

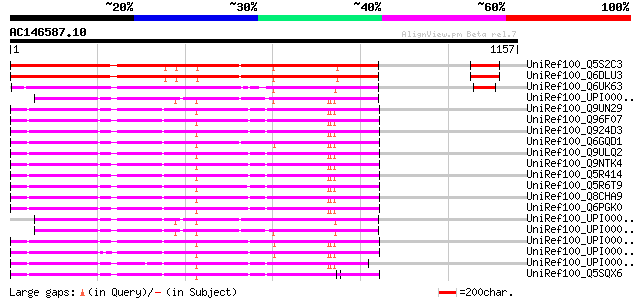
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146587.10 + phase: 0 /pseudo
(1157 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q5S2C3 PIR [Arabidopsis thaliana] 1386 0.0
UniRef100_Q6DLU3 PIROGI [Arabidopsis thaliana] 1386 0.0
UniRef100_Q6UK63 Rac-binding component of SCAR regulatory comple... 395 e-108
UniRef100_UPI000035F078 UPI000035F078 UniRef100 entry 350 1e-94
UniRef100_Q9UN29 P53 inducible protein [Homo sapiens] 349 2e-94
UniRef100_Q96F07 P53 inducible protein [Homo sapiens] 349 2e-94
UniRef100_Q924D3 CYFIP2 [Mus musculus] 348 5e-94
UniRef100_Q6GQD1 MGC80158 protein [Xenopus laevis] 348 7e-94
UniRef100_Q9ULQ2 KIAA1168 protein [Homo sapiens] 348 7e-94
UniRef100_Q9NTK4 Hypothetical protein DKFZp761I12121 [Homo sapiens] 348 7e-94
UniRef100_Q5R414 Hypothetical protein DKFZp459I162 [Pongo pygmaeus] 347 9e-94
UniRef100_Q5R6T9 Hypothetical protein DKFZp459P191 [Pongo pygmaeus] 347 1e-93
UniRef100_Q8CHA9 MKIAA1168 protein [Mus musculus] 347 2e-93
UniRef100_Q6PGK0 Cyfip2 protein [Mus musculus] 347 2e-93
UniRef100_UPI00003A9E55 UPI00003A9E55 UniRef100 entry 345 5e-93
UniRef100_UPI00003A9E54 UPI00003A9E54 UniRef100 entry 345 5e-93
UniRef100_UPI00003AAAFA UPI00003AAAFA UniRef100 entry 344 1e-92
UniRef100_UPI00003AAAFB UPI00003AAAFB UniRef100 entry 343 1e-92
UniRef100_UPI00002929FB UPI00002929FB UniRef100 entry 335 5e-90
UniRef100_Q5SQX6 Cytoplasmic FMR1 interacting protein 2 [Mus mus... 320 2e-85
>UniRef100_Q5S2C3 PIR [Arabidopsis thaliana]
Length = 1283
Score = 1386 bits (3587), Expect = 0.0
Identities = 714/915 (78%), Positives = 775/915 (84%), Gaps = 87/915 (9%)
Query: 1 MAVPVEEAIAALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLSEDTKA 60
MAVPVEEAIAALSTFSLEDEQPEVQGP V VS ER AT+SPIEY DVAAYRLSLSEDTKA
Sbjct: 1 MAVPVEEAIAALSTFSLEDEQPEVQGPAVMVSAERAATDSPIEYSDVAAYRLSLSEDTKA 60
Query: 61 LNQLSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSRLREI 120
LNQL++L QEGKEMAS+LYTYRSCVKALPQLP+SMK SQADLYLETYQVLDLEMSRLREI
Sbjct: 61 LNQLNTLIQEGKEMASILYTYRSCVKALPQLPESMKHSQADLYLETYQVLDLEMSRLREI 120
Query: 121 QRWQASASSKLATDMQRFSRPERRINGPTISHLWSMLKLFDVLVQLDHLKNAKASIPNDF 180
QRWQ+SAS+KLA DMQRFSRPERRINGPT++HLWSMLKL DVLVQLDHLKNAKASIPNDF
Sbjct: 121 QRWQSSASAKLAADMQRFSRPERRINGPTVTHLWSMLKLLDVLVQLDHLKNAKASIPNDF 180
Query: 181 SWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPVYLLHIFS 240
SWYKRTFTQVS QWQDTD+MREELDDLQIFLSTRWAILLNLHVEMFRVN
Sbjct: 181 SWYKRTFTQVSAQWQDTDTMREELDDLQIFLSTRWAILLNLHVEMFRVN----------- 229
Query: 241 LSTVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSEKDSESLYKRVKI 300
VEDILQVLIVF VESLELDFALLFPER+ILLRVLPVLVVL T SEKD+E+LYKRVK+
Sbjct: 230 --NVEDILQVLIVFIVESLELDFALLFPERYILLRVLPVLVVLATPSEKDTEALYKRVKL 287
Query: 301 NRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELPP------ 354
NRLINIFKN+ VIPAFPDLHLSPAAI+KELS YF KFSSQTRLLTL APHELPP
Sbjct: 288 NRLINIFKNDPVIPAFPDLHLSPAAILKELSVYFQKFSSQTRLLTLPAPHELPPREALEY 347
Query: 355 ------LNHIGAVRAEHDDFTIRFASAMNQ-----STDGSDVDWSKEVKGNMYDMIVEGF 403
+NHIGA+RAEHDDFTIRFAS+MNQ S DG+ +W +EVKGNMYDM+VEGF
Sbjct: 348 QRHYLIVNHIGALRAEHDDFTIRFASSMNQLLLLKSNDGAYTEWCREVKGNMYDMVVEGF 407
Query: 404 QLLSRWTARIWEQCAWKFSRPCKD------ASPSFSDYEKVVRYNYSAEERKALVELVSC 457
QLLSRWTARIWEQCAWKFSRPC+D AS S+SDYEKVVR+NY+AEERKALVELV
Sbjct: 408 QLLSRWTARIWEQCAWKFSRPCRDAGETPEASGSYSDYEKVVRFNYTAEERKALVELVGY 467
Query: 458 IKSVGSMMQRCDTLVADALWETIHAEVQDFVQNTLASMLRTTFRKKKDLSRILSDMRTLS 517
IKSVGSM+QRCDTLVADALWETIHAEVQDFVQNTLA+MLRTTFRKKKDLSRILSDMRTLS
Sbjct: 468 IKSVGSMLQRCDTLVADALWETIHAEVQDFVQNTLATMLRTTFRKKKDLSRILSDMRTLS 527
Query: 518 ADWMANTNKSESELQSSQHGGEESKANIFYPRAVAPTAAQVHCLQFLIYEVVSGGNLRRP 577
ADWMANT + E E+ SSQHG +ES+ N FYPR VAPTAAQVHCLQFLIYEVVSGGNLRRP
Sbjct: 528 ADWMANT-RPEHEMPSSQHGNDESRGNFFYPRPVAPTAAQVHCLQFLIYEVVSGGNLRRP 586
Query: 578 GGLFGNSGSEVPVNDLKQLETF----------VMLAVTVATLTDLGFLWFREFYLESSRV 627
GG FGN+GSE+PVNDLKQLETF + + ++ LTDLGFLWFREFYLESSRV
Sbjct: 587 GGFFGNNGSEIPVNDLKQLETFFYKLSFFLHILDYSASIGILTDLGFLWFREFYLESSRV 646
Query: 628 IQFPIECSLPWMLVDCVLESPNSGLLESVLMPFDIYNDSAQQALVLLKQRFLYDEIEAEV 687
IQFPIECSLPWML+D +LE+ NSGLLESVL+PFDIYNDSAQQALV+L+QRFLYDEIEAEV
Sbjct: 647 IQFPIECSLPWMLIDYILEAQNSGLLESVLLPFDIYNDSAQQALVVLRQRFLYDEIEAEV 706
Query: 688 DHCFDIFVARLCETIFTYYKSWAASELLDPTFLFASENAEKYAVQPMRLNMLLKMTRVK- 746
DH FDIFV+RL E+IFTYYKSW+ASELLDP+FLFA +N EK+++QP+R L KMT+VK
Sbjct: 707 DHGFDIFVSRLSESIFTYYKSWSASELLDPSFLFALDNGEKFSIQPVRFTALFKMTKVKI 766
Query: 747 ---------------------------------------ELEKLLDVLKHSHELLSIDLS 767
ELEKL+D+LKHSHELLS DLS
Sbjct: 767 LGRTINLRSLIAQRMNRIFRENLEFLFDRFESQDLCAVVELEKLIDILKHSHELLSQDLS 826
Query: 768 VDSFSLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFILCNTTQRFIRSSKTVPVQK 827
+D FSLMLNEMQENISLVSFSSRLA+QIWSEMQSDFLPNFILCNTTQRF+RSSK P QK
Sbjct: 827 IDPFSLMLNEMQENISLVSFSSRLATQIWSEMQSDFLPNFILCNTTQRFVRSSKVPPTQK 886
Query: 828 PSIPSAKPSFYCGTQ 842
PS+PSAKPSFYCGTQ
Sbjct: 887 PSVPSAKPSFYCGTQ 901
Score = 97.8 bits (242), Expect = 2e-18
Identities = 45/64 (70%), Positives = 51/64 (79%)
Query: 1053 GYLEESAQVQSSSPERLGDSVAWGGCTIIYLLGQQLHFELFDFSYQILNIAEVEAASVVQ 1112
GYLEE QS+ E LGDS+AWGGCTIIYLLGQQLHFELFDFSYQ+LN++EVE S
Sbjct: 1149 GYLEEITAPQSAQHEVLGDSIAWGGCTIIYLLGQQLHFELFDFSYQVLNVSEVETVSASH 1208
Query: 1113 TQKN 1116
T +N
Sbjct: 1209 THRN 1212
>UniRef100_Q6DLU3 PIROGI [Arabidopsis thaliana]
Length = 1282
Score = 1386 bits (3587), Expect = 0.0
Identities = 714/915 (78%), Positives = 775/915 (84%), Gaps = 87/915 (9%)
Query: 1 MAVPVEEAIAALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLSEDTKA 60
MAVPVEEAIAALSTFSLEDEQPEVQGP V VS ER AT+SPIEY DVAAYRLSLSEDTKA
Sbjct: 1 MAVPVEEAIAALSTFSLEDEQPEVQGPAVMVSAERAATDSPIEYSDVAAYRLSLSEDTKA 60
Query: 61 LNQLSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSRLREI 120
LNQL++L QEGKEMAS+LYTYRSCVKALPQLP+SMK SQADLYLETYQVLDLEMSRLREI
Sbjct: 61 LNQLNTLIQEGKEMASILYTYRSCVKALPQLPESMKHSQADLYLETYQVLDLEMSRLREI 120
Query: 121 QRWQASASSKLATDMQRFSRPERRINGPTISHLWSMLKLFDVLVQLDHLKNAKASIPNDF 180
QRWQ+SAS+KLA DMQRFSRPERRINGPT++HLWSMLKL DVLVQLDHLKNAKASIPNDF
Sbjct: 121 QRWQSSASAKLAADMQRFSRPERRINGPTVTHLWSMLKLLDVLVQLDHLKNAKASIPNDF 180
Query: 181 SWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPVYLLHIFS 240
SWYKRTFTQVS QWQDTD+MREELDDLQIFLSTRWAILLNLHVEMFRVN
Sbjct: 181 SWYKRTFTQVSAQWQDTDTMREELDDLQIFLSTRWAILLNLHVEMFRVN----------- 229
Query: 241 LSTVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSEKDSESLYKRVKI 300
VEDILQVLIVF VESLELDFALLFPER+ILLRVLPVLVVL T SEKD+E+LYKRVK+
Sbjct: 230 --NVEDILQVLIVFIVESLELDFALLFPERYILLRVLPVLVVLATPSEKDTEALYKRVKL 287
Query: 301 NRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELPP------ 354
NRLINIFKN+ VIPAFPDLHLSPAAI+KELS YF KFSSQTRLLTL APHELPP
Sbjct: 288 NRLINIFKNDPVIPAFPDLHLSPAAILKELSVYFQKFSSQTRLLTLPAPHELPPREALEY 347
Query: 355 ------LNHIGAVRAEHDDFTIRFASAMNQ-----STDGSDVDWSKEVKGNMYDMIVEGF 403
+NHIGA+RAEHDDFTIRFAS+MNQ S DG+ +W +EVKGNMYDM+VEGF
Sbjct: 348 QRHYLIVNHIGALRAEHDDFTIRFASSMNQLLLLKSNDGAYTEWCREVKGNMYDMVVEGF 407
Query: 404 QLLSRWTARIWEQCAWKFSRPCKD------ASPSFSDYEKVVRYNYSAEERKALVELVSC 457
QLLSRWTARIWEQCAWKFSRPC+D AS S+SDYEKVVR+NY+AEERKALVELV
Sbjct: 408 QLLSRWTARIWEQCAWKFSRPCRDAGETPEASGSYSDYEKVVRFNYTAEERKALVELVGY 467
Query: 458 IKSVGSMMQRCDTLVADALWETIHAEVQDFVQNTLASMLRTTFRKKKDLSRILSDMRTLS 517
IKSVGSM+QRCDTLVADALWETIHAEVQDFVQNTLA+MLRTTFRKKKDLSRILSDMRTLS
Sbjct: 468 IKSVGSMLQRCDTLVADALWETIHAEVQDFVQNTLATMLRTTFRKKKDLSRILSDMRTLS 527
Query: 518 ADWMANTNKSESELQSSQHGGEESKANIFYPRAVAPTAAQVHCLQFLIYEVVSGGNLRRP 577
ADWMANT + E E+ SSQHG +ES+ N FYPR VAPTAAQVHCLQFLIYEVVSGGNLRRP
Sbjct: 528 ADWMANT-RPEHEMPSSQHGNDESRGNFFYPRPVAPTAAQVHCLQFLIYEVVSGGNLRRP 586
Query: 578 GGLFGNSGSEVPVNDLKQLETF----------VMLAVTVATLTDLGFLWFREFYLESSRV 627
GG FGN+GSE+PVNDLKQLETF + + ++ LTDLGFLWFREFYLESSRV
Sbjct: 587 GGFFGNNGSEIPVNDLKQLETFFYKLSFFLHILDYSASIGILTDLGFLWFREFYLESSRV 646
Query: 628 IQFPIECSLPWMLVDCVLESPNSGLLESVLMPFDIYNDSAQQALVLLKQRFLYDEIEAEV 687
IQFPIECSLPWML+D +LE+ NSGLLESVL+PFDIYNDSAQQALV+L+QRFLYDEIEAEV
Sbjct: 647 IQFPIECSLPWMLIDYILEAQNSGLLESVLLPFDIYNDSAQQALVVLRQRFLYDEIEAEV 706
Query: 688 DHCFDIFVARLCETIFTYYKSWAASELLDPTFLFASENAEKYAVQPMRLNMLLKMTRVK- 746
DH FDIFV+RL E+IFTYYKSW+ASELLDP+FLFA +N EK+++QP+R L KMT+VK
Sbjct: 707 DHGFDIFVSRLSESIFTYYKSWSASELLDPSFLFALDNGEKFSIQPVRFTALFKMTKVKI 766
Query: 747 ---------------------------------------ELEKLLDVLKHSHELLSIDLS 767
ELEKL+D+LKHSHELLS DLS
Sbjct: 767 LGRTINLRSLIAQRMNRIFRENLEFLFDRFESQDLCAVVELEKLIDILKHSHELLSQDLS 826
Query: 768 VDSFSLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFILCNTTQRFIRSSKTVPVQK 827
+D FSLMLNEMQENISLVSFSSRLA+QIWSEMQSDFLPNFILCNTTQRF+RSSK P QK
Sbjct: 827 IDPFSLMLNEMQENISLVSFSSRLATQIWSEMQSDFLPNFILCNTTQRFVRSSKVPPTQK 886
Query: 828 PSIPSAKPSFYCGTQ 842
PS+PSAKPSFYCGTQ
Sbjct: 887 PSVPSAKPSFYCGTQ 901
Score = 97.8 bits (242), Expect = 2e-18
Identities = 45/64 (70%), Positives = 51/64 (79%)
Query: 1053 GYLEESAQVQSSSPERLGDSVAWGGCTIIYLLGQQLHFELFDFSYQILNIAEVEAASVVQ 1112
GYLEE QS+ E LGDS+AWGGCTIIYLLGQQLHFELFDFSYQ+LN++EVE S
