

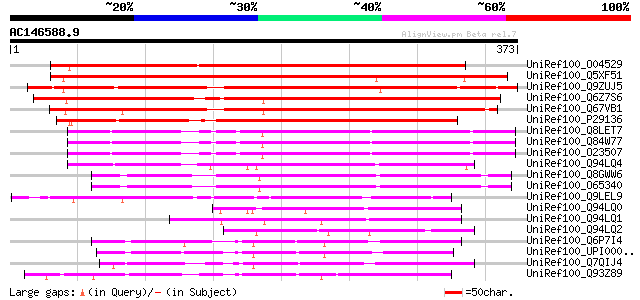
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146588.9 + phase: 0
(373 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O04529 F20P5.11 protein [Arabidopsis thaliana] 387 e-106
UniRef100_Q5XF51 At1g24140 [Arabidopsis thaliana] 377 e-103
UniRef100_Q9ZUJ5 T2K10.2 protein [Arabidopsis thaliana] 350 3e-95
UniRef100_Q6Z7S6 Putative zinc metalloproteinase [Oryza sativa] 334 2e-90
UniRef100_Q67VB1 Putative metalloproteinase [Oryza sativa] 334 2e-90
UniRef100_P29136 Metalloendoproteinase 1 precursor [Glycine max] 301 3e-80
UniRef100_Q8LET7 Proteinase like protein [Arabidopsis thaliana] 267 3e-70
UniRef100_Q84W77 Hypothetical protein At4g16640 [Arabidopsis tha... 267 4e-70
UniRef100_O23507 Proteinase like protein [Arabidopsis thaliana] 267 4e-70
UniRef100_Q94LQ4 Putative metalloproteinase [Oryza sativa] 254 3e-66
UniRef100_Q8GWW6 Putative metalloproteinase [Arabidopsis thaliana] 253 8e-66
UniRef100_O65340 Metalloproteinase [Arabidopsis thaliana] 252 1e-65
UniRef100_Q9LEL9 Matrix metalloproteinase [Cucumis sativus] 224 4e-57
UniRef100_Q94LQ0 Putative metalloproteinase [Oryza sativa] 166 1e-39
UniRef100_Q94LQ1 Putative metalloproteinase [Oryza sativa] 151 3e-35
UniRef100_Q94LQ2 Putative metalloproteinase [Oryza sativa] 139 1e-31
UniRef100_Q6P7I4 MGC68506 protein [Xenopus laevis] 136 1e-30
UniRef100_UPI0000361AF8 UPI0000361AF8 UniRef100 entry 135 1e-30
UniRef100_Q7QIJ4 ENSANGP00000021627 [Anopheles gambiae str. PEST] 135 2e-30
UniRef100_Q93Z89 Matrix metalloproteinase MMP2 [Glycine max] 133 7e-30
>UniRef100_O04529 F20P5.11 protein [Arabidopsis thaliana]
Length = 378
Score = 387 bits (993), Expect = e-106
Identities = 186/315 (59%), Positives = 235/315 (74%), Gaps = 11/315 (3%)
Query: 31 SWIPPGTLPVGP---------WDAFRNFTGCRQGENYNGLSNLKNYFQHFGYIPRSPKSN 81
+W P + V P WDAF NFTGC G+N +GL +K YFQ FGYIP + N
Sbjct: 20 AWFFPNSTAVPPSLRNTTRVFWDAFSNFTGCHHGQNVDGLYRIKKYFQRFGYIPETFSGN 79
Query: 82 FSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVADIINGTTTMNAGKDTE 141
F+DDFDD L+ A++ YQ NFNLNVTGELD +T++ +++PRCG D++NGT+ M+ G+ +
Sbjct: 80 FTDDFDDILKAAVELYQTNFNLNVTGELDALTIQHIVIPRCGNPDVVNGTSLMHGGR-RK 138
Query: 142 TTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTYAFFPGNELTETVKSVFATAFARWSE 201
T + S+ H V +T+FPG+PRWP +++LTYAF P N LTE VKSVF+ AF RWS+
Sbjct: 139 TFEVNFSRTHLHAVKRYTLFPGEPRWPRNRRDLTYAFDPKNPLTEEVKSVFSRAFGRWSD 198
Query: 202 VTTLKFTETTLYSGADIKIGFFNGDHGDGEPFDGSLGTLAHAFSPRNGRFHLDAAEDWVV 261
VT L FT + +S +DI IGF+ GDHGDGEPFDG LGTLAHAFSP +G+FHLDA E+WVV
Sbjct: 199 VTALNFTLSESFSTSDITIGFYTGDHGDGEPFDGVLGTLAHAFSPPSGKFHLDADENWVV 258
Query: 262 SGDV-SKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPTISSRMKKVVLADDDVRG 320
SGD+ S S+ AVDLESVAVHEIGHLLGLGHSS EE+IM+PTI++ +KV L +DDV G
Sbjct: 259 SGDLDSFLSVTAAVDLESVAVHEIGHLLGLGHSSVEESIMYPTITTGKRKVDLTNDDVEG 318
Query: 321 IQYLYGTNPSFNGST 335
IQYLYG NP+FNG+T
Sbjct: 319 IQYLYGANPNFNGTT 333
>UniRef100_Q5XF51 At1g24140 [Arabidopsis thaliana]
Length = 384
Score = 377 bits (969), Expect = e-103
Identities = 191/344 (55%), Positives = 242/344 (69%), Gaps = 8/344 (2%)
Query: 31 SWIPPGTL---PVGPWDAFRNFTGCRQGENYNGLSNLKNYFQHFGYIPRSPKS-NFSDDF 86
S IPP L PW++F NFTGC G+ Y+GL LK YFQHFGYI + S NF+DDF
Sbjct: 28 SAIPPQLLRNATGNPWNSFLNFTGCHAGKKYDGLYMLKQYFQHFGYITETNLSGNFTDDF 87
Query: 87 DDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVADIINGTTTMNAGKDTETTSNS 146
