

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146630.11 + phase: 0
(265 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
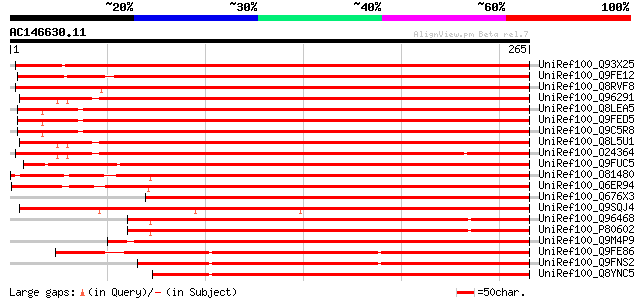
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q93X25 2-Cys peroxiredoxin [Pisum sativum] 475 e-133
UniRef100_Q9FE12 Peroxiredoxin precursor [Phaseolus vulgaris] 436 e-121
UniRef100_Q8RVF8 Thioredoxin peroxidase [Nicotiana tabacum] 412 e-114
UniRef100_Q96291 2-cys peroxiredoxin BAS1, chloroplast precursor... 400 e-110
UniRef100_Q8LEA5 2-cys peroxiredoxin-like protein [Arabidopsis t... 397 e-109
UniRef100_Q9FED5 Putative 2-cys peroxiredoxin protein [Arabidops... 396 e-109
UniRef100_Q9C5R8 AT5g06290 [Arabidopsis thaliana] 394 e-108
UniRef100_Q8L5U1 Putative 2-cys peroxiredoxin BAS1 [Arabidopsis ... 393 e-108
UniRef100_O24364 2-cys peroxiredoxin BAS1, chloroplast precursor... 392 e-108
UniRef100_Q9FUC5 2-Cys peroxiredoxin [Brassica napus] 391 e-108
UniRef100_O81480 Thioredoxin peroxidase [Secale cereale] 380 e-104
UniRef100_Q6ER94 Putative thioredoxin peroxidase [Oryza sativa] 379 e-104
UniRef100_Q676X3 2-cys peroxiredoxin-like protein [Hyacinthus or... 375 e-103
UniRef100_Q9SQJ4 2Cys-peroxiredoxin [Brassica campestris] 367 e-100
UniRef100_Q96468 2-cys peroxiredoxin BAS1, chloroplast precursor... 365 e-100
UniRef100_P80602 2-cys peroxiredoxin BAS1, chloroplast precursor... 364 1e-99
UniRef100_Q9M4P9 2-Cys-peroxiredoxin precursor [Riccia fluitans] 354 1e-96
UniRef100_Q9FE86 Peroxiredoxin precursor [Chlamydomonas reinhard... 326 3e-88
UniRef100_Q9FNS2 Peroxiredoxin [Chlamydomonas reinhardtii] 317 2e-85
UniRef100_Q8YNC5 Peroxiredoxin [Anabaena sp.] 302 7e-81
>UniRef100_Q93X25 2-Cys peroxiredoxin [Pisum sativum]
Length = 263
Score = 475 bits (1223), Expect = e-133
Identities = 242/262 (92%), Positives = 249/262 (94%), Gaps = 1/262 (0%)
Query: 4 CSAPSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRRSF 63
CSAP ASLL SNPN LFSPK SSP LSSLSIPNA NSLPKLRTSLPLSLNR +SSRR+F
Sbjct: 3 CSAPFASLLYSNPNTLFSPKFSSP-RLSSLSIPNAPNSLPKLRTSLPLSLNRSSSSRRTF 61
Query: 64 VVRASSELPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEIT 123
VVRAS ELPLVGN+APDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEIT
Sbjct: 62 VVRASGELPLVGNSAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEIT 121
Query: 124 AFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYG 183
AFSDRHAEF +NTEILGVSVDSVFSHLAWVQ+DRKSGGLGDL YPLVSDVTKSIS+SYG
Sbjct: 122 AFSDRHAEFDAINTEILGVSVDSVFSHLAWVQSDRKSGGLGDLKYPLVSDVTKSISESYG 181
Query: 184 VLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVCPAG 243
VLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVCPAG
Sbjct: 182 VLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVCPAG 241
Query: 244 WKPGEKSMKPDPKLSKEYFSAV 265
WKPGEKSMKPDPK SKEYF+AV
Sbjct: 242 WKPGEKSMKPDPKGSKEYFAAV 263
>UniRef100_Q9FE12 Peroxiredoxin precursor [Phaseolus vulgaris]
Length = 260
Score = 436 bits (1121), Expect = e-121
Identities = 228/262 (87%), Positives = 236/262 (90%), Gaps = 6/262 (2%)
Query: 5 SAPSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPK-LRTSLPLSLNRFTSSRRSF 63
SAP ASL+SSNPNILFSPK S SSLS PN+ NSL K LRTSL N + R+F