Sbjct: 1148 GYLEEITAPQSAQHEVLGDSIAWGGCTIIYLLGQQLHFELFDFSYQVLNVSEVETVSASH 1207
Query: 1113 TQKN 1116
T +N
Sbjct: 1208 THRN 1211
>UniRef100_Q6UK63 Rac-binding component of SCAR regulatory complex [Dictyostelium
discoideum]
Length = 1336
Score = 395 bits (1016), Expect = e-108
Identities = 258/906 (28%), Positives = 474/906 (51%), Gaps = 100/906 (11%)
Query: 5 VEEAIAALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLSEDTKALNQL 64
V E L +F D+Q E++ + + A + + Y D AY SE+T + ++
Sbjct: 13 VFERCDVLESFPFHDDQSEIEEQSPSIGYD--AYDKSLNYTDRGAYETQWSEETIGMEKM 70
Query: 65 SSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSRLREIQRWQ 124
+ ++G +++YTYRSC KALP + + + ++ +Y ++VL+ E+ +L++ +Q
Sbjct: 71 EEVLKQGDSFINMVYTYRSCSKALPTVKTAEQVNKTQIYEGNFEVLEPEIKKLKDFMYFQ 130
Query: 125 ASASSKLATDMQRFSRP---ERRINGPTISHLWSMLKLFDVLVQLDHLKNAKASIPNDFS 181
+++ + ++ + S + ++++ D+L LD LKN KA + NDFS
Sbjct: 131 KDTIKLFCDHIKKLASTYDKKKETISASESFINYLVRILDLLAILDALKNMKACLNNDFS 190
Query: 182 WYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPVYLLHIFSL 241
++KR + Q T+ +E L +FL+ + +I +L +E+ ++K+
Sbjct: 191 FFKRATGFLRKQMSGTEDQTQENHTLYLFLANQNSITSSLKLELHNIDKF---------- 240
Query: 242 STVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSE-KDSESLYKRVKI 300
+DIL +++ + LE + +L E+H LLRV+P ++ L+ ++ K + + K + I
Sbjct: 241 ---DDILPMIVNQCADYLEQEKYILPSEKHCLLRVMPFVLFLIDENDSKHNINKNKNLNI 297
Query: 301 NRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELPPLNHIGA 360
+R IFK V+P + D+ ++ +++K S +F + + T L + + ++ +
Sbjct: 298 SRYAKIFKKNPVVPLYGDMQITLESLVKR-SPHFDEKAWGTSTLDSKSALDYEIIHVLDQ 356
Query: 361 VRAEHDDFTIRFASAMNQSTDGSDVDWSKE----VKGNMYDMIVEGFQLLSRWTARIWEQ 416
R+ ++++ RFA+ +N+ + KE + ++ + + G ++LS WT+R+ +Q
Sbjct: 357 TRSLYNEYMARFANMVNEIR-AAKARGPKEPLPLAESDIQAITLMGLRILSDWTSRVLQQ 415
Query: 417 CAWKFSRPCKDASPSFS-DYEKVVRYNYSAEERKALVELVSCIKSVGSMMQRCDTLVADA 475
AWK+S+P D + S + DYE+VV++NY+ EER ALV+L++ IKS+ S+M + +TL+
Sbjct: 416 SAWKYSKPNNDPTISATFDYERVVKFNYTKEERTALVQLIAMIKSLASLMMKSETLLQPI 475
Query: 476 LWETIHAEVQDFVQ----NTLASMLRTTFRKKKDLSRILSDMRTLSADWMANTNKSESEL 531
L +TIH E+Q+FVQ T+ S ++ +KK ++ +S ++ +S DW + +E+ +
Sbjct: 476 LRKTIHQELQEFVQINLKETIKSFVKNNPKKKDNIKLEMSQLKNISVDWFSGFEPAEA-V 534
Query: 532 QSSQHGGEESKANIFYPRAVAPTAAQVHCLQFLIYEVVSGGNLRRPGGLFGNSGSEVPVN 591
+ + E K I RAV P+ Q+ + L+ ++ + + +
Sbjct: 535 PNKKSKEVEEKVQI-PARAVPPSPTQLELILTLVSSLM-------------DKKKDFSSD 580
Query: 592 DLKQLETF----------VMLAVTVATLTDLGFLWFREFYLESSRVIQFPIECSLPWMLV 641
+ E F + L+ ++ ++TDL LW+REFYLE + +QFPIE SLPW+L
Sbjct: 581 QYRDFEAFSSKAFFYRYLLSLSSSIISITDLADLWYREFYLELNNRVQFPIETSLPWILT 640
Query: 642 DCVLESPNSGLLESVLMPFDIYNDSAQQALVLLKQRFLYDEIEAEVDHCFDIFVARLCET 701
D +LES + L+E + P +YND+AQ+AL+ L QRFLYDEIEAE++ CFD + +L
Sbjct: 641 DHILESDDPSLIEHLFYPLGLYNDTAQRALLSLNQRFLYDEIEAELNLCFDQLLYKLSGK 700
Query: 702 IFTYYKSWAASELLDPTFLFASENAE---KYAVQPMRLNMLLK----------------- 741
++T++K+ A+S LLD + E A K R ++LL+
Sbjct: 701 VYTHFKTQASSILLDKPYKTQLELAHFNGKLHTPKSRFDVLLRQKHITLLGRSIDLCGLL 760
Query: 742 -----------------------MTRVKELEKLLDVLKHSHELLSIDLSVDSFSLMLNEM 778
+T + ELE + +K +H+LLS +D F + NE+
Sbjct: 761 AQRQNNTIRQNLDYAISRFEACDLTSIVELETQIANIKLTHKLLSEYFDIDPFESIFNEI 820
Query: 779 QENISLVSFSSRLASQIWSEMQSDFLPNFILCNTTQRFIRS--SKTVPVQKPSIPSAKPS 836
E+ SLVS+ R+ I E+ +DF PN+ + TQRFI++ + T +++ ++P P
Sbjct: 821 NESTSLVSYHGRIVLHIIFELVADFAPNYTFNSITQRFIKAPYTFTEELKRDALPKTNPV 880
Query: 837 FYCGTQ 842
F G +
Sbjct: 881 FLFGNK 886
Score = 55.8 bits (133), Expect = 7e-06
Identities = 25/50 (50%), Positives = 31/50 (62%)
Query: 1058 SAQVQSSSPERLGDSVAWGGCTIIYLLGQQLHFELFDFSYQILNIAEVEA 1107
S + S E GD + W GC++I+ LGQQ FELFDF Y ILN+ E A
Sbjct: 1146 SNENDKSFHELFGDGLMWAGCSLIHFLGQQYRFELFDFCYHILNVEEAAA 1195
>UniRef100_UPI000035F078 UPI000035F078 UniRef100 entry
Length = 1256
Score = 350 bits (898), Expect = 1e-94
Identities = 252/863 (29%), Positives = 441/863 (50%), Gaps = 107/863 (12%)
Query: 56 EDTKALNQLSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMS 115
E + ++ + +EG E A +LYT+RSC +A+PQ+ + + ++ ++Y +T +VL+ E++
Sbjct: 64 EQATVHSSMNEMLEEGHEYAIMLYTWRSCSRAIPQVKCNEQPNRVEIYEKTVEVLEPEVT 123
Query: 116 RLREIQRWQASASSKLATDMQRFSRPERRINGPTISHLWSMLKLFDVLVQLDHLKNAKAS 175
+L +Q +A + +++R ERR + + ++L ++ K ++ LD LKN K S
Sbjct: 124 KLMNFMHFQRTAIDRFCGEVRRLCHTERRKDFVSEAYLLTLGKFINMFAVLDELKNMKCS 183
Query: 176 IPNDFSWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPVYL 235
+ ND S YKR Q + + S++E +L +FL+ I +L ++ +N Y
Sbjct: 184 VKNDHSAYKRA-AQFLRKMSEPSSIQES-QNLSMFLANHNKITQSLQQQLEVINGY---- 237
Query: 236 LHIFSLSTVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSEKDSESL- 294
E++L ++ V+ E L E+H+LL+V+ + L+ + + L
Sbjct: 238 ---------EELLADIVNLCVDYYENKMYLTPSEKHMLLKVMGFGLYLMDGNSSNIYKLD 288
Query: 295 -YKRVKINRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELP 353
KR+ + ++ FK V+P F D+ + + +K S +F + S+ ++++ +
Sbjct: 289 AKKRINLTKIDKFFKQLQVVPLFGDMQIELSRYIKT-SAHFEENKSRWTCTSISSSPQYN 347
Query: 354 PLNHIGAVRAEHDDFTIRFASAMN---------QSTDGSDVDWSKEVKGNMYDMIVEGFQ 404
+ +R +H F A N Q + +D+++ K ++D+ ++G Q
Sbjct: 348 ICEQMIQIREDHMRFISELARYSNSEVVTGSGRQESQKTDIEYRK-----LFDLSLQGMQ 402
Query: 405 LLSRWTARIWEQCAWKFSRPC-----KDASPSFSDYEKVVRYNYSAEERKALVELVSCIK 459
LLS+W+A + E +WK P K+ + +YE+ RYNY++EE+ ALVE+++ IK
Sbjct: 403 LLSQWSAHVMEVYSWKLVHPTDKYSNKECPDNAEEYERATRYNYTSEEKFALVEVMAMIK 462
Query: 460 SVGSMMQRCDTLVADALWETIHAEVQDFVQNTLASMLRTTFRKKKD-LSRILSDMRTLSA 518
+ +M R +++ A+ TI++ +QDF Q TL LR +KKK+ + +L +R
Sbjct: 463 GLQVLMGRMESVFNHAIRHTIYSALQDFAQVTLRDPLRQAIKKKKNVIQSVLQAIRKTIC 522
Query: 519 DWMANTNKSESELQSSQHGGEESKA--NIFYPR-AVAPTAAQVHCLQFLIYEVV---SGG 572
DW E + G ++ K +I PR AV P++ Q++ ++ ++ +V SG
Sbjct: 523 DWETGR---EPHNDPALRGEKDPKGGFDIKVPRRAVGPSSTQLYMVRTMLESLVADKSGS 579
Query: 573 NLRRPGGLFGNSGSEVPVNDLKQL--ETF-----VMLAVTVATLTDLGFLWFREFYLESS 625
L G + + D+++ E+F + + T+ DL LWFREF+LE +
Sbjct: 580 KKTLRSSLEGPT-----ILDIEKFHRESFFYTHLLNFSETLQHCCDLSQLWFREFFLELT 634
Query: 626 --RVIQFPIECSLPWMLVDCVLESPNSGLLESVLMPFDIYNDSAQQALVLLKQRFLYDEI 683
R IQFPIE S+PW+L D +LE+ + ++E VL P D+YNDSA AL+ K++FLYDEI
Sbjct: 635 MGRRIQFPIEMSMPWILTDHILETKEASMMEYVLYPLDLYNDSAHYALITFKKQFLYDEI 694
Query: 684 EAEVDHCFDIFVARLCETIFTYYKSWAASELLDPTFLFASEN---------AEKYAV--- 731
EAEV+ CFD FV +L + IF YYK A S LLD +N + +Y
Sbjct: 695 EAEVNLCFDQFVYKLADQIFAYYKILAGSLLLDKRLRAECKNQGANIPWPSSNRYETLLK 754
Query: 732 --------QPMRLNMLL----------------------KMTRVKELEKLLDVLKHSHEL 761
+ + LN L+ +T + ELE LLD+ + +H+L
Sbjct: 755 QRHVQLLGRSIDLNRLITQRVSAALYKSLELAINRFESEDLTSIMELEGLLDINRMTHKL 814
Query: 762 LSIDLSVDSFSLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFILCNTTQRFIRS-- 819
LS L++DSF+ M E N+S + R+ ++ E+ DFLPN+ +T RF+R+
Sbjct: 815 LSKFLTLDSFNAMFREANHNVS--APYGRITLHVFWELNYDFLPNYCYNGSTNRFVRTVL 872
Query: 820 SKTVPVQKPSIPSAKPSFYCGTQ 842
+ Q+ P+A+P + G++
Sbjct: 873 PFSQEFQRDKPPNAQPHYLYGSK 895
Score = 45.8 bits (107), Expect = 0.008
Identities = 18/33 (54%), Positives = 23/33 (69%)
Query: 1070 GDSVAWGGCTIIYLLGQQLHFELFDFSYQILNI 1102
GD + W GC II LLGQQ F++ DFSY +L +
Sbjct: 1157 GDGLHWAGCMIIVLLGQQRRFDILDFSYHLLKV 1189
>UniRef100_Q9UN29 P53 inducible protein [Homo sapiens]
Length = 1253
Score = 349 bits (896), Expect = 2e-94
Identities = 263/914 (28%), Positives = 447/914 (48%), Gaps = 99/914 (10%)
Query: 3 VPVEEAIA---ALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLS---E 56
V +E+A++ L L D+QP ++ P + + + + D A+ ++ E
Sbjct: 5 VTLEDALSNVDLLEELPLPDQQPCIEPPPSSIMYQANFDTN---FEDRNAFVTGIARYIE 61
Query: 57 DTKALNQLSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSR 116
+ ++ + +EG E A +LYT+RSC +A+PQ+ + + ++ ++Y +T +VL+ E+++
Sbjct: 62 QATVHSSMNEMLEEGHEYAVMLYTWRSCSRAIPQVKCNEQPNRVEIYEKTVEVLEPEVTK 121
Query: 117 LREIQRWQASASSKLATDMQRFSRPERRINGPTISHLWSMLKLFDVLVQLDHLKNAKASI 176
L + +Q A + ++++R ERR + + ++L ++ K ++ LD LKN K S+
Sbjct: 122 LMKFMYFQRKAIERFCSEVKRLCHAERRKDFVSEAYLLTLGKFINMFAVLDELKNMKCSV 181
Query: 177 PNDFSWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPVYLL 236
ND S YKR Q + D S++E +L +FL+ I LH ++ + Y
Sbjct: 182 KNDHSAYKRA-AQFLRKMADPQSIQES-QNLSMFLANHNRITQCLHQQLEVIPGY----- 234
Query: 237 HIFSLSTVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSEKDSESL-- 294
E++L ++ V+ E L E+H+LL+V+ + L+ + + L
Sbjct: 235 --------EELLADIVNICVDYYENKMYLTPSEKHMLLKVMGFGLYLMDGNVSNIYKLDA 286
Query: 295 YKRVKINRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELPP 354
KR+ ++++ FK V+P F D+ + A +K + Y S T + +P +
Sbjct: 287 KKRINLSKIDKFFKQLQVVPLFGDMQIELARYIKTSAHYEENKSKWTCTQSSISP-QYNI 345
Query: 355 LNHIGAVRAEHDDFTIRFASAMNQST---DGSDVDWSKEVKGNMYDMIVEGFQLLSRWTA 411
+ +R +H F A N G D S E ++D+ + G QLLS+W+A
Sbjct: 346 CEQMVQIRDDHIRFISELARYSNSEVVTGSGLDSQKSDEEYRELFDLALRGLQLLSKWSA 405
Query: 412 RIWEQCAWKFSRPC-----KDASPSFSDYEKVVRYNYSAEERKALVELVSCIKSVGSMMQ 466
+ E +WK P KD + +YE+ RYNY++EE+ A VE+++ IK + +M
Sbjct: 406 HVMEVYSWKLVHPTDKFCNKDCPGTAEEYERATRYNYTSEEKFAFVEVIAMIKGLQVLMG 465
Query: 467 RCDTLVADALWETIHAEVQDFVQNTLASMLRTTFRKKKD-LSRILSDMRTLSADWMANTN 525
R +++ A+ TI+A +QDF Q TL LR RKKK+ L +L +R DW
Sbjct: 466 RMESVFNQAIRNTIYAALQDFAQVTLREPLRQAVRKKKNVLISVLQAIRKTICDWEGGRE 525