DD L+ A++ YQ+NF LNVTG LD++TL+ V++PRCG D++NGT+TM++G+ T S +
Sbjct: 88 DDILKNAVEMYQRNFQLNVTGVLDELTLKHVVIPRCGNPDVVNGTSTMHSGRKTFEVSFA 147
Query: 147 DSKLRFHTVSHFTVFPGQPRWPEGKQELTYAFFPGNELTETVKSVFATAFARWSEVTTLK 206
RFH V H++ FPG+PRWP +++LTYAF P N LTE VKSVF+ AF RW EVT L
Sbjct: 148 GRGQRFHAVKHYSFFPGEPRWPRNRRDLTYAFDPRNALTEEVKSVFSRAFTRWEEVTPLT 207
Query: 207 FTETTLYSGADIKIGFFNGDHGDGEPFDGSLGTLAHAFSPRNGRFHLDAAEDWVVSGDVS 266
FT +S +DI IGF++G+HGDGEPFDG + TLAHAFSP G FHLD E+W+VSG+
Sbjct: 208 FTRVERFSTSDISIGFYSGEHGDGEPFDGPMRTLAHAFSPPTGHFHLDGEENWIVSGEGG 267
Query: 267 KS--SLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPTISSRMKKVVLADDDVRGIQYL 324
S+ AVDLESVAVHEIGHLLGLGHSS E +IM+PTI + +KV L DDV G+QYL
Sbjct: 268 DGFISVSEAVDLESVAVHEIGHLLGLGHSSVEGSIMYPTIRTGRRKVDLTTDDVEGVQYL 327
Query: 325 YGTNPSFNG--STVISSPERNIGNGGCSLTSLWSPWRLFSLLTF 366
YG NP+FNG S S+ +R+ G+ G S S L +LL +
Sbjct: 328 YGANPNFNGSRSPPPSTQQRDTGDSGAPGRSDGSRSVLTNLLQY 371
>UniRef100_Q9ZUJ5 T2K10.2 protein [Arabidopsis thaliana]
Length = 360
Score = 350 bits (899), Expect = 3e-95
Identities = 192/366 (52%), Positives = 234/366 (63%), Gaps = 24/366 (6%)
Query: 14 FSLTLLIVSARLFPDVPSWIPPGTL---PVGPWDAFRNFTGCRQGENYNGLSNLKNYFQH 70
F T+ +SA+ + +V S IPP W+ F GC GEN NGLS LK YF+
Sbjct: 11 FFFTVNPISAKFYTNVSS-IPPLQFLNATQNAWETFSKLAGCHIGENINGLSKLKQYFRR 69
Query: 71 FGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVADIING 130
FGYI + N +DDFDD LQ AI TYQKNFNL VTG+LD TLRQ++ PRCG D+I+G
Sbjct: 70 FGYITTT--GNCTDDFDDVLQSAINTYQKNFNLKVTGKLDSSTLRQIVKPRCGNPDLIDG 127
Query: 131 TTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTYAFFPGNELTETVKS 190
+ MN GK TT ++ FPG+PRWP+ K++LTYAF P N LT+ VK
Sbjct: 128 VSEMNGGKILRTTEK------------YSFFPGKPRWPKRKRDLTYAFAPQNNLTDEVKR 175
Query: 191 VFATAFARWSEVTTLKFTETTLYSGADIKIGFFNGDHGDGEPFDGSLGTLAHAFSPRNGR 250
VF+ AF RW+EVT L FT + ADI IGFF+G+HGDGEPFDG++GTLAHA SP G
Sbjct: 176 VFSRAFTRWAEVTPLNFTRSESILRADIVIGFFSGEHGDGEPFDGAMGTLAHASSPPTGM 235
Query: 251 FHLDAAEDWVVS-GDVSKSSLP--TAVDLESVAVHEIGHLLGLGHSSEEEAIMFPTISSR 307
HLD EDW++S G++S+ LP T VDLESVAVHEIGHLLGLGHSS E+AIMFP IS
Sbjct: 236 LHLDGDEDWLISNGEISRRILPVTTVVDLESVAVHEIGHLLGLGHSSVEDAIMFPAISGG 295
Query: 308 MKKVVLADDDVRGIQYLYGTNPSFNGSTVISSPERNIGNGGCSLTSLWSPWRLFSLLTFV 367
+KV LA DD+ GIQ+LYG NP NG S P R + G W W + SL +
Sbjct: 296 DRKVELAKDDIEGIQHLYGGNP--NGDGGGSKPSRESQSTGGDSVRRWRGW-MISLSSIA 352
Query: 368 LSHFLL 373
FL+
Sbjct: 353 TCIFLI 358
>UniRef100_Q6Z7S6 Putative zinc metalloproteinase [Oryza sativa]
Length = 372
Score = 334 bits (857), Expect = 2e-90
Identities = 185/356 (51%), Positives = 231/356 (63%), Gaps = 21/356 (5%)
Query: 18 LLIVSARLFPDV-PSWIPPGTLPV------GPWDAFRNFTGCRQGENYNGLSNLKNYFQH 70
L++V+A F V P+ P LP PW AF+N +GC GE GL LK+Y H
Sbjct: 12 LVVVAAAAFVFVSPAMAFPMGLPATANPFPNPWSAFQNLSGCHAGEEREGLGRLKDYLSH 71
Query: 71 FGYIPRSPKSN-FSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVADIIN 129
FGY+P P S+ +SD FDD L+ AI YQ+NF LN TGELD T+ Q++ PRCGVAD+IN
Sbjct: 72 FGYLPPPPSSSPYSDAFDDSLEAAIAAYQRNFGLNATGELDTDTVDQMVAPRCGVADVIN 131
Query: 130 GTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTYAFFPGNELT---E 186
GT+TM+ ++S + LR + ++ FPG P WP ++ L YA + +
Sbjct: 132 GTSTMDR-------NSSAAALRGRHL--YSYFPGGPMWPPFRRNLRYAITATSATSIDRA 182
Query: 187 TVKSVFATAFARWSEVTTLKFTETTLYSGADIKIGFFNGDHGDGEPFDGSLGTLAHAFSP 246
T+ +VFA AF+RW+ T L+FTE + S ADI IGF++GDHGDGE FDG LGTLAHAFSP
Sbjct: 183 TLSAVFARAFSRWAAATRLQFTEVSSASNADITIGFYSGDHGDGEAFDGPLGTLAHAFSP 242