Sbjct: 4 SAPCASLISSNPNILFSPKFPSSS-FSSLSFPNSPNSLFKPLRTSL----NPSSPPLRTF 58
Query: 64 VVRASSELPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEIT 123
V RASSELPLVGN APDFEAEAVFDQEFIKVKLS+YIGKKYVILFFYPLDFTFVCPTEIT
Sbjct: 59 VARASSELPLVGNTAPDFEAEAVFDQEFIKVKLSDYIGKKYVILFFYPLDFTFVCPTEIT 118
Query: 124 AFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYG 183
AFSDR+AEF LNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPL+SDVTKSISKSY
Sbjct: 119 AFSDRYAEFEALNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLISDVTKSISKSYD 178
Query: 184 VLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVCPAG 243
VLIPDQGIALRGLFIIDKEGVIQHSTINNL IGRSVDETKRTLQALQYVQENPDEVCPAG
Sbjct: 179 VLIPDQGIALRGLFIIDKEGVIQHSTINNLAIGRSVDETKRTLQALQYVQENPDEVCPAG 238
Query: 244 WKPGEKSMKPDPKLSKEYFSAV 265
WKPGEKSMKPDPKLSKEYFSA+
Sbjct: 239 WKPGEKSMKPDPKLSKEYFSAI 260
>UniRef100_Q8RVF8 Thioredoxin peroxidase [Nicotiana tabacum]
Length = 271
Score = 412 bits (1058), Expect = e-114
Identities = 216/269 (80%), Positives = 229/269 (84%), Gaps = 7/269 (2%)
Query: 4 CSAPSASLLSSNPNIL-FSPKLSSPPHLSS-LSIPNASNSLPKL-----RTSLPLSLNRF 56
CSA S +LLSSNP SPK S +S LS+P++ N L R + LS
Sbjct: 3 CSASSTALLSSNPKAASISPKSSFQAPISQCLSVPSSFNGLRNCKPFVSRVARSLSTRVA 62
Query: 57 TSSRRSFVVRASSELPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTF 116
S RR FVVRASSELPLVGN APDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTF
Sbjct: 63 QSQRRRFVVRASSELPLVGNQAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTF 122
Query: 117 VCPTEITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTK 176
VCPTEITAFSDR+ EF +LNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPL+SDVTK
Sbjct: 123 VCPTEITAFSDRYGEFEKLNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLISDVTK 182
Query: 177 SISKSYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENP 236
SISKSY VLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDET RTLQALQYVQ+NP
Sbjct: 183 SISKSYNVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETLRTLQALQYVQDNP 242
Query: 237 DEVCPAGWKPGEKSMKPDPKLSKEYFSAV 265
DEVCPAGWKPGEKSMKPDPK SKEYF+++
Sbjct: 243 DEVCPAGWKPGEKSMKPDPKGSKEYFASI 271
>UniRef100_Q96291 2-cys peroxiredoxin BAS1, chloroplast precursor [Arabidopsis
thaliana]
Length = 266
Score = 400 bits (1027), Expect = e-110
Identities = 210/265 (79%), Positives = 231/265 (86%), Gaps = 8/265 (3%)
Query: 6 APSASLLSSNPNILFSPK--LSSPP--HLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRR 61
A S +L+SS + +F K LSSP L +LS P+AS SL R+ + ++SRR
Sbjct: 5 ASSTTLISSPSSRVFPAKSSLSSPSVSFLRTLSSPSASASL---RSGFARRSSLSSTSRR 61
Query: 62 SFVVRASSE-LPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPT 120
SF V+A ++ LPLVGN APDFEAEAVFDQEFIKVKLS+YIGKKYVILFFYPLDFTFVCPT
Sbjct: 62 SFAVKAQADDLPLVGNKAPDFEAEAVFDQEFIKVKLSDYIGKKYVILFFYPLDFTFVCPT 121
Query: 121 EITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISK 180
EITAFSDRH+EF +LNTE+LGVSVDSVFSHLAWVQTDRKSGGLGDLNYPL+SDVTKSISK
Sbjct: 122 EITAFSDRHSEFEKLNTEVLGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLISDVTKSISK 181
Query: 181 SYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVC 240
