

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146630.4 + phase: 0
(100 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
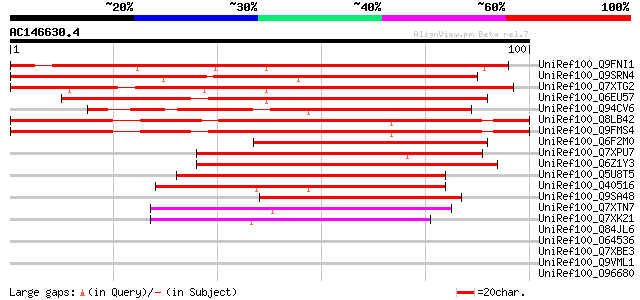
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FNI1 Gb|AAF02129.1 [Arabidopsis thaliana] 125 2e-28
UniRef100_Q9SRN4 T19F11.1 protein [Arabidopsis thaliana] 116 1e-25
UniRef100_Q7XTG2 OJ991214_12.5 protein [Oryza sativa] 99 2e-20
UniRef100_Q6EU57 Hypothetical protein OJ1112_G07.32 [Oryza sativa] 91 7e-18
UniRef100_Q94CV6 P0423B08.15 protein [Oryza sativa] 84 8e-16
UniRef100_Q8LB42 Hypothetical protein [Arabidopsis thaliana] 79 3e-14
UniRef100_Q9FMS4 Gb|AAF02129.1 [Arabidopsis thaliana] 78 4e-14
UniRef100_Q6F2M0 Hypothetical protein OSJNBa0020H14.5 [Oryza sat... 71 5e-12
UniRef100_Q7XPU7 OSJNBa0088H09.9 protein [Oryza sativa] 69 4e-11
UniRef100_Q6Z1Y3 Hypothetical protein B1147B12.27 [Oryza sativa] 56 2e-07
UniRef100_Q5U8T5 Cyclin B-like protein [Nicotiana tabacum] 54 9e-07
UniRef100_Q40516 B-type cyclin [Nicotiana tabacum] 51 6e-06
UniRef100_Q9SA48 F3O9.30 [Arabidopsis thaliana] 50 1e-05
UniRef100_Q7XTN7 OSJNBa0093O08.10 protein [Oryza sativa] 50 2e-05
UniRef100_Q7XK21 OSJNBa0044K18.24 protein [Oryza sativa] 46 2e-04
UniRef100_Q84JL6 Hypothetical protein At1g79160 [Arabidopsis tha... 42 0.004
UniRef100_O64536 YUP8H12R.23 protein [Arabidopsis thaliana] 42 0.004
UniRef100_Q7XBE3 Humj1 [Humulus japonicus] 39 0.023
UniRef100_Q9VML1 CG9019-PA [Drosophila melanogaster] 39 0.039
UniRef100_O96680 Dissatisfaction [Drosophila melanogaster] 39 0.039
>UniRef100_Q9FNI1 Gb|AAF02129.1 [Arabidopsis thaliana]
Length = 122
Score = 125 bits (315), Expect = 2e-28
Identities = 74/110 (67%), Positives = 82/110 (74%), Gaps = 17/110 (15%)
Query: 1 MSRRNGSGPKLDLKLNLSPPRVN-RRMESSPTRSASVSP---PSSCVSSENG-------- 48
MSRR+ PKL+LKLNLSPP + RRM SP+RSA+ SP PSSCVSSE
Sbjct: 1 MSRRS---PKLELKLNLSPPTSSQRRMVRSPSRSATTSPTSPPSSCVSSEMNQDEPSVRY 57
Query: 49 SSSPEATSMLLVGCPRCLMYVMLSENDPKCPKCHSTVLLDFL--NAENNN 96
S+SPE TSM+LVGCPRCLMYVMLSE+DPKCPKC STVLLDFL NA N N
Sbjct: 58 STSPETTSMVLVGCPRCLMYVMLSEDDPKCPKCKSTVLLDFLHENATNAN 107
>UniRef100_Q9SRN4 T19F11.1 protein [Arabidopsis thaliana]
Length = 117
