAVQPAIITSEEIIPVASP 865
Query: 454 TTQPTTPEEQHAHSPPSSSPNPSPSTTATPSPPYQPTPISVPKPVTRSQHGIFKPKRQLN 513
P+ + H H PP + T PP + +RS H ++ ++
Sbjct: 866 ---PSAVSDDHLHPPPERR-----RSYRTGKPPIWQKDF-ITTSTSRSNHCLYPISDNID 916
Query: 514 LNTS-------VPRSPLPRNP---VSALRDPNWKMAMDDEFNALIKNKTWELVPRPPDVN 563
+ + S + P A D W AM +E AL NKTWE+V P
Sbjct: 917 YSCLSSTYQCYIASSSVETEPQFYYQAANDCRWVHAMKEEIQALEDNKTWEVVSLPKGKK 976
Query: 564 VIRSMWIFTHKEKSDGVFERYKARLVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALS 623
I W++ K K+ G ER+KARLV G Q+ G+D ETFSPVVK T+RTVL+LA+S
Sbjct: 977 AIGCKWVYKIKYKASGEIERFKARLVAKGYNQKEGLDYQETFSPVVKMVTLRTVLTLAVS 1036
Query: 624 KAWSIHQLDVKNAFLHGELKETVYMHQPMGFR-DPNLPNHVCLLKKSLYGLKQAPRAWYK 682
K W I Q+DV NAFL G+L E VYM P GF+ D VC L KSLYGLKQA R W
Sbjct: 1037 KGWDIQQMDVYNAFLQGDLIEEVYMQLPQGFQYDKTGDPKVCRLLKSLYGLKQASRQWNV 1096
Query: 683 RFADYVSTIGFSHSTSDHSLFIYRKGTAMAYILLYVDDIILTASSDALRVTIISLLSTEF 742
+ + GF S D+SL + R + +L+YVDD+++T SS L +L F
Sbjct: 1097 KLTTALLAAGFQQSHLDYSLMLKRTADGIVIVLIYVDDLLITGSSLQLIDDAKQVLKANF 1156
Query: 743 AMKDLGSLHYFLGIAVTHHTGGLFLSQRKYAAEIIERAGMAACKSSSTPVDTKPKLSANS 802
+KDLG+L YFLG+ + G+ + QRKYA E+I G+ K S TPV+ KL+
Sbjct: 1157 KIKDLGTLRYFLGMEFARNASGMLMHQRKYALELISDLGLGGSKPSVTPVELHLKLTTRE 1216
Query: 803 SAPH----------ADPSHYRSLAGALQYLTFTRPDIAYAVQQVCLFMHDPREEHMHALK 852
H ADP+ Y+ L G L YLT TRPDI++AVQ + FMH P+ HM A
Sbjct: 1217 FDLHVGSSGADSLLADPTEYQRLVGRLLYLTITRPDISFAVQHLSQFMHAPKVSHMEAAI 1276
Query: 853 RIVRYIQGTLDHGLHLYPSSTSTLISYTDADWGGCPDTRRSTSGYCVFLGDNLISWSAKR 912
R+V+Y++ GL++ + TL +Y DADWG C +TR+S +GY + G L+SW +K+
Sbjct: 1277 RVVKYVKQAPGLGLYMAVQTADTLQAYCDADWGSCINTRKSITGYMIQFGSALLSWKSKK 1336
Query: 913 QATLSRSSAEAEYRGVANVVSESCWLRNLLLELHCPIRKASLVYCDNVSAIYLSGNPVQH 972
Q T+SRSSAEAEYR +A+ V+E WL L EL P+ +YCD+ +AI ++ NPV H
Sbjct: 1337 QPTISRSSAEAEYRSLASTVAELVWLTGLFKELDMPLSLPVSLYCDSKAAIQIAANPVFH 1396
Query: 973 QRTKHIEMDIHSVREKVARGEVRVLHVPSRYQIADIFTKGL 1013
+RTKHI++D H +REKV G V + ++P++ Q ADI TKGL
Sbjct: 1397 ERTKHIDIDCHFIREKVQAGLVMIHYLPTQEQPADILTKGL 1437
>UniRef100_Q9SLF0 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1333
Score = 608 bits (1567), Expect = e-172
Identities = 369/1022 (36%), Positives = 545/1022 (53%), Gaps = 45/1022 (4%)
Query: 45 LTLKNVLHSPQLIKNLVSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESR--GDLY 102
L + +V + + +L+S+ + +N ++ V+D +T +M R G +
Sbjct: 313 LVMISVFYVEEFQSDLISIGQLMDENRCVLQMSDRFLVVQD-RTSRMVMGAGRRVGGTFH 371
Query: 103 PITTSQAISPSTFAALAPSLWHARLGHPGAPVVDSLRKNKFIECNRASGSHICHSCSLGK 162