Query: 526 KSESEL---QSSQHGGEESKANIFYPRAVAPTAAQVHCLQFLIYEVV---SGGNLRRPGG 579
+ GG + K RAV P++ Q++ ++ ++ ++ SG
Sbjct: 526 PPNDPCLRGEKDPKGGFDIKVP---RRAVGPSSTQLYMVRTMLESLIADKSGSKKTLRSS 582
Query: 580 LFGNSGSEVPVNDLKQLETF----VMLAVTVATLTDLGFLWFREFYLESS--RVIQFPIE 633
L G + + D + F + ++ + DL LWFREF+LE + R IQFPIE
Sbjct: 583 LDGPI--VLAIEDFHKQSFFFTHLLNISEALQQCCDLSQLWFREFFLELTMGRRIQFPIE 640
Query: 634 CSLPWMLVDCVLESPNSGLLESVLMPFDIYNDSAQQALVLLKQRFLYDEIEAEVDHCFDI 693
S+PW+L D +LE+ ++E VL P D+YNDSA AL K++FLYDEIEAEV+ CFD
Sbjct: 641 MSMPWILTDHILETKEPSMMEYVLYPLDLYNDSAYYALTKFKKQFLYDEIEAEVNLCFDQ 700
Query: 694 FVARLCETIFTYYKSWAASELLDPTFLFASEN---------AEKYAV-----------QP 733
FV +L + IF YYK+ A S LLD F +N + +Y +
Sbjct: 701 FVYKLADQIFAYYKAMAGSVLLDKRFRAECKNYGVIIPYPPSNRYETLLKQRHVQLLGRS 760
Query: 734 MRLNMLL----------------------KMTRVKELEKLLDVLKHSHELLSIDLSVDSF 771
+ LN L+ +T + ELE LL++ + +H LL +++DSF
Sbjct: 761 IDLNRLITQRISAAMYKSLDQAISRFESEDLTSIVELEWLLEINRLTHRLLCKHMTLDSF 820
Query: 772 SLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFILCNTTQRFIRSS--KTVPVQKPS 829
M E N+S + R+ ++ E+ DFLPN+ +T RF+R++ T Q+
Sbjct: 821 DAMFREANHNVS--APYGRITLHVFWELNFDFLPNYCYNGSTNRFVRTAIPFTQEPQRDK 878
Query: 830 IPSAKPSFYCGTQP 843
+ +P + G++P
Sbjct: 879 PANVQPYYLYGSKP 892
Score = 47.8 bits (112), Expect = 0.002
Identities = 19/33 (57%), Positives = 24/33 (72%)
Query: 1070 GDSVAWGGCTIIYLLGQQLHFELFDFSYQILNI 1102
GD + W GC+II LLGQQ F+LFDF Y +L +
Sbjct: 1153 GDGLNWAGCSIIVLLGQQRRFDLFDFCYHLLKV 1185
>UniRef100_Q96F07 P53 inducible protein [Homo sapiens]
Length = 1253
Score = 349 bits (896), Expect = 2e-94
Identities = 263/914 (28%), Positives = 447/914 (48%), Gaps = 99/914 (10%)
Query: 3 VPVEEAIA---ALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLS---E 56
V +E+A++ L L D+QP ++ P + + + + D A+ ++ E
Sbjct: 5 VTLEDALSNVDLLEELPLPDQQPCIEPPPSSIMYQANFDTN---FEDRNAFVTGIARYIE 61
Query: 57 DTKALNQLSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSR 116
+ ++ + +EG E A +LYT+RSC +A+PQ+ + + ++ ++Y +T +VL+ E+++
Sbjct: 62 QATVHSSMNEMLEEGHEYAVMLYTWRSCSRAIPQVKCNEQPNRVEIYEKTVEVLEPEVTK 121
Query: 117 LREIQRWQASASSKLATDMQRFSRPERRINGPTISHLWSMLKLFDVLVQLDHLKNAKASI 176
L + +Q A + ++++R ERR + + ++L ++ K ++ LD LKN K S+
Sbjct: 122 LMKFMYFQRKAIERFCSEVKRLCHAERRKDFVSEAYLLTLGKFINMFAVLDELKNMKCSV 181
Query: 177 PNDFSWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPVYLL 236
ND S YKR Q + D S++E +L +FL+ I LH ++ + Y
Sbjct: 182 KNDHSAYKRA-AQFLRKMADPQSIQES-QNLSMFLANHNRITQCLHQQLEVIPGY----- 234
Query: 237 HIFSLSTVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSEKDSESL-- 294
E++L ++ V+ E L E+H+LL+V+ + L+ + + L
Sbjct: 235 --------EELLADIVNICVDYYENKMYLTPSEKHMLLKVMGFGLYLMDGNVSNIYKLDA 286
Query: 295 YKRVKINRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELPP 354
KR+ ++++ FK V+P F D+ + A +K + Y S T + +P +
Sbjct: 287 KKRINLSKIDKFFKQLQVVPLFGDMQIELARYIKTSAHYEENKSKWTCTQSSISP-QYNI 345
Query: 355 LNHIGAVRAEHDDFTIRFASAMNQST---DGSDVDWSKEVKGNMYDMIVEGFQLLSRWTA 411
+ +R +H F A N G D S E ++D+ + G QLLS+W+A
Sbjct: 346 CEQMVQIRDDHIRFISELARYSNSEVVTGSGLDSQKSDEEYRELFDLALRGLQLLSKWSA 405
Query: 412 RIWEQCAWKFSRPC-----KDASPSFSDYEKVVRYNYSAEERKALVELVSCIKSVGSMMQ 466
+ E +WK P KD + +YE+ RYNY++EE+ A VE+++ IK + +M
Sbjct: 406 HVMEVYSWKLVHPTDKFCNKDCPGTAEEYERATRYNYTSEEKFAFVEVIAMIKGLQVLMG 465
Query: 467 RCDTLVADALWETIHAEVQDFVQNTLASMLRTTFRKKKD-LSRILSDMRTLSADWMANTN 525
R +++ A+ TI+A +QDF Q TL LR RKKK+ L +L +R DW
Sbjct: 466 RMESVFNQAIRNTIYAALQDFAQVTLREPLRQAVRKKKNVLISVLQAIRKTICDWEGGRE 525
Query: 526 KSESEL---QSSQHGGEESKANIFYPRAVAPTAAQVHCLQFLIYEVV---SGGNLRRPGG 579
+ GG + K RAV P++ Q++ ++ ++ ++ SG
Sbjct: 526 PPNDPCLRGEKDPKGGFDIKVP---RRAVGPSSTQLYMVRTMLESLIADKSGSKKTLRSS 582
Query: 580 LFGNSGSEVPVNDLKQLETF----VMLAVTVATLTDLGFLWFREFYLESS--RVIQFPIE 633
L G + + D + F + ++ + DL LWFREF+LE + R IQFPIE
Sbjct: 583 LDGPI--VLAIEDFHKQSFFFTHLLNISEALQQCCDLSQLWFREFFLELTMGRRIQFPIE 640
Query: 634 CSLPWMLVDCVLESPNSGLLESVLMPFDIYNDSAQQALVLLKQRFLYDEIEAEVDHCFDI 693
S+PW+L D +LE+ ++E VL P D+YNDSA AL K++FLYDEIEAEV+ CFD
Sbjct: 641 MSMPWILTDHILETKEPSMMEYVLYPLDLYNDSAYYALTKFKKQFLYDEIEAEVNLCFDQ 700
Query: 694 FVARLCETIFTYYKSWAASELLDPTFLFASEN---------AEKYAV-----------QP 733
FV +L + IF YYK+ A S LLD F +N + +Y +
Sbjct: 701 FVYKLADQIFAYYKAMAGSVLLDKRFRAECKNYGVIIPYPPSNRYETLLKQRHVQLLGRS 760
Query: 734 MRLNMLL----------------------KMTRVKELEKLLDVLKHSHELLSIDLSVDSF 771
+ LN L+ +T + ELE LL++ + +H LL +++DSF
Sbjct: 761 IDLNRLITQRTSAAMYKSLDQAISRFESEDLTSIVELEWLLEINRLTHRLLCKHMTLDSF 820
Query: 772 SLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFILCNTTQRFIRSS--KTVPVQKPS 829
M E N+S + R+ ++ E+ DFLPN+ +T RF+R++ T Q+
Sbjct: 821 DAMFREANHNVS--APYGRITLHVFWELNFDFLPNYCYNGSTNRFVRTAIPFTQEPQRDK 878
Query: 830 IPSAKPSFYCGTQP 843
+ +P + G++P
Sbjct: 879 PANVQPYYLYGSKP 892
Score = 47.8 bits (112), Expect = 0.002
Identities = 19/33 (57%), Positives = 24/33 (72%)
Query: 1070 GDSVAWGGCTIIYLLGQQLHFELFDFSYQILNI 1102
GD + W GC+II LLGQQ F+LFDF Y +L +
Sbjct: 1153 GDGLNWAGCSIIVLLGQQRRFDLFDFCYHLLKV 1185
>UniRef100_Q924D3 CYFIP2 [Mus musculus]
Length = 1253
Score = 348 bits (893), Expect = 5e-94
Identities = 262/914 (28%), Positives = 447/914 (48%), Gaps = 99/914 (10%)
Query: 3 VPVEEAIA---ALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLS---E 56
V +E+A++ L L D+QP ++ P + + + + D A+ ++ E
Sbjct: 5 VTLEDALSNVDLLEELPLPDQQPCIEPPPSSIMYQANFDTN---FEDRNAFVTGIARYIE 61
Query: 57 DTKALNQLSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSR 116
+ ++ + +EG + A +LYT+RSC +A+PQ+ + + ++ ++Y +T +VL+ E+++
Sbjct: 62 QATVHSSMNEMLEEGHDYAVMLYTWRSCSRAIPQVKCNEQPNRVEIYEKTVEVLEPEVTK 121
Query: 117 LREIQRWQASASSKLATDMQRFSRPERRINGPTISHLWSMLKLFDVLVQLDHLKNAKASI 176
L + +Q A + ++++R ERR + + ++L ++ K ++ LD LKN K S+
Sbjct: 122 LMKFMYFQRKAIERFCSEVKRLCHAERRKDFVSEAYLLTLGKFINMFAVLDELKNMKCSV 181
Query: 177 PNDFSWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPVYLL 236
ND S YKR Q + D S++E +L +FL+ I LH ++ + Y
Sbjct: 182 KNDHSAYKRA-AQFLRKMADPQSIQES-QNLSMFLANHNRITQCLHQQLEVIPGY----- 234
Query: 237 HIFSLSTVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSEKDSESL-- 294
E++L ++ V+ E L E+H+LL+V+ + L+ + + L
Sbjct: 235 --------EELLADIVNICVDYYENKMYLTPSEKHMLLKVMGFGLYLMDGNVSNIYKLDA 286
Query: 295 YKRVKINRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELPP 354
KR+ ++++ FK V+P F D+ + A +K + Y S T + +P +
Sbjct: 287 KKRINLSKIDKFFKQLQVVPLFGDMQIELARYIKTSAHYEENKSKWTCTQSSISP-QYNI 345
Query: 355 LNHIGAVRAEHDDFTIRFASAMNQST---DGSDVDWSKEVKGNMYDMIVEGFQLLSRWTA 411
+ +R +H F A N G D S E ++D+ + G QLLS+W+A
Sbjct: 346 CEQMVQIRDDHIRFISELARYSNSEVVTGSGLDSQKSDEEYRELFDLALRGLQLLSKWSA 405
Query: 412 RIWEQCAWKFSRPC-----KDASPSFSDYEKVVRYNYSAEERKALVELVSCIKSVGSMMQ 466
+ E +WK P KD + +YE+ RYNY++EE+ A VE+++ IK + +M
Sbjct: 406 HVMEVYSWKLVHPTDKFCNKDCPGTAEEYERATRYNYTSEEKFAFVEVIAMIKGLQVLMG 465
Query: 467 RCDTLVADALWETIHAEVQDFVQNTLASMLRTTFRKKKD-LSRILSDMRTLSADWMANTN 525
R +++ A+ TI+A +QDF Q TL LR RKKK+ L +L +R DW
Sbjct: 466 RMESVFNQAIRNTIYAALQDFAQVTLREPLRQAVRKKKNVLISVLQAIRKTICDWEGGRE 525
Query: 526 KSESEL---QSSQHGGEESKANIFYPRAVAPTAAQVHCLQFLIYEVV---SGGNLRRPGG 579
+ GG + K RAV P++ Q++ ++ ++ ++ SG
Sbjct: 526 PPNDPCLRGEKDPKGGFDIKVP---RRAVGPSSTQLYMVRTMLESLIADKSGSKKTLRSS 582
Query: 580 LFGNSGSEVPVNDLKQLETF----VMLAVTVATLTDLGFLWFREFYLESS--RVIQFPIE 633
L G + + D + F + ++ + DL LWFREF+LE + R IQFPIE
Sbjct: 583 LDGPI--VLAIEDFHKQSFFFTHLLNISEALQQCCDLSQLWFREFFLELTMGRRIQFPIE 640
Query: 634 CSLPWMLVDCVLESPNSGLLESVLMPFDIYNDSAQQALVLLKQRFLYDEIEAEVDHCFDI 693
S+PW+L D +LE+ ++E VL P D+YNDSA AL K++FLYDEIEAEV+ CFD
Sbjct: 641 MSMPWILTDHILETKEPSMMEYVLYPLDLYNDSAYYALTKFKKQFLYDEIEAEVNLCFDQ 700
Query: 694 FVARLCETIFTYYKSWAASELLDPTFLFASEN---------AEKYAV-----------QP 733
FV +L + IF YYK+ A S LLD F +N + +Y +
Sbjct: 701 FVYKLADQIFAYYKAMAGSVLLDKRFRAECKNYGVIIPYPPSNRYETLLKQRHVQLLGRS 760
Query: 734 MRLNMLL----------------------KMTRVKELEKLLDVLKHSHELLSIDLSVDSF 771
+ LN L+ +T + ELE LL++ + +H LL +++DSF
Sbjct: 761 IDLNRLITQRISAAMYKSLDQAISRFESEDLTSIVELEWLLEINRLTHRLLCKHMTLDSF 820
Query: 772 SLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFILCNTTQRFIRSS--KTVPVQKPS 829
M E N+S + R+ ++ E+ DFLPN+ +T RF+R++ T Q+
Sbjct: 821 DAMFREANHNVS--APYGRITLHVFWELNFDFLPNYCYNGSTNRFVRTAIPFTQEPQRDK 878
Query: 830 IPSAKPSFYCGTQP 843
+ +P + G++P
Sbjct: 879 PANVQPYYLYGSKP 892
Score = 47.8 bits (112), Expect = 0.002
Identities = 19/33 (57%), Positives = 24/33 (72%)
Query: 1070 GDSVAWGGCTIIYLLGQQLHFELFDFSYQILNI 1102
GD + W GC+II LLGQQ F+LFDF Y +L +
Sbjct: 1153 GDGLNWAGCSIIVLLGQQRRFDLFDFCYHLLKV 1185
>UniRef100_Q6GQD1 MGC80158 protein [Xenopus laevis]
Length = 1253
Score = 348 bits (892), Expect = 7e-94
Identities = 262/912 (28%), Positives = 450/912 (48%), Gaps = 95/912 (10%)
Query: 3 VPVEEAIA---ALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLS---E 56
V +E+A++ L L D+QP ++ P + + + + D A+ ++ E
Sbjct: 5 VTLEDALSNVDLLEELPLPDQQPCIEPPPSSIMYQANFDTN---FEDRNAFVTGIARYIE 61
Query: 57 DTKALNQLSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSR 116
+ ++ + +EG+E A +LYT+RSC +A+PQ+ + + ++ ++Y +T +VL+ E+++
