

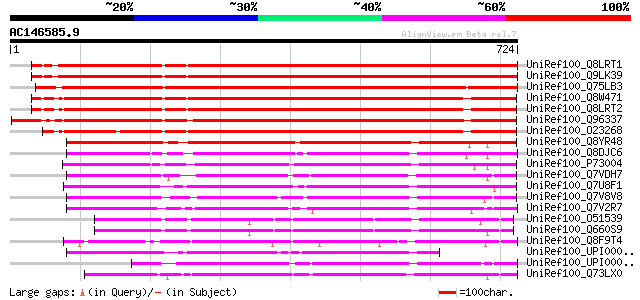
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146585.9 - phase: 0
(724 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LRT1 Acyl-CoA synthetase-like protein [Arabidopsis t... 949 0.0
UniRef100_Q9LK39 Long-chain-fatty-acid CoA ligase [Arabidopsis t... 949 0.0
UniRef100_Q75LB3 Putative AMP-binding protein [Oryza sativa] 938 0.0
UniRef100_Q8W471 A6 anther-specific protein [Arabidopsis thaliana] 927 0.0
UniRef100_Q8LRT2 Acyl-CoA synthetase-like protein [Arabidopsis t... 925 0.0
UniRef100_Q96337 AMP-binding protein [Brassica napus] 919 0.0
UniRef100_O23268 AMP-binding protein [Arabidopsis thaliana] 911 0.0
UniRef100_Q8YR48 Alr3602 protein [Anabaena sp.] 482 e-134
UniRef100_Q8DJC6 Long-chain-fatty-acid CoA ligase [Synechococcus... 473 e-132
UniRef100_P73004 Long-chain-fatty-acid CoA ligase [Synechocystis... 449 e-124
UniRef100_Q7VDH7 Long-chain acyl-CoA synthetase [Prochlorococcus... 392 e-107
UniRef100_Q7U8F1 Putative long-chain-fatty-acid--CoA ligase [Syn... 388 e-106
UniRef100_Q7V8V8 Putative long-chain-fatty-acid--CoA ligase [Pro... 384 e-105
UniRef100_Q7V2R7 Putative long-chain-fatty-acid--CoA ligase [Pro... 371 e-101
UniRef100_O51539 Long-chain-fatty-acid CoA ligase [Borrelia burg... 345 4e-93
UniRef100_Q660S9 Long-chain-fatty-acid CoA ligase [Borrelia gari... 341 4e-92
UniRef100_Q8F9T4 Long-chain-fatty-acid CoA ligase [Leptospira in... 337 6e-91
UniRef100_UPI0000322607 UPI0000322607 UniRef100 entry 333 1e-89
UniRef100_UPI0000323D49 UPI0000323D49 UniRef100 entry 327 1e-87
UniRef100_Q73LX0 AMP-binding enzyme family protein [Treponema de... 320 9e-86
>UniRef100_Q8LRT1 Acyl-CoA synthetase-like protein [Arabidopsis thaliana]
Length = 722
Score = 949 bits (2452), Expect = 0.0
Identities = 468/694 (67%), Positives = 564/694 (80%), Gaps = 14/694 (2%)
Query: 32 RCFRVFSQSNTETVQIIRKCSPFLESSMLVGNNNNNNNAAVDSSHEWKVVPDIWRSSAEK 91
R FRV +S + ++ R+CSPFLE L A SS+EWK VPDIWRSS EK
Sbjct: 42 RGFRVRCESKIQEKEL-RRCSPFLERLSLP-------REAALSSNEWKSVPDIWRSSVEK 93
Query: 92 YGDKVALVDQYHHPPSTITYNQLEQAILDYAEGLRVIGVRPDEKLALFADNSCRWLVADQ 151
YGD+VA+VD YH PPST TY QLEQ ILD+ EGLRV+GV+ DEK+ALFADNSCRWLVADQ
Sbjct: 94 YGDRVAVVDPYHDPPSTFTYRQLEQEILDFVEGLRVVGVKADEKIALFADNSCRWLVADQ 153
Query: 152 GMMASGAINVVRGSRSSVEELLQIYNHSESVALAVDGPEMFNRIAKPFYSKTGMRFIILL 211
G+MA+GA+NVVRGSRSSVEELLQIY HSESVAL VD PE FNRIA+ F K +F+ILL
Sbjct: 154 GIMATGAVNVVRGSRSSVEELLQIYCHSESVALVVDNPEFFNRIAESFSYKAAPKFVILL 213
Query: 212 WGEKSDLNLIAEENKEVPIFSFMEVIDLGRESRMALSDSHEASQRYVYEAINSDDIATLI 271
WGEKS L + P++S+ E+ G+E R + S++ S +Y YE I+ DDIAT++
Sbjct: 214 WGEKSSL---VTAGRHTPVYSYNEIKKFGQERRAKFARSND-SGKYEYEYIDPDDIATIM 269
Query: 272 YTSGTTGNPKGVMLTHRNLLHQIKNLWDTVPAEVGDRFLSMLPPWHAYERACEYFIFTCG 331
YTSGTTGNPKGVMLTH+NLLHQI+NL D VPAE G+RFLSMLP WHAYERACEYFIFTCG
Sbjct: 270 YTSGTTGNPKGVMLTHQNLLHQIRNLSDFVPAEAGERFLSMLPSWHAYERACEYFIFTCG 329
Query: 332 IEQVYTTVRNLKDDLGRYQPHYMISVPLVFETLYSGIQKQISTSPPVRKLVALTFIRVSL 391
+EQ YT++R LKDDL RYQPHY+ISVPLV+ETLYSGIQKQIS S P RK +ALT I+VSL
Sbjct: 330 VEQKYTSIRFLKDDLKRYQPHYLISVPLVYETLYSGIQKQISASSPARKFLALTLIKVSL 389
Query: 392 AYMEYKRIYEGKCLTRNVKQPSIVNSMLDCLWARIIATILFPIHLLAIKFVYSKIHSAIG 451
AY E KR+YEG CLT+N K P + S++D LWAR++A L+P+H+LA K V+ KI S+IG
Sbjct: 390 AYTEMKRVYEGLCLTKNQKPPMYIVSLVDWLWARVVAFFLWPLHMLAEKLVHRKIRSSIG 449
Query: 452 LSKAGISGGGSLPLEVDKFFEAIGVKVQNGYGLTETSPVIAARRPRCNVIGSVGHPVQHT 511
++KAG+SGGGSLP+ VDKFFEAIGV VQNGYGLTETSPV++ARR RCNV+GSVGHP++ T
Sbjct: 450 ITKAGVSGGGSLPMHVDKFFEAIGVNVQNGYGLTETSPVVSARRLRCNVLGSVGHPIKDT 509
Query: 512 EFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPH 571
EFK+VD ETG VLPPGSKGI+KVRGPPVM GYYKNPLAT Q +D DGW NTGD+GWI P
Sbjct: 510 EFKIVDHETGTVLPPGSKGIVKVRGPPVMKGYYKNPLATKQVIDDDGWFNTGDMGWITPQ 569
Query: 572 HSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLG 631
HSTGRSR+ GVIV++GRAKDTIVLSTGENVEP E+EEAAMRS++IQQIVVIGQD+RRLG
Sbjct: 570 HSTGRSRSCGGVIVLEGRAKDTIVLSTGENVEPLEIEEAAMRSNLIQQIVVIGQDQRRLG 629
Query: 632 AIIVPNSEEVLKVARE-LSIIDSISSNVVSEEKVLNLIYKELKTWMSESPFQIGPILLVN 690
AI++PN E A++ +S +DS N +S+E + +++Y+EL+ W S+ FQ+GP+L+V+
Sbjct: 630 AIVIPNKEAAEGAAKQKISPVDS-EVNELSKETITSMVYEELRKWTSQCSFQVGPVLIVD 688
Query: 691 EPFTIDNGLMTPTMKIRRDRVVAKYKEQIDDLYK 724
EPFTIDNGLMTPTMKIRRD+VV +YK +I+ LYK
Sbjct: 689 EPFTIDNGLMTPTMKIRRDKVVDQYKNEIERLYK 722
>UniRef100_Q9LK39 Long-chain-fatty-acid CoA ligase [Arabidopsis thaliana]
Length = 722
Score = 949 bits (2452), Expect = 0.0
Identities = 468/694 (67%), Positives = 564/694 (80%), Gaps = 14/694 (2%)
Query: 32 RCFRVFSQSNTETVQIIRKCSPFLESSMLVGNNNNNNNAAVDSSHEWKVVPDIWRSSAEK 91
R FRV +S + ++ R+CSPFLE L A SS+EWK VPDIWRSS EK
Sbjct: 42 RGFRVRCESKIQEKEL-RRCSPFLERLSLP-------REAALSSNEWKSVPDIWRSSVEK 93
Query: 92 YGDKVALVDQYHHPPSTITYNQLEQAILDYAEGLRVIGVRPDEKLALFADNSCRWLVADQ 151
YGD+VA+VD YH PPST TY QLEQ ILD+ EGLRV+GV+ DEK+ALFADNSCRWLVADQ
Sbjct: 94 YGDRVAVVDPYHDPPSTFTYRQLEQEILDFVEGLRVVGVKADEKIALFADNSCRWLVADQ 153
Query: 152 GMMASGAINVVRGSRSSVEELLQIYNHSESVALAVDGPEMFNRIAKPFYSKTGMRFIILL 211
G+MA+GA+NVVRGSRSSVEELLQIY HSESVAL VD PE FNRIA+ F K +F+ILL
Sbjct: 154 GIMATGAVNVVRGSRSSVEELLQIYCHSESVALVVDNPEFFNRIAESFSYKAAPKFVILL 213
Query: 212 WGEKSDLNLIAEENKEVPIFSFMEVIDLGRESRMALSDSHEASQRYVYEAINSDDIATLI 271
WGEKS L + P++S+ E+ G+E R + S++ S +Y YE I+ DDIAT++
Sbjct: 214 WGEKSSL---VTAGRHTPVYSYNEIKKFGQERRAKFARSND-SGKYEYEYIDPDDIATIM 269
Query: 272 YTSGTTGNPKGVMLTHRNLLHQIKNLWDTVPAEVGDRFLSMLPPWHAYERACEYFIFTCG 331
YTSGTTGNPKGVMLTH+NLLHQI+NL D VPAE G+RFLSMLP WHAYERACEYFIFTCG
Sbjct: 270 YTSGTTGNPKGVMLTHQNLLHQIRNLSDFVPAEAGERFLSMLPSWHAYERACEYFIFTCG 329
Query: 332 IEQVYTTVRNLKDDLGRYQPHYMISVPLVFETLYSGIQKQISTSPPVRKLVALTFIRVSL 391
+EQ YT++R LKDDL RYQPHY+ISVPLV+ETLYSGIQKQIS S P RK +ALT I+VSL
Sbjct: 330 VEQKYTSIRFLKDDLKRYQPHYLISVPLVYETLYSGIQKQISASSPARKFLALTLIKVSL 389
Query: 392 AYMEYKRIYEGKCLTRNVKQPSIVNSMLDCLWARIIATILFPIHLLAIKFVYSKIHSAIG 451
AY E KR+YEG CLT+N K P + S++D LWAR++A L+P+H+LA K V+ KI S+IG
Sbjct: 390 AYTEMKRVYEGLCLTKNQKPPMYIVSLVDWLWARVVAFFLWPLHMLAEKLVHRKIRSSIG 449
Query: 452 LSKAGISGGGSLPLEVDKFFEAIGVKVQNGYGLTETSPVIAARRPRCNVIGSVGHPVQHT 511
++KAG+SGGGSLP+ VDKFFEAIGV VQNGYGLTETSPV++ARR RCNV+GSVGHP++ T
Sbjct: 450 ITKAGVSGGGSLPMHVDKFFEAIGVNVQNGYGLTETSPVVSARRLRCNVLGSVGHPIKDT 509
Query: 512 EFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPH 571
EFK+VD ETG VLPPGSKGI+KVRGPPVM GYYKNPLAT Q +D DGW NTGD+GWI P
Sbjct: 510 EFKIVDHETGTVLPPGSKGIVKVRGPPVMKGYYKNPLATKQVIDDDGWFNTGDMGWITPQ 569
Query: 572 HSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLG 631
HSTGRSR+ GVIV++GRAKDTIVLSTGENVEP E+EEAAMRS++IQQIVVIGQD+RRLG
Sbjct: 570 HSTGRSRSCGGVIVLEGRAKDTIVLSTGENVEPLEIEEAAMRSNLIQQIVVIGQDQRRLG 629
Query: 632 AIIVPNSEEVLKVARE-LSIIDSISSNVVSEEKVLNLIYKELKTWMSESPFQIGPILLVN 690
AI++PN E A++ +S +DS N +S+E + +++Y+EL+ W S+ FQ+GP+L+V+