Score = 116 bits (290), Expect = 1e-25
Identities = 63/94 (67%), Positives = 70/94 (74%), Gaps = 5/94 (5%)
Query: 1 MSRRNGSGPKLDLKLNLSPPRVNRRMES---SPTRSASVSPPSSCVSSENGSSSPEA-TS 56
MSRRN +GPKL+L+LNLSPP S SP RS + SP SSCVSSE E TS
Sbjct: 1 MSRRNKNGPKLELRLNLSPPPSQASQMSLVRSPNRSNTTSP-SSCVSSETNQEENETITS 59
Query: 57 MLLVGCPRCLMYVMLSENDPKCPKCHSTVLLDFL 90
M+LVGCPRCLMYVMLS++DPKCPKC STVLLDFL
Sbjct: 60 MVLVGCPRCLMYVMLSDDDPKCPKCKSTVLLDFL 93
>UniRef100_Q7XTG2 OJ991214_12.5 protein [Oryza sativa]
Length = 116
Score = 99.4 bits (246), Expect = 2e-20
Identities = 56/106 (52%), Positives = 71/106 (66%), Gaps = 12/106 (11%)
Query: 1 MSRRNGSGPK-LDLKLNLSPPRVNRRMESSPTRSASV---SPPSSCVSSENG-----SSS 51
MSR N + +DLKLNLS P R +SS R+ + S PSSC+SSEN S+S
Sbjct: 1 MSRSNKKSSRGIDLKLNLSLPA---RGDSSSRRAMAADEESSPSSCLSSENEHGLQWSNS 57
Query: 52 PEATSMLLVGCPRCLMYVMLSENDPKCPKCHSTVLLDFLNAENNNN 97
PEATSM+L CPRC +YVML ++DP+CP+C S V+LDFL +N NN
Sbjct: 58 PEATSMVLAACPRCFIYVMLPQDDPRCPQCKSPVILDFLQQDNGNN 103
>UniRef100_Q6EU57 Hypothetical protein OJ1112_G07.32 [Oryza sativa]
Length = 123
Score = 90.9 bits (224), Expect = 7e-18
Identities = 50/96 (52%), Positives = 61/96 (63%), Gaps = 17/96 (17%)
Query: 11 LDLKLNLSPPRVNRRMESSPTRSASVSPPSSCVSSENG--------------SSSPEATS 56
LDLKLNLS P V R + + +S PSSC+SSE+ S SPEATS
Sbjct: 15 LDLKLNLSLPAVARAVSPAADDESS---PSSCLSSESELRQQHGGGGGQLQWSDSPEATS 71
Query: 57 MLLVGCPRCLMYVMLSENDPKCPKCHSTVLLDFLNA 92
M+L CPRC +YVML+E DP+CPKC S V+LDFL+A
Sbjct: 72 MVLAACPRCFLYVMLAEADPRCPKCRSPVILDFLHA 107
>UniRef100_Q94CV6 P0423B08.15 protein [Oryza sativa]
Length = 129
Score = 84.0 bits (206), Expect = 8e-16
Identities = 46/80 (57%), Positives = 54/80 (67%), Gaps = 15/80 (18%)
Query: 16 NLSPPRVNRRMESSPTRSASVSPPSSCVSSENGSSSPEATS------MLLVGCPRCLMYV 69
N SP RR+ SS S++ SPPSSCVSSE SP+A M+L GCPRC+MYV
Sbjct: 47 NTSP----RRLSSSS--SSTASPPSSCVSSEG---SPDAAGGGAGPPMVLAGCPRCMMYV 97
Query: 70 MLSENDPKCPKCHSTVLLDF 89
MLS DP+CP+CHS VLLDF
Sbjct: 98 MLSREDPRCPRCHSAVLLDF 117
>UniRef100_Q8LB42 Hypothetical protein [Arabidopsis thaliana]
Length = 93
Score = 79.0 bits (193), Expect = 3e-14
Identities = 47/101 (46%), Positives = 64/101 (62%), Gaps = 11/101 (10%)
Query: 1 MSRRNGSGPKLDLKLNLSPPRVNRRMESSPTRSASVSPPSSCVSSENGSSSPEATSMLLV 60