Sbjct: 62 QATVHSSMNEMLEEGQEYAVMLYTWRSCSRAIPQVKCNEQPNRVEIYEKTVEVLEPEVTK 121
Query: 117 LREIQRWQASASSKLATDMQRFSRPERRINGPTISHLWSMLKLFDVLVQLDHLKNAKASI 176
L + +Q A + +++R ERR + + ++L ++ K ++ LD LKN K S+
Sbjct: 122 LMKFMYFQRKAIERFCNEVKRLCHTERRKDFVSEAYLLTLGKFINMFAVLDELKNMKCSV 181
Query: 177 PNDFSWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPVYLL 236
ND S YKR Q + D S++E +L +FL+ I LH ++ + Y
Sbjct: 182 KNDHSAYKRA-AQFLRKMADPQSIQES-QNLSMFLANHNRITQCLHQQLEVIPGY----- 234
Query: 237 HIFSLSTVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSEKDSESL-- 294
E++L ++ V+ E L E+H+LL+V+ + L+ + + L
Sbjct: 235 --------EELLADIVNICVDYYENKMYLTPSEKHMLLKVMGFGLYLMDGNVSNIYKLDA 286
Query: 295 YKRVKINRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELPP 354
KR+ ++++ FK V+P F D+ + A ++ + Y S T + +P +
Sbjct: 287 KKRINLSKIDKFFKQLQVVPLFGDMQIELARYIETSAHYEENKSKWTCTQSSISP-QYNI 345
Query: 355 LNHIGAVRAEHDDFTIRFASAMNQST---DGSDVDWSKEVKGNMYDMIVEGFQLLSRWTA 411
+ +R +H F A N G D S E ++D+ + G QLLS+W+A
Sbjct: 346 CEQMVQIRDDHIRFISELARYSNSEVVTGSGLDSQKSDEEYRELFDLALRGLQLLSKWSA 405
Query: 412 RIWEQCAWKFSRPC-----KDASPSFSDYEKVVRYNYSAEERKALVELVSCIKSVGSMMQ 466
+ E +WK P KD + +YE+ RYNY++EE+ A VE+++ +K + +M
Sbjct: 406 HVMEVYSWKLVHPTDKFCNKDCPGTAEEYERATRYNYTSEEKFAFVEVIAMVKGLQVLMG 465
Query: 467 RCDTLVADALWETIHAEVQDFVQNTLASMLRTTFRKKKD-LSRILSDMRTLSADWMANTN 525
R +++ A+ TI+A +QDF Q++L LR RKKK+ L +L +R DW A
Sbjct: 466 RMESVFNQAIRNTIYAALQDFAQSSLREPLRQAVRKKKNVLISVLQAIRKTVCDWEAGR- 524
Query: 526 KSESELQSSQHGGEESKA--NIFYPR-AVAPTAAQVHCLQFLIYEVVSGGNLRRPGGLFG 582
E G ++ K +I PR AV P++ Q++ ++ ++ +++ + +
Sbjct: 525 --EPPNDPCLRGEKDPKGGFDINVPRRAVGPSSTQLYMVRTMLESLIADKSGSKKTLRSS 582
Query: 583 NSGSEVPVNDLKQLETFVM-----LAVTVATLTDLGFLWFREFYLESS--RVIQFPIECS 635
G V + ++F + + DL LWFREF+LE + R IQFPIE S
Sbjct: 583 LDGPIVQAIEEFHKQSFFFTHLLNFSEALQQCCDLSQLWFREFFLELTMGRRIQFPIEMS 642
Query: 636 LPWMLVDCVLESPNSGLLESVLMPFDIYNDSAQQALVLLKQRFLYDEIEAEVDHCFDIFV 695
+PW+L D +LE+ ++E VL P D+YNDSA AL K++FLYDEIEAEV+ CFD FV
Sbjct: 643 MPWILTDHILETKEPSMMEYVLYPLDLYNDSAYYALTKFKKQFLYDEIEAEVNLCFDQFV 702
Query: 696 ARLCETIFTYYKSWAASELLDPTFLFASEN---------AEKYAV-----------QPMR 735
+L + IF YYK+ + S LLD F +N + +Y + +
Sbjct: 703 YKLSDQIFAYYKAMSGSVLLDKRFRAECKNYGVIIPYPPSNRYETLLKQRHVQLLGRSID 762
Query: 736 LNMLL----------------------KMTRVKELEKLLDVLKHSHELLSIDLSVDSFSL 773
LN L+ +T + ELE LLD+ + +H LLS L++DSF
Sbjct: 763 LNRLITQRISAAMYKSLDQAISRFESEDLTSIVELEWLLDINRLTHRLLSKHLTLDSFDA 822
Query: 774 MLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFILCNTTQRFIRSS--KTVPVQKPSIP 831
M E N+S + R ++ E+ DFLPN+ +T RF+R++ T Q+
Sbjct: 823 MFREANHNVS--APYGRNTLHVFWELNFDFLPNYCYNGSTNRFVRTAIPFTQEPQRDKPA 880
Query: 832 SAKPSFYCGTQP 843
+ +P + G++P
Sbjct: 881 NVQPYYLYGSKP 892
Score = 47.4 bits (111), Expect = 0.003
Identities = 19/33 (57%), Positives = 23/33 (69%)
Query: 1070 GDSVAWGGCTIIYLLGQQLHFELFDFSYQILNI 1102
GD + W GC II LLGQQ F+LFDF Y +L +
Sbjct: 1153 GDGLNWAGCAIIVLLGQQRRFDLFDFCYHLLKV 1185
>UniRef100_Q9ULQ2 KIAA1168 protein [Homo sapiens]
Length = 1281
Score = 348 bits (892), Expect = 7e-94
Identities = 262/914 (28%), Positives = 447/914 (48%), Gaps = 99/914 (10%)
Query: 3 VPVEEAIA---ALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLS---E 56
V +E+A++ L L D+QP ++ P + + + + D A+ ++ E
Sbjct: 33 VTLEDALSNVDLLEELPLPDQQPCIEPPPSSIMYQANFDTN---FEDRNAFVTGIARYIE 89
Query: 57 DTKALNQLSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSR 116
+ ++ + +EG E A +LYT+RSC +A+PQ+ + + ++ ++Y +T +VL+ E+++
Sbjct: 90 QATVHSSMNEMLEEGHEYAVMLYTWRSCSRAIPQVKCNEQPNRVEIYEKTVEVLEPEVTK 149
Query: 117 LREIQRWQASASSKLATDMQRFSRPERRINGPTISHLWSMLKLFDVLVQLDHLKNAKASI 176
L + +Q A + ++++R ERR + + ++L ++ K ++ LD LKN K S+
Sbjct: 150 LMKFMYFQRKAIERFCSEVKRLCHAERRKDFVSEAYLLTLGKFINMFAVLDELKNMKCSV 209
Query: 177 PNDFSWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPVYLL 236
ND S YKR Q + D S++E +L +FL+ I LH ++ + Y
Sbjct: 210 KNDHSAYKRA-AQFLRKMADPQSIQES-QNLSMFLANHNRITQCLHQQLEVIPGY----- 262
Query: 237 HIFSLSTVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSEKDSESL-- 294
E++L ++ V+ E L E+H+LL+V+ + L+ + + L
Sbjct: 263 --------EELLADIVNICVDYYENKMYLTPSEKHMLLKVMGFGLYLMDGNVSNIYKLDA 314
Query: 295 YKRVKINRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELPP 354
KR+ ++++ FK V+P F D+ + A ++ + Y S T + +P +
Sbjct: 315 KKRINLSKIDKFFKQLQVVPLFGDMQIELARYIETSAHYEENKSKWTCTQSSISP-QYNI 373
Query: 355 LNHIGAVRAEHDDFTIRFASAMNQST---DGSDVDWSKEVKGNMYDMIVEGFQLLSRWTA 411
+ +R +H F A N G D S E ++D+ + G QLLS+W+A
Sbjct: 374 CEQMVQIRDDHIRFISELARYSNSEVVTGSGLDSQKSDEEYRELFDLALRGLQLLSKWSA 433
Query: 412 RIWEQCAWKFSRPC-----KDASPSFSDYEKVVRYNYSAEERKALVELVSCIKSVGSMMQ 466
+ E +WK P KD + +YE+ RYNY++EE+ A VE+++ IK + +M
Sbjct: 434 HVMEVYSWKLVHPTDKFCNKDCPGTAEEYERATRYNYTSEEKFAFVEVIAMIKGLQVLMG 493
Query: 467 RCDTLVADALWETIHAEVQDFVQNTLASMLRTTFRKKKD-LSRILSDMRTLSADWMANTN 525
R +++ A+ TI+A +QDF Q TL LR RKKK+ L +L +R DW
Sbjct: 494 RMESVFNQAIRNTIYAALQDFAQVTLREPLRQAVRKKKNVLISVLQAIRKTICDWEGGRE 553
Query: 526 KSESEL---QSSQHGGEESKANIFYPRAVAPTAAQVHCLQFLIYEVV---SGGNLRRPGG 579
+ GG + K RAV P++ Q++ ++ ++ ++ SG
Sbjct: 554 PPNDPCLRGEKDPKGGFDIKVP---RRAVGPSSTQLYMVRTMLESLIADKSGSKKTLRSS 610
Query: 580 LFGNSGSEVPVNDLKQLETF----VMLAVTVATLTDLGFLWFREFYLESS--RVIQFPIE 633
L G + + D + F + ++ + DL LWFREF+LE + R IQFPIE
Sbjct: 611 LDGPI--VLAIEDFHKQSFFFTHLLNISEALQQCCDLSQLWFREFFLELTMGRRIQFPIE 668
Query: 634 CSLPWMLVDCVLESPNSGLLESVLMPFDIYNDSAQQALVLLKQRFLYDEIEAEVDHCFDI 693
S+PW+L D +LE+ ++E VL P D+YNDSA AL K++FLYDEIEAEV+ CFD
Sbjct: 669 MSMPWILTDHILETKEPSMMEYVLYPLDLYNDSAYYALTKFKKQFLYDEIEAEVNLCFDQ 728
Query: 694 FVARLCETIFTYYKSWAASELLDPTFLFASEN---------AEKYAV-----------QP 733
FV +L + IF YYK+ A S LLD F +N + +Y +
Sbjct: 729 FVYKLADQIFAYYKAMAGSVLLDKRFRAECKNYGVIIPYPPSNRYETLLKQRHVQLLGRS 788
Query: 734 MRLNMLL----------------------KMTRVKELEKLLDVLKHSHELLSIDLSVDSF 771
+ LN L+ +T + ELE LL++ + +H LL +++DSF
Sbjct: 789 IDLNRLITQRISAAMYKSLDQAISRFESEDLTSIVELEWLLEINRLTHRLLCKHMTLDSF 848
Query: 772 SLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFILCNTTQRFIRSS--KTVPVQKPS 829
M E N+S + R+ ++ E+ DFLPN+ +T RF+R++ T Q+
Sbjct: 849 DAMFREANHNVS--APYGRITLHVFWELNFDFLPNYCYNGSTNRFVRTAIPFTQEPQRDK 906
Query: 830 IPSAKPSFYCGTQP 843
+ +P + G++P
Sbjct: 907 PANVQPYYLYGSKP 920
Score = 46.6 bits (109), Expect = 0.004
Identities = 19/33 (57%), Positives = 24/33 (72%)
Query: 1070 GDSVAWGGCTIIYLLGQQLHFELFDFSYQILNI 1102
GD + W GC+II LLGQQ F+LFDF Y +L +
Sbjct: 1181 GDGLNWVGCSIIVLLGQQRRFDLFDFCYHLLKV 1213
>UniRef100_Q9NTK4 Hypothetical protein DKFZp761I12121 [Homo sapiens]
Length = 1253
Score = 348 bits (892), Expect = 7e-94
Identities = 262/914 (28%), Positives = 447/914 (48%), Gaps = 99/914 (10%)
Query: 3 VPVEEAIA---ALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLS---E 56
V +E+A++ L L D+QP ++ P + + + + D A+ ++ E
Sbjct: 5 VTLEDALSNVDLLEELPLPDQQPCIEPPPSSIMYQANFDTN---FEDRNAFVTGIARYIE 61
Query: 57 DTKALNQLSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSR 116
+ ++ + +EG E A +LYT+RSC +A+PQ+ + + ++ ++Y +T +VL+ E+++
Sbjct: 62 QATVHSSMNEMLEEGHEYAVMLYTWRSCSRAIPQVKCNEQPNRVEIYEKTVEVLEPEVTK 121
Query: 117 LREIQRWQASASSKLATDMQRFSRPERRINGPTISHLWSMLKLFDVLVQLDHLKNAKASI 176
L + +Q A + ++++R ERR + + ++L ++ K ++ LD LKN K S+
Sbjct: 122 LMKFMYFQRKAIERFCSEVKRLCHAERRKDFVSEAYLLTLGKFINMFAVLDELKNMKCSV 181
Query: 177 PNDFSWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPVYLL 236
ND S YKR Q + D S++E +L +FL+ I LH ++ + Y
Sbjct: 182 KNDHSAYKRA-AQFLRKMADPQSIQES-QNLSMFLANHNRITQCLHQQLEVIPGY----- 234
Query: 237 HIFSLSTVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSEKDSESL-- 294
E++L ++ V+ E L E+H+LL+V+ + L+ + + L
Sbjct: 235 --------EELLADIVNICVDYYENKMYLTPSEKHMLLKVMGFGLYLMDGNVSNIYKLDA 286
Query: 295 YKRVKINRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELPP 354
KR+ ++++ FK V+P F D+ + A ++ + Y S T + +P +
Sbjct: 287 KKRINLSKIDKFFKQLQVVPLFGDMQIELARYIETSAHYEENKSKWTCTQSSISP-QYNI 345
Query: 355 LNHIGAVRAEHDDFTIRFASAMNQST---DGSDVDWSKEVKGNMYDMIVEGFQLLSRWTA 411
+ +R +H F A N G D S E ++D+ + G QLLS+W+A
Sbjct: 346 CEQMVQIRDDHIRFISELARYSNSEVVTGSGLDSQKSDEEYRELFDLALRGLQLLSKWSA 405
Query: 412 RIWEQCAWKFSRPC-----KDASPSFSDYEKVVRYNYSAEERKALVELVSCIKSVGSMMQ 466
+ E +WK P KD + +YE+ RYNY++EE+ A VE+++ IK + +M
Sbjct: 406 HVMEVYSWKLVHPTDKFCNKDCPGTAEEYERATRYNYTSEEKFAFVEVIAMIKGLQVLMG 465
Query: 467 RCDTLVADALWETIHAEVQDFVQNTLASMLRTTFRKKKD-LSRILSDMRTLSADWMANTN 525
R +++ A+ TI+A +QDF Q TL LR RKKK+ L +L +R DW
Sbjct: 466 RMESVFNQAIRNTIYAALQDFAQVTLREPLRQAVRKKKNVLISVLQAIRKTICDWEGGRE 525
Query: 526 KSESEL---QSSQHGGEESKANIFYPRAVAPTAAQVHCLQFLIYEVV---SGGNLRRPGG 579
+ GG + K RAV P++ Q++ ++ ++ ++ SG
Sbjct: 526 PPNDPCLRGEKDPKGGFDIKVP---RRAVGPSSTQLYMVRTMLESLIADKSGSKKTLRSS 582
Query: 580 LFGNSGSEVPVNDLKQLETF----VMLAVTVATLTDLGFLWFREFYLESS--RVIQFPIE 633
L G + + D + F + ++ + DL LWFREF+LE + R IQFPIE
Sbjct: 583 LDGPI--VLAIEDFHKQSFFFTHLLNISEALQQCCDLSQLWFREFFLELTMGRRIQFPIE 640
Query: 634 CSLPWMLVDCVLESPNSGLLESVLMPFDIYNDSAQQALVLLKQRFLYDEIEAEVDHCFDI 693
S+PW+L D +LE+ ++E VL P D+YNDSA AL K++FLYDEIEAEV+ CFD