Query: 247 RNGRFHLDAAEDWVVSGDVS-KSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPTIS 305
+GRFHLDAAE WV SGDVS SS TAVDLESVAVHEIGHLLGLGHSS ++IM+PTI
Sbjct: 243 TDGRFHLDAAEAWVASGDVSTSSSFGTAVDLESVAVHEIGHLLGLGHSSVPDSIMYPTIR 302
Query: 306 SRMKKVVLADDDVRGIQYLYGTNPSFNGSTVISSPERNIGNGGCSLTSLWSPWRLF 361
+ +KV L DDV GIQ LYGTNP+F G T S + G + + PW F
Sbjct: 303 TGTRKVDLESDDVLGIQSLYGTNPNFKGVTPTSPSTSSREMDGSAAAAGIRPWSGF 358
>UniRef100_Q67VB1 Putative metalloproteinase [Oryza sativa]
Length = 371
Score = 334 bits (857), Expect = 2e-90
Identities = 175/342 (51%), Positives = 217/342 (63%), Gaps = 26/342 (7%)
Query: 30 PSWIPPGTLP-----VGPWDAFRNFTGCRQGENYNGLSNLKNYFQHFGYIPRSPKSN--- 81
P++ P LP PW AF+N +GC G+ GL LK+Y HFGY+ S+
Sbjct: 26 PAFALPAGLPDIKSLTNPWSAFKNLSGCHFGDERQGLGKLKDYLWHFGYLSYPSSSSLSP 85
Query: 82 -FSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVADIINGTTTMNAGKDT 140
F+D FD D++ AIK YQ NF L+VTG+LD T+ Q+M PRCGVAD++NGT+TM G
Sbjct: 86 SFNDLFDADMELAIKMYQGNFGLDVTGDLDAATVSQMMAPRCGVADVVNGTSTMGGGGGV 145
Query: 141 ETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTYAFFPGNELT---ETVKSVFATAFA 197
++ FPG PRWP + L YA ++ + T+ VFA+AFA
Sbjct: 146 RGRGL------------YSYFPGSPRWPRSRTTLRYAITATSQTSIDRATLSKVFASAFA 193
Query: 198 RWSEVTTLKFTETTLYSGADIKIGFFNGDHGDGEPFDGSLGTLAHAFSPRNGRFHLDAAE 257
RWS TTL FTE + ADI IGF+ GDHGDGE FDG LGTLAHAFSP NGR HLDA+E
Sbjct: 194 RWSAATTLNFTEAASAADADITIGFYGGDHGDGEAFDGPLGTLAHAFSPTNGRLHLDASE 253
Query: 258 DWVVSGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPTISSRMKKVVLADDD 317
WV GDV+++S AVDLESVAVHEIGH+LGLGHSS ++IMFPT++SR KKV LA DD
Sbjct: 254 AWVAGGDVTRASSNAAVDLESVAVHEIGHILGLGHSSAADSIMFPTLTSRTKKVNLATDD 313
Query: 318 VRGIQYLYGTNPSFNGSTVISSPERNIGNGGCSLTSLWSPWR 359
V GIQ LYG NP+F G T ++ R + + G L PWR
Sbjct: 314 VAGIQGLYGNNPNFKGVTPPATSSREMDSAGAG--ELSRPWR 353
>UniRef100_P29136 Metalloendoproteinase 1 precursor [Glycine max]
Length = 305
Score = 301 bits (770), Expect = 3e-80
Identities = 162/299 (54%), Positives = 194/299 (64%), Gaps = 21/299 (7%)
Query: 35 PGTLPVGP--WD--AFRNFTGCRQGENYNGLSNLKNYFQHFGYIPRSPKSNFSDDFDDDL 90
P GP WD A FT G+NY GLSN+KNYF H GYIP +P +F D+FDD L
Sbjct: 24 PSVSAHGPYAWDGEATYKFTTYHPGQNYKGLSNVKNYFHHLGYIPNAP--HFDDNFDDTL 81
Query: 91 QEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVADIINGTTTMNAGKDTETTSNSDSKL 150
AIKTYQKN+NLNVTG+ D TL+Q+M PRCGV DII T +TTS
Sbjct: 82 VSAIKTYQKNYNLNVTGKFDINTLKQIMTPRCGVPDIIINTN--------KTTS------ 127
Query: 151 RFHTVSHFTVFPGQPRWPEGKQELTYAFFPGNELTETVKSVFATAFARWSEVTTLKFTET 210
F +S +T F PRW G +LTYAF P L +T KS A AF++W+ V + F ET
Sbjct: 128 -FGMISDYTFFKDMPRWQAGTTQLTYAFSPEPRLDDTFKSAIARAFSKWTPVVNIAFQET 186
Query: 211 TLYSGADIKIGFFNGDHGDGEPFDGSLGTLAHAFSPRNGRFHLDAAEDWVVSGDVSKSSL 270
T Y A+IKI F + +HGD PFDG G L HAF+P +GR H DA E WV SGDV+KS +
Sbjct: 187 TSYETANIKILFASKNHGDPYPFDGPGGILGHAFAPTDGRCHFDADEYWVASGDVTKSPV 246
Query: 271 PTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPTISSRMKKVVLADDDVRGIQYLYGTNP 329
+A DLESVAVHEIGHLLGLGHSS+ AIM+P+I R +KV LA DD+ GI+ LYG NP
Sbjct: 247 TSAFDLESVAVHEIGHLLGLGHSSDLRAIMYPSIPPRTRKVNLAQDDIDGIRKLYGINP 305
>UniRef100_Q8LET7 Proteinase like protein [Arabidopsis thaliana]
Length = 364
Score = 267 bits (683), Expect = 3e-70
Identities = 154/334 (46%), Positives = 196/334 (58%), Gaps = 25/334 (7%)
Query: 43 WDAFRNFTGCRQGENYNGLSNLKNYFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFN 102
W F + G + +G+S LK Y FGY+ + FSD FD L+ AI YQ+N
Sbjct: 52 WHDFSRLVDVQIGSHVSGVSELKRYLHRFGYV-KDGSEIFSDVFDGPLESAISLYQENLG 110