S+GVLI DQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDET RTLQALQY+QENPDEVC
Sbjct: 182 SFGVLIHDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETMRTLQALQYIQENPDEVC 241
Query: 241 PAGWKPGEKSMKPDPKLSKEYFSAV 265
PAGWKPGEKSMKPDPKLSKEYFSA+
Sbjct: 242 PAGWKPGEKSMKPDPKLSKEYFSAI 266
>UniRef100_Q8LEA5 2-cys peroxiredoxin-like protein [Arabidopsis thaliana]
Length = 271
Score = 397 bits (1020), Expect = e-109
Identities = 210/268 (78%), Positives = 230/268 (85%), Gaps = 9/268 (3%)
Query: 5 SAPSASLLSSN------PNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTS 58
S+ S +LLSS+ + L SP +S P L S S +S+SL +SL ++
Sbjct: 6 SSSSTTLLSSSRVLLPSKSSLLSPTVSVPRTLHSSSA--SSSSLCSGFSSLGSLTTSRSA 63
Query: 59 SRRSFVVRASSE-LPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFV 117
SRR+F V+A ++ LPLVGN APDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFV
Sbjct: 64 SRRNFAVKAQADDLPLVGNKAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFV 123
Query: 118 CPTEITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKS 177
CPTEITAFSDR+ EF +LNTE+LGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSD+TKS
Sbjct: 124 CPTEITAFSDRYEEFEKLNTEVLGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDITKS 183
Query: 178 ISKSYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPD 237
ISKS+GVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDET RTLQALQYVQENPD
Sbjct: 184 ISKSFGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETMRTLQALQYVQENPD 243
Query: 238 EVCPAGWKPGEKSMKPDPKLSKEYFSAV 265
EVCPAGWKPGEKSMKPDPKLSKEYFSA+
Sbjct: 244 EVCPAGWKPGEKSMKPDPKLSKEYFSAI 271
>UniRef100_Q9FED5 Putative 2-cys peroxiredoxin protein [Arabidopsis thaliana]
Length = 273
Score = 396 bits (1018), Expect = e-109
Identities = 209/268 (77%), Positives = 230/268 (84%), Gaps = 9/268 (3%)
Query: 5 SAPSASLLSSN------PNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTS 58
S+ S +LLSS+ + L SP +S P + S S +S+SL +SL ++
Sbjct: 8 SSSSTTLLSSSRVLLPSKSSLLSPTVSFPRIIPSSSA--SSSSLCSGFSSLGSLTTNRSA 65
Query: 59 SRRSFVVRASSE-LPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFV 117
SRR+F V+A ++ LPLVGN APDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFV
Sbjct: 66 SRRNFAVKAQADDLPLVGNKAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFV 125
Query: 118 CPTEITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKS 177
CPTEITAFSDR+ EF +LNTE+LGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSD+TKS
Sbjct: 126 CPTEITAFSDRYEEFEKLNTEVLGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDITKS 185
Query: 178 ISKSYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPD 237
ISKS+GVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDET RTLQALQYVQENPD
Sbjct: 186 ISKSFGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETMRTLQALQYVQENPD 245
Query: 238 EVCPAGWKPGEKSMKPDPKLSKEYFSAV 265
EVCPAGWKPGEKSMKPDPKLSKEYFSA+
Sbjct: 246 EVCPAGWKPGEKSMKPDPKLSKEYFSAI 273
>UniRef100_Q9C5R8 AT5g06290 [Arabidopsis thaliana]
Length = 271
Score = 394 bits (1012), Expect = e-108
Identities = 208/268 (77%), Positives = 229/268 (84%), Gaps = 9/268 (3%)
Query: 5 SAPSASLLSSN------PNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTS 58
S+ S +LLSS+ + L SP +S P + S S +S+SL +SL ++