Sbjct: 641 MSMPWILTDHILETKEPSMMEYVLYPLDLYNDSAYYALTKFKKQFLYDEIEAEVNLCFDQ 700
Query: 694 FVARLCETIFTYYKSWAASELLDPTFLFASEN---------AEKYAV-----------QP 733
FV +L + IF YYK+ A S LLD F +N + +Y +
Sbjct: 701 FVYKLADQIFAYYKAMAGSVLLDKRFRAECKNYGVIIPYPPSNRYETLLKQRHVQLLGRS 760
Query: 734 MRLNMLL----------------------KMTRVKELEKLLDVLKHSHELLSIDLSVDSF 771
+ LN L+ +T + ELE LL++ + +H LL +++DSF
Sbjct: 761 IDLNRLITQRISAAMYKSLDQAISRFESEDLTSIVELEWLLEINRLTHRLLCKHMTLDSF 820
Query: 772 SLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFILCNTTQRFIRSS--KTVPVQKPS 829
M E N+S + R+ ++ E+ DFLPN+ +T RF+R++ T Q+
Sbjct: 821 DAMFREANHNVS--APYGRITLHVFWELNFDFLPNYCYNGSTNRFVRTAIPFTQEPQRDK 878
Query: 830 IPSAKPSFYCGTQP 843
+ +P + G++P
Sbjct: 879 PANVQPYYLYGSKP 892
Score = 47.8 bits (112), Expect = 0.002
Identities = 19/33 (57%), Positives = 24/33 (72%)
Query: 1070 GDSVAWGGCTIIYLLGQQLHFELFDFSYQILNI 1102
GD + W GC+II LLGQQ F+LFDF Y +L +
Sbjct: 1153 GDGLNWAGCSIIVLLGQQRRFDLFDFCYHLLKV 1185
>UniRef100_Q5R414 Hypothetical protein DKFZp459I162 [Pongo pygmaeus]
Length = 1253
Score = 347 bits (891), Expect = 9e-94
Identities = 262/914 (28%), Positives = 447/914 (48%), Gaps = 99/914 (10%)
Query: 3 VPVEEAIA---ALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLS---E 56
V +E+A++ L L D+QP ++ P + + + + D A+ ++ E
Sbjct: 5 VTLEDALSNVDLLEELPLPDQQPCIEPPPSSIMYQANFDTN---FEDRNAFVTGIARYIE 61
Query: 57 DTKALNQLSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSR 116
+ ++ + +EG E A +LYT+RSC +A+PQ+ + + ++ ++Y +T +VL+ E+++
Sbjct: 62 QATVHSSMNEMLEEGHEYAVMLYTWRSCSRAIPQVKCNEQPNRVEIYEKTVEVLEPEVTK 121
Query: 117 LREIQRWQASASSKLATDMQRFSRPERRINGPTISHLWSMLKLFDVLVQLDHLKNAKASI 176
L + +Q A + ++++R ERR + + ++L ++ K ++ LD LKN K S+
Sbjct: 122 LMKFMYFQRKAIERFCSEVKRLCHAERRKDFVSEAYLLTLGKFINMFAVLDELKNMKCSV 181
Query: 177 PNDFSWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPVYLL 236
ND S YKR Q + D S++E +L +FL+ I LH ++ + Y
Sbjct: 182 KNDHSAYKRA-AQFLRKMADPQSIQES-QNLSMFLANHNRITQCLHQQLEVIPGY----- 234
Query: 237 HIFSLSTVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSEKDSESL-- 294
E++L ++ V+ E L E+H+LL+V+ + L+ + + L
Sbjct: 235 --------EELLADIVNICVDYYENKMYLTPSEKHMLLKVMGFGLYLMDGNVSNIYKLDA 286
Query: 295 YKRVKINRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELPP 354
KR+ ++++ FK V+P F D+ + A ++ + Y S T + +P +
Sbjct: 287 KKRINLSKIDKFFKQLQVVPLFGDMQIELARYIETSAHYEENKSKWTCTQSSISP-QYNI 345
Query: 355 LNHIGAVRAEHDDFTIRFASAMNQST---DGSDVDWSKEVKGNMYDMIVEGFQLLSRWTA 411
+ +R +H F A N G D S E ++D+ + G QLLS+W+A
Sbjct: 346 CEQMVQIRDDHIRFISELARYSNSEVVTGSGLDSQKSDEEYRELFDLALRGLQLLSKWSA 405
Query: 412 RIWEQCAWKFSRPC-----KDASPSFSDYEKVVRYNYSAEERKALVELVSCIKSVGSMMQ 466
+ E +WK P KD + +YE+ RYNY++EE+ A VE+++ IK + +M
Sbjct: 406 HVMEVYSWKLVHPTDKFCNKDCPGTAEEYERATRYNYTSEEKFAFVEVIAMIKGLQVLMG 465
Query: 467 RCDTLVADALWETIHAEVQDFVQNTLASMLRTTFRKKKD-LSRILSDMRTLSADWMANTN 525
R +++ A+ TI+A +QDF Q TL LR RKKK+ L +L +R DW
Sbjct: 466 RMESVFNQAIRNTIYAALQDFAQVTLREPLRQAVRKKKNVLISVLQAIRKTICDWEGGRE 525
Query: 526 KSESEL---QSSQHGGEESKANIFYPRAVAPTAAQVHCLQFLIYEVV---SGGNLRRPGG 579
+ GG + K RAV P++ Q++ ++ ++ ++ SG
Sbjct: 526 PPNDPCLRGEKDPKGGFDIKVP---RRAVGPSSTQLYMVRTMLESLIADKSGSKKTLRSS 582
Query: 580 LFGNSGSEVPVNDLKQLETF----VMLAVTVATLTDLGFLWFREFYLESS--RVIQFPIE 633
L G + + D + F + ++ + DL LWFREF+LE + R IQFPIE
Sbjct: 583 LDGPI--VLAIEDFHKQSFFFTHLLNISEALQQCCDLSQLWFREFFLELTMGRRIQFPIE 640
Query: 634 CSLPWMLVDCVLESPNSGLLESVLMPFDIYNDSAQQALVLLKQRFLYDEIEAEVDHCFDI 693
S+PW+L D +LE+ ++E VL P D+YNDSA AL K++FLYDEIEAEV+ CFD
Sbjct: 641 MSMPWILTDHILETKEPSMMEYVLYPLDLYNDSAYYALTKFKKQFLYDEIEAEVNLCFDQ 700
Query: 694 FVARLCETIFTYYKSWAASELLDPTFLFASEN---------AEKYAV-----------QP 733
FV +L + IF YYK+ A S LLD F +N + +Y +
Sbjct: 701 FVYKLADQIFAYYKAMAGSVLLDKRFRAECKNYGVIIPYPPSNRYETLLKQRHVQLLGRS 760
Query: 734 MRLNMLL----------------------KMTRVKELEKLLDVLKHSHELLSIDLSVDSF 771
+ LN L+ +T + ELE LL++ + +H LL +++DSF
Sbjct: 761 IDLNRLITQRISAAMYKSLDQAISRFESEDLTSIVELEWLLEINRLTHRLLCKHMTLDSF 820
Query: 772 SLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFILCNTTQRFIRSS--KTVPVQKPS 829
M E N+S + R+ ++ E+ DFLPN+ +T RF+R++ T Q+
Sbjct: 821 DAMFREANHNVS--APYGRITLHVFWELNFDFLPNYCYNGSTNRFVRTAIPFTQEPQRDK 878
Query: 830 IPSAKPSFYCGTQP 843
+ +P + G++P
Sbjct: 879 PANIQPYYLYGSKP 892
Score = 47.8 bits (112), Expect = 0.002
Identities = 19/33 (57%), Positives = 24/33 (72%)
Query: 1070 GDSVAWGGCTIIYLLGQQLHFELFDFSYQILNI 1102
GD + W GC+II LLGQQ F+LFDF Y +L +
Sbjct: 1153 GDGLNWAGCSIIVLLGQQRRFDLFDFCYHLLKV 1185
>UniRef100_Q5R6T9 Hypothetical protein DKFZp459P191 [Pongo pygmaeus]
Length = 1253
Score = 347 bits (890), Expect = 1e-93
Identities = 263/915 (28%), Positives = 448/915 (48%), Gaps = 101/915 (11%)
Query: 3 VPVEEAIA---ALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLS---E 56
V +E+A++ L L D+QP ++ P + + + + D A+ ++ E
Sbjct: 5 VTLEDALSNVDLLEELPLPDQQPCIEPPPSSIMYQANFDTN---FEDRNAFVTGIARYIE 61
Query: 57 DTKALNQLSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSR 116
+ ++ + +EG E A +LYT+RSC +A+PQ+ + + ++ ++Y +T +VL+ E+++
Sbjct: 62 QATVHSSMNEMLEEGHEYAVMLYTWRSCSRAIPQVKCNEQPNRVEIYEKTVEVLEPEVTK 121
Query: 117 LREIQRWQASASSKLATDMQRFSRPERRINGPTISHLWSMLKLFDVLVQLDHLKNAKASI 176
L + +Q A + ++++R ERR + + ++L ++ K ++ LD LKN K S+
Sbjct: 122 LMKFMYFQRKAIERFCSEVKRLCHAERRKDFVSEAYLLTLGKFINMFAVLDELKNMKCSV 181
Query: 177 PNDFSWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPVYLL 236
ND S YKR Q + D S++E +L +FL+ I LH ++ + Y
Sbjct: 182 KNDHSAYKRA-AQFLRKMADPQSIQES-QNLSMFLANHNRITQCLHQQLEVIPGY----- 234
Query: 237 HIFSLSTVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSEKDSESL-- 294
E++L ++ V+ E L E+H+LL+V+ + L+ + + L
Sbjct: 235 --------EELLADIVNICVDYYENKMYLTPSEKHMLLKVMGFGLYLMDGNVSNIYKLDA 286
Query: 295 YKRVKINRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELPP 354
KR+ ++++ FK V+P F D+ + A ++ + Y S T + +P +
Sbjct: 287 KKRINLSKIDKFFKQLQVVPLFGDMQIELARYIETSAHYEENKSKWTCTQSSISP-QYNI 345
Query: 355 LNHIGAVRAEHDDFTIRFASAMNQST---DGSDVDWSKEVKGNMYDMIVEGFQLLSRWTA 411
+ +R +H F A N G D S E ++D+ + G QLLS+W+A
Sbjct: 346 CEQMVQIRDDHIRFISELARYSNSEVVTGSGLDSQKSDEEYRELFDLALRGLQLLSKWSA 405
Query: 412 RIWEQCAWKFSRPC-----KDASPSFSDYEKVVRYNYSAEERKALVELVSCIKSVGSMMQ 466
+ E +WK P KD + +YE+ RYNY++EE+ A VE+++ IK + +M
Sbjct: 406 HVMEVYSWKLVHPTDKFCNKDCPGTAEEYERATRYNYTSEEKFAFVEVIAMIKGLQVLMG 465
Query: 467 RCDTLVADALWETIHAEVQDFVQNTLASMLRTTFRKKKD-LSRILSDMRTLSADWMANTN 525
R +++ A+ TI+A +QDF Q TL LR RKKK+ L +L +R DW
Sbjct: 466 RTESVFNQAIRNTIYAALQDFAQVTLREPLRQAVRKKKNVLISVLQAIRKTICDWEGGRE 525
Query: 526 KSESEL---QSSQHGGEESKANIFYPR-AVAPTAAQVHCLQFLIYEVV---SGGNLRRPG 578
+ GG + K PR AV P++ Q++ ++ ++ ++ SG
Sbjct: 526 PPNDPFLRGEKDPKGGFDIKV----PRCAVGPSSTQLYMVRTMLESLIADKSGSKKTLRS 581
Query: 579 GLFGNSGSEVPVNDLKQLETF----VMLAVTVATLTDLGFLWFREFYLESS--RVIQFPI 632
L G + + D + F + ++ + DL LWFREF+LE + R IQFPI
Sbjct: 582 SLDGPI--VLAIEDFHKQSFFFTHLLNISEALQQCCDLSQLWFREFFLELTMGRRIQFPI 639
Query: 633 ECSLPWMLVDCVLESPNSGLLESVLMPFDIYNDSAQQALVLLKQRFLYDEIEAEVDHCFD 692
E S+PW+L D +LE+ ++E VL P D+YNDSA AL K++FLYDEIEAEV+ CFD
Sbjct: 640 EMSMPWILTDHILETKEPSMMEYVLYPLDLYNDSAYYALTKFKKQFLYDEIEAEVNLCFD 699
Query: 693 IFVARLCETIFTYYKSWAASELLDPTFLFASEN---------AEKYAV-----------Q 732
FV +L + IF YYK+ A S LLD F +N + +Y +
Sbjct: 700 QFVYKLADQIFAYYKAMAGSVLLDKRFRAECKNYGVIIPYPPSNRYETLLKQRHVQLLGR 759
Query: 733 PMRLNMLL----------------------KMTRVKELEKLLDVLKHSHELLSIDLSVDS 770
+ LN L+ +T + ELE LL++ + +H LL +++DS
Sbjct: 760 SIDLNRLITQRISAAMYKSLDQAISRFESEDLTSIVELEWLLEINRLTHRLLCKHMTLDS 819
Query: 771 FSLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFILCNTTQRFIRSS--KTVPVQKP 828
F M E N+S + R+ ++ E+ DFLPN+ +T RF+R++ T Q+
Sbjct: 820 FDAMFREANHNVS--APYGRITLHVFWELNFDFLPNYCYNGSTNRFVRTAIPFTQEPQRD 877
Query: 829 SIPSAKPSFYCGTQP 843
+ +P + G++P
Sbjct: 878 KPANIQPYYLYGSKP 892
Score = 47.8 bits (112), Expect = 0.002
Identities = 19/33 (57%), Positives = 24/33 (72%)
Query: 1070 GDSVAWGGCTIIYLLGQQLHFELFDFSYQILNI 1102
GD + W GC+II LLGQQ F+LFDF Y +L +
Sbjct: 1153 GDGLNWAGCSIIVLLGQQRRFDLFDFCYHLLKV 1185
>UniRef100_Q8CHA9 MKIAA1168 protein [Mus musculus]
Length = 1259
Score = 347 bits (889), Expect = 2e-93
Identities = 261/914 (28%), Positives = 447/914 (48%), Gaps = 99/914 (10%)
Query: 3 VPVEEAIA---ALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLS---E 56
V +E+A++ L L D+QP ++ P + + + + D A+ ++ E
Sbjct: 11 VTLEDALSNVDLLEELPLPDQQPCIEPPPSSIMYQANFDTN---FEDRNAFVTGIARYIE 67
Query: 57 DTKALNQLSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSR 116
+ ++ + +EG + A +LYT+RSC +A+PQ+ + + ++ ++Y +T +VL+ E+++
Sbjct: 68 QATVHSSMNEMLEEGHDYAVMLYTWRSCSRAIPQVKCNEQPNRVEIYEKTVEVLEPEVTK 127
Query: 117 LREIQRWQASASSKLATDMQRFSRPERRINGPTISHLWSMLKLFDVLVQLDHLKNAKASI 176
L + +Q A + ++++R ERR + + ++L ++ K ++ LD LKN K S+
Sbjct: 128 LMKFMYFQRKAIERFCSEVKRLCHAERRKDFVSEAYLLTLGKFINMFAVLDELKNMKCSV 187
Query: 177 PNDFSWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPVYLL 236
ND S YKR Q + D S++E +L +FL+ I LH ++ + Y
Sbjct: 188 KNDHSAYKRA-AQFLRKMADPQSIQES-QNLSMFLANHNRITQCLHQQLEVIPGY----- 240