+T A S + LWH+R+GHP A VV SL + + + C C K
Sbjct: 372 FRSTEIAASVTVKEEKNYELWHSRMGHPAARVV-SLIPESSVSVSSTHLNKACDVCHRAK 430
Query: 163 HIKLPFVSSNSCTIMPFDIIHSDIWTS-PVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQ 221
+ F S + T+ F++I+ D+W S +G RY++ +DDYS +W + L+ KS+
Sbjct: 431 QTRNSFPLSINKTLRIFELIYCDLWGPYRTPSHTGARYFLTIIDDYSRGVWLYLLNDKSE 490
Query: 222 VFSIFLSFRTFIRTQFEREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGK 281
+F QF ++K ++ DNG EF +F +E GV SC T +N +
Sbjct: 491 APCHLKNFFAMTDRQFNVKIKTVRSDNGTEF--LCLTKFFQEQGVIHERSCVATPERNDR 548
Query: 282 AERKIRTINNIIRTLLVHASLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQKEPS 341
ERK R + N+ R L A+LP FW + A YLIN P+ L +P + L++K+P
Sbjct: 549 VERKHRHLLNVARALRFQANLPIQFWGECVLTAAYLINRTPSSVLNDSTPYERLHKKQPR 608
Query: 342 YSHLRVFGCLCYPLFPSTTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIF 401
+ HLRVFG LCY + +K RS CVF+GYP + ++ +DL + +SR V+F
Sbjct: 609 FDHLRVFGSLCYAHNRNRGGDKFAERSRRCVFVGYPHGQKGWRLFDLEQNEFFVSRDVVF 668
Query: 402 DETQFPFAKLHNPQPYTYGFMDDGPSPYVIHHLTSQPSLGQPAQHDLP---NTQPTTQPT 458
E +FPF ++ + Q + +P V + + LGQ A P ++ + T
Sbjct: 669 SELEFPF-RISHEQNVIEEEEEALWAPIVDGLIEEEVHLGQNAGPTPPICVSSPISPSAT 727
Query: 459 TPEEQHAHSPPSSS---PNPSPSTTATPSPPYQPT-----PISVPKPVTR---SQHGIFK 507
+ +H+ S P + P P+ STT+ SP PT P+S KP T + +
Sbjct: 728 SSRSEHSTSSPLDTEVVPTPATSTTSASSPS-SPTNLQFLPLSRAKPTTAQAVAPPAVPP 786
Query: 508 PKRQLNLNTSVP--------RSPLPRNPVSALRDPNWKMAMDDE-------------FNA 546
P+RQ N + P + + + S L +++ D+ +A
Sbjct: 787 PRRQSTRNKAPPVTLKDFVVNTTVCQESPSKLNSILYQLQKRDDTRRFSASHTTYVAIDA 846
Query: 547 LIKNKTWELVPRPPDVNVIRSMWIFTHKEKSDGVFERYKARLVGDGKTQQVGVDCGETFS 606
+N TW + PP I S W++ K SDG ERYKARLV G Q+ G D GETF+
Sbjct: 847 QEENHTWTIEDLPPGKRAIGSQWVYKVKHNSDGSVERYKARLVALGNKQKEGEDYGETFA 906
Query: 607 PVVKPATIRTVLSLALSKAWSIHQLDVKNAFLHGELKETVYMHQPMGFRDPNLPNHVCLL 666
PV K AT+R L +A+ + W IHQ+DV NAFLHG+L+E VYM P GF + + PN VC L
Sbjct: 907 PVAKMATVRLFLDVAVKRNWEIHQMDVHNAFLHGDLREEVYMKLPPGF-EASHPNKVCRL 965
Query: 667 KKSLYGLKQAPRAWYKRFADYVSTIGFSHSTSDHSLFIYRKGTAMAYILLYVDDIILTAS 726
+K+LYGLKQAPR W+++ + GF S +D+SLF KG+ IL+YVDD+I+T +
Sbjct: 966 RKALYGLKQAPRCWFEKLTTALKRYGFQQSLADYSLFTLVKGSVRIKILIYVDDLIITGN 1025
Query: 727 SDALRVTIISLLSTEFAMKDLGSLHYFLGIAVTHHTGGLFLSQRKYAAEIIERAGMAACK 786
S L++ F MKDLG L YFLGI V T G+++ QRKYA +II G+ K
Sbjct: 1026 SQRATQQFKEYLASCFHMKDLGPLKYFLGIEVARSTTGIYICQRKYALDIISETGLLGVK 1085
Query: 787 SSSTPVDTKPKLSANSSAPHADPSHYRSLAGALQYLTFTRPDIAYAVQQVCLFMHDPREE 846