Sbjct: 630 AIVIPNKEAAEGAAKQKISPVDS-EVNELSKETITSMVYEELRKWTSQCSFQVGPVLIVD 688
Query: 691 EPFTIDNGLMTPTMKIRRDRVVAKYKEQIDDLYK 724
EPFTIDNGLMTPTMKIRRD+VV +YK +I+ LYK
Sbjct: 689 EPFTIDNGLMTPTMKIRRDKVVDQYKNEIERLYK 722
>UniRef100_Q75LB3 Putative AMP-binding protein [Oryza sativa]
Length = 750
Score = 938 bits (2425), Expect = 0.0
Identities = 460/687 (66%), Positives = 555/687 (79%), Gaps = 11/687 (1%)
Query: 38 SQSNTETVQIIRKCSPFLESSMLVGNNNNNNNAAVDSSHEWKVVPDIWRSSAEKYGDKVA 97
S +T + RKCSP LES++L G N ++H+W VPDIWR++AEKY D+VA
Sbjct: 75 SAGSTLENPVRRKCSPLLESALLPGGNGL-------TTHDWMAVPDIWRTAAEKYADRVA 127
Query: 98 LVDQYHHPPSTITYNQLEQAILDYAEGLRVIGVRPDEKLALFADNSCRWLVADQGMMASG 157
LVD YH PPS +TY QLEQ ILD+++GLR IGV PDEK+ALFADNSCRWLVADQG+MA+G
Sbjct: 128 LVDPYHEPPSELTYKQLEQEILDFSQGLRAIGVAPDEKIALFADNSCRWLVADQGIMATG 187
Query: 158 AINVVRGSRSSVEELLQIYNHSESVALAVDGPEMFNRIAKPFYSKTGMRFIILLWGEKSD 217
AINVVRG+RSS EEL QIY HSES+AL VD P+ FNR+A+ F S+ RFI+LLWGEKS
Sbjct: 188 AINVVRGTRSSDEELFQIYTHSESIALVVDSPQFFNRLAESFISRINARFIVLLWGEKSC 247
Query: 218 LNLIAEENKEVPIFSFMEVIDLGRESRMALSDSHEASQRYVYEAINSDDIATLIYTSGTT 277
LN +E +P++ F ++ LGRESR L SHE Q+ V+E I DD+ATLIYTSGT+
Sbjct: 248 LN--SEVVNGIPLYDFKDITQLGRESRNTLRHSHEQGQQVVFETITPDDVATLIYTSGTS 305
Query: 278 GNPKGVMLTHRNLLHQIKNLWDTVPAEVGDRFLSMLPPWHAYERACEYFIFTCGIEQVYT 337
G PKGVMLTHRNLLHQIKNLWD VPA GDRFLSMLPPWHAYERA EYFIFT GI+QVYT
Sbjct: 306 GTPKGVMLTHRNLLHQIKNLWDFVPAVPGDRFLSMLPPWHAYERASEYFIFTYGIQQVYT 365
Query: 338 TVRNLKDDLGRYQPHYMISVPLVFETLYSGIQKQISTSPPVRKLVALTFIRVSLAYMEYK 397
TV+ LK+DL RYQP Y++SVPLV+E LYS IQ+QIS+S RK VAL I++SL YME K
Sbjct: 366 TVKYLKEDLQRYQPQYIVSVPLVYEILYSSIQRQISSSSTARKFVALALIKISLLYMEAK 425
Query: 398 RIYEGKCLTRNVKQPSIVNSMLDCLWARIIATILFPIHLLAIKFVYSKIHSAIGLSKAGI 457
RIYEG L+ N +PS + M++ L ARI+A +L+P+H LA VY KIHSAIG+SKAGI
Sbjct: 426 RIYEGTVLSNNPVKPSFIVYMVNWLSARIVAALLWPLHNLAKTLVYKKIHSAIGISKAGI 485
Query: 458 SGGGSLPLEVDKFFEAIGVKVQNGYGLTETSPVIAARRPRCNVIGSVGHPVQHTEFKVVD 517
SGGGSLP+ VDKFFEAIGVKVQNGYGLTETSPV+AARRP CNV+G+VGHPV+HTE KVVD
Sbjct: 486 SGGGSLPMHVDKFFEAIGVKVQNGYGLTETSPVVAARRPFCNVLGTVGHPVKHTEIKVVD 545
Query: 518 SETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRS 577
ETGEVLP GSKG++KVRGP VM GYYKNP ATN+ LD++GW +TGD+GWIAPH TG S
Sbjct: 546 METGEVLPDGSKGVVKVRGPQVMKGYYKNPSATNKVLDQEGWFDTGDIGWIAPHCPTGPS 605
Query: 578 RNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIVPN 637
R G++V++GRAKDTIVL+TGENVEPAE+EEAA RS +I QIVV+GQDKRR+GA+IVPN
Sbjct: 606 RKCGGMLVLEGRAKDTIVLTTGENVEPAEIEEAASRSDLINQIVVVGQDKRRIGALIVPN 665
Query: 638 SEEVLKVARELSIIDSISSNVVSEEKVLNLIYKELKTWMSESPFQIGPILLVNEPFTIDN 697
+EVL A+ SI+D +N ++++KVLNL+Y EL+TWM + FQIGPIL+V+EPFT+DN
Sbjct: 666 YDEVLATAKRKSILD--GNNELAKDKVLNLLYDELRTWMVDCSFQIGPILIVDEPFTVDN 723
Query: 698 GLMTPTMKIRRDRVVAKYKEQIDDLYK 724
GL+TPT+K+RRD+V AKY +ID LYK
Sbjct: 724 GLLTPTLKLRRDKVTAKYHREIDALYK 750
>UniRef100_Q8W471 A6 anther-specific protein [Arabidopsis thaliana]
Length = 727
Score = 927 bits (2395), Expect = 0.0
Identities = 463/692 (66%), Positives = 555/692 (79%), Gaps = 25/692 (3%)
Query: 32 RCFRVFSQSNTETVQIIRKCSPFLESSMLVGNNNNNNNAAVDSSHEWKVVPDIWRSSAEK 91
R FRV +S + V + SPFLESS G+ AA+ SS EWK VPDIWRSSAEK
Sbjct: 60 RRFRVHCESKEKEV---KPSSPFLESSSFSGD------AALRSS-EWKAVPDIWRSSAEK 109
Query: 92 YGDKVALVDQYHHPPSTITYNQLEQAILDYAEGLRVIGVRPDEKLALFADNSCRWLVADQ 151
YGD+VALVD YH PP +TY QLEQ ILD+AEGLRV+GV+ DEK+ALFADNSCRWLV+DQ
Sbjct: 110 YGDRVALVDPYHDPPLKLTYKQLEQEILDFAEGLRVLGVKADEKIALFADNSCRWLVSDQ 169
Query: 152 GMMASGAINVVRGSRSSVEELLQIYNHSESVALAVDGPEMFNRIAKPFYSKTGMRFIILL 211
G+MA+GA+NVVRGSRSSVEELLQIY HSESVA+ VD PE FNRIA+ F SK +RF+ILL
Sbjct: 170 GIMATGAVNVVRGSRSSVEELLQIYRHSESVAIVVDNPEFFNRIAESFTSKASLRFLILL 229
Query: 212 WGEKSDLNLIAEENKEVPIFSFMEVIDLGRESRMALSDSHEASQRYVYEAINSDDIATLI 271
WGEKS L + ++P++S+ E+I+ G+ESR LS S++ ++ Y + I+SDD A ++
Sbjct: 230 WGEKSSL---VTQGMQIPVYSYAEIINQGQESRAKLSASND-TRSYRNQFIDSDDTAAIM 285
Query: 272 YTSGTTGNPKGVMLTHRNLLHQIKNLWDTVPAEVGDRFLSMLPPWHAYERACEYFIFTCG 331
YTSGTTGNPKGVMLTHRNLLHQIK+L VPA+ GD+FLSMLP WHAYERA EYFIFTCG
Sbjct: 286 YTSGTTGNPKGVMLTHRNLLHQIKHLSKYVPAQAGDKFLSMLPSWHAYERASEYFIFTCG 345
Query: 332 IEQVYTTVRNLKDDLGRYQPHYMISVPLVFETLYSGIQKQISTSPPVRKLVALTFIRVSL 391
+EQ+YT++R LKDDL RYQP+Y++SVPLV+ETLYSGIQKQIS S RK +ALT I+VS+
Sbjct: 346 VEQMYTSIRYLKDDLKRYQPNYIVSVPLVYETLYSGIQKQISASSAGRKFLALTLIKVSM 405
Query: 392 AYMEYKRIYEGKCLTRNVKQPSIVNSMLDCLWARIIATILFPIHLLAIKFVYSKIHSAIG 451
AYME KRIYEG CLT+ K P + + +D LWAR+IA +L+P+H+LA K +Y KIHS+IG
Sbjct: 406 AYMEMKRIYEGMCLTKEQKPPMYIVAFVDWLWARVIAALLWPLHMLAKKLIYKKIHSSIG 465
Query: 452 LSKAGISGGGSLPLEVDKFFEAIGVKVQNGYGLTETSPVIAARRPRCNVIGSVGHPVQHT 511
+SKAGISGGGSLP+ VDKFFEAIGV +QNGYGLTETSPV+ AR CNV+GS GHP+ T
Sbjct: 466 ISKAGISGGGSLPIHVDKFFEAIGVILQNGYGLTETSPVVCARTLSCNVLGSAGHPMHGT 525
Query: 512 EFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPH 571
EFK+VD ET VLPPGSKGI+KVRGP VM GYYKNP T Q L++ GW NTGD GWIAPH
Sbjct: 526 EFKIVDPETNNVLPPGSKGIIKVRGPQVMKGYYKNPSTTKQVLNESGWFNTGDTGWIAPH 585
Query: 572 HSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLG 631
HS GRSR+ GVIV++GRAKDTIVLSTGENVEP E+EEAAMRS +I+QIVVIGQD+RRLG
Sbjct: 586 HSKGRSRHCGGVIVLEGRAKDTIVLSTGENVEPLEIEEAAMRSRVIEQIVVIGQDRRRLG 645
Query: 632 AIIVPNSEEVLKVARELSIIDSISSNVVSEEKVLNLIYKELKTWMSESPFQIGPILLVNE 691
AII+PN EE +V E S+E + +L+Y+EL+ W SE FQ+GP+L+V++
Sbjct: 646 AIIIPNKEEAQRVDPE-----------TSKETLKSLVYQELRKWTSECSFQVGPVLIVDD 694
Query: 692 PFTIDNGLMTPTMKIRRDRVVAKYKEQIDDLY 723
PFTIDNGLMTPTMKIRRD VVAKYKE+ID LY
Sbjct: 695 PFTIDNGLMTPTMKIRRDMVVAKYKEEIDQLY 726
>UniRef100_Q8LRT2 Acyl-CoA synthetase-like protein [Arabidopsis thaliana]
Length = 709
Score = 925 bits (2391), Expect = 0.0
Identities = 463/692 (66%), Positives = 555/692 (79%), Gaps = 25/692 (3%)
Query: 32 RCFRVFSQSNTETVQIIRKCSPFLESSMLVGNNNNNNNAAVDSSHEWKVVPDIWRSSAEK 91
R FRV +S + V + SPFLESS G+ AA+ SS EWK VPDIWRSSAEK
Sbjct: 42 RRFRVHCESKEKEV---KPSSPFLESSSFSGD------AALRSS-EWKAVPDIWRSSAEK 91
Query: 92 YGDKVALVDQYHHPPSTITYNQLEQAILDYAEGLRVIGVRPDEKLALFADNSCRWLVADQ 151
YGD+VALVD YH PP +TY QLEQ ILD+AEGLRV+GV+ DEK+ALFADNSCRWLV+DQ
Sbjct: 92 YGDRVALVDPYHDPPLKLTYKQLEQEILDFAEGLRVLGVKADEKIALFADNSCRWLVSDQ 151
Query: 152 GMMASGAINVVRGSRSSVEELLQIYNHSESVALAVDGPEMFNRIAKPFYSKTGMRFIILL 211
G+MA+GA+NVVRGSRSSVEELLQIY HSESVA+ VD PE FNRIA+ F SK +RF+ILL
Sbjct: 152 GIMATGAVNVVRGSRSSVEELLQIYRHSESVAIVVDNPEFFNRIAESFTSKASLRFLILL 211
Query: 212 WGEKSDLNLIAEENKEVPIFSFMEVIDLGRESRMALSDSHEASQRYVYEAINSDDIATLI 271
WGEKS L + ++P++S+ E+I+ G+ESR LS S++ ++ Y + I+SDD A ++
Sbjct: 212 WGEKSSL---VTQGMQIPVYSYAEIINQGQESRAKLSASND-TRSYRNQFIDSDDTAAIM 267
Query: 272 YTSGTTGNPKGVMLTHRNLLHQIKNLWDTVPAEVGDRFLSMLPPWHAYERACEYFIFTCG 331
YTSGTTGNPKGVMLTHRNLLHQIK+L VPA GD+FLSMLP WHAYERA EYFIFTCG
Sbjct: 268 YTSGTTGNPKGVMLTHRNLLHQIKHLSKYVPALAGDKFLSMLPSWHAYERASEYFIFTCG 327
Query: 332 IEQVYTTVRNLKDDLGRYQPHYMISVPLVFETLYSGIQKQISTSPPVRKLVALTFIRVSL 391