Sbjct: 6 SSSSTTLLSSSRVLLPSKSSLLSPTVSFPRIIPSSSA--SSSSLCSGFSSLGSLTTNRSA 63
Query: 59 SRRSFVVRASSE-LPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFV 117
SRR+F V+A ++ LPLVGN APDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFV
Sbjct: 64 SRRNFAVKAQADDLPLVGNKAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFV 123
Query: 118 CPTEITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKS 177
CPTEITAFSDR+ EF +LNTE+LGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSD+TKS
Sbjct: 124 CPTEITAFSDRYEEFEKLNTEVLGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDITKS 183
Query: 178 ISKSYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPD 237
ISKS+GVLIPDQGIALRGLFIIDKEGVIQHS INNLGIGRSVDET RTLQALQYVQENPD
Sbjct: 184 ISKSFGVLIPDQGIALRGLFIIDKEGVIQHSPINNLGIGRSVDETMRTLQALQYVQENPD 243
Query: 238 EVCPAGWKPGEKSMKPDPKLSKEYFSAV 265
EVCPAGWKPGEKSMKPDPKLSKEYFSA+
Sbjct: 244 EVCPAGWKPGEKSMKPDPKLSKEYFSAI 271
>UniRef100_Q8L5U1 Putative 2-cys peroxiredoxin BAS1 [Arabidopsis thaliana]
Length = 266
Score = 393 bits (1009), Expect = e-108
Identities = 207/265 (78%), Positives = 229/265 (86%), Gaps = 8/265 (3%)
Query: 6 APSASLLSSNPNILFSPK--LSSPP--HLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRR 61
A S +L+SS + +F K LSSP L +LS P+AS SL R+ + ++SRR
Sbjct: 5 ASSTTLISSPSSRVFPAKSSLSSPSVSFLRTLSSPSASASL---RSGFARRSSLSSTSRR 61
Query: 62 SFVVRASSE-LPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPT 120
SF V+A ++ LPLVGN APDFEAEAVFDQEFIKVKLS+YIGKKYVILFFYPLDFTFVCPT
Sbjct: 62 SFAVKAQADDLPLVGNKAPDFEAEAVFDQEFIKVKLSDYIGKKYVILFFYPLDFTFVCPT 121
Query: 121 EITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISK 180
EITAFSDRH+EF +LNTE+LGVSVDSVFSHLAWVQTDRKSGGLGDLNYPL+S TKSISK
Sbjct: 122 EITAFSDRHSEFEKLNTEVLGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLISYFTKSISK 181
Query: 181 SYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVC 240
S+GVLI DQGIALRGLFIIDKEGVIQHSTINNLGIG+SVDET RTLQALQY+QENPDEVC
Sbjct: 182 SFGVLIHDQGIALRGLFIIDKEGVIQHSTINNLGIGQSVDETMRTLQALQYIQENPDEVC 241
Query: 241 PAGWKPGEKSMKPDPKLSKEYFSAV 265
PAGWKPGEKSMKPDPKLSKEYFSA+
Sbjct: 242 PAGWKPGEKSMKPDPKLSKEYFSAI 266
>UniRef100_O24364 2-cys peroxiredoxin BAS1, chloroplast precursor [Spinacia oleracea]
Length = 265
Score = 392 bits (1007), Expect = e-108
Identities = 209/267 (78%), Positives = 229/267 (85%), Gaps = 9/267 (3%)
Query: 4 CSAPSASLLSSNPNILFSPK--LSSPP--HLSSLSIPNASNSLPKLRTSLPLSLNRFTSS 59
C A S +L+SS + +F K LSSP L +LS P+AS SL R+ + ++S
Sbjct: 3 CVASSTTLISSPSSRVFPAKSSLSSPSVSFLRTLSSPSASASL---RSGFARRSSLSSTS 59
Query: 60 RRSFVVRASSE-LPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVC 118
RRSF V+A ++ LPLVGN APDFEAEAVFDQEFIKVKLS+YIGKKYVILFFYPLDFTFVC
Sbjct: 60 RRSFAVKAQADDLPLVGNKAPDFEAEAVFDQEFIKVKLSDYIGKKYVILFFYPLDFTFVC 119
Query: 119 PTEITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSI 178
PTEITAFSDRH+EF +LNTE+LGVSVDSVFSHLAWVQTDRKSGGLGDLNYPL+SDVTKSI
Sbjct: 120 PTEITAFSDRHSEFEKLNTEVLGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLISDVTKSI 179
Query: 179 SKSYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDE 238
SKS+GVLI DQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDET RTLQALQY NPDE