Query: 103 LNVTGELDDMTLRQVMLPRCGVADIINGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFP 162
L +TG LD T+ + LPRCGV D T T N+D HT +H+T F
Sbjct: 111 LPITGRLDTSTVTLMSLPRCGVXD-------------THMTINNDF---LHTTAHYTYFN 154
Query: 163 GQPRWPEGKQELTYAFFPGNEL----TETVKSVFATAFARWSEVTTLKFTETTLYSGADI 218
G+P+W + LTYA ++L +E VK+VF AF++WS V + F E ++ AD+
Sbjct: 155 GKPKW--NRDTLTYAISKTHKLDYLTSEDVKTVFRRAFSQWSSVIPVSFEEVDDFTTADL 212
Query: 219 KIGFFNGDHGDGEPFDGSLGTLAHAFSPRNGRFHLDAAEDWVVSGDVSKSSLPTAVDLES 278
KIGF+ GDHGDG PFDG LGTLAHAF+P NGR HLDAAE W+V D+ K S AVDLES
Sbjct: 213 KIGFYAGDHGDGLPFDGVLGTLAHAFAPENGRLHLDAAETWIVDDDL-KGSSEVAVDLES 271
Query: 279 VAVHEIGHLLGLGHSSEEEAIMFPTISSRMKKVVLADDDVRGIQYLYGTNPSFNGSTVIS 338
VA HEIGHLLGLGHSS+E A+M+P++ R KKV L DDV G+ LYG NP ++
Sbjct: 272 VATHEIGHLLGLGHSSQESAVMYPSLRPRTKKVDLTVDDVAGVLKLYGPNPKLRLDSLTQ 331
Query: 339 SPERNIGNGGCSLTSLWSPWRLFSLLTFVLSHFL 372
S E +I NG S L + + LL L FL
Sbjct: 332 S-EDSIKNGTVSHRFLSGNFIGYVLLVVGLILFL 364
>UniRef100_Q84W77 Hypothetical protein At4g16640 [Arabidopsis thaliana]
Length = 364
Score = 267 bits (682), Expect = 4e-70
Identities = 154/334 (46%), Positives = 196/334 (58%), Gaps = 25/334 (7%)
Query: 43 WDAFRNFTGCRQGENYNGLSNLKNYFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFN 102
W F + G + +G+S LK Y FGY+ FSD FD L+ AI YQ+N
Sbjct: 52 WHDFSRLVDVQIGSHVSGVSELKRYLHRFGYV-NDGSEIFSDVFDGPLESAISLYQENLG 110
Query: 103 LNVTGELDDMTLRQVMLPRCGVADIINGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFP 162
L +TG LD T+ + LPRCGV+D T T N+D HT +H+T F
Sbjct: 111 LPITGRLDTSTVTLMSLPRCGVSD-------------THMTINNDF---LHTTAHYTYFN 154
Query: 163 GQPRWPEGKQELTYAFFPGNEL----TETVKSVFATAFARWSEVTTLKFTETTLYSGADI 218
G+P+W + LTYA ++L +E VK+VF AF++WS V + F E ++ AD+
Sbjct: 155 GKPKW--NRDTLTYAISKTHKLDYLTSEDVKTVFRRAFSQWSSVIPVSFEEVDDFTTADL 212
Query: 219 KIGFFNGDHGDGEPFDGSLGTLAHAFSPRNGRFHLDAAEDWVVSGDVSKSSLPTAVDLES 278
KIGF+ GDHGDG PFDG LGTLAHAF+P NGR HLDAAE W+V D+ K S AVDLES
Sbjct: 213 KIGFYAGDHGDGLPFDGVLGTLAHAFAPENGRLHLDAAETWIVDDDL-KGSSEVAVDLES 271
Query: 279 VAVHEIGHLLGLGHSSEEEAIMFPTISSRMKKVVLADDDVRGIQYLYGTNPSFNGSTVIS 338
VA HEIGHLLGLGHSS+E A+M+P++ R KKV L DDV G+ LYG NP ++
Sbjct: 272 VATHEIGHLLGLGHSSQESAVMYPSLRPRTKKVDLTVDDVAGVLKLYGPNPKLRLDSLTQ 331
Query: 339 SPERNIGNGGCSLTSLWSPWRLFSLLTFVLSHFL 372
S E +I NG S L + + LL L FL
Sbjct: 332 S-EDSIKNGTVSHRFLSGNFIGYVLLVVGLILFL 364
>UniRef100_O23507 Proteinase like protein [Arabidopsis thaliana]
Length = 364
Score = 267 bits (682), Expect = 4e-70
Identities = 154/334 (46%), Positives = 196/334 (58%), Gaps = 25/334 (7%)
Query: 43 WDAFRNFTGCRQGENYNGLSNLKNYFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFN 102
W F + G + +G+S LK Y FGY+ FSD FD L+ AI YQ+N
Sbjct: 52 WHDFSRLVDVQIGSHVSGVSELKRYLHRFGYV-NDGSEIFSDVFDGPLESAISLYQENLG 110
Query: 103 LNVTGELDDMTLRQVMLPRCGVADIINGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFP 162
L +TG LD T+ + LPRCGV+D T T N+D HT +H+T F
Sbjct: 111 LPITGRLDTSTVTLMSLPRCGVSD-------------THMTINNDF---LHTTAHYTYFN 154
Query: 163 GQPRWPEGKQELTYAFFPGNEL----TETVKSVFATAFARWSEVTTLKFTETTLYSGADI 218
G+P+W + LTYA ++L +E VK+VF AF++WS V + F E ++ AD+
Sbjct: 155 GKPKW--NRDTLTYAISKTHKLDYLTSEDVKTVFRRAFSQWSSVIPVSFEEVDDFTTADL 212
Query: 219 KIGFFNGDHGDGEPFDGSLGTLAHAFSPRNGRFHLDAAEDWVVSGDVSKSSLPTAVDLES 278
KIGF+ GDHGDG PFDG LGTLAHAF+P NGR HLDAAE W+V D+ K S AVDLES
Sbjct: 213 KIGFYAGDHGDGLPFDGVLGTLAHAFAPENGRLHLDAAETWIVDDDL-KGSSEVAVDLES 271