+EQ+YT++R LKDDL RYQP+Y++SVPLV+ETLYSGIQKQIS S RK +ALT I+VS+
Sbjct: 328 VEQMYTSIRYLKDDLKRYQPNYIVSVPLVYETLYSGIQKQISASSAGRKFLALTLIKVSM 387
Query: 392 AYMEYKRIYEGKCLTRNVKQPSIVNSMLDCLWARIIATILFPIHLLAIKFVYSKIHSAIG 451
AYME KRIYEG CLT+ K P + + +D LWAR+IA +L+P+H+LA K +Y KIHS+IG
Sbjct: 388 AYMEMKRIYEGMCLTKEQKPPMYIVAFVDWLWARVIAALLWPLHMLAKKLIYKKIHSSIG 447
Query: 452 LSKAGISGGGSLPLEVDKFFEAIGVKVQNGYGLTETSPVIAARRPRCNVIGSVGHPVQHT 511
+SKAGISGGGSLP+ VDKFFEAIGV +QNGYGLTETSPV+ AR CNV+GS GHP+ T
Sbjct: 448 ISKAGISGGGSLPIHVDKFFEAIGVILQNGYGLTETSPVVCARTLSCNVLGSAGHPMHGT 507
Query: 512 EFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPH 571
EFK+VD ET VLPPGSKGI+KVRGP VM GYYKNP T Q L++ GW NTGD GWIAPH
Sbjct: 508 EFKIVDPETNNVLPPGSKGIIKVRGPQVMKGYYKNPSTTKQVLNESGWFNTGDTGWIAPH 567
Query: 572 HSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLG 631
HS GRSR+ +GVIV++GRAKDTIVLSTGENVEP E+EEAAMRS +I+QIVVIGQD+RRLG
Sbjct: 568 HSKGRSRHCAGVIVLEGRAKDTIVLSTGENVEPLEIEEAAMRSRVIEQIVVIGQDRRRLG 627
Query: 632 AIIVPNSEEVLKVARELSIIDSISSNVVSEEKVLNLIYKELKTWMSESPFQIGPILLVNE 691
AII+PN EE +V E S+E + +L+Y+EL+ W SE FQ+GP+L+V++
Sbjct: 628 AIIIPNKEEAQRVDPE-----------TSKETLKSLVYQELRKWTSECSFQVGPVLIVDD 676
Query: 692 PFTIDNGLMTPTMKIRRDRVVAKYKEQIDDLY 723
PFTIDNGLMTPTMKIRRD VVAKYKE+ID LY
Sbjct: 677 PFTIDNGLMTPTMKIRRDMVVAKYKEEIDQLY 708
>UniRef100_Q96337 AMP-binding protein [Brassica napus]
Length = 701
Score = 919 bits (2374), Expect = 0.0
Identities = 459/721 (63%), Positives = 562/721 (77%), Gaps = 31/721 (4%)
Query: 3 IIPFPSNTFSYHFFTTTTITLPRTNYARPRCFRVFSQSNTETVQIIRKCSPFLESSMLVG 62
++ + S++ +H I+ PR R FR+ S + V +P +ESS
Sbjct: 11 LLSYGSSSRDFHDLRFRLISGPRVRVPSFRRFRIHCVSKGKDV------TPIIESS---- 60
Query: 63 NNNNNNNAAVDSSHEWKVVPDIWRSSAEKYGDKVALVDQYHHPPSTITYNQLEQAILDYA 122
+++ +AA+ S EWK VPDIWRSSAEKYGDK+ALVD YH PP +TYNQLEQ ILD+A
Sbjct: 61 -SSSGGDAALSYS-EWKAVPDIWRSSAEKYGDKIALVDPYHDPPLKLTYNQLEQEILDFA 118
Query: 123 EGLRVIGVRPDEKLALFADNSCRWLVADQGMMASGAINVVRGSRSSVEELLQIYNHSESV 182
EGLR +GV+ DEK+ALFADNSCRWLV+DQG+M++GA+NVVRGSRSSVEELLQIY HSESV
Sbjct: 119 EGLRAVGVKADEKIALFADNSCRWLVSDQGIMSTGAVNVVRGSRSSVEELLQIYQHSESV 178
Query: 183 ALAVDGPEMFNRIAKPFYSKTGMRFIILLWGEKSDLNLIAEENKEVPIFSFMEVIDLGRE 242
AL VD PE FNRIA F SK +RF+ILLWGEKS L + ++P++S+ ++ +LG+E
Sbjct: 179 ALVVDNPEFFNRIADTFTSKVSLRFLILLWGEKSSL---VTQGMQIPVYSYTDIKNLGQE 235
Query: 243 SRMALSDSHEASQRYVYEAINSDDIATLIYTSGTTGNPKGVMLTHRNLLHQIKNLWDTVP 302
R +D+ ++ IN DD A ++YTSGTTGNPKGVMLTHRNLLHQIK+L VP
Sbjct: 236 KRAGSNDTRKS--------INPDDTAAIMYTSGTTGNPKGVMLTHRNLLHQIKHLSAYVP 287
Query: 303 AEVGDRFLSMLPPWHAYERACEYFIFTCGIEQVYTTVRNLKDDLGRYQPHYMISVPLVFE 362
AE GDRFLSMLP WHAYERACEYFIFTCG+EQ+YT++R LK+DL RYQPHY+ISVPLV+E
Sbjct: 288 AEAGDRFLSMLPSWHAYERACEYFIFTCGVEQMYTSIRFLKEDLKRYQPHYLISVPLVYE 347
Query: 363 TLYSGIQKQISTSPPVRKLVALTFIRVSLAYMEYKRIYEGKCLTRNVKQPSIVNSMLDCL 422
TLYSGIQKQISTS RK +ALT I++SLAYME KRIYEG CLT+ K P + S++D L
Sbjct: 348 TLYSGIQKQISTSSAARKYLALTLIKISLAYMEMKRIYEGMCLTKEQKPPMYIVSLVDWL 407
Query: 423 WARIIATILFPIHLLAIKFVYSKIHSAIGLSKAGISGGGSLPLEVDKFFEAIGVKVQNGY 482
AR++A +L+P+H+LA +Y KIH++IG+SKAGISGGGSLP+ +DKFFEAIGV +QNGY
Sbjct: 408 RARVVAALLWPLHMLAKILIYKKIHASIGISKAGISGGGSLPIHIDKFFEAIGVILQNGY 467
Query: 483 GLTETSPVIAARRPRCNVIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVRGPPVMNG 542
GLTETSPVI AR CNVIGS G+P+ TEFK+VD ET VLPPGSKGI+KVRGP +M G
Sbjct: 468 GLTETSPVICARTLSCNVIGSAGYPMHGTEFKIVDPETNTVLPPGSKGIVKVRGPQIMKG 527
Query: 543 YYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGENV 602
YYKNP T Q L++ GW NTGD+GWIAPHHSTGRSR G+IV++GRAKDTIVLSTGENV
Sbjct: 528 YYKNPTTTKQVLNESGWFNTGDMGWIAPHHSTGRSRRCGGLIVLEGRAKDTIVLSTGENV 587
Query: 603 EPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIVPNSEEVLKVARELSIIDSISSNVVSEE 662
EP E+EEAAMRS +I QIVVIGQD+RRLGAII+PN EE K+ E S +S E
Sbjct: 588 EPLEIEEAAMRSRLIDQIVVIGQDQRRLGAIIMPNKEEAEKLDPETS--------QLSSE 639
Query: 663 KVLNLIYKELKTWMSESPFQIGPILLVNEPFTIDNGLMTPTMKIRRDRVVAKYKEQIDDL 722
K+ +L+Y+EL+ W SE FQ+GP+L+V+EPFTIDNGLMTPTMKIRRD+VVAKYK++I+ L
Sbjct: 640 KLKSLVYQELRKWTSECSFQVGPVLIVDEPFTIDNGLMTPTMKIRRDKVVAKYKDEINQL 699
Query: 723 Y 723
Y
Sbjct: 700 Y 700
>UniRef100_O23268 AMP-binding protein [Arabidopsis thaliana]
Length = 698
Score = 911 bits (2355), Expect = 0.0
Identities = 454/676 (67%), Positives = 543/676 (80%), Gaps = 27/676 (3%)
Query: 48 IRKCSPFLESSMLVGNNNNNNNAAVDSSHEWKVVPDIWRSSAEKYGDKVALVDQYHHPPS 107
++ SPFLESS G+ AA+ SS EWK VPDIWRSSAEKYGD+VALVD YH PP
Sbjct: 49 VKPSSPFLESSSFSGD------AALRSS-EWKAVPDIWRSSAEKYGDRVALVDPYHDPPL 101
Query: 108 TITYNQLEQAILDYAEGLRVIGVRPDEKLALFADNSCRWLVADQGMMASGAINVVRGSRS 167
+TY QLEQ ILD+AEGLRV+GV+ DEK+ALFADNSCRWLV+DQG A+NVVRGSRS
Sbjct: 102 KLTYKQLEQEILDFAEGLRVLGVKADEKIALFADNSCRWLVSDQG-----AVNVVRGSRS 156
Query: 168 SVEELLQIYNHSESVALAVDGPEMFNRIAKPFYSKTGMRFIILLWGEKSDLNLIAEENKE 227
SVEELLQIY HSESVA+ VD PE FNRIA+ F SK +RF+ILLWGEKS L + +
Sbjct: 157 SVEELLQIYRHSESVAIVVDNPEFFNRIAESFTSKASLRFLILLWGEKSSL---VTQGMQ 213
Query: 228 VPIFSFMEVIDLGRESRMALSDSHEASQRYVYEAINSDDIATLIYTSGTTGNPKGVMLTH 287
+P++S+ E+I+ G+ESR LS S++ ++ Y + I+SDD A ++YTSGTTGNPKGVMLTH
Sbjct: 214 IPVYSYAEIINQGQESRAKLSASND-TRSYRNQFIDSDDTAAIMYTSGTTGNPKGVMLTH 272
Query: 288 RNLLHQIKNLWDTVPAEVGDRFLSMLPPWHAYERACEYFIFTCGIEQVYTTVRNLKDDLG 347
RNLLHQIK+L VPA+ GD+FLSMLP WHAYERA EYFIFTCG+EQ+YT++R LKDDL
Sbjct: 273 RNLLHQIKHLSKYVPAQAGDKFLSMLPSWHAYERASEYFIFTCGVEQMYTSIRYLKDDLK 332
Query: 348 RYQPHYMISVPLVFETLYSGIQKQISTSPPVRKLVALTFIRVSLAYMEYKRIYEGKCLTR 407
RYQP+Y++SVPLV+ETLYSGIQKQIS S RK +ALT I+VS+AYME KRIYEG CLT+
Sbjct: 333 RYQPNYIVSVPLVYETLYSGIQKQISASSAGRKFLALTLIKVSMAYMEMKRIYEGMCLTK 392
Query: 408 NVKQPSIVNSMLDCLWARIIATILFPIHLLAIKFVYSKIHSAIGLSKAGISGGGSLPLEV 467
K P + + +D LWAR+IA +L+P+H+LA K +Y KIHS+IG+SKAGISGGGSLP+ V
Sbjct: 393 EQKPPMYIVAFVDWLWARVIAALLWPLHMLAKKLIYKKIHSSIGISKAGISGGGSLPIHV 452
Query: 468 DKFFEAIGVKVQNGYGLTETSPVIAARRPRCNVIGSVGHPVQHTEFKVVDSETGEVLPPG 527
DKFFEAIGV +QNGYGLTETSPV+ AR CNV+GS GHP+ TEFK+VD ET VLPPG
Sbjct: 453 DKFFEAIGVILQNGYGLTETSPVVCARTLSCNVLGSAGHPMHGTEFKIVDPETNNVLPPG 512
Query: 528 SKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVD 587
SKGI+KVRGP VM GYYKNP T Q L++ GW NTGD GWIAPHHS GRSR+ GVIV++
Sbjct: 513 SKGIIKVRGPQVMKGYYKNPSTTKQVLNESGWFNTGDTGWIAPHHSKGRSRHCGGVIVLE 572
Query: 588 GRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIVPNSEEVLKVARE 647
GRAKDTIVLSTGENVEP E+EEAAMRS +I+QIVVIGQD+RRLGAII+PN EE +V E
Sbjct: 573 GRAKDTIVLSTGENVEPLEIEEAAMRSRVIEQIVVIGQDRRRLGAIIIPNKEEAQRVDPE 632
Query: 648 LSIIDSISSNVVSEEKVLNLIYKELKTWMSESPFQIGPILLVNEPFTIDNGLMTPTMKIR 707
S+E + +L+Y+EL+ W SE FQ+GP+L+V++PFTIDNGLMTPTMKIR
Sbjct: 633 -----------TSKETLKSLVYQELRKWTSECSFQVGPVLIVDDPFTIDNGLMTPTMKIR 681
Query: 708 RDRVVAKYKEQIDDLY 723
RD VVAKYKE+ID LY
Sbjct: 682 RDMVVAKYKEEIDQLY 697
>UniRef100_Q8YR48 Alr3602 protein [Anabaena sp.]
