

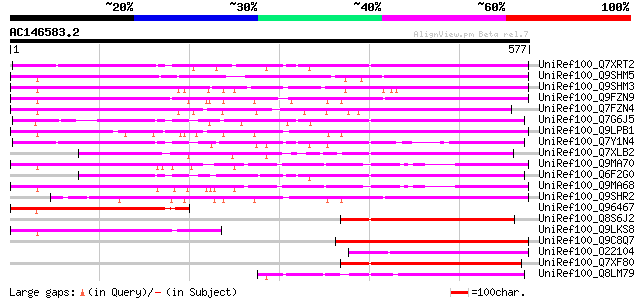
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146583.2 + phase: 0 /pseudo
(577 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XRT2 OSJNBa0042F21.11 protein [Oryza sativa] 325 2e-87
UniRef100_Q9SHM5 F7F22.15 [Arabidopsis thaliana] 293 7e-78
UniRef100_Q9SHM3 F7F22.17 [Arabidopsis thaliana] 281 4e-74
UniRef100_Q9FZN9 Retroelement pol polyprotein-like [Arabidopsis ... 280 6e-74
UniRef100_Q7FZN4 T17A2.3 protein [Arabidopsis thaliana] 277 5e-73
UniRef100_Q7G6J5 Putative polyprotein [Oryza sativa] 260 9e-68
UniRef100_Q9LPB1 T32E20.9 [Arabidopsis thaliana] 253 1e-65
UniRef100_Q7Y1N4 Putative retrotransposon gag protein [Oryza sat... 246 1e-63
UniRef100_Q7XLB2 OSJNBa0011K22.17 protein [Oryza sativa] 230 7e-59
UniRef100_Q9MA70 F2J6.11 protein [Arabidopsis thaliana] 227 8e-58
UniRef100_Q6F2G0 Hypothetical protein OSJNBa0083K01.26 [Oryza sa... 225 2e-57
UniRef100_Q9MA68 F2J6.13 protein [Arabidopsis thaliana] 223 1e-56
UniRef100_Q9SHR2 F28L22.6 protein [Arabidopsis thaliana] 167 6e-40
UniRef100_Q96467 Hypothetical protein [Hordeum vulgare] 162 2e-38
UniRef100_Q8S6J2 Hypothetical protein OSJNBa0019N10.34 [Oryza sa... 153 1e-35
UniRef100_Q9LKS8 Hypothetical protein T15F17.m [Arabidopsis thal... 150 7e-35
UniRef100_Q9C8Q7 Athila ORF 1, putative; 43045-40843 [Arabidopsi... 150 7e-35
UniRef100_O22104 Vicia faba mRNA expressed from retrotransposon-... 150 1e-34
UniRef100_Q7XF80 Hypothetical protein [Oryza sativa] 149 3e-34
UniRef100_Q8LM79 Hypothetical protein OSJNAa0082N11.11 [Oryza sa... 148 4e-34
>UniRef100_Q7XRT2 OSJNBa0042F21.11 protein [Oryza sativa]
Length = 920
Score = 325 bits (834), Expect = 2e-87
Identities = 218/636 (34%), Positives = 324/636 (50%), Gaps = 79/636 (12%)
Query: 4 FSGSPTEDPNLHISSFLRLSG--TIKE-NQEAVRLHLFPFSLRDRASAWFHSLEVGSITS 60
F G P ED N H+ FL + TIK + +AVRL LFPFSL RA WF++ +I +
Sbjct: 268 FCGKPNEDANAHLLQFLEICSMYTIKGVSPDAVRLRLFPFSLLRRAKQWFYA-NCAAINT 326
Query: 61 WDDMRRAFLARFFPPSKTAKLRDQITRFNQKDGESLYEAWEHFKEMLRLCPHHGLEKWLI 120
WD +FL++FFP KT LR +I+RF Q ES+ EAWE +E + CPHHG++ WLI
Sbjct: 327 WDKCSTSFLSKFFPIGKTNALRGRISRFQQTRDESIPEAWERLQEYVAACPHHGMDDWLI 386
Query: 121 VHTFYNGLSDTTKMSVDAAAGGALMNKNYTEAYALIEDMAQNHYQWTNERAVTTPTPSKK 180
+ FYNGL+ ++ +DAAAGGA +K A LIE M N W+ ER T +
Sbjct: 387 LQNFYNGLTLMSRDHLDAAAGGAFFSKTVQGAVELIEKMVSNT-GWSKERLQT------R 439
Query: 181 EAGIHEVSEYNHLAAKVEQLNYA------------------------------------Q 204
+ G+H V E LAAK++ L