Query: 279 VAVHEIGHLLGLGHSSEEEAIMFPTISSRMKKVVLADDDVRGIQYLYGTNPSFNGSTVIS 338
VA HEIGHLLGLGHSS+E A+M+P++ R KKV L DDV G+ LYG NP ++
Sbjct: 272 VATHEIGHLLGLGHSSQESAVMYPSLRPRTKKVDLTVDDVAGVLKLYGPNPKLRLDSLTQ 331
Query: 339 SPERNIGNGGCSLTSLWSPWRLFSLLTFVLSHFL 372
S E +I NG S L + + LL L FL
Sbjct: 332 S-EDSIKNGTVSHRFLSGNFIGYVLLVVGLILFL 364
>UniRef100_Q94LQ4 Putative metalloproteinase [Oryza sativa]
Length = 355
Score = 254 bits (649), Expect = 3e-66
Identities = 143/314 (45%), Positives = 184/314 (58%), Gaps = 28/314 (8%)
Query: 43 WDAFRNFTGCRQGENYNGLSNLKNYFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFN 102
W +F+ +G + GL+ LK Y FGY+ + P + +D FD+ L+ A++ YQ F+
Sbjct: 36 WHSFKQLLDAGRGSHVTGLAELKRYLARFGYMAK-PGRDTTDAFDEHLEVAVRRYQTRFS 94
Query: 103 LNVTGELDDMTLRQVMLPRCGVADIINGTTTMNAGKDTETTSNS--DSKLRFHTVSHFTV 160
L VTG LD+ TL Q+M PRCGV D D E + + VS FT
Sbjct: 95 LPVTGRLDNATLDQIMSPRCGVGD-----------DDVERPVSVALSPGAQGGVVSRFTF 143
Query: 161 FPGQPRWPEGKQE--LTYAFFP----GNELTETVKSVFATAFARWSEVTTLKFTETTLYS 214
F G+PRW L+YA P G V++VF AFARW+ + F ET Y
Sbjct: 144 FKGEPRWTRSDPPIVLSYAVSPTATVGYLPPAAVRAVFQRAFARWARTIPVGFVETDDYE 203
Query: 215 GADIKIGFFNGDHGDGEPFDGSLGTLAHAFSPRNGRFHLDAAEDWVVSGDVSKSSLPTAV 274
ADIK+GF+ G+HGDG PFDG LG L HAFSP+NGR HLDA+E W V DV ++ +A+
Sbjct: 204 AADIKVGFYAGNHGDGVPFDGPLGILGHAFSPKNGRLHLDASEHWAVDFDVDATA--SAI 261
Query: 275 DLESVAVHEIGHLLGLGHSSEEEAIMFPTISSRMKKVVLADDDVRGIQYLYGTNPSFNGS 334
DLESVA HEIGH+LGLGHS+ A+M+P+I R KKV L DDV G+Q LYG+NP F+ S
Sbjct: 262 DLESVATHEIGHVLGLGHSASPRAVMYPSIKPREKKVRLTVDDVEGVQALYGSNPQFSLS 321
Query: 335 ------TVISSPER 342
T SSP R
Sbjct: 322 SLSEQGTSSSSPRR 335
>UniRef100_Q8GWW6 Putative metalloproteinase [Arabidopsis thaliana]
Length = 342
Score = 253 bits (645), Expect = 8e-66
Identities = 145/315 (46%), Positives = 188/315 (59%), Gaps = 34/315 (10%)
Query: 61 LSNLKNYFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLP 120
+ +K + Q +GY+P++ +S+ D ++A+ YQKN L +TG+ D TL Q++LP
Sbjct: 50 IPEIKRHLQQYGYLPQNKESD-----DVSFEQALVRYQKNLGLPITGKPDSDTLSQILLP 104
Query: 121 RCGVADIINGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQ-ELTYAFF 179
RCG D + T FHT + FPG+PRW +LTYAF
Sbjct: 105 RCGFPDDVEPKTAP-----------------FHTGKKYVYFPGRPRWTRDVPLKLTYAFS 147
Query: 180 PGN---ELTET-VKSVFATAFARWSEVTTLKFTETTLYSGADIKIGFFNGDHGDGEPFDG 235
N L T ++ VF AF +W+ V + F ET Y ADIKIGFFNGDHGDGEPFDG
Sbjct: 148 QENLTPYLAPTDIRRVFRRAFGKWASVIPVSFIETEDYVIADIKIGFFNGDHGDGEPFDG 207
Query: 236 SLGTLAHAFSPRNGRFHLDAAEDWVVSGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSSE 295
LG LAH FSP NGR HLD AE W V D KSS+ AVDLESVAVHEIGH+LGLGHSS
Sbjct: 208 VLGVLAHTFSPENGRLHLDKAETWAVDFDEEKSSV--AVDLESVAVHEIGHVLGLGHSSV 265
Query: 296 EEAIMFPTISSRMKKVVLADDDVRGIQYLYGTNPSFN-GSTVISSPERNIGNGGCSLTSL 354
++A M+PT+ R KKV L DDV G+Q LYGTNP+F S + S N+ +G + +
Sbjct: 266 KDAAMYPTLKPRSKKVNLNMDDVVGVQSLYGTNPNFTLNSLLASETSTNLADG----SRI 321
Query: 355 WSPWRLFSLLTFVLS 369
S ++S L+ V++
Sbjct: 322 RSQGMIYSTLSTVIA 336
>UniRef100_O65340 Metalloproteinase [Arabidopsis thaliana]
Length = 341
Score = 252 bits (644), Expect = 1e-65
Identities = 145/314 (46%), Positives = 185/314 (58%), Gaps = 33/314 (10%)
Query: 61 LSNLKNYFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLP 120
+ +K + Q +GY+P++ +S+ D ++A+ YQKN L +TG+ D TL Q++LP
Sbjct: 50 IPEIKRHLQQYGYLPQNKESD-----DVSFEQALVRYQKNLGLPITGKPDSDTLSQILLP 104
Query: 121 RCGVADIINGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQ-ELTYAFF 179
RCG D + T FHT + FPG+PRW +LTYAF