Sbjct: 180 SKSFGVLIHDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETMRTLQALQYT-GNPDE 238
Query: 239 VCPAGWKPGEKSMKPDPKLSKEYFSAV 265
VCPAGWKPGEKSMKPDPKLSKEYFSA+
Sbjct: 239 VCPAGWKPGEKSMKPDPKLSKEYFSAI 265
>UniRef100_Q9FUC5 2-Cys peroxiredoxin [Brassica napus]
Length = 270
Score = 391 bits (1005), Expect = e-108
Identities = 205/259 (79%), Positives = 224/259 (86%), Gaps = 3/259 (1%)
Query: 8 SASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRRSFVVRA 67
S+ LL S P+ FSP S L S S+ +S+ + PL+ R +SSR SF V+A
Sbjct: 14 SSVLLPSKPSP-FSPAASFLRTLPSTSVSTSSSLRSCFSSISPLTCIR-SSSRPSFAVKA 71
Query: 68 SSE-LPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEITAFS 126
++ LPLVGN APDFEAEAVFDQEFIKVKLSEYIGKKYVILF YPLDFTFVCPTEITAFS
Sbjct: 72 QADDLPLVGNKAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFLYPLDFTFVCPTEITAFS 131
Query: 127 DRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYGVLI 186
DR+ EF +LNTE+LGVSVDSVFSHLAWVQT+RKSGGLGDLNYPLVSD+TKSISKS+GVLI
Sbjct: 132 DRYEEFEKLNTEVLGVSVDSVFSHLAWVQTERKSGGLGDLNYPLVSDITKSISKSFGVLI 191
Query: 187 PDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVCPAGWKP 246
PDQGIALRGLFIIDK+GVIQHSTINNLGIGRSVDET RTLQALQYVQENPDEVCPAGWKP
Sbjct: 192 PDQGIALRGLFIIDKKGVIQHSTINNLGIGRSVDETMRTLQALQYVQENPDEVCPAGWKP 251
Query: 247 GEKSMKPDPKLSKEYFSAV 265
GEKSMKPDPKLSKEYFSA+
Sbjct: 252 GEKSMKPDPKLSKEYFSAI 270
>UniRef100_O81480 Thioredoxin peroxidase [Secale cereale]
Length = 258
Score = 380 bits (977), Expect = e-104
Identities = 204/268 (76%), Positives = 222/268 (82%), Gaps = 13/268 (4%)
Query: 1 MACCSAPSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSR 60
MAC A SAS +S+ ++ SPK + P S+ P LR + R +R
Sbjct: 1 MAC--AFSASTVSTAAALVASPKPAGAP--SACRFPALRRGRAGLRCA------RLEDAR 50
Query: 61 -RSFVVRASSE--LPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFV 117
RSFV RA++E LPLVGN APDF AEAVFDQEFI VKLS+YIGKKYVILFFYPLDFTFV
Sbjct: 51 ARSFVARAAAEYDLPLVGNKAPDFAAEAVFDQEFINVKLSDYIGKKYVILFFYPLDFTFV 110
Query: 118 CPTEITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKS 177
CPTEITAFSDRH EF ++NTEILGVSVDSVFSHLAWVQT+RKSGGLGDL YPLVSDVTKS
Sbjct: 111 CPTEITAFSDRHEEFEKINTEILGVSVDSVFSHLAWVQTERKSGGLGDLKYPLVSDVTKS 170
Query: 178 ISKSYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPD 237
ISKS+GVLIPDQGIALRGLF+IDKEGVIQHSTINNLGIGRSVDET RTLQALQYVQENPD
Sbjct: 171 ISKSFGVLIPDQGIALRGLFMIDKEGVIQHSTINNLGIGRSVDETLRTLQALQYVQENPD 230
Query: 238 EVCPAGWKPGEKSMKPDPKLSKEYFSAV 265
EVCPAGWKPGEKSMKPDPK SKEYF+A+
Sbjct: 231 EVCPAGWKPGEKSMKPDPKGSKEYFAAI 258
>UniRef100_Q6ER94 Putative thioredoxin peroxidase [Oryza sativa]
Length = 261
Score = 379 bits (972), Expect = e-104
Identities = 199/267 (74%), Positives = 222/267 (82%), Gaps = 11/267 (4%)
Query: 2 ACCSAPSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRR 61
ACCS+ + ++ SS+ L ++P SLS+P A + P L LS + S+R
Sbjct: 3 ACCSSLATAVSSSSAKPLAGIPPAAP---HSLSLPRAPAARP-----LRLSASSSRSARA 54
Query: 62 S-FVVRASS--ELPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVC 118
S FV RA + PLVGN APDF+AEAVFDQEFI VKLS+YIGKKYVILFFYPLDFTFVC
Sbjct: 55 SSFVARAGGVDDAPLVGNKAPDFDAEAVFDQEFINVKLSDYIGKKYVILFFYPLDFTFVC 114