Length = 683
Score = 482 bits (1240), Expect = e-134
Identities = 260/659 (39%), Positives = 403/659 (60%), Gaps = 51/659 (7%)
Query: 81 VPDIWRSSAEKYGDKVALVDQYHHPPSTITYNQLEQAILDYAEGLRVIGVRPDEKLALFA 140
+P+IW + +K+G+ +AL + + P ITY QL I +A GL+ +GV+ ++++L A
Sbjct: 59 LPEIWPIAGKKFGETLALYNPHAKPEVKITYQQLADKIQLFASGLQALGVQAGDRISLIA 118
Query: 141 DNSCRWLVADQGMMASGAINVVRGSRSSVEELLQIYNHSESVALAVDGPEMFNRIAKPFY 200
DNS RW +ADQG+M +GA++ VR S++ EELL I HS S AL V+ + FN++ +
Sbjct: 119 DNSPRWFIADQGIMTAGAVDAVRSSQAEKEELLYIVAHSGSTALVVEDLKTFNKLQDRLH 178
Query: 201 SKTGMRFIILLWGEKSDLNLIAEENKEVPIFSFMEVIDLGRESRMALSDSHEASQRYVYE 260
++ +++L E + + + +F+++I++G+ + +V
Sbjct: 179 DLP-IQVVVILTDETPPTE------QSLKVLNFLQLIEVGK------------NHTFVPV 219
Query: 261 AINSDDIATLIYTSGTTGNPKGVMLTHRNLLHQIKNLWDTVPAEVGDRFLSMLPPWHAYE 320
+++ATLIYTSGTTG PKGVML++ NLLHQ+ V +VGD LS+LP WH+YE
Sbjct: 220 KRKRNELATLIYTSGTTGKPKGVMLSYSNLLHQVTTFRVVVQPKVGDTALSILPSWHSYE 279
Query: 321 RACEYFIFTCGIEQVYTTVRNLKDDLGRYQPHYMISVPLVFETLYSGIQKQISTSPPVRK 380
R EY++ + G Q+YT +R++K DL +Y+P+YM++VP ++E++Y G+QKQ P ++
Sbjct: 280 RTVEYYLLSQGCTQIYTNLRSVKADLKQYKPNYMVAVPRLWESIYEGVQKQFREQPANKQ 339
Query: 381 LVALTFIRVSLAYMEYKRIYEGKCLTRNVKQPSIVNSMLDCLWARIIATILFPIHLLAIK 440
+ + +S Y++ +RI +G L + S L+ L A+++ P H+L K
Sbjct: 340 RLIKFLLGMSEKYIKAQRIAQGTSLDH------LHASSLERLIAKVLEFAFLPFHVLGQK 393
Query: 441 FVYSKIHSAIG-LSKAGISGGGSLPLEVDKFFEAIGVKVQNGYGLTETSPVIAARRPRCN 499
VY K+ A G K ISGGG+LP +D FFE IGV++ GYGLTETSPV RRP N
Sbjct: 394 LVYGKVKEATGGRFKQVISGGGALPKHIDTFFEIIGVQILQGYGLTETSPVTNVRRPWRN 453
Query: 500 VIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGW 559
IG+ G P+ TE K+VD ET + LP G +G++ ++GP VM GYY+NP AT +A+D GW
Sbjct: 454 FIGTSGQPIPGTEVKIVDPETRQPLPVGKRGLVLLKGPQVMQGYYQNPEATAKAIDPQGW 513
Query: 560 LNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQ 619
++GDLGW+ P + +V+ GRAKDTIVLS GEN+EP +E+A +RS I Q
Sbjct: 514 FDSGDLGWVTPDNE----------LVLTGRAKDTIVLSNGENIEPQPIEDACLRSPYIDQ 563
Query: 620 IVVIGQDKRRLGAIIVPNSEEVLKVARELSIIDSI---------SSNVVSEEKVL-NLIY 669
I+++GQD+R +GA+IVPN E + K A +++ S+ S + E K++ L
Sbjct: 564 IMLVGQDQRSIGALIVPNVEALAKWAESQNLVLSVEDDNLTSSSSQKINLESKMIQGLFR 623
Query: 670 KELKTWMSESP-----FQIGPILLVNEPFTIDNGLMTPTMKIRRDRVVAKYKEQIDDLY 723
+EL + P ++GP L+ EPF+I+NGLMT T+K+RR V +Y++ I+ ++
Sbjct: 624 QELNREVQNRPGYRPDDRVGPFKLILEPFSIENGLMTQTLKLRRHVVTERYRDIINAMF 682
>UniRef100_Q8DJC6 Long-chain-fatty-acid CoA ligase [Synechococcus elongatus]
Length = 658
Score = 473 bits (1217), Expect = e-132
Identities = 275/660 (41%), Positives = 391/660 (58%), Gaps = 50/660 (7%)
Query: 81 VPDIWRSSAEKYGDKVALVDQYHHPPSTITYNQLEQAILDYAEGLRVIGVRPDEKLALFA 140
+P++W A+++GD VAL Y PP+ ITY++L Q I +A GL+ +GV +++ALF
Sbjct: 33 LPEMWPLLAQRHGDVVALDAPYEDPPTRITYSELYQRIQRFAAGLQALGVAAGDRVALFP 92
Query: 141 DNSCRWLVADQGMMASGAINVVRGSRSSVEELLQIYNHSESVALAVDGPEMFNRIAKPFY 200
DNS RWL+ADQG M +GAINVVR + +ELL I S + L ++ ++ +P
Sbjct: 93 DNSPRWLIADQGSMMAGAINVVRSGTADAQELLYILRDSGATLLLIENLATLGKLQEPLV 152
Query: 201 SKTGMRFIILLWGEKSDLNLIAEENKEVPIFSFMEVIDLGRESRMALSDSHEASQRYVYE 260
TG++ ++LL GE +L + + +F +V G+ +
Sbjct: 153 D-TGVKTVVLLSGESPEL-----AGFPLRLLNFGQVFTEGQYGTVRAV------------ 194
Query: 261 AINSDDIATLIYTSGTTGNPKGVMLTHRNLLHQIKNLWDTVPAEVGDRFLSMLPPWHAYE 320
AI D++ATL+YTSGTTG PKGVM+TH LL QI NLW V +VGDR LS+LP WHAYE
Sbjct: 195 AITPDNLATLMYTSGTTGQPKGVMVTHGGLLSQIVNLWAIVQPQVGDRVLSILPIWHAYE 254
Query: 321 RACEYFIFTCGIEQVYTTVRNLKDDLGRYQPHYMISVPLVFETLYSGIQKQISTSPPVRK 380
R EYF+F CG Q YT +R+ K+DL R +PHYMI+VP ++E+ Y G+QKQ+ SP ++
Sbjct: 255 RVAEYFLFACGCSQTYTNLRHFKNDLKRCKPHYMIAVPRIWESFYEGVQKQLRDSPATKR 314
Query: 381 LVALTFIRVSLAYMEYKRIYEGKCLTRNVKQPSIVNSMLDCLWARIIATILFPIHLLAIK 440
+A F+ V Y+ +R+ G LT P L AR+ +L P++ L K
Sbjct: 315 RLAQFFLSVGQQYILQRRLLTGLSLTN--PHPRGWQKWL----ARVQTLLLKPLYELGEK 368
Query: 441 FVYSKIHSAIGLS-KAGISGGGSLPLEVDKFFEAIGVKVQNGYGLTETSPVIAARRPRCN 499
+YSKI A G K ISGGG+L +D F+E I ++V GYGLTET+ V+ ARR N
Sbjct: 369 RLYSKIREATGGEIKQVISGGGALAPHLDTFYEVINLEVLVGYGLTETAVVLTARRSWAN 428
Query: 500 VIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGW 559
+ GS G P+ T K+VD ET L G KG++ +GP VM GYY P AT + LD +GW
Sbjct: 429 LRGSAGRPIPDTAIKIVDPETKAPLEFGQKGLVMAKGPQVMRGYYNQPEATAKVLDAEGW 488
Query: 560 LNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQ 619
+TGDLG++ P +G +V+ GR KDTIVLS GEN+EP +E+A +RS I Q
Sbjct: 489 FDTGDLGYLTP----------NGDLVLTGRQKDTIVLSNGENIEPQPIEDACVRSPYIDQ 538
Query: 620 IVVIGQDKRRLGAIIVPNSE--EVLKVARELSI------IDSISSNVVSEEK--VLNLIY 669
I+++GQD++ LGA+IVPN E E VA+ + + S VV+ E +++L
Sbjct: 539 IMLVGQDQKALGALIVPNLEALEAWVVAKGYRLELPNRPAQAGSGEVVTLESKVIIDLYR 598
Query: 670 KELKTWMSESP-----FQIGPILLVNEPFTIDNGLMTPTMKIRRDRVVAKYKEQIDDLYK 724
+EL + P +I V EPFTI+NGL+T T+KIRR V +Y++ I+ +++
Sbjct: 599 QELLREVQNRPGYRPDDRIATFRFVLEPFTIENGLLTQTLKIRRHVVSDRYRDMINAMFE 658
>UniRef100_P73004 Long-chain-fatty-acid CoA ligase [Synechocystis sp.]