Query: 237 HIFSLSTVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSEKDSESL-- 294
E++L ++ V+ E L E+H+LL+V+ + L+ + + L
Sbjct: 241 --------EELLADIVNICVDYYENKMYLTPSEKHMLLKVMGFGLYLMDGNVSNIYKLDA 292
Query: 295 YKRVKINRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELPP 354
KR+ ++++ FK V+P F D+ + A ++ + Y S T + +P +
Sbjct: 293 KKRINLSKIDKFFKQLQVVPLFGDMQIELARYIETSAHYEENKSKWTCTQSSISP-QYNI 351
Query: 355 LNHIGAVRAEHDDFTIRFASAMNQST---DGSDVDWSKEVKGNMYDMIVEGFQLLSRWTA 411
+ +R +H F A N G D S E ++D+ + G QLLS+W+A
Sbjct: 352 CEQMVQIRDDHIRFISELARYSNSEVVTGSGLDSQKSDEEYRELFDLALRGLQLLSKWSA 411
Query: 412 RIWEQCAWKFSRPC-----KDASPSFSDYEKVVRYNYSAEERKALVELVSCIKSVGSMMQ 466
+ E +WK P KD + +YE+ RYNY++EE+ A VE+++ IK + +M
Sbjct: 412 HVMEVYSWKLVHPTDKFCNKDCPGTAEEYERATRYNYTSEEKFAFVEVIAMIKGLQVLMG 471
Query: 467 RCDTLVADALWETIHAEVQDFVQNTLASMLRTTFRKKKD-LSRILSDMRTLSADWMANTN 525
R +++ A+ TI+A +QDF Q TL LR RKKK+ L +L +R DW
Sbjct: 472 RMESVFNQAIRNTIYAALQDFAQVTLREPLRQAVRKKKNVLISVLQAIRKTICDWEGGRE 531
Query: 526 KSESEL---QSSQHGGEESKANIFYPRAVAPTAAQVHCLQFLIYEVV---SGGNLRRPGG 579
+ GG + K RAV P++ Q++ ++ ++ ++ SG
Sbjct: 532 PPNDPCLRGEKDPKGGFDIKVP---RRAVGPSSTQLYMVRTMLESLIADKSGSKKTLRSS 588
Query: 580 LFGNSGSEVPVNDLKQLETF----VMLAVTVATLTDLGFLWFREFYLESS--RVIQFPIE 633
L G + + D + F + ++ + DL LWFREF+LE + R IQFPIE
Sbjct: 589 LDGPI--VLAIEDFHKQSFFFTHLLNISEALQQCCDLSQLWFREFFLELTMGRRIQFPIE 646
Query: 634 CSLPWMLVDCVLESPNSGLLESVLMPFDIYNDSAQQALVLLKQRFLYDEIEAEVDHCFDI 693
S+PW+L D +LE+ ++E VL P D+YNDSA AL K++FLYDEIEAEV+ CFD
Sbjct: 647 MSMPWILTDHILETKEPSMMEYVLYPLDLYNDSAYYALTKFKKQFLYDEIEAEVNLCFDQ 706
Query: 694 FVARLCETIFTYYKSWAASELLDPTFLFASEN---------AEKYAV-----------QP 733
FV +L + IF YYK+ A S LLD F +N + +Y +
Sbjct: 707 FVYKLADQIFAYYKAMAGSVLLDKRFRAECKNYGVIIPYPPSNRYETLLKQRHVQLLGRS 766
Query: 734 MRLNMLL----------------------KMTRVKELEKLLDVLKHSHELLSIDLSVDSF 771
+ LN L+ +T + ELE LL++ + +H LL +++DSF
Sbjct: 767 IDLNRLITQRISAAMYKSLDQAISRFESEDLTSIVELEWLLEINRLTHRLLCKHMTLDSF 826
Query: 772 SLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFILCNTTQRFIRSS--KTVPVQKPS 829
M E N+S + R+ ++ E+ DFLPN+ +T RF+R++ T Q+
Sbjct: 827 DAMFREANHNVS--APYGRITLHVFWELNFDFLPNYCYNGSTNRFVRTAIPFTQEPQRDK 884
Query: 830 IPSAKPSFYCGTQP 843
+ +P + G++P
Sbjct: 885 PANVQPYYLYGSKP 898
Score = 47.8 bits (112), Expect = 0.002
Identities = 19/33 (57%), Positives = 24/33 (72%)
Query: 1070 GDSVAWGGCTIIYLLGQQLHFELFDFSYQILNI 1102
GD + W GC+II LLGQQ F+LFDF Y +L +
Sbjct: 1159 GDGLNWAGCSIIVLLGQQRRFDLFDFCYHLLKV 1191
>UniRef100_Q6PGK0 Cyfip2 protein [Mus musculus]
Length = 894
Score = 347 bits (889), Expect = 2e-93
Identities = 262/915 (28%), Positives = 447/915 (48%), Gaps = 99/915 (10%)
Query: 3 VPVEEAIA---ALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLS---E 56
V +E+A++ L L D+QP ++ P + + + + D A+ ++ E
Sbjct: 5 VTLEDALSNVDLLEELPLPDQQPCIEPPPSSIMYQANFDTN---FEDRNAFVTGIARYIE 61
Query: 57 DTKALNQLSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSR 116
+ ++ + +EG + A +LYT+RSC +A+PQ+ + + ++ ++Y +T +VL+ E+++
Sbjct: 62 QATVHSSMNEMLEEGHDYAVMLYTWRSCSRAIPQVKCNEQPNRVEIYEKTVEVLEPEVTK 121
Query: 117 LREIQRWQASASSKLATDMQRFSRPERRINGPTISHLWSMLKLFDVLVQLDHLKNAKASI 176
L + +Q A + ++++R ERR + + ++L ++ K ++ LD LKN K S+
Sbjct: 122 LMKFMYFQRKAIERFCSEVKRLCHAERRKDFVSEAYLLTLGKFINMFAVLDELKNMKCSV 181
Query: 177 PNDFSWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPVYLL 236
ND S YKR Q + D S++E +L +FL+ I LH ++ + Y
Sbjct: 182 KNDHSAYKRA-AQFLRKMADPQSIQES-QNLSMFLANHNRITQCLHQQLEVIPGY----- 234
Query: 237 HIFSLSTVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSEKDSESL-- 294
E++L ++ V+ E L E+H+LL+V+ + L+ + + L
Sbjct: 235 --------EELLADIVNICVDYYENKMYLTPSEKHMLLKVMGFGLYLMDGNVSNIYKLDA 286
Query: 295 YKRVKINRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELPP 354
KR+ ++++ FK V+P F D+ + A +K + Y S T + +P +
Sbjct: 287 KKRINLSKIDKFFKQLQVVPPFGDMQIELARYIKTSAHYEENKSKWTCTQSSISP-QYNI 345
Query: 355 LNHIGAVRAEHDDFTIRFASAMNQST---DGSDVDWSKEVKGNMYDMIVEGFQLLSRWTA 411
+ +R +H F A N G D S E ++D+ + G QLLS+W+A
Sbjct: 346 CEQMVQIRDDHIRFISELARYSNSEVVTGSGLDSQKSDEEYRELFDLALRGLQLLSKWSA 405
Query: 412 RIWEQCAWKFSRPC-----KDASPSFSDYEKVVRYNYSAEERKALVELVSCIKSVGSMMQ 466
+ E +WK P KD + +YE+ RYNY++EE+ A VE+++ IK + +M
Sbjct: 406 HVMEVYSWKLVHPTDKFCNKDCPGTAEEYERATRYNYTSEEKFAFVEVIAMIKGLQVLMG 465
Query: 467 RCDTLVADALWETIHAEVQDFVQNTLASMLRTTFRKKKD-LSRILSDMRTLSADWMANTN 525
R +++ A+ TI+A +QDF Q TL LR RKKK+ L +L +R DW
Sbjct: 466 RMESVFNQAIRNTIYAALQDFAQVTLREPLRQAVRKKKNVLISVLQAIRKTICDWEGGRE 525
Query: 526 KSESEL---QSSQHGGEESKANIFYPRAVAPTAAQVHCLQFLIYEVV---SGGNLRRPGG 579
+ GG + K RAV P++ Q++ ++ ++ ++ SG
Sbjct: 526 PPNDPCLRGEKDPKGGFDIKVP---RRAVGPSSTQLYMVRTMLESLIADKSGSKKTLRSS 582
Query: 580 LFGNSGSEVPVNDLKQLETF----VMLAVTVATLTDLGFLWFREFYLESS--RVIQFPIE 633
L G + + D + F + ++ + DL LWFREF+LE + R IQFPIE
Sbjct: 583 LDGPI--VLAIEDFHKQSFFFTHLLNISEALQQCCDLSQLWFREFFLELTMGRRIQFPIE 640
Query: 634 CSLPWMLVDCVLESPNSGLLESVLMPFDIYNDSAQQALVLLKQRFLYDEIEAEVDHCFDI 693
S+PW+L D +LE+ ++E VL P D+YNDSA AL K++FLYDEIEAEV+ CFD
Sbjct: 641 MSMPWILTDHILETKEPSMMEYVLYPLDLYNDSAYYALTKFKKQFLYDEIEAEVNLCFDQ 700
Query: 694 FVARLCETIFTYYKSWAASELLDPTFLFASEN---------AEKYAV-----------QP 733
FV +L + IF YYK+ A S LLD F +N + +Y +
Sbjct: 701 FVYKLADQIFAYYKAMAGSVLLDKRFRAECKNYGVIIPYPPSNRYETLLKQRHVQLLGRS 760
Query: 734 MRLNMLL----------------------KMTRVKELEKLLDVLKHSHELLSIDLSVDSF 771
+ LN L+ +T + ELE LL++ + +H LL +++DSF
Sbjct: 761 IDLNRLITQRISAAMYKSLDQAISRFESEDLTSIVELEWLLEINRLTHRLLCKHMTLDSF 820
Query: 772 SLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFILCNTTQRFIRSS--KTVPVQKPS 829
M E N+S + R+ ++ E+ DFLPN+ +T RF+R++ T Q+
Sbjct: 821 DAMFREANHNVS--APYGRITLHVFWELNFDFLPNYCYNGSTNRFVRTAIPFTQEPQRDK 878
Query: 830 IPSAKPSFYCGTQPS 844
+ +P + G++ S
Sbjct: 879 PANVQPYYLYGSKAS 893
>UniRef100_UPI00003A9E55 UPI00003A9E55 UniRef100 entry
Length = 1251
Score = 345 bits (885), Expect = 5e-93
Identities = 254/856 (29%), Positives = 430/856 (49%), Gaps = 95/856 (11%)
Query: 56 EDTKALNQLSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMS 115
E + ++ + +EG+E A +LYT+RSC +A+PQ+ + + ++ ++Y +T +VL+ E++
Sbjct: 61 EQATVHSSMNEMLEEGQEYAVMLYTWRSCSRAIPQVKCNEQPNRVEIYEKTVEVLEPEVT 120
Query: 116 RLREIQRWQASASSKLATDMQRFSRPERRINGPTISHLWSMLKLFDVLVQLDHLKNAKAS 175
+L +Q +A + +++R ERR + + ++L ++ K ++ LD LKN K S
Sbjct: 121 KLMNFMYFQRNAIERFCGEVRRLCHAERRKDFVSEAYLITLGKFINMFAVLDELKNMKCS 180
Query: 176 IPNDFSWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPVYL 235
+ ND S YKR Q + D S++E +L +FL+ I +L ++ + Y
Sbjct: 181 VKNDHSAYKRA-AQFLRKMADPQSIQES-QNLSMFLANHNKITQSLQQQLEVIVGY---- 234
Query: 236 LHIFSLSTVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSEKDSESL- 294
E++L ++ V+ E L E+H+LL+V+ + L+ S + L
Sbjct: 235 ---------EELLADIVNLCVDYYENKMYLTPSEKHMLLKVMGFGLYLMDGSVSNIYKLD 285
Query: 295 -YKRVKINRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELP 353
KR+ + ++ FK V+P F D+ + A +K + Y S T + ++P +
Sbjct: 286 AKKRINLAKIDKFFKQLQVVPLFGDMQIELARYIKTSAHYEENKSRWTCTSSSSSP-QYN 344
Query: 354 PLNHIGAVRAEHDDFTIRFASAMN---------QSTDGSDVDWSKEVKGNMYDMIVEGFQ 404
+ +R +H F A N Q +D ++ K ++D+ ++G Q
Sbjct: 345 ICEQMIQIREDHMRFISELARYSNSEVVTGSGRQEAQKTDAEYRK-----LFDLSLQGLQ 399
Query: 405 LLSRWTARIWEQCAWKFSRPC-----KDASPSFSDYEKVVRYNYSAEERKALVELVSCIK 459
LLS+W+A + E +WK P KD + +YE+ RYNY++EE+ ALVE+++ IK
Sbjct: 400 LLSQWSAHVMEVYSWKLVHPTDKYSNKDCPDNAEEYERATRYNYTSEEKFALVEVIAMIK 459
Query: 460 SVGSMMQRCDTLVADALWETIHAEVQDFVQNTLASMLRTTFRKKKD-LSRILSDMRTLSA 518
+ +M R +++ A+ TI+A +QDF Q TL LR +KKK+ + +L +R
Sbjct: 460 GLQVLMGRMESVFNHAIRHTIYAALQDFAQVTLREPLRQAIKKKKNVIQSVLQAIRKTVC 519
Query: 519 DWMANTNKSESELQSSQHGGEESKA--NIFYPR-AVAPTAAQVHCLQFLIYEVVSGGNLR 575
DW A E + G ++ K+ +I PR AV P++ Q++ ++ + + S L+
Sbjct: 520 DWEAG---HEPFNDPALRGEKDPKSGFDIKVPRRAVGPSSTQLYMVRTMTESLNSAELLK 576
Query: 576 RPGGLFGNSGSEVPVNDLKQLETFVML---AVTVATLTDLGFLWFREFYLESS--RVIQF 630
+ L ++ L+Q + L T+ DL LWFREF+LE + R IQF
Sbjct: 577 QLKALGLEKLLQMTHKFLRQSYIYPPLLNFGETLQQCCDLSQLWFREFFLELTMGRRIQF 636
Query: 631 PIECSLPWMLVDCVLESPNSGLLESVLMPFDIYNDSAQQALVLLKQRFLYDEIEAEVDHC 690
PIE S+PW+L D +LE+ + ++E VL D+YNDSA AL K++FLYDEIEAEV+ C
Sbjct: 637 PIEMSMPWILTDHILETKEASMMEYVLYSLDLYNDSAHYALTKFKKQFLYDEIEAEVNLC 696
Query: 691 FDIFVARLCETIFTYYKSWAASELLDPTFLFASEN--AEKYAVQPMRLNMLLK------- 741
FD FV +L + IF YYK+ A S LLD +N A +Q R LLK
Sbjct: 697 FDQFVYKLADQIFAYYKAMAGSLLLDKRLRSECKNQGATIQLLQSNRYETLLKQRHVQLL 756
Query: 742 ---------------------------------MTRVKELEKLLDVLKHSHELLSIDLSV 768
+T + EL+ L+++ K +H+LLS +++
Sbjct: 757 GRSIDLNRLITQRISAAMYRSMELAIGRFESEDLTSIVELDGLIEINKMTHKLLSRYMTL 816
Query: 769 DSFSLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFILCNTTQRFIRS--SKTVPVQ 826
DSF M E N+S + R+ ++ E+ DFLPN+ +T RF+R+ + Q
Sbjct: 817 DSFDAMFREANHNVS--APYGRITLHVFWELNYDFLPNYCYNGSTNRFVRTVLPFSQEFQ 874
Query: 827 KPSIPSAKPSFYCGTQ 842
+ P+A+P + G++
Sbjct: 875 RDKQPNAQPQYLYGSK 890
Score = 43.5 bits (101), Expect = 0.037
Identities = 17/33 (51%), Positives = 22/33 (66%)
Query: 1070 GDSVAWGGCTIIYLLGQQLHFELFDFSYQILNI 1102
GD + W GC II LLGQQ F++ DF Y +L +
Sbjct: 1152 GDGLHWAGCMIIVLLGQQRRFDVLDFCYHLLKV 1184