Sbjct: 105 RCGFPDDVEPKTAP-----------------FHTGKKYVYFPGRPRWTRDVPLKLTYAFS 147
Query: 180 PGN---ELTETVKSVFATAFARWSEVTTLKFTETTLYSGADIKIGFFNGDHGDGEPFDGS 236
N L T VF AF W+ V + F ET Y ADIKIGFFNGDHGDGEPFDG
Sbjct: 148 QENLTPYLAPTDIRVFRRAFGEWASVIPVSFIETEDYVIADIKIGFFNGDHGDGEPFDGV 207
Query: 237 LGTLAHAFSPRNGRFHLDAAEDWVVSGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSSEE 296
LG LAH FSP NGR HLD AE W V D KSS+ AVDLESVAVHEIGH+LGLGHSS +
Sbjct: 208 LGVLAHTFSPENGRLHLDKAETWAVDFDEEKSSV--AVDLESVAVHEIGHVLGLGHSSVK 265
Query: 297 EAIMFPTISSRMKKVVLADDDVRGIQYLYGTNPSFN-GSTVISSPERNIGNGGCSLTSLW 355
+A M+PT+ R KKV L DDV G+Q LYGTNP+F S + S N+ +G + +
Sbjct: 266 DAAMYPTLKPRSKKVNLNMDDVVGVQSLYGTNPNFTLNSLLASETSTNLADG----SRIR 321
Query: 356 SPWRLFSLLTFVLS 369
S ++S L+ V++
Sbjct: 322 SQGMIYSALSTVIA 335
>UniRef100_Q9LEL9 Matrix metalloproteinase [Cucumis sativus]
Length = 320
Score = 224 bits (570), Expect = 4e-57
Identities = 136/336 (40%), Positives = 183/336 (53%), Gaps = 34/336 (10%)
Query: 2 KKQQLIFPLTISFSLTLLIVSARLFPDVPSWIPPGTLPVGPWDA--------FRNFTGCR 53
K Q+IFP T+ F LFP+ P+ P L + +N GC
Sbjct: 5 KALQIIFPFTLLF--------LSLFPN-PNTSSPIILKHSSQNMNSSNSLMFLKNLQGCH 55
Query: 54 QGENYNGLSNLKNYFQHFGYIPRSPKSN----FSDDFDDDLQEAIKTYQKNFNLNVTGEL 109
G+ G+ +K Y Q FGYI + + + F D FD L+ A+KTYQ N NL +G L
Sbjct: 56 LGDTKQGIHQIKKYLQRFGYITTNIQKHSNPIFDDTFDHILESALKTYQTNHNLAPSGIL 115
Query: 110 DDMTLRQVMLPRCGVADIINGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPE 169
D T+ Q+ +PRCGV D+I T ++ T+N + FH VSHFT F G +WP
Sbjct: 116 DSNTIAQIAMPRCGVQDVIKNKKTKK--RNQNFTNNGHT--HFHKVSHFTFFEGNLKWPS 171
Query: 170 GKQELTYAFFPGNELTETVKSVFATAFARWSEVTTLKFTETTLYSGADIKIGFFNGDHGD 229
K L+Y F P N + +K V + AF++WS T KF+ Y ADIKI F G+HGD
Sbjct: 172 SKLHLSYGFLP-NYPIDAIKPV-SRAFSKWSLNTHFKFSHVADYRKADIKISFERGEHGD 229
Query: 230 GEPFDGSLGTLAHAFSPRNGRFHLDAAEDWVVSGDVSKSSLPTAVDLESVAVHEIGHLLG 289
PFDG G LAHA++P +GR H D + W S ++ D+E+VA+HEIGH+LG
Sbjct: 230 NAPFDGVGGVLAHAYAPTDGRLHFDGDDAW------SVGAISGYFDVETVALHEIGHILG 283
Query: 290 LGHSSEEEAIMFPTISSRMKKVVLADDDVRGIQYLY 325
L HS+ EEAIMFP+I + K L DD+ GI+ LY
Sbjct: 284 LQHSTIEEAIMFPSIPEGVTK-GLHGDDIAGIKALY 318
>UniRef100_Q94LQ0 Putative metalloproteinase [Oryza sativa]
Length = 261
Score = 166 bits (419), Expect = 1e-39
Identities = 94/201 (46%), Positives = 121/201 (59%), Gaps = 24/201 (11%)
Query: 150 LRFH--TVSHFTVFPGQPRWPEGKQE--LTYA--------FFPGNELTETVKSVFATAFA 197
LR H VS + + G+PRW + LTYA + PG + V +VF +AFA
Sbjct: 38 LRHHGGVVSRYAFWTGKPRWTRHGRPMVLTYAVSHTDAVGYLPG----DAVLAVFRSAFA 93
Query: 198 RWSEVTTLKFTETTLYSGA-----DIKIGFFN-GDHGDGEPFDGSLGTLAHAFSPRNGRF 251
RW+EV + F E T A DI++GF+ G+HGDG PFDG L AHA P +GR
Sbjct: 94 RWAEVIPVSFAEITTEDDAAAAEADIRVGFYGAGEHGDGHPFDGPLNVYAHATGPEDGRI 153
Query: 252 HLDAAEDWVVSGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPTISSRMKKV 311
DAAE W V D++ + P AVDLE+VA HEIGH LGL HS+ E ++M+P + +R +KV
Sbjct: 154 DFDAAERWAV--DLAADASPAAVDLETVATHEIGHALGLDHSTSESSVMYPYVGTRERKV 211
Query: 312 VLADDDVRGIQYLYGTNPSFN 332
L DDV GIQ LYG NPSF+
Sbjct: 212 RLTVDDVEGIQELYGVNPSFS 232
>UniRef100_Q94LQ1 Putative metalloproteinase [Oryza sativa]
Length = 289
Score = 151 bits (381), Expect = 3e-35
Identities = 92/236 (38%), Positives = 123/236 (51%), Gaps = 23/236 (9%)
Query: 118 MLPRCGVADIINGTTTMNAGKDTETTSNSDSKLRFHT----VSHFTVFPGQPRWPEGKQE 173
M+ C D+ T T A T T+ + + FT FPG+PRW +
Sbjct: 1 MMSLCRSGDVARLTATQPALAQTTMTTTLQPRPEDNNNDSGAGPFTFFPGKPRWTRPDRV 60