Length = 696
Score = 449 bits (1154), Expect = e-124
Identities = 255/664 (38%), Positives = 383/664 (57%), Gaps = 51/664 (7%)
Query: 76 HEWKVVPDIWRSSAEKYGDKVALVDQYHHPPSTITYNQLEQAILDYAEGLRVIGVRPDEK 135
H + DIW + E + D VAL D++ HPP T+TY QL + I +A GL+ +GV P +
Sbjct: 68 HSIHCLADIWAITGENFADIVALNDRHSHPPVTLTYAQLREEITAFAAGLQSLGVTPHQH 127
Query: 136 LALFADNSCRWLVADQGMMASGAINVVRGSRSSVEELLQIYNHSESVALAVDGPEMFNRI 195
LA+FADNS RW +ADQG M +GA+N VR +++ +ELL I S S L + + +++
Sbjct: 128 LAIFADNSPRWFIADQGSMLAGAVNAVRSAQAERQELLYILEDSNSRTLIAENRQTLSKL 187
Query: 196 AKPFYSKTGMRFIILLWGEKSDLNLIAEENKEVPIFSFMEVIDLGRESRMALSDSHEASQ 255
A + ++ IILL E+ E+ +P ++F +V+ LG E
Sbjct: 188 ALDGET-IDLKLIILLTDEE------VAEDSAIPQYNFAQVMALGAGKIPTPVPRQE--- 237
Query: 256 RYVYEAINSDDIATLIYTSGTTGNPKGVMLTHRNLLHQIKNLWDTVPAEVGDRFLSMLPP 315
+D+ATLIYTSGTTG PKGVML+H NLLHQ++ L + GD+ LS+LP
Sbjct: 238 ---------EDLATLIYTSGTTGQPKGVMLSHGNLLHQVRELDSVIIPRPGDQVLSILPC 288
Query: 316 WHAYERACEYFIFTCGIEQVYTTVRNLKDDLGRYQPHYMISVPLVFETLYSGIQKQISTS 375
WH+ ER+ EYF+ + G YT++R+ K D+ +PH+++ VP ++E+LY G+QK
Sbjct: 289 WHSLERSAEYFLLSRGCTMNYTSIRHFKGDVKDIKPHHIVGVPRLWESLYEGVQKTFREK 348
Query: 376 PPVRKLVALTFIRVSLAYMEYKRIYEGKCLTRNVKQPSIVNSMLDCLWARIIATILFPIH 435
P ++ + F +S Y+ KRI N+ + S + L AR A +L P+H
Sbjct: 349 SPGQQKLINFFFGISQKYILAKRI------ANNLSLNHLHASAIARLVARCQALVLSPLH 402
Query: 436 LLAIKFVYSKIHSAI-GLSKAGISGGGSLPLEVDKFFEAIGVKVQNGYGLTETSPVIAAR 494
L K VY K+ A G + ISGGG+L +D F+E + V GYGLTET+PV AR
Sbjct: 403 YLGDKIVYHKVRQAAGGRLETLISGGGALARHLDDFYEITSIPVLVGYGLTETAPVTNAR 462
Query: 495 RPRCNVIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQAL 554
+ N+ S G P+ TE ++VD ET E LPP ++G++ +RGP VM GYY P AT + L
Sbjct: 463 VHKHNLRYSSGRPIPFTEIRIVDMETKEDLPPETQGLVLIRGPQVMQGYYNKPEATAKVL 522
Query: 555 DKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRS 614
D++GW ++GDLGW+ P + +++ GRAKDTIVLS GENVEP +E+A +RS
Sbjct: 523 DQEGWFDSGDLGWVTPQND----------LILTGRAKDTIVLSNGENVEPQPIEDACLRS 572
Query: 615 SIIQQIVVIGQDKRRLGAIIVPNSEEVLKVARELSIIDSISSNVVSEE----------KV 664
+ I QI+++GQD++ LGA+IVPN + + K A ++ ++ S E +V
Sbjct: 573 AYIDQIMLVGQDQKSLGALIVPNFDALQKWAETKNLQITVPEPSASSEGMQASGLYDPQV 632
Query: 665 LNLIYKELKTWMSESP-----FQIGPILLVNEPFTIDNGLMTPTMKIRRDRVVAKYKEQI 719
+ L+ EL + + P QI + PF+++NG+MT T+K++R V Y+ I
Sbjct: 633 VGLMRSELHREVRDRPGYRADDQIKDFRFIPAPFSLENGMMTQTLKLKRPVVTQTYQHLI 692
Query: 720 DDLY 723
D+++
Sbjct: 693 DEMF 696
>UniRef100_Q7VDH7 Long-chain acyl-CoA synthetase [Prochlorococcus marinus]
Length = 675
Score = 392 bits (1008), Expect = e-107
Identities = 243/658 (36%), Positives = 370/658 (55%), Gaps = 58/658 (8%)
Query: 81 VPDIWRSSAEKYGDKVALVDQYHHPPSTITYNQLEQAILDYAEGLRVIGVRPDEKLALFA 140
V IW E +G+ +A+ ++ P Y +L I A +GV + +ALF+
Sbjct: 59 VDQIWPWLKENHGNVLAVNSPHYIFPERYNYEELADKIAVAAAAFESLGVVSGDVVALFS 118
Query: 141 DNSCRWLVADQGMMASGAINVVRGSRSSVEELLQIYNHSESVALAVDGPEMFNRIAKPFY 200
+NS RWL+ADQG+M GA N VRGS + VEEL I S SV L V E++ +A
Sbjct: 119 ENSPRWLIADQGLMRLGASNAVRGSSAPVEELRYILADSRSVGLVVQSSELWKALALSED 178
Query: 201 SKTGMRFIILLWGEKSDLNLIAEEN------KEVPIFSFMEVIDLGRESRMALSDSHEAS 254
K +F++ + G+ + NL+ E + PI +F E + S
Sbjct: 179 DKKQFKFVLQIEGQPEN-NLLGWEEFLLKGANKSPINAFKEATGITANS----------- 226
Query: 255 QRYVYEAINSDDIATLIYTSGTTGNPKGVMLTHRNLLHQIKNLWDTVPAEVGDRFLSMLP 314
IAT++YTSGTTG PKGV LTH NLLHQ+++L G LS+LP
Sbjct: 227 -----------SIATILYTSGTTGKPKGVPLTHDNLLHQMRSLACIASPPPGTPLLSVLP 275
Query: 315 PWHAYERACEYFIFTCGIEQVYTTVRNLKDDLGRYQPHYMISVPLVFETLYSGIQKQIST 374
WH+YER+ EY+ F+C Q YTT+++ K DL +P M +VP ++E + +G + +
Sbjct: 276 IWHSYERSAEYYFFSCACSQHYTTIKHFKQDLQTVRPVVMATVPRLWEAVKTGFDEALKK 335
Query: 375 SPPVRKLVALTFIRVSLAYMEYKRIYEGKCLTRNVKQPSIVNSMLDCLWARIIATILFPI 434
P R++V + S + R +RN+ I + ++ ++ +PI
Sbjct: 336 MPRSRQMVLKAALNNSRLFKAALR------KSRNLLLLEINKTNRAIAFSEVV--FRWPI 387
Query: 435 HLLAIKFVYSKIHSAI--GLSKAGISGGGSLPLEVDKFFEAIGVKVQNGYGLTETSPVIA 492
H LA K ++ K+ + G + I+GGG++ VD FFEA+GV++ GYGLTETSPV++
Sbjct: 388 HALASKLLWPKVLQQLSGGRLRFPINGGGAIAPHVDTFFEALGVELLVGYGLTETSPVLS 447
Query: 493 ARRPRCNVIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQ 552
RR N+ GS G P+ TEF++VD G L +G + VRGP VMNGY + P T++
Sbjct: 448 CRRTWRNIRGSSGLPLPETEFRIVDPLNGGPLKFSQQGRVLVRGPQVMNGYLRKPEETSK 507
Query: 553 ALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAM 612
LD+ GW +TGDLG + + G IV+ GRAKDTIVLS GENVEP LEE +
Sbjct: 508 VLDQKGWFDTGDLGMLL----------ADGSIVLTGRAKDTIVLSNGENVEPGPLEEFLL 557
Query: 613 RSSIIQQIVVIGQDKRRLGAIIVPNSEEVLKVA--RELSIIDSISSNVVSEEKVLNLIYK 670
S +I+QI++IGQD+R+LGA+IVP+ +++LK A R L + + + + + K+ L+ +
Sbjct: 558 GSPLIEQIIMIGQDERQLGALIVPDIDQILKWAHQRNLFLKEDLGGG-MGDLKLRRLLQR 616
Query: 671 ELKTWMSESP-----FQIGPILLVNEPFTIDNGLMTPTMKIRRDRVVAKYKEQIDDLY 723
E+ +S+ P ++ I LV +PF+I+NGL+T T+K RR+++V + ++ I+D++
Sbjct: 617 EVNDILSKRPGSRSDERVAGIALV-KPFSIENGLLTQTLKQRRNKIVERDQKAIEDIF 673
>UniRef100_Q7U8F1 Putative long-chain-fatty-acid--CoA ligase [Synechococcus sp.]