>UniRef100_UPI00003A9E54 UPI00003A9E54 UniRef100 entry
Length = 1254
Score = 345 bits (885), Expect = 5e-93
Identities = 255/863 (29%), Positives = 435/863 (49%), Gaps = 107/863 (12%)
Query: 56 EDTKALNQLSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMS 115
E + ++ + +EG+E A +LYT+RSC +A+PQ+ + + ++ ++Y +T +VL+ E++
Sbjct: 62 EQATVHSSMNEMLEEGQEYAVMLYTWRSCSRAIPQVKCNEQPNRVEIYEKTVEVLEPEVT 121
Query: 116 RLREIQRWQASASSKLATDMQRFSRPERRINGPTISHLWSMLKLFDVLVQLDHLKNAKAS 175
+L +Q +A + +++R ERR + + ++L ++ K ++ LD LKN K S
Sbjct: 122 KLMNFMYFQRNAIERFCGEVRRLCHAERRKDFVSEAYLITLGKFINMFAVLDELKNMKCS 181
Query: 176 IPNDFSWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPVYL 235
+ ND S YKR Q + D S++E +L +FL+ I +L ++ + Y
Sbjct: 182 VKNDHSAYKRA-AQFLRKMADPQSIQES-QNLSMFLANHNKITQSLQQQLEVIVGY---- 235
Query: 236 LHIFSLSTVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSEKDSESL- 294
E++L ++ V+ E L E+H+LL+V+ + L+ S + L
Sbjct: 236 ---------EELLADIVNLCVDYYENKMYLTPSEKHMLLKVMGFGLYLMDGSVSNIYKLD 286
Query: 295 -YKRVKINRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELP 353
KR+ + ++ FK V+P F D+ + A +K + Y S T + ++P +
Sbjct: 287 AKKRINLAKIDKFFKQLQVVPLFGDMQIELARYIKTSAHYEENKSRWTCTSSSSSP-QYN 345
Query: 354 PLNHIGAVRAEHDDFTIRFASAMN---------QSTDGSDVDWSKEVKGNMYDMIVEGFQ 404
+ +R +H F A N Q +D ++ K ++D+ ++G Q
Sbjct: 346 ICEQMIQIREDHMRFISELARYSNSEVVTGSGRQEAQKTDAEYRK-----LFDLSLQGLQ 400
Query: 405 LLSRWTARIWEQCAWKFSRPC-----KDASPSFSDYEKVVRYNYSAEERKALVELVSCIK 459
LLS+W+A + E +WK P KD + +YE+ RYNY++EE+ ALVE+++ IK
Sbjct: 401 LLSQWSAHVMEVYSWKLVHPTDKYSNKDCPDNAEEYERATRYNYTSEEKFALVEVIAMIK 460
Query: 460 SVGSMMQRCDTLVADALWETIHAEVQDFVQNTLASMLRTTFRKKKD-LSRILSDMRTLSA 518
+ +M R +++ A+ TI+A +QDF Q TL LR +KKK+ + +L +R
Sbjct: 461 GLQVLMGRMESVFNHAIRHTIYAALQDFAQVTLREPLRQAIKKKKNVIQSVLQAIRKTVC 520
Query: 519 DWMANTNKSESELQSSQHGGEESKA--NIFYPR-AVAPTAAQVHCLQFLIYEVV---SGG 572
DW A E + G ++ K+ +I PR AV P++ Q++ ++ ++ ++ SG
Sbjct: 521 DWEAG---HEPFNDPALRGEKDPKSGFDIKVPRRAVGPSSTQLYMVRTMLESLIADKSGS 577
Query: 573 NLRRPGGLFGNSGSEVPVNDLKQL--ETF-----VMLAVTVATLTDLGFLWFREFYLESS 625
L G + + D+++ E+F + + T+ DL LWFREF+LE +
Sbjct: 578 KKTLRSSLEGPT-----ILDIEKFHRESFFYTHLINFSETLQQCCDLSQLWFREFFLELT 632
Query: 626 --RVIQFPIECSLPWMLVDCVLESPNSGLLESVLMPFDIYNDSAQQALVLLKQRFLYDEI 683
R IQFPIE S+PW+L D +LE+ + ++E VL D+YNDSA AL K++FLYDEI
Sbjct: 633 MGRRIQFPIEMSMPWILTDHILETKEASMMEYVLYSLDLYNDSAHYALTKFKKQFLYDEI 692
Query: 684 EAEVDHCFDIFVARLCETIFTYYKSWAASELLDPTFLFASEN--AEKYAVQPMRLNMLLK 741
EAEV+ CFD FV +L + IF YYK+ A S LLD +N A +Q R LLK
Sbjct: 693 EAEVNLCFDQFVYKLADQIFAYYKAMAGSLLLDKRLRSECKNQGATIQLLQSNRYETLLK 752
Query: 742 ----------------------------------------MTRVKELEKLLDVLKHSHEL 761
+T + EL+ L+++ K +H+L
Sbjct: 753 QRHVQLLGRSIDLNRLITQRISAAMYRSMELAIGRFESEDLTSIVELDGLIEINKMTHKL 812
Query: 762 LSIDLSVDSFSLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFILCNTTQRFIRS-- 819
LS +++DSF M E N+S + R+ ++ E+ DFLPN+ +T RF+R+
Sbjct: 813 LSRYMTLDSFDAMFREANHNVS--APYGRITLHVFWELNYDFLPNYCYNGSTNRFVRTVL 870
Query: 820 SKTVPVQKPSIPSAKPSFYCGTQ 842
+ Q+ P+A+P + G++
Sbjct: 871 PFSQEFQRDKQPNAQPQYLYGSK 893
Score = 43.5 bits (101), Expect = 0.037
Identities = 17/33 (51%), Positives = 22/33 (66%)
Query: 1070 GDSVAWGGCTIIYLLGQQLHFELFDFSYQILNI 1102
GD + W GC II LLGQQ F++ DF Y +L +
Sbjct: 1155 GDGLHWAGCMIIVLLGQQRRFDVLDFCYHLLKV 1187
>UniRef100_UPI00003AAAFA UPI00003AAAFA UniRef100 entry
Length = 1257
Score = 344 bits (882), Expect = 1e-92
Identities = 260/913 (28%), Positives = 446/913 (48%), Gaps = 96/913 (10%)
Query: 3 VPVEEAIA---ALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLS---E 56
V +E+A++ L L D+QP ++ P + + + + D A+ ++ E
Sbjct: 8 VTLEDALSNVDLLEELPLPDQQPCIEPPPSSIMYQANFDTN---FEDRNAFVTGIARYIE 64
Query: 57 DTKALNQLSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSR 116
+ ++ + +EG E A +LYT+RSC +A+PQ+ + + ++ ++Y +T +VL+ E+++
Sbjct: 65 QATVHSSMNEMLEEGHEYAVMLYTWRSCSRAIPQVKCNEQPNRVEIYEKTVEVLEPEVTK 124
Query: 117 LREIQRWQASASSKLATDMQRFSRPERRINGPTISHLWSMLKLFDVLVQLDHLKNAKASI 176
L + +Q A + ++++R ERR + + ++L ++ K ++ LD LKN K S+
Sbjct: 125 LMKFMYFQRKAIERFCSEVKRLCHAERRKDFVSEAYLLTLGKFINMFAVLDELKNMKCSV 184
Query: 177 PNDFSWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAIL-LNLHVEMFRVNKYPVYL 235
ND S YKR Q + D S++E +L +FL+ I LH ++ + Y
Sbjct: 185 KNDHSAYKRA-AQFLRKMADPQSIQES-QNLSMFLANHNRITQAMLHQQLEVIPGY---- 238
Query: 236 LHIFSLSTVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSEKDSESL- 294
E++L ++ V+ E L E+H+LL+V+ + L+ + + L
Sbjct: 239 ---------EELLADIVNICVDYYENKMYLTPSEKHMLLKVMGFGLYLMDGNVSNIYKLD 289
Query: 295 -YKRVKINRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELP 353
KR+ ++++ FK V+P F D+ + A +K + Y S T + +P +
Sbjct: 290 AKKRINLSKIDKFFKQLQVVPLFGDMQIELARYIKTSAHYEENKSKWTCTQSSISP-QYN 348
Query: 354 PLNHIGAVRAEHDDFTIRFASAMNQST---DGSDVDWSKEVKGNMYDMIVEGFQLLSRWT 410
+ +R +H F A N G D S E ++D+ + G QLLS+W+
Sbjct: 349 ICEQMVQIRDDHIRFISELARYSNSEVVTGSGLDSQKSDEEYRELFDLALRGLQLLSKWS 408
Query: 411 ARIWEQCAWKFSRPC-----KDASPSFSDYEKVVRYNYSAEERKALVELVSCIKSVGSMM 465
A + E +WK P KD + +YE+ RYNY++EE+ A VE+++ IK + +M
Sbjct: 409 AHVMEVYSWKLVHPTDKFCNKDCPGTAEEYERATRYNYTSEEKFAFVEVIAMIKGLQVLM 468
Query: 466 QRCDTLVADALWETIHAEVQDFVQNTLASMLRTTFRKKKD-LSRILSDMRTLSADWMANT 524
R +++ A+ TI+A +QDF Q TL LR RKKK+ L +L +R DW
Sbjct: 469 GRMESVFNQAIRNTIYAALQDFAQVTLREPLRQAVRKKKNVLISVLQAIRKTICDWEGGR 528
Query: 525 NKSESEL---QSSQHGGEESKANIFYPRAVAPTAAQVHCLQFLIYEVVSGGNLRRPGGLF 581
+ GG + K RAV P++ Q++ ++ ++ +++ + +
Sbjct: 529 EPPNDPCLRGEKDPKGGFDIKVP---RRAVGPSSTQLYMVRTMLESLIADKSGSKKTLRS 585
Query: 582 GNSGSEVPVNDLKQLETFVM-----LAVTVATLTDLGFLWFREFYLESS--RVIQFPIEC 634
G V + ++F ++ + DL LWFREF+LE + R IQFPIE
Sbjct: 586 SLDGPIVLAIEEFHKQSFFFTHLLNISEALQQCCDLSQLWFREFFLELTMGRRIQFPIEM 645
Query: 635 SLPWMLVDCVLESPNSGLLESVLMPFDIYNDSAQQALVLLKQRFLYDEIEAEVDHCFDIF 694
S+PW+L D +LE+ ++E VL P D+YNDSA AL K++FLYDEIEAEV+ CFD F
Sbjct: 646 SMPWILTDHILETKEPSMMEYVLYPLDLYNDSAYYALTKFKKQFLYDEIEAEVNLCFDQF 705
Query: 695 VARLCETIFTYYKSWAASELLDPTFLFASEN---------AEKYAV-----------QPM 734
V +L + IF YYK+ A S LLD F +N + +Y + +
Sbjct: 706 VYKLADQIFAYYKAMAGSVLLDKRFRAECKNYGVIIPYPPSNRYETLLKQRHVQLLGRSI 765
Query: 735 RLNMLL----------------------KMTRVKELEKLLDVLKHSHELLSIDLSVDSFS 772
LN L+ +T + ELE LL++ + +H LL +++DSF
Sbjct: 766 DLNRLITQRISAAMYKSLDQAISRFESEDLTSIVELEWLLEINRLTHRLLCKHMTLDSFD 825
Query: 773 LMLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFILCNTTQRFIRSS--KTVPVQKPSI 830
M E N+S + R+ ++ E+ DFLPN+ +T RF+R++ T Q+
Sbjct: 826 AMFREANHNVS--APYGRITLHVFWELNFDFLPNYCYNGSTNRFVRTAIPFTQEPQRDKP 883
Query: 831 PSAKPSFYCGTQP 843
+ +P + G++P
Sbjct: 884 ANVQPYYLYGSKP 896
Score = 47.8 bits (112), Expect = 0.002
Identities = 19/33 (57%), Positives = 24/33 (72%)
Query: 1070 GDSVAWGGCTIIYLLGQQLHFELFDFSYQILNI 1102
GD + W GC+II LLGQQ F+LFDF Y +L +
Sbjct: 1157 GDGLNWAGCSIIVLLGQQRRFDLFDFCYHLLKV 1189
>UniRef100_UPI00003AAAFB UPI00003AAAFB UniRef100 entry
Length = 1244
Score = 343 bits (881), Expect = 1e-92
Identities = 259/912 (28%), Positives = 444/912 (48%), Gaps = 97/912 (10%)
Query: 3 VPVEEAIA---ALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLS---E 56
V +E+A++ L L D+QP ++ P + + + + D A+ ++ E
Sbjct: 1 VTLEDALSNVDLLEELPLPDQQPCIEPPPSSIMYQANFDTN---FEDRNAFVTGIARYIE 57
Query: 57 DTKALNQLSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSR 116
+ ++ + +EG E A +LYT+RSC +A+PQ+ + + ++ ++Y +T +VL+ E+++
Sbjct: 58 QATVHSSMNEMLEEGHEYAVMLYTWRSCSRAIPQVKCNEQPNRVEIYEKTVEVLEPEVTK 117
Query: 117 LREIQRWQASASSKLATDMQRFSRPERRINGPTISHLWSMLKLFDVLVQLDHLKNAKASI 176
L + +Q A + ++++R ERR + + ++L ++ K ++ LD LKN K S+
Sbjct: 118 LMKFMYFQRKAIERFCSEVKRLCHAERRKDFVSEAYLLTLGKFINMFAVLDELKNMKCSV 177
Query: 177 PNDFSWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPVYLL 236
ND S YKR Q + D S++E +L +FL+ N + + P Y
Sbjct: 178 KNDHSAYKRA-AQFLRKMADPQSIQES-QNLSMFLANH-----NRITQASPIQVIPGY-- 228
Query: 237 HIFSLSTVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSEKDSESL-- 294
E++L ++ V+ E L E+H+LL+V+ + L+ + + L
Sbjct: 229 --------EELLADIVNICVDYYENKMYLTPSEKHMLLKVMGFGLYLMDGNVSNIYKLDA 280
Query: 295 YKRVKINRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELPP 354
KR+ ++++ FK V+P F D+ + A +K + Y S T + +P +
Sbjct: 281 KKRINLSKIDKFFKQLQVVPLFGDMQIELARYIKTSAHYEENKSKWTCTQSSISP-QYNI 339
Query: 355 LNHIGAVRAEHDDFTIRFASAMNQST---DGSDVDWSKEVKGNMYDMIVEGFQLLSRWTA 411
+ +R +H F A N G D S E ++D+ + G QLLS+W+A
Sbjct: 340 CEQMVQIRDDHIRFISELARYSNSEVVTGSGLDSQKSDEEYRELFDLALRGLQLLSKWSA 399
Query: 412 RIWEQCAWKFSRPC-----KDASPSFSDYEKVVRYNYSAEERKALVELVSCIKSVGSMMQ 466
+ E +WK P KD + +YE+ RYNY++EE+ A VE+++ IK + +M
Sbjct: 400 HVMEVYSWKLVHPTDKFCNKDCPGTAEEYERATRYNYTSEEKFAFVEVIAMIKGLQVLMG 459
Query: 467 RCDTLVADALWETIHAEVQDFVQNTLASMLRTTFRKKKD-LSRILSDMRTLSADWMANTN 525
R +++ A+ TI+A +QDF Q TL LR RKKK+ L +L +R DW
Sbjct: 460 RMESVFNQAIRNTIYAALQDFAQVTLREPLRQAVRKKKNVLISVLQAIRKTICDWEGGRE 519
Query: 526 KSESEL---QSSQHGGEESKANIFYPRAVAPTAAQVHCLQFLIYEVVSGGNLRRPGGLFG 582
+ GG + K RAV P++ Q++ ++ ++ +++ + +