++ P++ KL ++S DP YR L G L YL TR D+A++V + FM +PRE+
Sbjct: 1086 PANFPLEQNHKLGLSTSPLLTDPQRYRRLVGRLIYLAVTRLDLAFSVHILARFMQEPRED 1145
Query: 847 HMHALKRIVRYIQGTLDHGLHLYPSSTSTLISYTDADWGGCPDTRRSTSGYCVFLGDNLI 906
H A R+VRY++ G+ L S + + D+DW G P +RRS +GY V GD+ I
Sbjct: 1146 HWAAALRVVRYLKADPGQGVFLRRSGDFQITGWCDSDWAGDPMSRRSVTGYFVQFGDSPI 1205
Query: 907 SWSAKRQATLSRSSAEAEYRGVANVVSESCWLRNLLLELHCPIRKASLVYCDNVSAIYLS 966
SW K+Q T+S+SSAEAEYR ++ + SE WL+ LL L + ++ CD+ SAIY++
Sbjct: 1206 SWKTKKQDTVSKSSAEAEYRAMSFLASELLWLKQLLFSLGVSHVQPMIMCCDSKSAIYIA 1265
Query: 967 GNPVQHQRTKHIEMDIHSVREKVARGEVRVLHVPSRYQIADIFTKGLPLVLFEDFRNNLS 1026
NPV H+RTKHIE+D H VR++ +G + HV + Q+ADIFTK L F FR L
Sbjct: 1266 TNPVFHERTKHIEIDYHFVRDEFVKGVITPRHVGTTSQLADIFTKPLGRDCFSAFRIKLG 1325
Query: 1027 VR 1028
+R
Sbjct: 1326 IR 1327
>UniRef100_Q5XWK9 Gag-pol polyprotein-like [Solanum tuberosum]
Length = 1212
Score = 599 bits (1545), Expect = e-169
Identities = 350/916 (38%), Positives = 503/916 (54%), Gaps = 54/916 (5%)
Query: 1 MTSEQGNLSSYFNLSNNRGIIVGNGHSIPIRGYSHTNLSFPNPPLTLKNVLHSPQLIKNL 60
MT+ L + I + NG ++PI T + P T KNV SP+L +L
Sbjct: 330 MTNSTSILKNVRKYQGPSQIQIANGSNLPI-----TKVGDITP--TFKNVFVSPKLSTSL 382
Query: 61 VSVRKFTTDNSVSVEFDPFGFSVKDFQTGMRLMRCESRGDLYPITTSQAISP-----STF 115
+SV + DN+ V F G V+D +G + + G L+PI S I P T
Sbjct: 383 ISVGQLV-DNNCDVNFSRNGCLVQDQVSGTIIAKGPKVGRLFPIHFS--IPPVLSFACTS 439
Query: 116 AALAPSLWHARLGHPGAPVVDSL-------RKNKFIECNRASGSHICHSCSLGKHIKLPF 168
A +WH RLGHP + V+ + KNKF + S C +C LGK LPF
Sbjct: 440 TASKTEVWHKRLGHPNSVVLSHISNSGLLGNKNKF-----SVASIDCSTCKLGKSKTLPF 494
Query: 169 VSSNSCTIMPFDIIHSDIW-TSPVLSSSGHRYYVLFVDDYSNFLWTFPLSKKSQVFSIFL 227
+ S FD+IHSD+W SP++S + +Y++ F+DDYS F W + L KS+VFS+F
Sbjct: 495 PNFGSRATKCFDVIHSDVWGISPIISHAHFKYFMTFIDDYSRFTWVYFLRSKSEVFSMFK 554
Query: 228 SFRTFIRTQFEREVKNIQCDNGKEFDNRHFWEFCKENGVAFRLSCPHTSSQNGKAERKIR 287
+F +I TQF +K ++ D+G E+ + F +F + G+ + SCP+T QNG AERK R
Sbjct: 555 TFLAYIETQFSTCIKLLRSDSGGEYMSYEFKKFLLDKGIVSQHSCPYTPQQNGVAERKNR 614
Query: 288 TINNIIRTLLVHASLPPSFWHHALQMATYLINILPNKQLAYQSPLKILYQKEPSYSHLRV 347
+ ++ RTLL+ +S+P +W AL A YLIN LP+K L +SP LY + P+YS
Sbjct: 615 HLLDVTRTLLIESSVPSKYWVEALSTAVYLINRLPSKVLNLESPYFRLYHQNPNYSDFHT 674
Query: 348 FGCLCYPLFPSTTINKLQARSTPCVFLGYPSNHRSYKCYDLSSRKIIISRHVIFDETQFP 407
FGC+C+ P + NKL +ST C F+GY ++ + + CYD S K ISR+V+F E Q+
Sbjct: 675 FGCVCFVHLPPSQCNKLSVQSTKCAFMGYSTSQKGFICYDPCSHKFRISRNVVFFENQYF 734
Query: 408 FAKLHNPQPYTYGFMDDGPSPYVIHHLTSQPSLGQPAQHDLPNTQPTTQPTTPEEQHAHS 467