Length = 637
Score = 388 bits (997), Expect = e-106
Identities = 240/655 (36%), Positives = 360/655 (54%), Gaps = 50/655 (7%)
Query: 77 EWKVVPDIWRSSAEKYGDKVALVDQYHHPPSTITYNQLEQAILDYAEGLRVIGVRPDEKL 136
E + V +W AE++G A+ + ++ +L Q I A + GV+ + +
Sbjct: 23 ELERVDAVWPWLAERHGTITAVDAPHAAHAERFSFAELAQRIATAAAAFQRQGVQEGDVV 82
Query: 137 ALFADNSCRWLVADQGMMASGAINVVRGSRSSVEELLQIYNHSESVALAVDGPEMFNRIA 196
ALFA+NS RWLVADQG+M GA + VRG+ + VEEL I + + AL V +++ R+
Sbjct: 83 ALFAENSPRWLVADQGLMRCGAADAVRGASAPVEELRYILDDCNATALVVQNADLWRRLD 142
Query: 197 KPFYSKTGMRFIILLWGEKSDLNLIAEENKEVPIFSFMEVIDLGRESRMALSDSHEASQR 256
+ G+R ++ L GE E LG E+ +A A Q+
Sbjct: 143 LTASQRQGLRLVLQLEGEP-------------------EQGVLGWEAFLASG----AGQQ 179
Query: 257 YVYEAINSDDIATLIYTSGTTGNPKGVMLTHRNLLHQIKNLWDTVPAEVGDRFLSMLPPW 316
V IAT++YTSGTTG PKGV LTH NLLHQ+++L + G LS+LP W
Sbjct: 180 SVTPTSARTAIATVLYTSGTTGQPKGVPLTHANLLHQMQSLACVAHPQPGAPVLSVLPIW 239
Query: 317 HAYERACEYFIFTCGIEQVYTTVRNLKDDLGRYQPHYMISVPLVFETLYSGIQKQISTSP 376
HAYER+ Y+ +C Q YT ++ LK DL R +P M +VP ++E++ +G + + T P
Sbjct: 240 HAYERSASYYFLSCACTQTYTNIKQLKKDLPRVRPIAMATVPRLWESVQAGFEDVVKTFP 299
Query: 377 PVRKLVALTFIRVSLAYMEYKRIYEGKCLTRNVKQPSIVNSMLDCLWARIIATILFPIHL 436
P R+ + + S A+ + R L QP + + A +A + +P+H
Sbjct: 300 PSRQRLLRAALANSAAHRKAVRTARNLLL-----QPVALPGRMT---AAAVAALRWPLHA 351
Query: 437 LAIKFVYSKIHSAIGLSKAG--ISGGGSLPLEVDKFFEAIGVKVQNGYGLTETSPVIAAR 494
LA ++ K+ + + ISGGG++ +D FFEA+G+++ GYGLTETSPV++ R
Sbjct: 352 LASALIWPKLRLQLSGGRLAYPISGGGAIAPHIDAFFEAVGIELLVGYGLTETSPVVSCR 411
Query: 495 RPRCNVIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQAL 554
RP N+ GS G P+ TEF++VD E+G L +G + VRGP VM GY P A+ + L
Sbjct: 412 RPWRNIRGSSGLPMPDTEFRIVDQESGASLGFRERGRVLVRGPQVMGGYLGKPEASAKVL 471
Query: 555 DKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRS 614
DGW +TGDLG + P G + + GRAKDTIVLS+GEN+EP LEEA + S
Sbjct: 472 SADGWFDTGDLGMLLP----------DGSVALTGRAKDTIVLSSGENIEPGPLEEALVAS 521
Query: 615 SIIQQIVVIGQDKRRLGAIIVPNSEEVLKVARE--LSIIDSISSNVVSEEKVLNLIYKEL 672
+I+Q++++GQD+R+LGA++VP E + A E LS+ + + + +LNL+ +E
Sbjct: 522 PLIEQVMLVGQDERQLGALLVPRVEPIRAWASEQGLSVGEDLGGR-PGDSVLLNLLMREC 580
Query: 673 KTWMSESPFQIGPILLVN----EPFTIDNGLMTPTMKIRRDRVVAKYKEQIDDLY 723
+ P G L EPF+I+NGL+T T+K RRDR+ + I+ +Y
Sbjct: 581 NLVLRLRPGARGDERLCGVGLVEPFSIENGLLTQTLKQRRDRISRRDAAVIERIY 635
>UniRef100_Q7V8V8 Putative long-chain-fatty-acid--CoA ligase [Prochlorococcus
marinus]
Length = 664
Score = 384 bits (985), Expect = e-105
Identities = 239/653 (36%), Positives = 366/653 (55%), Gaps = 48/653 (7%)
Query: 81 VPDIWRSSAEKYGDKVALVDQYHHPPSTITYNQLEQAILDYAEGLRVIGVRPDEKLALFA 140
V IW A+K+G +A+ + P Y +L + I A +G+ + +A FA
Sbjct: 47 VDQIWLWLAQKHGQILAVDAPHATHPERFRYGELAEQIALAAAAFSHLGIGAGDVVAFFA 106
Query: 141 DNSCRWLVADQGMMASGAINVVRGSRSSVEELLQIYNHSESVALAVDGPEMFNRIAKPFY 200
+NS RWL+ADQG+M +GA N VRG+ + +EEL I S +VAL V E+++++A
Sbjct: 107 ENSPRWLIADQGLMRTGAANAVRGATAPLEELRYILADSRAVALVVQNAELWHKLALTDE 166
Query: 201 SKTGMRFIILLWGEKSDLNLIAEENKEVPIFSFMEVI--DLGRESRMALSDSHEASQRYV 258
+ +R ++ L GE +D + + E++ G+ + L D S V
Sbjct: 167 QRRPLRLVLQLEGEPAD-----------GVMGWQELLAAGAGQPAPDPLKDRDSGSAATV 215
Query: 259 YEAINSDDIATLIYTSGTTGNPKGVMLTHRNLLHQIKNLWDTVPAEVGDRFLSMLPPWHA 318
AT++YTSGTTG PKGV L+H NLLHQ++ L G LS+LP WHA
Sbjct: 216 --------TATILYTSGTTGQPKGVPLSHANLLHQMRCLACVAFPSPGAPVLSVLPIWHA 267
Query: 319 YERACEYFIFTCGIEQVYTTVRNLKDDLGRYQPHYMISVPLVFETLYSGIQKQISTSPPV 378
YER+ EY+ F+C Q YTT++ LK DL R +P M +VP ++E + +G + + P
Sbjct: 268 YERSAEYYFFSCACTQSYTTIKQLKRDLPRVRPIVMATVPRLWEAIQAGFDEAVKGMPTG 327
Query: 379 RKLVALTFIRVSLAYMEYKRIYEGKCLTRNVKQPSIVNSMLDCLWARIIATILFPIHLLA 438
R+ + +R++LA +R + ++ + L A + AT+ +P+H LA
Sbjct: 328 RQRL----LRMALANSGSQRKAWRRSRNLLLEPLPLTTRTL----ALLEATLRWPLHGLA 379
Query: 439 IKFVYSKIHSAI--GLSKAGISGGGSLPLEVDKFFEAIGVKVQNGYGLTETSPVIAARRP 496
++ K+ + + G + I+GGG++ VD FFEA+G+++ GYGLTETSPV++ RRP
Sbjct: 380 GALLWPKVLNQLCGGRLRFPINGGGAIAPHVDAFFEAVGLELLVGYGLTETSPVVSCRRP 439
Query: 497 RCNVIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDK 556
N+ GS G P+ TEF++VD E+G L +G + VRGP VM GY + P AT + LD
Sbjct: 440 WRNIRGSSGPPLPETEFRIVDPESGVDLMFRQRGRVLVRGPQVMAGYLRKPEATAKVLDG 499
Query: 557 DGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSI 616
GW +TGDLG + P G +V+ GRAKDTIVLS+GEN+EP LE A + S +
Sbjct: 500 QGWFDTGDLGMLLP----------DGSLVLTGRAKDTIVLSSGENIEPGPLEAALVASPL 549
Query: 617 IQQIVVIGQDKRRLGAIIVPNSEEVLKVARELSIIDSIS-SNVVSEEKVLNLIYKELKTW 675
++Q++++GQD+R+L A++VP EE+L A + ++ S +E + L+ EL
Sbjct: 550 LEQVMLVGQDERQLAALVVPREEEMLAWAEDQGLLMQTGLSGSPGDEALRRLLRGELNRL 609
Query: 676 M-----SESPFQIGPILLVNEPFTIDNGLMTPTMKIRRDRVVAKYKEQIDDLY 723
+ S + ++ + LV E FTI+NGL+T T+K RRDR+ + I LY
Sbjct: 610 LAQRSGSRADERLAGVALV-EAFTIENGLLTQTLKQRRDRITLRDGALIAALY 661
>UniRef100_Q7V2R7 Putative long-chain-fatty-acid--CoA ligase [Prochlorococcus marinus
subsp. pastoris]
Length = 641
Score = 371 bits (952), Expect = e-101
Identities = 239/654 (36%), Positives = 352/654 (53%), Gaps = 55/654 (8%)
Query: 81 VPDIWRSSAEKYGDKVALVDQYHHPPSTITYNQLEQAILDYAEGLRVIGVRPDEKLALFA 140
V IW + G+ +A+ D I+Y++L I + + G+ + + L +
Sbjct: 32 VDQIWEYLKLRCGNTIAINDLRGKNKEKISYSELADLITKVSNSFKNYGLVKGDVVTLIS 91
Query: 141 DNSCRWLVADQGMMASGAINVVRGSRSSVEELLQIYNHSESVALAVDGPEMFNRIAKPFY 200
+NS RWL+ADQG+M GAIN VRG S EL I HS SV L V E++ ++ K
Sbjct: 92 ENSPRWLIADQGLMRLGAINAVRGINSPSVELEYIIEHSNSVGLIVQSKEVWLKLNKKNE 151
Query: 201 SKTGMRFIILLWGEKSDLNLIAEENKEVPIFSFMEVIDLGRESRMALSDSHEASQRYVYE 260
K +FII L E+ + + ++ E I G E+ + + + ++Y ++
Sbjct: 152 LKKRFKFIINL-----------EDEQFEDLINWPEFIKAGEENLL----KNNSFEKYNHQ 196
Query: 261 AINSDDIATLIYTSGTTGNPKGVMLTHRNLLHQIKNLWDTVPAEVGDRFLSMLPPWHAYE 320
+DIA+++YTSGTTG PKGV LTH N LHQI NL + E G LS+LP WH+YE
Sbjct: 197 I---NDIASILYTSGTTGKPKGVPLTHANFLHQIINLANIADPEPGTSVLSVLPIWHSYE 253
Query: 321 RACEYFIFTCGIEQVYTTVRNLKDDLGRYQPHYMISVPLVFETLYSGIQKQISTSPPVRK 380
R+ EYF F+CG Q YT + LKDD+ + +P M +VP ++E ++ G + PP ++
Sbjct: 254 RSAEYFFFSCGCTQFYTNPKFLKDDIKQVKPVVMATVPRLWEAIHDGFFLALKKMPPKKQ 313
Query: 381 LVALTFIRVSLAYMEYKRIYEGKCLTRNVKQPSIVNSMLDCLWARIIATI----LFPIHL 436
+ I+ S + R K + Q S + A+I TI F IH
Sbjct: 314 KIIKILIKNSSIFKRNLR----KIRNLEINQVSSL--------AKISLTISVIGRFFIHK 361
Query: 437 LAIKFVYSKIHSAI--GLSKAGISGGGSLPLEVDKFFEAIGVKVQNGYGLTETSPVIAAR 494
++ F++ I + K I+GGG+LP VD FFE++G+ V GYGLTETSPV+ R
Sbjct: 362 ISSTFLWPSILKQLCGEKLKFPINGGGALPEHVDLFFESLGIDVLVGYGLTETSPVLTCR 421
Query: 495 RPRCNVIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQAL 554
R NV GS G P+ TE K+++ E ++L G + V+GP VM GY N AT L
Sbjct: 422 RRELNVRGSSGQPLAFTEIKIMNEEKDKILKFKEIGKILVKGPQVMKGYLNNISATKDVL 481
Query: 555 DKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRS 614
KDGW +TGDLG++ P +G +V+ GRAKDTIVLS+GEN+EP LE + S
Sbjct: 482 SKDGWFDTGDLGFLIP----------NGSLVITGRAKDTIVLSSGENIEPNPLETEILSS 531
Query: 615 SIIQQIVVIGQDKRRLGAIIVPNSEEVLKVARELSIIDSISSN----VVSEEKVLNLIYK 670
I Q+ ++GQDK+ L A++ PN E V E + I ++SN + + ++ NL+
Sbjct: 532 EFINQVQLVGQDKKCLTALVAPNIELVENKFFE-NDISKLNSNKKIGLFFKSQINNLLKN 590
Query: 671 ELKTWMSESPFQIGPILLVNEPFTIDNGLMTPTMKIRRDRVVAKYKEQIDDLYK 724
L E QI V + FT++NGL+T T+K +R +V Y I+++YK
Sbjct: 591 RLGARFEE---QILECYFV-DAFTLENGLLTQTLKQKRREIVDLYSTHIEEMYK 640