M++ S LDLKLN+SP ++SS +S+S S S E EA SM++V
Sbjct: 1 MNQEAASNLGLDLKLNISPS-----LDSSLLTESSLS---SLCSEEAEGGGGEAKSMVVV 52
Query: 61 GCPRCLMYVMLS-ENDPKCPKCHSTVLLDFLNAENNNNKKS 100
GCP C+MY++ S ENDP+CP+C+S VLLDFL N++KKS
Sbjct: 53 GCPNCIMYIITSLENDPRCPRCNSHVLLDFLT--GNHSKKS 91
>UniRef100_Q9FMS4 Gb|AAF02129.1 [Arabidopsis thaliana]
Length = 93
Score = 78.2 bits (191), Expect = 4e-14
Identities = 47/101 (46%), Positives = 63/101 (61%), Gaps = 11/101 (10%)
Query: 1 MSRRNGSGPKLDLKLNLSPPRVNRRMESSPTRSASVSPPSSCVSSENGSSSPEATSMLLV 60
M++ S LDLKLN+SP ++SS +S SS S E EA SM++V
Sbjct: 1 MNQEAASNLGLDLKLNISPS-----LDSSLLTESS---SSSLCSEEAEGGGGEAKSMVVV 52
Query: 61 GCPRCLMYVMLS-ENDPKCPKCHSTVLLDFLNAENNNNKKS 100
GCP C+MY++ S ENDP+CP+C+S VLLDFL N++KKS
Sbjct: 53 GCPNCIMYIITSLENDPRCPRCNSHVLLDFLT--GNHSKKS 91
>UniRef100_Q6F2M0 Hypothetical protein OSJNBa0020H14.5 [Oryza sativa]
Length = 621
Score = 71.2 bits (173), Expect = 5e-12
Identities = 30/45 (66%), Positives = 35/45 (77%)
Query: 48 GSSSPEATSMLLVGCPRCLMYVMLSENDPKCPKCHSTVLLDFLNA 92
G ++ M+L GCPRC+MYVMLS DPKCPKCHSTVLLDF +A
Sbjct: 136 GGGGGGSSGMILAGCPRCMMYVMLSREDPKCPKCHSTVLLDFNDA 180
>UniRef100_Q7XPU7 OSJNBa0088H09.9 protein [Oryza sativa]
Length = 81
Score = 68.6 bits (166), Expect = 4e-11
Identities = 33/57 (57%), Positives = 41/57 (71%), Gaps = 2/57 (3%)
Query: 37 SPPSSCVSSENGSSSPEATSMLLVGCPRCLMYVMLSEND--PKCPKCHSTVLLDFLN 91
SP SSCVSS+ + A M++ GCP+CLMYVMLSE + PKCP+C S VLL FL+
Sbjct: 7 SPASSCVSSDAEEEAAVAKPMVVAGCPQCLMYVMLSEEEQQPKCPRCKSPVLLHFLH 63
>UniRef100_Q6Z1Y3 Hypothetical protein B1147B12.27 [Oryza sativa]
Length = 213
Score = 56.2 bits (134), Expect = 2e-07
Identities = 27/58 (46%), Positives = 37/58 (63%)
Query: 37 SPPSSCVSSENGSSSPEATSMLLVGCPRCLMYVMLSENDPKCPKCHSTVLLDFLNAEN 94
SP SCVSSE A +M++ GCPRCLMYVML+ ++ K P+C S D+++ N
Sbjct: 3 SPARSCVSSEAEDQQAAAAAMVVAGCPRCLMYVMLAVSNTKQPRCPSGFGDDWISQVN 60
>UniRef100_Q5U8T5 Cyclin B-like protein [Nicotiana tabacum]
Length = 473
Score = 53.9 bits (128), Expect = 9e-07
Identities = 23/52 (44%), Positives = 37/52 (70%)
Query: 33 SASVSPPSSCVSSENGSSSPEATSMLLVGCPRCLMYVMLSENDPKCPKCHST 84
S+SVS S ++ +++ ++ M++VGC RC MYVM++E DP+CP+C ST