Query: 174 LTYAFFPGNELTE----TVKSVFATAFARWSEVTTLKFTETTLYSGADIKIGFFNGDHG- 228
LTYA P V++ +AFARW++V ++F E Y ADIK+GF+ G
Sbjct: 61 LTYAVSPTATADHLPPSAVRAALRSAFARWADVIPMRFLEAERYDAADIKVGFYLYTDGR 120
Query: 229 -----------DGEPFDGSLGTLAHAFSP-RNGRFHLDAAEDWVVSGDVSKSSLPTAVDL 276
D + D G LAH+ P ++G+ HL AA W V+ ++ + P AVDL
Sbjct: 121 CDGCACIDSDDDDDDGDDCEGVLAHSSMPEKSGQIHLHAAHRWTVN--LAADTAPLAVDL 178
Query: 277 ESVAVHEIGHLLGLGHSSEEEAIMFPTISSRMKKVVLADDDVRGIQYLYGTNPSFN 332
ESVA HEIGH+LGL HSS ++M+P IS R +KV L DDV GIQ LYG NP F+
Sbjct: 179 ESVAAHEIGHVLGLDHSSSRSSMMYPFISCRERKVRLTTDDVHGIQELYGANPHFS 234
>UniRef100_Q94LQ2 Putative metalloproteinase [Oryza sativa]
Length = 266
Score = 139 bits (350), Expect = 1e-31
Identities = 86/198 (43%), Positives = 107/198 (53%), Gaps = 17/198 (8%)
Query: 158 FTVFPGQPRWPEGKQELTYAFFP----GNELTETVKSVFATAFARWSEVTTLKFTETTLY 213
FT G+PRW LTYA P + + V+ F +A ARW+EVT L+F E Y
Sbjct: 30 FTFLWGRPRWNRPDMRLTYAVSPLATADHLPRDAVREAFRSALARWAEVTPLRFAEAARY 89
Query: 214 SGADIKIGFFNGDHGDGEPF-------DGSLGTLAHAFSPRNGRFHLDAAEDWVVSG--D 264
ADI++GF+ DG+ G LAHA P++GR HL AA W V+
Sbjct: 90 EEADIRVGFYLHT-ADGKCDACGCVCKGGGEEALAHAHPPQDGRIHLHAARKWAVTNVAG 148
Query: 265 VSKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIMFPTISSRMKKVVLADDDVRGIQYL 324
+ P AVDLESVAVHEIGH LGLGHSS E ++M+ KV L DDDV+G+Q L
Sbjct: 149 AGGDAPPLAVDLESVAVHEIGHALGLGHSSSESSMMYRHYRG---KVSLTDDDVKGVQEL 205
Query: 325 YGTNPSFNGSTVISSPER 342
YG P T S P+R
Sbjct: 206 YGAKPPRVDLTADSKPKR 223
>UniRef100_Q6P7I4 MGC68506 protein [Xenopus laevis]
Length = 497
Score = 136 bits (342), Expect = 1e-30
Identities = 96/278 (34%), Positives = 135/278 (48%), Gaps = 37/278 (13%)
Query: 61 LSNLKNYFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLP 120
L + + + +GY+ + K + L A++ +Q +L V+G+LD +T++Q++ P
Sbjct: 29 LQTAQVFLEKYGYLEETTKQHNGKQ----LASAVREFQWLSHLPVSGQLDTITVQQMVQP 84
Query: 121 RCGVADI--INGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTYAF 178
RCG+ DI + + + G+ S +K +W K+ LTY
Sbjct: 85 RCGMKDIESLELVKSHHHGQQRRKRYTSKNK----------------KWY--KRHLTYQI 126
Query: 179 --FPGNELTETVKSVFATAFARWSEVTTLKFTETTLYSGADIKIGFFNGDHGDG--EPFD 234
+P V+ AF WS V++L F+E L ADI++ F+ GDH DG FD
Sbjct: 127 VNWPWYLSKHQVRQAVKAAFQLWSNVSSLTFSEA-LRDPADIRLAFYEGDHNDGAGNAFD 185
Query: 235 GSLGTLAHAFSPRNGRFHLDAAEDWVVSGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSS 294
G G LAHAF PR G H D+AE W + G +L V HEIGH LGL HSS
Sbjct: 186 GPGGALAHAFFPRRGEAHFDSAEHWSLDGK--------GRNLFVVLAHEIGHTLGLQHSS 237
Query: 295 EEEAIMFPTISSRMKKVVLADDDVRGIQYLYGTNPSFN 332
+ A+M P K VL DDV IQ LYG PS N
Sbjct: 238 FKNALMSPYYKKLNKDYVLNFDDVLAIQNLYGAPPSGN 275
>UniRef100_UPI0000361AF8 UPI0000361AF8 UniRef100 entry
Length = 468
Score = 135 bits (341), Expect = 1e-30
Identities = 95/266 (35%), Positives = 131/266 (48%), Gaps = 28/266 (10%)
Query: 65 KNYFQHFGYIPRSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGV 124
+ + + +GY+ + +D+ + AI +Q+ L VTG LD TLRQ+ PRCGV
Sbjct: 1 QTFLEKYGYLHHDTHVHNADE----IHSAISDFQRISRLPVTGRLDSATLRQMSEPRCGV 56
Query: 125 ADIINGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTYAF--FPGN 182
+D G+ A + T + K R T +W K+ LTY +P +
Sbjct: 57 SD--EGSQKNWAQRVKATFTRQRRKARSATQDR--------KWY--KRHLTYQIINWPRH 104
Query: 183 ELTETVKSVFATAFARWSEVTTLKFTETTLYSGADIKIGFFNGDHGDG--EPFDGSLGTL 240
+V+ V AF WS V+ L F E + ADI++ F+ GDH DG FDG GTL
Sbjct: 105 LPLSSVRLVVHAAFQLWSNVSNLVFREAS-EGPADIRLAFYEGDHNDGTGNAFDGPGGTL 163
Query: 241 AHAFSPRNGRFHLDAAEDWVVSGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIM 300