Query: 119 PTEITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSI 178
PTEITAFSDR+ EF +LNTEILGVS+DSVFSHLAWVQTDRKSGGLGDL YPL+SDVTKSI
Sbjct: 115 PTEITAFSDRYDEFEKLNTEILGVSIDSVFSHLAWVQTDRKSGGLGDLKYPLISDVTKSI 174
Query: 179 SKSYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDE 238
SKS+GVLIPDQGIALRGLFIIDKEGVIQHSTINNL IGRSVDET RTLQALQYVQ+NPDE
Sbjct: 175 SKSFGVLIPDQGIALRGLFIIDKEGVIQHSTINNLAIGRSVDETMRTLQALQYVQDNPDE 234
Query: 239 VCPAGWKPGEKSMKPDPKLSKEYFSAV 265
VCPAGWKPG+KSMKPDPK SKEYF+A+
Sbjct: 235 VCPAGWKPGDKSMKPDPKGSKEYFAAI 261
>UniRef100_Q676X3 2-cys peroxiredoxin-like protein [Hyacinthus orientalis]
Length = 196
Score = 375 bits (964), Expect = e-103
Identities = 183/196 (93%), Positives = 192/196 (97%)
Query: 70 ELPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEITAFSDRH 129
ELPLVGN+AP FEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEITAFSDR+
Sbjct: 1 ELPLVGNSAPGFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEITAFSDRY 60
Query: 130 AEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYGVLIPDQ 189
+EF ++NTE+LGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYGVLIPDQ
Sbjct: 61 SEFEKVNTEVLGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYGVLIPDQ 120
Query: 190 GIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVCPAGWKPGEK 249
GIALRGLFIIDKEGVIQHSTINNLGIGRSVDET RTLQALQYVQENPDEVCPAGWKPGEK
Sbjct: 121 GIALRGLFIIDKEGVIQHSTINNLGIGRSVDETMRTLQALQYVQENPDEVCPAGWKPGEK 180
Query: 250 SMKPDPKLSKEYFSAV 265
SMKPDPK SKEYF+++
Sbjct: 181 SMKPDPKRSKEYFASI 196
>UniRef100_Q9SQJ4 2Cys-peroxiredoxin [Brassica campestris]
Length = 273
Score = 367 bits (943), Expect = e-100
Identities = 200/269 (74%), Positives = 222/269 (82%), Gaps = 9/269 (3%)
Query: 6 APSASLLSSNPNILFSPKLSSPPHLSSLSIPNASNSLPK--LRTSLPL-SLNRFTSSRRS 62
A S +L+SS+ ++L + K S P S +P S+ P LR+ +PL S +SSRRS
Sbjct: 5 ASSTTLISSSASVLPATKSSLLPSPSLSFLPTLSSPSPSASLRSLVPLPSPQSASSSRRS 64
Query: 63 FVVRASSE-LPLVGNAAPDFEAEAVFDQEFIK---VKLSEYIGKKYVILFFYPLDFTFVC 118
F V+ ++ LPLVGN APDFEAE VFDQEFIK VKLS+YIGKKYVILFF PLDFTFVC
Sbjct: 65 FAVKGQTDDLPLVGNKAPDFEAEGVFDQEFIKFIKVKLSDYIGKKYVILFFLPLDFTFVC 124
Query: 119 PTEITAFSDRHAEFAELNTEILGVSVDSV--FSHLAWVQTDRKSGGLGDLNYPLVSDVTK 176
PTEITAFSDR+AEF +LNTE+LGVSVDSV FSHLA VQTDRK GGLGDLNYPL+SDVTK
Sbjct: 125 PTEITAFSDRYAEFEKLNTEVLGVSVDSVSVFSHLAGVQTDRKFGGLGDLNYPLISDVTK 184
Query: 177 SISKSYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENP 236
SISKS+GVLI DQGIALRGLFIIDKEGVIQHSTI NLGIGRSVDET RTLQALQY+QE P
Sbjct: 185 SISKSFGVLIHDQGIALRGLFIIDKEGVIQHSTIXNLGIGRSVDETMRTLQALQYIQEGP 244
Query: 237 DEVCPAGWKPGEKSMKPDPKLSKEYFSAV 265
EVCPAGWKPGEKSMKPDPKLSKE FSA+
Sbjct: 245 GEVCPAGWKPGEKSMKPDPKLSKELFSAI 273
>UniRef100_Q96468 2-cys peroxiredoxin BAS1, chloroplast precursor [Hordeum vulgare]
Length = 210
Score = 365 bits (938), Expect = e-100
Identities = 184/207 (88%), Positives = 195/207 (93%), Gaps = 3/207 (1%)
Query: 61 RSFVVRASSE--LPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVC 118
RSFV RA++E LPLVGN APDF AEAVFDQEFI VKLS+YIGKKYVILFFYPLDFTFVC
Sbjct: 5 RSFVARAAAEYDLPLVGNKAPDFAAEAVFDQEFINVKLSDYIGKKYVILFFYPLDFTFVC 64
Query: 119 PTEITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSI 178