Sbjct: 416 SSSVSSGRSDLAKSTKAAATSSSPMVVVGCQRCHMYVMVTEADPRCPQCKST 467
>UniRef100_Q40516 B-type cyclin [Nicotiana tabacum]
Length = 473
Score = 51.2 bits (121), Expect = 6e-06
Identities = 27/62 (43%), Positives = 39/62 (62%), Gaps = 6/62 (9%)
Query: 29 SPTRSASVSPPSSCVSSE---NGSSSPEATS---MLLVGCPRCLMYVMLSENDPKCPKCH 82
+P +S + SS +S + S+ ATS M++VGC RC MYVM++E DP+CP+C
Sbjct: 406 TPAKSLLAASSSSVLSEQADLRKSTEAAATSSSKMVVVGCQRCHMYVMVTEADPRCPQCK 465
Query: 83 ST 84
ST
Sbjct: 466 ST 467
>UniRef100_Q9SA48 F3O9.30 [Arabidopsis thaliana]
Length = 259
Score = 50.1 bits (118), Expect = 1e-05
Identities = 20/39 (51%), Positives = 28/39 (71%)
Query: 49 SSSPEATSMLLVGCPRCLMYVMLSENDPKCPKCHSTVLL 87
+S+ A S + GCP CL YV +++N+PKCP+CHS V L
Sbjct: 207 ASATVAASQVAAGCPGCLSYVFVAKNNPKCPRCHSFVPL 245
>UniRef100_Q7XTN7 OSJNBa0093O08.10 protein [Oryza sativa]
Length = 238
Score = 49.7 bits (117), Expect = 2e-05
Identities = 24/65 (36%), Positives = 36/65 (54%), Gaps = 7/65 (10%)
Query: 28 SSPTRSASVSPPSSCVSSENGS-------SSPEATSMLLVGCPRCLMYVMLSENDPKCPK 80
+S + SAS S SS + + S ++P + SM CP CL YV+++E DP+CP+
Sbjct: 140 TSSSSSASTSSSSSSIGKRHRSPPSGGAVATPASPSMRAAACPSCLTYVLIAEADPRCPR 199
Query: 81 CHSTV 85
C V
Sbjct: 200 CAGNV 204
>UniRef100_Q7XK21 OSJNBa0044K18.24 protein [Oryza sativa]
Length = 748
Score = 45.8 bits (107), Expect = 2e-04
Identities = 23/63 (36%), Positives = 35/63 (55%), Gaps = 9/63 (14%)
Query: 28 SSPTRSASVSPPSSCVSS---------ENGSSSPEATSMLLVGCPRCLMYVMLSENDPKC 78
+SP S+S S +S SS G+++P + SM P CL YV+++E DP+C
Sbjct: 137 ASPATSSSSSASTSSSSSIGKCHRSPPAGGAATPASPSMRAAAYPSCLTYVLIAEADPRC 196
Query: 79 PKC 81
P+C
Sbjct: 197 PRC 199
>UniRef100_Q84JL6 Hypothetical protein At1g79160 [Arabidopsis thaliana]
Length = 239
Score = 42.0 bits (97), Expect = 0.004
Identities = 16/32 (50%), Positives = 23/32 (71%)
Query: 56 SMLLVGCPRCLMYVMLSENDPKCPKCHSTVLL 87
S ++ GCP CL YV++ N+PKCP+C + V L
Sbjct: 191 SPVVAGCPGCLSYVLVMMNNPKCPRCDTIVPL 222
>UniRef100_O64536 YUP8H12R.23 protein [Arabidopsis thaliana]
Length = 204
Score = 42.0 bits (97), Expect = 0.004
Identities = 16/32 (50%), Positives = 23/32 (71%)
Query: 56 SMLLVGCPRCLMYVMLSENDPKCPKCHSTVLL 87
S ++ GCP CL YV++ N+PKCP+C + V L
Sbjct: 156 SPVVAGCPGCLSYVLVMMNNPKCPRCDTIVPL 187