F P ++ + P L P DL ++ +P E+ +
Sbjct: 735 F-------------------PTIVDLSSVSPLL--PTFEDLSSSFKRFKPGFVYERRRPT 773
Query: 468 PPSSSPNPSPSTTATPSPPYQPTPISVPKPVTRSQHGIFKPKRQLNLNTSVPRSPLPRNP 527
P + +P P T P + + S P TR + + ++++ +P
Sbjct: 774 LPYPNTDPPPETA--PQLESENSSRSGPLEPTRRSTRVSRTPNWYGFSSTLSNISVPSCY 831
Query: 528 VSALRDPNWKMAMDDEFNALIKNKTWELVPRPPDVNVIRSMWIFTHKEKSDGVFERYKAR 587
A + W+ AM++E AL +N TW++V P +V I W+++ K SDG +RYKAR
Sbjct: 832 SQASKHECWQKAMEEELLALKENDTWDIVSCPSNVRPIGCKWVYSIKLHSDGTLDRYKAR 891
Query: 588 LVGDGKTQQVGVDCGETFSPVVKPATIRTVLSLALSKAWSIHQLDVKNAFLHGELKETVY 647
LV G Q+ GVD ETF+PV K T+RT++++A S+ WS++Q DVKNAFLHG+LKE +Y
Sbjct: 892 LVVLGNRQEYGVDYEETFAPVAKMTTVRTIIAIAASQNWSLYQKDVKNAFLHGDLKEDIY 951
Query: 648 MHQPMG-FRDPNLPNHVCLLKKSLYGLKQAPRAWYKRFADYVSTIGFSHSTSDHSLFIYR 706
M P F P + VC LK+SLYGLKQAPRAW+ +F + F S D SLF+ +
Sbjct: 952 MKPPPDLFSSPT--SDVCKLKRSLYGLKQAPRAWFDKFRSTLLQFSFELSKYDSSLFLRK 1009
Query: 707 KGTAMAYILLYVDDIILTASSDALRVTIISLLSTEFAMKDLGSLHYFLGIAVTHHTGGLF 766
T+ +L+YVDDII+T + +L + L F MKDLG+L YFLG+ V + G+F
Sbjct: 1010 TSTSCVLLLVYVDDIIITGTDSSLITCLQQQLKDSFHMKDLGTLTYFLGLEVHNVASGVF 1069
Query: 767 LSQRKYAAEIIERAGMAACKSSSTPVDTKPKLSANSSAPHADPSHYRSLAGALQYLTFTR 826
L+Q KY ++I AG+ S TP++ K DP+ +R L G+L YLT TR
Sbjct: 1070 LNQHKYTQDLISLAGLQVSSSVDTPLEMNVKYRREEGDLLPDPTIFRQLVGSLNYLTITR 1129
Query: 827 PDIAYAVQQVCLFMHDPREEHMHALKRIVRYIQGTLDHGLHLYPSSTSTLISYTDADWGG 886
PDI++AVQQV FM PR H+ A+ I+RY+ GT GL S L +++D+DW G
Sbjct: 1130 PDISFAVQQVSQFMQAPRHLHLVAVCHIIRYLLGTSTRGLFFPSGSPIRLNAFSDSDWAG 1189
Query: 887 CPDTRRSTSGYCVFLG 902
CPDTRRS SG+C+FLG
Sbjct: 1190 CPDTRRSVSGWCMFLG 1205
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.135 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,831,103,304
Number of Sequences: 2790947
Number of extensions: 83263410
Number of successful extensions: 479000
Number of sequences better than 10.0: 5713
Number of HSP's better than 10.0 without gapping: 2497
Number of HSP's successfully gapped in prelim test: 3499
Number of HSP's that attempted gapping in prelim test: 395026
Number of HSP's gapped (non-prelim): 32458
length of query: 1038
length of database: 848,049,833
effective HSP length: 138
effective length of query: 900
effective length of database: 462,899,147
effective search space: 416609232300
effective search space used: 416609232300
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 80 (35.4 bits)
Medicago: description of AC146632.4