PTEITAFSDRH EF ++NTEILGVSVDSVFSHLAWVQT+RKSGGLGDL YPLVSDVTKSI
Sbjct: 65 PTEITAFSDRHEEFEKINTEILGVSVDSVFSHLAWVQTERKSGGLGDLKYPLVSDVTKSI 124
Query: 179 SKSYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDE 238
SKS+GVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDET RTLQALQYV++ PDE
Sbjct: 125 SKSFGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETLRTLQALQYVKK-PDE 183
Query: 239 VCPAGWKPGEKSMKPDPKLSKEYFSAV 265
VCPAGWKPGEKSMKPDPK SKEYF+A+
Sbjct: 184 VCPAGWKPGEKSMKPDPKGSKEYFAAI 210
>UniRef100_P80602 2-cys peroxiredoxin BAS1, chloroplast precursor [Triticum aestivum]
Length = 210
Score = 364 bits (934), Expect = 1e-99
Identities = 183/207 (88%), Positives = 195/207 (93%), Gaps = 3/207 (1%)
Query: 61 RSFVVRASSE--LPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVC 118
RSFV RA++E LPLVGN APDF AEAVFDQEFI VKLS+YIGKKYVILFFYPLDFTFVC
Sbjct: 5 RSFVARAAAEYDLPLVGNKAPDFAAEAVFDQEFINVKLSDYIGKKYVILFFYPLDFTFVC 64
Query: 119 PTEITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSI 178
PTEITAFSDRH EF ++NTEILGVSVDSVFSHLAWVQT+RKSGGLGDL YPLVSDVTKSI
Sbjct: 65 PTEITAFSDRHEEFEKINTEILGVSVDSVFSHLAWVQTERKSGGLGDLKYPLVSDVTKSI 124
Query: 179 SKSYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDE 238
SKS+GVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDET RTL+ALQYV++ PDE
Sbjct: 125 SKSFGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETLRTLRALQYVKK-PDE 183
Query: 239 VCPAGWKPGEKSMKPDPKLSKEYFSAV 265
VCPAGWKPGEKSMKPDPK SKEYF+A+
Sbjct: 184 VCPAGWKPGEKSMKPDPKGSKEYFAAI 210
>UniRef100_Q9M4P9 2-Cys-peroxiredoxin precursor [Riccia fluitans]
Length = 275
Score = 354 bits (908), Expect = 1e-96
Identities = 170/215 (79%), Positives = 192/215 (89%), Gaps = 3/215 (1%)
Query: 51 LSLNRFTSSRRSFVVRASSELPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFY 110
+SL +F+ + V A PLVGN APDFEAEAVFDQEF+K+KLSEYIGK+YV+LFFY
Sbjct: 64 VSLKQFSKGK---VASARCASPLVGNVAPDFEAEAVFDQEFVKIKLSEYIGKRYVVLFFY 120
Query: 111 PLDFTFVCPTEITAFSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPL 170
PLDFTFVCPTEITAFSD+H EF +LNTE++GVS DSVFSHLAW+QTDRKSGGLGDL YPL
Sbjct: 121 PLDFTFVCPTEITAFSDKHEEFEKLNTEVIGVSTDSVFSHLAWIQTDRKSGGLGDLKYPL 180
Query: 171 VSDVTKSISKSYGVLIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQ 230
VSD+TK I++ +GVLIPDQGIALRGLFIIDKEGVIQH+TINNL IGRSV+ET RTLQA+Q
Sbjct: 181 VSDLTKKIAEDFGVLIPDQGIALRGLFIIDKEGVIQHATINNLAIGRSVEETLRTLQAVQ 240
Query: 231 YVQENPDEVCPAGWKPGEKSMKPDPKLSKEYFSAV 265
YVQENPDEVCPAGWKPGEK+MKPD KLSKEYF+ V
Sbjct: 241 YVQENPDEVCPAGWKPGEKTMKPDTKLSKEYFAQV 275
>UniRef100_Q9FE86 Peroxiredoxin precursor [Chlamydomonas reinhardtii]
Length = 235
Score = 326 bits (836), Expect = 3e-88
Identities = 166/243 (68%), Positives = 197/243 (80%), Gaps = 12/243 (4%)
Query: 24 LSSPPHLSSLSIPNASNSLPKLRTSLPLSLNRFTSSRRSFVVRAS-SELPLVGNAAPDFE 82
L S S+++ + P++ S+ +RRS VVRAS +E PLVG+ APDF+
Sbjct: 4 LQSASRSSAVAFSRQARVAPRVAASV---------ARRSLVVRASHAEKPLVGSVAPDFK 54
Query: 83 AEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEITAFSDRHAEFAELNTEILGV 142
A+AVFDQEF ++ LS+Y GK YV+LFFYPLDFTFVCPTEITAFSDR+ EF ++NTE+LGV
Sbjct: 55 AQAVFDQEFQEITLSKYRGK-YVVLFFYPLDFTFVCPTEITAFSDRYKEFKDINTEVLGV 113