>UniRef100_Q7XBE3 Humj1 [Humulus japonicus]
Length = 155
Score = 39.3 bits (90), Expect = 0.023
Identities = 20/54 (37%), Positives = 28/54 (51%), Gaps = 1/54 (1%)
Query: 29 SPTRSASVSPPSSCVSSENGSSSPEATSMLLVG-CPRCLMYVMLSENDPKCPKC 81
S S+ SPP + S E+ E +L+VG C C MY M+ + +CPKC
Sbjct: 87 SEESSSESSPPPATTSRESQYRGAEKDHVLVVGGCKSCFMYFMVPKQVQECPKC 140
>UniRef100_Q9VML1 CG9019-PA [Drosophila melanogaster]
Length = 691
Score = 38.5 bits (88), Expect = 0.039
Identities = 32/97 (32%), Positives = 43/97 (43%), Gaps = 9/97 (9%)
Query: 4 RNGSGPKLDLKLNLSPPRVNRRMESSPTRSASVSPPSSCVSSENGSSSPEATSMLLVGCP 63
R+G G + L L PRVN E T S S SPP S GS SP T G P
Sbjct: 328 RSGGGEE-GLSLGSESPRVNVETE---TPSPSNSPPLSA-----GSISPAPTLTTSSGSP 378
Query: 64 RCLMYVMLSENDPKCPKCHSTVLLDFLNAENNNNKKS 100
+ S ++ P H+++++ N NNNN +
Sbjct: 379 QHRQMSRHSLSEATTPPSHASLMICASNNNNNNNNNN 415
>UniRef100_O96680 Dissatisfaction [Drosophila melanogaster]
Length = 693
Score = 38.5 bits (88), Expect = 0.039
Identities = 32/97 (32%), Positives = 43/97 (43%), Gaps = 9/97 (9%)
Query: 4 RNGSGPKLDLKLNLSPPRVNRRMESSPTRSASVSPPSSCVSSENGSSSPEATSMLLVGCP 63
R+G G + L L PRVN E T S S SPP S GS SP T G P
Sbjct: 328 RSGGGEE-GLSLGSESPRVNVETE---TPSPSNSPPLSA-----GSISPAPTLTTSSGSP 378
Query: 64 RCLMYVMLSENDPKCPKCHSTVLLDFLNAENNNNKKS 100
+ S ++ P H+++++ N NNNN +
Sbjct: 379 QHRQMSRHSLSEATTPPSHASLMICASNNNNNNNNNN 415
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.310 0.125 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 174,713,811
Number of Sequences: 2790947
Number of extensions: 6057763
Number of successful extensions: 24437
Number of sequences better than 10.0: 73
Number of HSP's better than 10.0 without gapping: 40
Number of HSP's successfully gapped in prelim test: 33
Number of HSP's that attempted gapping in prelim test: 24340
Number of HSP's gapped (non-prelim): 113
length of query: 100
length of database: 848,049,833
effective HSP length: 76
effective length of query: 24
effective length of database: 635,937,861
effective search space: 15262508664
effective search space used: 15262508664
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 68 (30.8 bits)
Medicago: description of AC146630.4