Sbjct: 520 PPNDPCLRGEKDPKGGFDIKVP---RRAVGPSSTQLYMVRTMLESLIADKSGSKKTLRSS 576
Query: 583 NSGSEVPVNDLKQLETFVM-----LAVTVATLTDLGFLWFREFYLESS--RVIQFPIECS 635
G V + ++F ++ + DL LWFREF+LE + R IQFPIE S
Sbjct: 577 LDGPIVLAIEEFHKQSFFFTHLLNISEALQQCCDLSQLWFREFFLELTMGRRIQFPIEMS 636
Query: 636 LPWMLVDCVLESPNSGLLESVLMPFDIYNDSAQQALVLLKQRFLYDEIEAEVDHCFDIFV 695
+PW+L D +LE+ ++E VL P D+YNDSA AL K++FLYDEIEAEV+ CFD FV
Sbjct: 637 MPWILTDHILETKEPSMMEYVLYPLDLYNDSAYYALTKFKKQFLYDEIEAEVNLCFDQFV 696
Query: 696 ARLCETIFTYYKSWAASELLDPTFLFASEN---------AEKYAV-----------QPMR 735
+L + IF YYK+ A S LLD F +N + +Y + +
Sbjct: 697 YKLADQIFAYYKAMAGSVLLDKRFRAECKNYGVIIPYPPSNRYETLLKQRHVQLLGRSID 756
Query: 736 LNMLL----------------------KMTRVKELEKLLDVLKHSHELLSIDLSVDSFSL 773
LN L+ +T + ELE LL++ + +H LL +++DSF
Sbjct: 757 LNRLITQRISAAMYKSLDQAISRFESEDLTSIVELEWLLEINRLTHRLLCKHMTLDSFDA 816
Query: 774 MLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFILCNTTQRFIRSS--KTVPVQKPSIP 831
M E N+S + R+ ++ E+ DFLPN+ +T RF+R++ T Q+
Sbjct: 817 MFREANHNVS--APYGRITLHVFWELNFDFLPNYCYNGSTNRFVRTAIPFTQEPQRDKPA 874
Query: 832 SAKPSFYCGTQP 843
+ +P + G++P
Sbjct: 875 NVQPYYLYGSKP 886
Score = 47.8 bits (112), Expect = 0.002
Identities = 19/33 (57%), Positives = 24/33 (72%)
Query: 1070 GDSVAWGGCTIIYLLGQQLHFELFDFSYQILNI 1102
GD + W GC+II LLGQQ F+LFDF Y +L +
Sbjct: 1147 GDGLNWAGCSIIVLLGQQRRFDLFDFCYHLLKV 1179
>UniRef100_UPI00002929FB UPI00002929FB UniRef100 entry
Length = 1319
Score = 335 bits (859), Expect = 5e-90
Identities = 255/886 (28%), Positives = 432/886 (47%), Gaps = 98/886 (11%)
Query: 3 VPVEEAIA---ALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLS---E 56
V +E+A++ L L D+QP ++ P + + + + D A+ ++ E
Sbjct: 6 VTLEDALSNVDLLEELPLPDQQPCIEPPPSSIMYQANFDTN---FEDRNAFVTGIARYIE 62
Query: 57 DTKALNQLSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSR 116
+ ++ + +EG E A +LYT+RSC +A+PQ+ + + ++ ++Y +T +VL+ E+++
Sbjct: 63 QATVHSSMNEMLEEGHEYAVMLYTWRSCSRAIPQVKCNEQPNRVEIYEKTVEVLEPEVTK 122
Query: 117 LREIQRWQASASSKLATDMQRFSRPERRINGPTISHLWSMLKLFDVLVQLDHLKNAKASI 176
L + +Q A + ++++R ERR + + ++L ++ K ++ LD LKN K S+
Sbjct: 123 LMKFMYFQRKAIERFCSEVKRLCHAERRKDFVSEAYLLTLGKFINMFAVLDELKNMKCSV 182
Query: 177 PNDFSWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPVYLL 236
ND S YKR Q + D S++E +L +FL+ I LH ++ + Y
Sbjct: 183 KNDHSAYKRA-AQFLRKMADPQSIQES-QNLSMFLANHNRITQCLHQQLEVIPGY----- 235
Query: 237 HIFSLSTVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSEKDSESL-- 294
E++L ++ V+ E L E+H+LL+V+ + L+ + + L
Sbjct: 236 --------EELLADIVNICVDYYENKMYLTPSEKHMLLKVMGFGLYLMDGNVSNIYKLDA 287
Query: 295 YKRVKINRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELPP 354
KR+ ++++ FK + V+P F D+ + + ++ + Y S T + +P +
Sbjct: 288 KKRINLSKIDKFFKLQ-VVPLFGDMQIELSRYIETSAHYEENKSKWTCTQSSISP-QYNL 345
Query: 355 LNHIGAVRAEHDDFTIRFASAMNQST---DGSDVDWSKEVKGNMYDMIVEGFQLLSRWTA 411
+ +R +H F A N G D S E ++D+ + G QLLS+W+
Sbjct: 346 CEQMVQIREDHIRFISELARYSNSEVVTGSGLDSQKSDEEYRELFDLALRGLQLLSKWST 405
Query: 412 RIWEQCAWKFSRPC-----KDASPSFSDYEKVVRYNYSAEERKALVELVSCIKSVGSMMQ 466
+ E +WK P KD + +YE+ RYNY++EE+ ALVE+++ IK + +M
Sbjct: 406 HVMEVYSWKLVHPTDKFCNKDCPGTAEEYERATRYNYTSEEKFALVEVIAMIKGLQVLMG 465
Query: 467 RCDTLVADALWETIHAEVQDFVQNTLASMLRTTFRKKKD-LSRILSDMRTLSADWMANTN 525
R +++ A+ TI+A +QDF Q TL LR RKKK+ L +L +R DW
Sbjct: 466 RMESVFNQAIRNTIYAALQDFAQMTLREPLRQAVRKKKNVLISVLQAIRKTVCDWEGARE 525
Query: 526 KSESEL---QSSQHGGEESKANIFYPRAVAPTAAQVHCLQFLIYEVV---SGGNLRRPGG 579
+ GG + K RAV P++ Q++ ++ ++ ++ SG
Sbjct: 526 PPNDPCLRGEKDPKGGFDIKVP---RRAVGPSSTQLYMVRTMLESLIADKSGSKKTLRSS 582
Query: 580 LFGNSGSEVPVNDLKQLETF----VMLAVTVATLTDLGFLWFREFYLESS--RVIQFPIE 633
L G V + D + F + + + DL LWFREF+LE + R IQFPIE
Sbjct: 583 LDGPI--VVAIEDFHKQSFFFTHLLNFSEALQQCCDLSQLWFREFFLELTMGRRIQFPIE 640
Query: 634 CSLPWMLVDCVLESPNSGLLESVLMPFDIYNDSAQQALVLLKQRFLYDEIEAEVDHCFDI 693
S+PW+L D +LE+ ++E VL P D+YNDS AL K++FLYDEIEAEV+ CFD
Sbjct: 641 MSMPWILTDHILETKEPSMMEYVLYPLDLYNDSGYYALTKFKKQFLYDEIEAEVNLCFDQ 700
Query: 694 FVARLCETIFTYYKSWAASELLDPTFLFASEN---------AEKYAV-----------QP 733
FV +L + IF YYK+ A S LLD F +N + +Y +
Sbjct: 701 FVYKLADQIFAYYKAMAGSVLLDKRFRAECKNYGVIIPYPPSNRYETLLKQRHVQLLGRS 760
Query: 734 MRLNMLL----------------------KMTRVKELEKLLDVLKHSHELLSIDLSVDSF 771
+ LN L+ +T + ELE LL++ + +H LLS +++DSF
Sbjct: 761 IDLNRLITQRISAAMYKSLDHAISRFESEDLTSIVELEWLLEINRLTHRLLSKHMTLDSF 820
Query: 772 SLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPNFILCNTTQRFI 817
M E N+S + R+ ++ E+ DFLPN+ +T R +
Sbjct: 821 DAMFREANHNVS--APYGRITLHVFWELNFDFLPNYCYNGSTNRSV 864
Score = 47.0 bits (110), Expect = 0.003
Identities = 18/33 (54%), Positives = 23/33 (69%)
Query: 1070 GDSVAWGGCTIIYLLGQQLHFELFDFSYQILNI 1102
GD + W GC +I LLGQQ F+LFDF Y +L +
Sbjct: 1181 GDGLNWAGCAVIVLLGQQRRFDLFDFCYHLLKV 1213
>UniRef100_Q5SQX6 Cytoplasmic FMR1 interacting protein 2 [Mus musculus]
Length = 1319
Score = 320 bits (820), Expect = 2e-85
Identities = 234/783 (29%), Positives = 392/783 (49%), Gaps = 55/783 (7%)
Query: 3 VPVEEAIA---ALSTFSLEDEQPEVQGPGVWVSTERGATESPIEYCDVAAYRLSLS---E 56
V +E+A++ L L D+QP ++ P + + + + D A+ ++ E
Sbjct: 5 VTLEDALSNVDLLEELPLPDQQPCIEPPPSSIMYQANFDTN---FEDRNAFVTGIARYIE 61
Query: 57 DTKALNQLSSLTQEGKEMASVLYTYRSCVKALPQLPDSMKQSQADLYLETYQVLDLEMSR 116
+ ++ + +EG + A +LYT+RSC +A+PQ+ + + ++ ++Y +T +VL+ E+++
Sbjct: 62 QATVHSSMNEMLEEGHDYAVMLYTWRSCSRAIPQVKCNEQPNRVEIYEKTVEVLEPEVTK 121
Query: 117 LREIQRWQASASSKLATDMQRFSRPERRINGPTISHLWSMLKLFDVLVQLDHLKNAKASI 176
L + +Q A + ++++R ERR + + ++L ++ K ++ LD LKN K S+
Sbjct: 122 LMKFMYFQRKAIERFCSEVKRLCHAERRKDFVSEAYLLTLGKFINMFAVLDELKNMKCSV 181
Query: 177 PNDFSWYKRTFTQVSGQWQDTDSMREELDDLQIFLSTRWAILLNLHVEMFRVNKYPVYLL 236
ND S YKR Q + D S++E +L +FL+ I LH ++ + Y
Sbjct: 182 KNDHSAYKRA-AQFLRKMADPQSIQES-QNLSMFLANHNRITQCLHQQLEVIPGY----- 234
Query: 237 HIFSLSTVEDILQVLIVFAVESLELDFALLFPERHILLRVLPVLVVLVTSSEKDSESL-- 294
E++L ++ V+ E L E+H+LL+V+ + L+ + + L
Sbjct: 235 --------EELLADIVNICVDYYENKMYLTPSEKHMLLKVMGFGLYLMDGNVSNIYKLDA 286
Query: 295 YKRVKINRLINIFKNEAVIPAFPDLHLSPAAIMKELSTYFPKFSSQTRLLTLAAPHELPP 354
KR+ ++++ FK V+P F D+ + A +K + Y S T + +P +
Sbjct: 287 KKRINLSKIDKFFKQLQVVPLFGDMQIELARYIKTSAHYEENKSKWTCTQSSISP-QYNI 345
Query: 355 LNHIGAVRAEHDDFTIRFASAMNQST---DGSDVDWSKEVKGNMYDMIVEGFQLLSRWTA 411
+ +R +H F A N G D S E ++D+ + G QLLS+W+A
Sbjct: 346 CEQMVQIRDDHIRFISELARYSNSEVVTGSGLDSQKSDEEYRELFDLALRGLQLLSKWSA 405
Query: 412 RIWEQCAWKFSRPC-----KDASPSFSDYEKVVRYNYSAEERKALVELVSCIKSVGSMMQ 466
+ E +WK P KD + +YE+ RYNY++EE+ A VE+++ IK + +M
Sbjct: 406 HVMEVYSWKLVHPTDKFCNKDCPGTAEEYERATRYNYTSEEKFAFVEVIAMIKGLQVLMG 465
Query: 467 RCDTLVADALWETIHAEVQDFVQNTLASMLRTTFRKKKD-LSRILSDMRTLSADWMANTN 525
R +++ A+ TI+A +QDF Q TL LR RKKK+ L +L +R DW
Sbjct: 466 RMESVFNQAIRNTIYAALQDFAQVTLREPLRQAVRKKKNVLISVLQAIRKTICDWEGGRE 525
Query: 526 KSESEL---QSSQHGGEESKANIFYPRAVAPTAAQVHCLQFLIYEVV---SGGNLRRPGG 579
+ GG + K RAV P++ Q++ ++ ++ ++ SG
Sbjct: 526 PPNDPCLRGEKDPKGGFDIKVP---RRAVGPSSTQLYMVRTMLESLIADKSGSKKTLRSS 582
Query: 580 LFGNSGSEVPVNDLKQLETF----VMLAVTVATLTDLGFLWFREFYLESS--RVIQFPIE 633
L G + + D + F + ++ + DL LWFREF+LE + R IQFPIE
Sbjct: 583 LDGPI--VLAIEDFHKQSFFFTHLLNISEALQQCCDLSQLWFREFFLELTMGRRIQFPIE 640
Query: 634 CSLPWMLVDCVLESPNSGLLESVLMPFDIYNDSAQQALVLLKQRFLYDEIEAEVDHCFDI 693
S+PW+L D +LE+ ++E VL P D+YNDSA AL K++FLYDEIEAEV+ CFD
Sbjct: 641 MSMPWILTDHILETKEPSMMEYVLYPLDLYNDSAYYALTKFKKQFLYDEIEAEVNLCFDQ 700
Query: 694 FVARLCETIFTYYKSWAASELLDPTFLFASENAEKYAVQP--MRLNMLLKMTRVKELEKL 751
FV +L + IF YYK+ A S LLD F +N P R LLK V+ L +
Sbjct: 701 FVYKLADQIFAYYKAMAGSVLLDKRFRAECKNYGVIIPYPPSNRYETLLKQRHVQLLGRS 760
Query: 752 LDV 754
+D+
Sbjct: 761 IDL 763
Score = 53.9 bits (128), Expect = 3e-05
Identities = 31/99 (31%), Positives = 55/99 (55%), Gaps = 4/99 (4%)
Query: 747 ELEKLLDVLKHSHELLSIDLSVDSFSLMLNEMQENISLVSFSSRLASQIWSEMQSDFLPN 806
ELE LL++ + +H LL +++DSF M E N+S + R+ ++ E+ DFLPN
Sbjct: 862 ELEWLLEINRLTHRLLCKHMTLDSFDAMFREANHNVS--APYGRITLHVFWELNFDFLPN 919
Query: 807 FILCNTTQRFIRSS--KTVPVQKPSIPSAKPSFYCGTQP 843
+ +T RF+R++ T Q+ + +P + G++P
Sbjct: 920 YCYNGSTNRFVRTAIPFTQEPQRDKPANVQPYYLYGSKP 958
Score = 47.8 bits (112), Expect = 0.002
Identities = 19/33 (57%), Positives = 24/33 (72%)
Query: 1070 GDSVAWGGCTIIYLLGQQLHFELFDFSYQILNI 1102
GD + W GC+II LLGQQ F+LFDF Y +L +
Sbjct: 1219 GDGLNWAGCSIIVLLGQQRRFDLFDFCYHLLKV 1251
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.331 0.140 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,750,174,162
Number of Sequences: 2790947
Number of extensions: 67624188
Number of successful extensions: 196485
Number of sequences better than 10.0: 87
Number of HSP's better than 10.0 without gapping: 63
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 196003
Number of HSP's gapped (non-prelim): 202
length of query: 1157
length of database: 848,049,833
effective HSP length: 139
effective length of query: 1018
effective length of database: 460,108,200
effective search space: 468390147600
effective search space used: 468390147600
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 81 (35.8 bits)
Medicago: description of AC146587.10