Query: 143 SVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYGVLIPDQGIALRGLFIIDKE 202
SVDS F+HLAW+QTDRK GGLGDL YPLV+D+ K ISK+YGVL D GI+LRGLFIIDKE
Sbjct: 114 SVDSQFTHLAWIQTDRKEGGLGDLAYPLVADLKKEISKAYGVLTED-GISLRGLFIIDKE 172
Query: 203 GVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVCPAGWKPGEKSMKPDPKLSKEYF 262
GV+QH+TINNL GRSVDETKR LQA+QYVQ NPDEVCPAGWKPG+K+MKPDPK SKEYF
Sbjct: 173 GVVQHATINNLAFGRSVDETKRVLQAIQYVQSNPDEVCPAGWKPGDKTMKPDPKGSKEYF 232
Query: 263 SAV 265
SAV
Sbjct: 233 SAV 235
>UniRef100_Q9FNS2 Peroxiredoxin [Chlamydomonas reinhardtii]
Length = 199
Score = 317 bits (812), Expect = 2e-85
Identities = 156/201 (77%), Positives = 179/201 (88%), Gaps = 3/201 (1%)
Query: 66 RAS-SELPLVGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEITA 124
RAS +E PLVG+ APDF+A+AVFDQEF ++ LS+Y GK YV+LFFYPLDFTFVCPTEITA
Sbjct: 1 RASHAEKPLVGSVAPDFKAQAVFDQEFQEITLSKYRGK-YVVLFFYPLDFTFVCPTEITA 59
Query: 125 FSDRHAEFAELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYGV 184
FSDR+ EF ++NTE+LGVSVDS F+HLAW+QTDRK GGLGDL YPLV+D+ K ISK+YGV
Sbjct: 60 FSDRYKEFKDINTEVLGVSVDSQFTHLAWIQTDRKEGGLGDLAYPLVADLKKEISKAYGV 119
Query: 185 LIPDQGIALRGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVCPAGW 244
L D GI+LRGLFIIDKEGV+QH+TINNL GRSVDETKR LQA+QYVQ NPDEVCPAGW
Sbjct: 120 LTED-GISLRGLFIIDKEGVVQHATINNLAFGRSVDETKRVLQAIQYVQSNPDEVCPAGW 178
Query: 245 KPGEKSMKPDPKLSKEYFSAV 265
KPG+K+MKPDPK SKEYFSAV
Sbjct: 179 KPGDKTMKPDPKGSKEYFSAV 199
>UniRef100_Q8YNC5 Peroxiredoxin [Anabaena sp.]
Length = 203
Score = 302 bits (773), Expect = 7e-81
Identities = 147/192 (76%), Positives = 165/192 (85%), Gaps = 1/192 (0%)
Query: 74 VGNAAPDFEAEAVFDQEFIKVKLSEYIGKKYVILFFYPLDFTFVCPTEITAFSDRHAEFA 133
VG APDF A AV DQEF +KLS+Y GK YV+LFFYPLDFTFVCPTEITAFSDR+ EF
Sbjct: 13 VGQQAPDFTATAVVDQEFKTIKLSDYRGK-YVVLFFYPLDFTFVCPTEITAFSDRYEEFK 71
Query: 134 ELNTEILGVSVDSVFSHLAWVQTDRKSGGLGDLNYPLVSDVTKSISKSYGVLIPDQGIAL 193
+LNTEILGVSVDS FSHLAW+QTDRKSGG+GDLNYPLVSD+ K +S +Y VL P GIAL
Sbjct: 72 KLNTEILGVSVDSEFSHLAWIQTDRKSGGVGDLNYPLVSDIKKEVSDAYNVLDPAAGIAL 131
Query: 194 RGLFIIDKEGVIQHSTINNLGIGRSVDETKRTLQALQYVQENPDEVCPAGWKPGEKSMKP 253
RGLFIIDK+G+IQH+TINNL GRSVDET RTLQA+QYVQ +PDEVCPAGW+PGEK+M P
Sbjct: 132 RGLFIIDKDGIIQHATINNLAFGRSVDETLRTLQAIQYVQSHPDEVCPAGWQPGEKTMTP 191
Query: 254 DPKLSKEYFSAV 265
DP SK YF+AV
Sbjct: 192 DPVKSKVYFAAV 203
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.134 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 435,389,738
Number of Sequences: 2790947
Number of extensions: 17390072
Number of successful extensions: 50201
Number of sequences better than 10.0: 1475
Number of HSP's better than 10.0 without gapping: 1205
Number of HSP's successfully gapped in prelim test: 270
Number of HSP's that attempted gapping in prelim test: 47572
Number of HSP's gapped (non-prelim): 1503
length of query: 265
length of database: 848,049,833
effective HSP length: 125
effective length of query: 140
effective length of database: 499,181,458
effective search space: 69885404120
effective search space used: 69885404120
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 73 (32.7 bits)
Medicago: description of AC146630.11