AHAF PR G H D AE W ++G +L VA HEIGH LGL HS A+M
Sbjct: 164 AHAFLPRRGEAHFDRAERWTLNGYKGH-------NLFMVAAHEIGHTLGLEHSPVRHALM 216
Query: 301 FPTISSRMKKVVLADDDVRGIQYLYG 326
P + +V + DD+ +Q LYG
Sbjct: 217 SPYYRKLGRSLVPSWDDIVAVQQLYG 242
>UniRef100_Q7QIJ4 ENSANGP00000021627 [Anopheles gambiae str. PEST]
Length = 465
Score = 135 bits (340), Expect = 2e-30
Identities = 100/282 (35%), Positives = 128/282 (44%), Gaps = 38/282 (13%)
Query: 67 YFQHFGYIP---RSPKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCG 123
Y FGY+ R+P S D D ++AI +Q LNVTGELD T++ + LPRCG
Sbjct: 2 YLSQFGYLSPKYRNPTSGNLLD-QDTWEKAIMEFQSFAGLNVTGELDGETMQLMSLPRCG 60
Query: 124 VADIINGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTYAF--FPG 181
V D + SD++ + + + RW ++LTY +P
Sbjct: 61 VKDKVG--------------FGSDTRSKRYALQG-------SRWKV--KDLTYRISKYPR 97
Query: 182 NELTETVKSVFATAFARWSEVTTLKFTETTLYSGADIKIGFFNGDHGDGEPFDGSLGTLA 241
V A AF WSE T L+FT I I F +HGDG+PFDG GTLA
Sbjct: 98 RLERTAVDKEIAKAFGVWSEYTDLRFTPKKT-GAVHIDIRFEENEHGDGDPFDGPGGTLA 156
Query: 242 HAFSP-RNGRFHLDAAEDWVVSGDVSKSSLPTAVDLESVAVHEIGHLLGLGHSSEEEAIM 300
HA+ P G H D AE W + P +L VA HE GH LGL HS A+M
Sbjct: 157 HAYFPVYGGDAHFDDAEQWTI-------DKPRGTNLFQVAAHEFGHSLGLSHSDVRSALM 209
Query: 301 FPTISSRMKKVVLADDDVRGIQYLYGTNPSFNGSTVISSPER 342
P L DD++GIQ LYGT G ++P R
Sbjct: 210 APFYRGYDPVFRLDSDDIQGIQTLYGTKTRNPGGGAGATPTR 251
>UniRef100_Q93Z89 Matrix metalloproteinase MMP2 [Glycine max]
Length = 357
Score = 133 bits (335), Expect = 7e-30
Identities = 110/325 (33%), Positives = 164/325 (49%), Gaps = 38/325 (11%)
Query: 12 ISFSLTLLIVSARL--FPDVPSWIPPGTLPVGPWDAFRNFTGCRQGENYNG--LSNLKNY 67
+SFS L + +L P + ++ P T +G NFT E + +S +K+Y
Sbjct: 27 VSFSSFLKQLKQKLEKSPTLKDFLKPTT--IGDIYYTLNFTEIFSSEERSAPPVSLIKDY 84
Query: 68 FQHFGYIPRS-PKSNFSDDFDDDLQEAIKTYQKNFNLNVTGELDDMTLRQVMLPRCGVAD 126
++GYI S P SN D + + AIKTYQ+ + L TG+L++ TL+Q+ RCGV D
Sbjct: 85 LSNYGYIESSGPLSNSMDQ--ETIISAIKTYQQYYCLQPTGKLNNETLQQMSFLRCGVPD 142
Query: 127 IINGTTTMNAGKDTETTSNSDSKLRFHTVSHFTVFPGQPRWPEGKQELTYAFFPGNELTE 186
I + +D + + H RW LTY F P N++
Sbjct: 143 I------------NIDYNFTDDNMSYPKAGH--------RWFP-HTNLTYGFLPENQIPA 181
Query: 187 TVKSVFATAFARWSEVT-TLKFTETTLYSGADIKIGFFNGDHG--DGEPFDGSLGTLAHA 243
+ VF +FARW++ + L TETT Y ADI++GF+N + D E + GSL L
Sbjct: 182 NMTKVFRDSFARWAQASGVLNLTETT-YDNADIQVGFYNFTYLGIDIEVYGGSLIFL-QP 239
Query: 244 FSPRNGRFHLDAAED-WVVSGDVSKSSLPTAV-DLESVAVHEIGHLLGLGHSSEEEAIMF 301
S + G LD W + + + S V DLES A+HEIGHLLGL HS++E+++M+
Sbjct: 240 DSTKKGVILLDGTNKLWALPSENGRLSWEEGVLDLESAAMHEIGHLLGLDHSNKEDSVMY 299
Query: 302 PTI-SSRMKKVVLADDDVRGIQYLY 325
P I S +KV L+ D +Q+ +
Sbjct: 300 PCILPSHQRKVQLSKSDKTNVQHQF 324
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.137 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 668,294,960
Number of Sequences: 2790947
Number of extensions: 29870548
Number of successful extensions: 73079
Number of sequences better than 10.0: 601
Number of HSP's better than 10.0 without gapping: 511
Number of HSP's successfully gapped in prelim test: 91
Number of HSP's that attempted gapping in prelim test: 71043
Number of HSP's gapped (non-prelim): 907
length of query: 373
length of database: 848,049,833
effective HSP length: 129
effective length of query: 244
effective length of database: 488,017,670
effective search space: 119076311480
effective search space used: 119076311480
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC146588.9