>UniRef100_O51539 Long-chain-fatty-acid CoA ligase [Borrelia burgdorferi]
Length = 645
Score = 345 bits (884), Expect = 4e-93
Identities = 221/608 (36%), Positives = 350/608 (57%), Gaps = 34/608 (5%)
Query: 122 AEGLRVIGVRPDEKLALFADNSCRWLVADQGMMASGAINVVRGSRSSVEELLQIYNHSES 181
A GL G++ EK+ + +D+ W++ D + G ++V RG+ SS +EL I NHSES
Sbjct: 48 ASGLLHCGIKRGEKVVIISDSRREWIIIDVATLGLGCVDVPRGNDSSEDELAYIINHSES 107
Query: 182 VALAVDGPEMFNRIAKPFYSKTGMRFIILLWGEKSDLNLIAEENKEVPIFSFMEVIDLGR 241
+ V+ + +++ + +R I+++ +KS E+ + +FS+ ++++LG
Sbjct: 108 TFIFVENNKQLHKVLSKKHDLRLVRCIVVIDDDKS----YEEKIGNITVFSYKKLLELGT 163
Query: 242 ESRMALSDSHEASQRYVYEAINSDDIATLIYTSGTTGNPKGVMLTHRNLLHQIKNLWDTV 301
E A S + E +S DIAT+IYTSGTTG PKGVML H + + Q+ L+D +
Sbjct: 164 EYLRANPKSFDME----IEKGSSKDIATIIYTSGTTGMPKGVMLRHESFIFQLDRLYDYL 219
Query: 302 PA-EVGDRFLSMLPPWHAYERACEYFIFTCGIEQVYTTVRN---LKDDLGRYQPHYMISV 357
P + G +S+LP WH++ERACEY + GI Y+ LKD L P ++SV
Sbjct: 220 PTLKPGKIMISILPLWHSFERACEYIVALKGIAIAYSKPIGPVLLKDFL-LLNPQMIVSV 278
Query: 358 PLVFETLYSGIQKQISTSPPVRKLVALTFIRVSLAYMEYKRIYEGKCLTRNVKQPSIVNS 417
P ++E + GI K++S S ++K V F++V + Y + K + G L+ K+ + + S
Sbjct: 279 PRIWEGIRIGIIKKVSESF-IKKFVFGGFLKVGIIYAKLKERFLG--LSPIYKKTNFLIS 335
Query: 418 MLDCLWARIIATILFPIHLLAIKFVYSKIHSAIGLS-KAGISGGGSLPLEVDKFFEAIGV 476
+ L+ I ++FPI LL V+ KI +A+G + + G+SGGG+L VD FF+A+G+
Sbjct: 336 LFSKLFLFIGIVLIFPIKLLGDILVFKKIKNALGQNFEFGVSGGGALVDYVDYFFKAVGI 395
Query: 477 KVQNGYGLTETSPVIAARRPRCNVIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVRG 536
KV GYGLTET P+++ RR + V +VG + E+KVV + G VLP G KG L VR
Sbjct: 396 KVLEGYGLTETGPILSVRRLKGPVAKTVGPILPDVEYKVVGID-GRVLPYGEKGELWVRS 454
Query: 537 PPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVL 596
P +M+GY+K+ T++ L +DGW NTGDL + ++ I + GR+KDTIVL
Sbjct: 455 PQIMSGYFKDKAKTSEVLTEDGWFNTGDLVRLTINNE----------ISIVGRSKDTIVL 504
Query: 597 STGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIVPNSEEVLKVARELSIIDSISS 656
GEN+EP LE +S I+ I+++GQD++ LGA+IVPN + + K A + S S
Sbjct: 505 RGGENIEPEPLERVLGKSLFIENIMIVGQDQKFLGAVIVPNFDNLEKWANSSGVSFSSRS 564
Query: 657 NVVSEEKVLNLIYKE----LKTWMSESPFQ-IGPILLVNEPFTIDNGLMTPTMKIRRDRV 711
++++ E V L K + T + F+ I +L+ +PFTI L T T+K++R +
Sbjct: 565 DLLANEDVNKLYSKHISDTINTKLGFKNFEKIVGFVLLQDPFTIGEEL-TNTLKLKRYYI 623
Query: 712 VAKYKEQI 719
KY+++I
Sbjct: 624 SKKYEDKI 631
>UniRef100_Q660S9 Long-chain-fatty-acid CoA ligase [Borrelia garinii]
Length = 645
Score = 341 bits (875), Expect = 4e-92
Identities = 220/608 (36%), Positives = 352/608 (57%), Gaps = 34/608 (5%)
Query: 122 AEGLRVIGVRPDEKLALFADNSCRWLVADQGMMASGAINVVRGSRSSVEELLQIYNHSES 181
A GL G++ EK+ + +D+ W++ D + G ++V RG+ SS +EL I NHSES
Sbjct: 48 ASGLLHYGIKRGEKVVIISDSRREWVIIDIAALGLGCVDVPRGNDSSEDELTYIINHSES 107
Query: 182 VALAVDGPEMFNRIAKPFYSKTGMRFIILLWGEKSDLNLIAEENKEVPIFSFMEVIDLGR 241
+ V+ + ++ + +++I+++ +KS E+ + +FS+ ++++LG
Sbjct: 108 TFIFVENNKQLQKVLSKKHDLRLVKYIVVIDDDKS----YEEKMGTITVFSYKKLLELGA 163
Query: 242 ESRMALSDSHEASQRYVYEAINSDDIATLIYTSGTTGNPKGVMLTHRNLLHQIKNLWDTV 301
E A S + E +S DIAT+IYTSGTTG PKGVML H + + Q+ L+D +
Sbjct: 164 EHLRANPKSFDIE----IEKGSSKDIATIIYTSGTTGMPKGVMLRHESFIFQLDRLYDYL 219
Query: 302 PA-EVGDRFLSMLPPWHAYERACEYFIFTCGIEQVYTTVRN---LKDDLGRYQPHYMISV 357
PA + G +S+LP WH++ERACEY + G+ Y+ LKD L P +ISV
Sbjct: 220 PAIKPGKIMISILPLWHSFERACEYIVALKGVAIAYSKPIGPVLLKDFL-LLNPQMIISV 278
Query: 358 PLVFETLYSGIQKQISTSPPVRKLVALTFIRVSLAYMEYKRIYEGKCLTRNVKQPSIVNS 417
P ++E + GI K++S S ++KLV F+++ + Y + + G + ++P++ S
Sbjct: 279 PRIWEGIRIGIIKKVSESL-IKKLVFGGFLKIGIIYAKLNEKFLG--FSPVYRKPNLFIS 335
Query: 418 MLDCLWARIIATILFPIHLLAIKFVYSKIHSAIGLS-KAGISGGGSLPLEVDKFFEAIGV 476
+ L+ I ++FPI LL V+ KI +A+G + + G+SGGG+L VD FF+AIG+
Sbjct: 336 IFSKLFLFIGIILIFPIKLLGDILVFKKIKNALGQNFEFGVSGGGALVDYVDYFFKAIGI 395
Query: 477 KVQNGYGLTETSPVIAARRPRCNVIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVRG 536
KV GYGLTET PV++ RR + V +VG + E+KVV + G+VLP G KG L V+
Sbjct: 396 KVLEGYGLTETGPVLSVRRLKGPVARTVGPILPDVEYKVVGID-GKVLPYGEKGELWVKS 454
Query: 537 PPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVL 596
P +M+GY+K+ T++ L +DGW NTGDL + ++ I + GR+KDTIVL
Sbjct: 455 PQIMSGYFKDKAKTSEVLTEDGWFNTGDLVRLTINNE----------ISIVGRSKDTIVL 504
Query: 597 STGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIVPNSEEVLKVARELSIIDSISS 656
GEN+EP LE +S I+ I+++GQD++ LGA+IVPN + + K A I S S
Sbjct: 505 RGGENIEPEPLERVLGKSLFIENIMIVGQDQKFLGAVIVPNFDNLEKWANSSGISFSSRS 564
Query: 657 NVVSEEKVLNLIYKELK-TWMSESPF----QIGPILLVNEPFTIDNGLMTPTMKIRRDRV 711
+++ E V L K + T ++S F +I +L+ + F+I L T T+K++R +
Sbjct: 565 DLLVNEGVNKLYSKHISDTINTKSGFKNFEKIVGFVLLQDSFSIGEEL-TNTLKLKRYYI 623
Query: 712 VAKYKEQI 719
KY+++I
Sbjct: 624 SQKYEDKI 631
>UniRef100_Q8F9T4 Long-chain-fatty-acid CoA ligase [Leptospira interrogans]
Length = 685
Score = 337 bits (865), Expect = 6e-91
Identities = 236/710 (33%), Positives = 364/710 (51%), Gaps = 93/710 (13%)
Query: 78 WKVVPDIWRSSAEKYGDKVAL----VDQYHHPPSTITYNQLEQAILDYAEGLRVIGVRPD 133
+K + D+ S EKYGD+ D+ HP S YNQL + AE L +G++
Sbjct: 4 YKNLADMLIQSTEKYGDRPVFWSKGEDKEFHPTS---YNQLYDMGIALAEALIQLGLKAR 60
Query: 134 EKLALFADNSCRWLVADQGMMASGAINVVRGSRSSVEELLQIYNHSESVALAVDGPEMFN 193
E + + ADN W++ D + SGA NV RG+ + EL I NHSE+ + ++ +M
Sbjct: 61 EHVGVLADNRLEWILTDYAVQFSGAANVPRGTDVTESELEYILNHSEAKIVFIENDKMLE 120
Query: 194 RIAKPFYSKTGMRFIILLWGEKSDLNLIAEENKEVPIFSFMEVIDLGRESRMALSDSHEA 253
+ Y+K + K + +I +++ + ++ DL E R + +
Sbjct: 121 K-----YNKVKSKV------PKVETIIIMDKSSSAKGKNIHKIYDLVEEGRSLRAKGSKK 169
Query: 254 SQRYVYEAINSDDIATLIYTSGTTGNPKGVMLTHRNLLHQIKNLWDTVPAEVG--DRFLS 311
+++ + E I +D+ TLIYTSGTTG PKGVML H N++HQ+ + + E+ D LS
Sbjct: 170 AEKRI-EEIKPEDLFTLIYTSGTTGMPKGVMLMHSNMIHQMVYVVPMLLTEIKPTDSMLS 228
Query: 312 MLPPWHAYERACEYFIFTCGIEQVYTTVRNLKDDLGRYQPHYMISVPLVFETLYSGIQKQ 371
+LP WH +ER EY + GI+ YT V +L++DL + +P +M S P V+E +Y+ I +
Sbjct: 229 ILPIWHIFERINEYGAISSGIQTYYTKVADLRNDLAKAKPSFMASAPRVWENVYANIYNK 288
Query: 372 IS---TSPPVRKLVALTFIRVSLAYMEYKRIYEGKCLTRNVKQPSIVNSMLDCLWARIIA 428
I+ +PP+R+++ S Y +R G L + + +I+ S++ + + I+
Sbjct: 289 INDPKQTPPIRRVLFKLAYFFSKHYNASRRFLNG--LEVDYENRNIMKSLVIGIRSLIVL 346
Query: 429 TILFPIHLLAIK------------------------------------FVYSKIHSAI-G 451
+ P L AI V SKI +A G
Sbjct: 347 LLTGPFTLSAISILAYLTLPVYGIHLPNWLFFSLVGLGLVFNAKTLDTIVLSKIRAATGG 406
Query: 452 LSKAGISGGGSLPLEVDKFFEAIGVKVQNGYGLTETSPVIAARRPRCNVIGSVGHPVQHT 511
KA +SGGG+L VD FF IG+ V GYG+TETSPVI+ R +IGSVG V +
Sbjct: 407 RLKASLSGGGALQSHVDNFFNDIGMLVLEGYGMTETSPVISVRPFVKPIIGSVGFLVPKS 466
Query: 512 EFKVVDSETGEVLPP------------GSKGILKVRGPPVMNGYYKNPLATNQALDKDGW 559
E ++ E G VL G KGI+ V+GP VM GYYKNP T + + DGW
Sbjct: 467 EL-IIKDENGNVLTHINDQYEVLAGKLGQKGIVFVKGPQVMKGYYKNPEVTKKTI-VDGW 524
Query: 560 LNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQ 619
+NTGD+G+I N + + GRAKDT+VL GENVEP +E S I+Q
Sbjct: 525 MNTGDIGFI----------NFKKTLTLTGRAKDTVVLLGGENVEPVPIENKMDESPFIKQ 574
Query: 620 IVVIGQDKRRLGAIIVPNSEEVLKVARELSIIDSISSNVVSEEKVLNLIYKELKTWMS-- 677
+VIGQD++ LGAIIVP+ E + +E I S ++ KV++ KE++++ S
Sbjct: 575 SMVIGQDQKVLGAIIVPDMEHLGVWCKENGIDSSKIDEIIKNPKVIDFYKKEVRSYNSTK 634
Query: 678 ---ESPFQIGPILLVNEPFTIDNGLMTPTMKIRRDRVVAKYKEQIDDLYK 724
+S Q+ ++L +PF + + L T +K++R + KY ++I +Y+
Sbjct: 635 TGFKSFEQVQHVILAKKPFEVGDEL-TDLLKMKRHVITEKYSKEIKKIYE 683
>UniRef100_UPI0000322607 UPI0000322607 UniRef100 entry
Length = 522
Score = 333 bits (853), Expect = 1e-89
Identities = 203/536 (37%), Positives = 299/536 (54%), Gaps = 43/536 (8%)
Query: 81 VPDIWRSSAEKYGDKVALVDQYHHPPSTITYNQLEQAILDYAEGLRVIGVRPDEKLALFA 140
V +W A ++G A+ + P ++ +LEQ I A R GV+P + +ALFA
Sbjct: 27 VDAVWPWLAAEHGTITAVDAPHAAHPERFSFAELEQRIATAAAAFRSQGVQPGDVVALFA 86
Query: 141 DNSCRWLVADQGMMASGAINVVRGSRSSVEELLQIYNHSESVALAVDGPEMFNRIAKPFY 200
+NS RWLVADQG+M GA + VRG+ + VEEL I + AL V +++ R+
Sbjct: 87 ENSPRWLVADQGLMRCGAADAVRGASAPVEELRYILEDCNATALVVQNADLWRRLDLTAS 146
Query: 201 SKTGMRFIILLWGEKSDLNLIAEENKEVPIFSFMEVIDLGRESRMALSDSHEASQRYVYE 260
+ +R ++ L GE ++ L G E+ +A A Q+ V
Sbjct: 147 QRQRLRLVLQLEGEPAEGVL-------------------GWEAFLASG----AGQQTVTA 183
Query: 261 AINSDDIATLIYTSGTTGNPKGVMLTHRNLLHQIKNLWDTVPAEVGDRFLSMLPPWHAYE 320
+ + IAT++YTSGTTG PKGV LTH NLLHQ+++L + G LS+LP WHAYE
Sbjct: 184 SRDRRAIATVLYTSGTTGQPKGVPLTHANLLHQMQSLACVAHPKPGSPVLSVLPIWHAYE 243
Query: 321 RACEYFIFTCGIEQVYTTVRNLKDDLGRYQPHYMISVPLVFETLYSGIQKQISTSPPVRK 380
R+ Y+ +C Q YTT++ LK DL R +P M +VP ++E++ +G + + T P R+
Sbjct: 244 RSASYYFLSCACTQTYTTIKQLKKDLPRVKPIAMATVPRLWESVQAGFEDVLKTFPASRQ 303
Query: 381 LVALTFIRVSLAYMEYKRIYEGKCLTRNVKQPSIVNSMLDCLWARIIATILFPIHLLAIK 440
+ +R +LA +R + RN+ + S+ + A A + +P+H LA
Sbjct: 304 RL----LRAALANSAAQR--KALRTARNLLLQQV--SLSGRITAAATAVLRWPLHALASA 355
Query: 441 FVYSKIHSAI--GLSKAGISGGGSLPLEVDKFFEAIGVKVQNGYGLTETSPVIAARRPRC 498
++ K+ + G ISGGG++ +D FFEA+G+++ GYGLTETSPV++ RRP
Sbjct: 356 LIWPKLRLQLSGGQLAYPISGGGAIAPHIDAFFEAVGIELLVGYGLTETSPVVSCRRPWR 415
Query: 499 NVIGSVGHPVQHTEFKVVDSETGEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDG 558
N+ GS G P+ TEF++VD E+G L +G + VRGP VM GY P A+++ L DG
Sbjct: 416 NIRGSSGLPMPDTEFRIVDPESGVALGFREQGRVLVRGPQVMGGYLGKPEASSKVLSADG 475
Query: 559 WLNTGDLGWIAPHHSTGRSRNSSGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRS 614
W +TGDLG + P G + + GRAKDTIVLS+GEN+EP LEEA + S
Sbjct: 476 WFDTGDLGMLLP----------DGSVALTGRAKDTIVLSSGENIEPGPLEEALVAS 521
>UniRef100_UPI0000323D49 UPI0000323D49 UniRef100 entry
Length = 524
Score = 327 bits (837), Expect = 1e-87
Identities = 211/558 (37%), Positives = 302/558 (53%), Gaps = 51/558 (9%)
Query: 175 IYNHSESVALAVDGPEMFNRIAKPFYSKTGMRFIILLWGEKSDLNLIAEENKEVPIFSFM 234
I HS SV L V E++ ++ K ++FII L E+ +
Sbjct: 3 IIEHSNSVGLIVQSKEIWLKLNNKEELKKRLKFIINLEDEQFE----------------- 45
Query: 235 EVIDLGRESRMALSDSHEASQRYVYEAINS--DDIATLIYTSGTTGNPKGVMLTHRNLLH 292
L S+ S+ E SQ Y +E N DD+AT++YTSGTTG PKGV LTH N LH
Sbjct: 46 ---SLISWSKFISSEEKENSQNYNFEKFNPEIDDVATILYTSGTTGKPKGVPLTHANFLH 102
Query: 293 QIKNLWDTVPAEVGDRFLSMLPPWHAYERACEYFIFTCGIEQVYTTVRNLKDDLGRYQPH 352
QI NL E G LS+LP WH+YER+ EYF F+CG Q YT + LKDD+ + +P
Sbjct: 103 QIINLAYIADPEPGTSVLSVLPIWHSYERSAEYFFFSCGCSQYYTIPKFLKDDITQIKPV 162
Query: 353 YMISVPLVFETLYSGIQKQISTSP-PVRKLVALTFIRVSLAYMEYKRIYEGKCLTRNVKQ 411
M +VP ++E ++ G + + P +KL+ S+ ++I RN+
Sbjct: 163 VMATVPRLWEAIHDGFFQALKKMPFKKQKLIKFLISNSSVFKRSLRKI-------RNIDI 215
Query: 412 PSIVNSMLDCLWARIIATILFPIHLLAIKFVYSKIHSAIGLSKAG--ISGGGSLPLEVDK 469
I L +I+ +P+H L+ F++ I + K I+GGG+LP VD
Sbjct: 216 NQITFKSKIPLMGSVISR--YPLHKLSTIFLWPNILRQLCGEKLKFPINGGGALPEHVDL 273
Query: 470 FFEAIGVKVQNGYGLTETSPVIAARRPRCNVIGSVGHPVQHTEFKVVDSETGEVLPPGSK 529
FFE++GV V GYGLTETSPV+ RR NV GS G P+ TE K+V+ + ++L
Sbjct: 274 FFESLGVDVLVGYGLTETSPVLTCRRRELNVRGSSGQPLSFTEIKIVNDDKKKILKFREV 333
Query: 530 GILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNSSGVIVVDGR 589
G + VRGP VM GY N +ATN L KDGW +TGDLG++ P +G + + GR
Sbjct: 334 GKILVRGPQVMKGYLNNAIATNDVLSKDGWFDTGDLGFLIP----------NGSLFITGR 383
Query: 590 AKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIVPNSEEVLK--VARE 647
AKDTIVLS+GEN+EP LE + S I QI ++GQDK+ L A++VPN E V + +
Sbjct: 384 AKDTIVLSSGENIEPNPLETEILSSEFINQIQLVGQDKKCLTALVVPNVELVKSKFLEED 443
Query: 648 LSIID-SISSNVVSEEKVLNLIYKELKTWMSESPFQIGPILLVNEPFTIDNGLMTPTMKI 706
LS ++ + + ++ NL+ L + S QI V + FT++NGL+T T+K
Sbjct: 444 LSKLNLNKKIGTFFKSQINNLLKSRLG---ARSEEQILDCYFV-DAFTLENGLLTQTLKQ 499
Query: 707 RRDRVVAKYKEQIDDLYK 724
+R + KY QI+++Y+
Sbjct: 500 KRKEIEKKYSLQIENMYE 517
>UniRef100_Q73LX0 AMP-binding enzyme family protein [Treponema denticola]
Length = 641
Score = 320 bits (820), Expect = 9e-86
Identities = 207/629 (32%), Positives = 334/629 (52%), Gaps = 35/629 (5%)
Query: 108 TITYNQLEQAILDYAEGLRVIGVRPDEKLALFADNSCRWLVADQGMMASGAINVVRGSRS 167
+++Y +L + LD+A GL IG RP + + L ADN WL A G+M GA +V RG +
Sbjct: 36 SLSYKELYETALDFAAGLLSIGARPKDHIGLIADNRKEWLHASFGIMNIGAADVPRGCDA 95
Query: 168 SVEELLQIYNHSESVALAVDGPEMFNRIAKPFYSKTGMRFIILLWGEKSDLNLIAEE--- 224
+ +E+ I + SE ++ ++ + II + + D + EE
Sbjct: 96 TEQEITHILSFSECKFAVLENEAQIRKVLVHLAEIPLLESIISI--DNVDFKKLEEEFNL 153
Query: 225 -NKEVPIFSFMEVIDLGRESRMALSDSHEASQRYVYEAINSDDIATLIYTSGTTGNPKGV 283
+++ ++ ++IDLG+ +R+ + + Y E +++DD+A++I+TSGTTGNPKGV
Sbjct: 154 KERKIEFHTYADIIDLGKSARI---EGKFKPEEYA-ENVDTDDLASIIFTSGTTGNPKGV 209
Query: 284 MLTHRNLLHQIKNLWDTVPAEVGDRFLSMLPPWHAYERACEYFIFTCGIEQVYTTVRN-- 341
+THRN + Q+ L D + + G + +S+LP WH++ERACEY I G Y+
Sbjct: 210 TMTHRNFMTQLMELPDRIILKPGQKAISVLPVWHSFERACEYVIIISGGTIAYSKPIGSV 269
Query: 342 LKDDLGRYQPHYMISVPLVFETLYSGIQKQISTSPPVRKLVALTFIRVSLAYMEYKRIYE 401
L D+ + P SVP ++E +Y GI K + + L FI V + M KR
Sbjct: 270 LLADMVKINPTLFPSVPRIWEAVYDGIFKAMKKRGRPLYYLFLFFIEVGIKTMRLKRRVT 329
Query: 402 GKCLTRNVKQPSIVNSMLDCLWARIIATILFPIHLLAIKFVYSKIHSAIG-LSKAGISGG 460
G+C ++ ++ +L L IA P++ + +Y I G +AG+SGG
Sbjct: 330 GQC-PHFQRKTKVIYPILAVLPLLCIA----PLYYIGDLLIYRAIRKKFGKCFRAGVSGG 384
Query: 461 GSLPLEVDKFFEAIGVKVQNGYGLTETSPVIAARRPRCNVIGSVGHPVQHTEFKVVDSET 520
G+LP VD+FF AI + V GYG+TET+PVI+ R V G++G P+ E K++D +
Sbjct: 385 GALPPNVDEFFWAIRINVMEGYGITETAPVISVRPMPKPVFGTLGKPLACFESKIID-KN 443
Query: 521 GEVLPPGSKGILKVRGPPVMNGYYKNPLATNQALDKDGWLNTGDLGWIAPHHSTGRSRNS 580
G+ LP KG+L VRG V GYYK+P T + +DKDGW +TGDL +
Sbjct: 444 GKELPHNRKGLLLVRGDAVTKGYYKDPERTAEVIDKDGWFDTGDLA----------IKTV 493
Query: 581 SGVIVVDGRAKDTIVLSTGENVEPAELEEAAMRSSIIQQIVVIGQDKRRLGAIIVPNSEE 640
G +++ GR K+TIVL GEN+EP +E S +I VV+GQD+R LGA+IV + E
Sbjct: 494 DGELILKGRKKNTIVLRGGENIEPVPIEVKLQESPLISLAVVLGQDQRSLGALIVVHKEN 553
Query: 641 VLKVARELSIIDSISSNVVSEEKVLNLIYKELKTWM-SESPF----QIGPILLVNEPFTI 695
+ A + + + ++ + V + E+ + S++ F +I I+L+ E F
Sbjct: 554 LQSWAANNGLRNVPVTELIHDSNVQKMYEAEVAELVNSKTGFKLFERINKIVLLPEEFQA 613
Query: 696 DNGLMTPTMKIRRDRVVAKYKEQIDDLYK 724
L + ++ R ++ Y+ +I +L+K
Sbjct: 614 GREL-SAKGEMMRHKINQLYRNEIYELFK 641
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,205,446,775
Number of Sequences: 2790947
Number of extensions: 51019942
Number of successful extensions: 201089
Number of sequences better than 10.0: 5663
Number of HSP's better than 10.0 without gapping: 4170
Number of HSP's successfully gapped in prelim test: 1493
Number of HSP's that attempted gapping in prelim test: 183998
Number of HSP's gapped (non-prelim): 11944
length of query: 724
length of database: 848,049,833
effective HSP length: 135
effective length of query: 589
effective length of database: 471,271,988
effective search space: 277579200932
effective search space used: 277579200932
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 79 (35.0 bits)
Medicago: description of AC146585.9