Sbjct: 440 QRGMHTVKETELLAAKLDLLMKCLDDHEKRPQGTVKALDSHVTCEVCGGTGHSGNDCLET 499
Query: 205 YKQGMRPNQNFYKNPQGSYGQTAP----PGYTNNQRVAQKSSLEILLENCMMNQNKQLQE 260
+++ M +N PQG G P PG NN + + SL L+ Q K
Sbjct: 500 HEEAMYMGKNNEYRPQGGEGWNQPRPYYPGGNNNGNFSNQPSLMDLV----FAQAKTTDA 555
Query: 261 LKNQTGSLNDSLSKLNTKVDSIAT-------HTKMLETQISQVAQQVAISS*TPGVFPGQ 313
L+ + + + L +N K+D A+ KM+ETQ++Q+A V + G PGQ
Sbjct: 556 LRKKLAANDKILENINVKLDGFASAFQNQLSFNKMIETQLAQLASLVPANE--SGRIPGQ 613
Query: 314 TETNPKAHVNAISLGGNK------------LEDTITKAKSVKGESVKLLGEKDAIKTLLD 361
+++ + +V AI+ G K + S + ++ +K + D
Sbjct: 614 PDSSIE-NVKAITTRGGKSTRDPPYPNPAGTNGMSKETPSTDSDDKEIQPDKTVPQEYCD 672
Query: 362 KNKALNPLRLTKLNLEAQFAKFLNILKRICIKIPFGEALSRMPLYTKFLKEIFSKKKAMD 421
P + K +++ QFA F+ ++++I I +P +A+ ++P Y ++LK+I + K+ +
Sbjct: 673 TRLLPFPQQSRKPSVDKQFACFVEVIQKIHINVPLLDAM-QVPTYARYLKDILNNKRPLP 731
Query: 422 HKETIALTRE-SSAIVKKQPPKLRDPGSFVIPCVIGKETVDKALCDLGASVSLLPLSLFK 480
E + LT S+ I+ K P K +DPG I C IG + D+ALCDLGASVS++P +F
Sbjct: 732 TTEVVKLTEHCSNLILHKLPEKKKDPGCPTITCSIGAQQFDQALCDLGASVSVMPKDVFD 791
Query: 481 RMGIGELKPTEMILKLADRSTIPLVGYIEDIPVKIEGIYIPTDFVVVDIEEDHDVPIILG 540
++ L PT M L+LAD S VG ED+P KI +IP DFVV+D++ + P+ILG
Sbjct: 792 KLNFTVLAPTPMRLQLADSSVHYPVGIAEDVPAKIRDFFIPVDFVVLDMDTGKETPLILG 851
Query: 541 RPFLATAGAIVDVQSGRIVFQASDAMIGFELENVMK 576
RPFL+TAGA +DV +G I F + FE + M+
Sbjct: 852 RPFLSTAGANIDVGTGSIRFHTNGKEEKFEFQPRME 887
>UniRef100_Q9SHM5 F7F22.15 [Arabidopsis thaliana]
Length = 1862
Score = 293 bits (751), Expect = 7e-78
Identities = 205/614 (33%), Positives = 316/614 (51%), Gaps = 66/614 (10%)
Query: 2 NQFSGSPTEDPNLHISSFLRLSGTIKEN---QEAVRLHLFPFSLRDRASAWFHSLEVGSI 58
N+F G P EDP H+ F RL K N ++ +L LFPFSL D+A W +L SI
Sbjct: 170 NKFHGLPMEDPLDHLDEFDRLCNLTKINGVSEDGFKLRLFPFSLGDKAHIWEKNLPHDSI 229
Query: 59 TSWDDMRRAFLARFFPPSKTAKLRDQITRFNQKDGESLYEAWEHFKEMLRLCPHHGLEKW 118
T+WDD ++AFL++FF ++TA+LR++I+ F+QK GES EAWE FK C HHG K
Sbjct: 230 TTWDDCKKAFLSKFFSNARTARLRNEISGFSQKTGESFCEAWERFKGYTNQCSHHGFTKA 289
Query: 119 LIVHTFYNGLSDTTKMSVDAAAGGALMNKNYTEAYALIEDMAQNHYQWTNERAVTTPTPS 178
++ T Y G+ +M +D A+ G NK+ E + L+E++AQ++ + NE T +
Sbjct: 290 SLLSTLYRGVLPRIRMLLDTASNGNFQNKDVEEGWELVENLAQSNGNY-NENCDRTVRGT 348
Query: 179 KKEAGIHEVSEYNHLAAKVEQLNYAQYKQ-GMRPNQNFYKNPQGSYGQTAPPGYTNNQRV 237
H E L K++++ +Q+K + Y+ G Q Y NN +
Sbjct: 349 ADSDDKHR-KEIKALNDKLDRILLSQHKHVHFLVDDEQYEVQDGEGNQLEEVSYINNNQG 407
Query: 238 AQKSSLEILLENCMMNQNKQLQELKNQTGSLNDSLSKLNTKVDSIATHTKMLETQISQVA 297
K N + N +LS +T V + + Q SQ
Sbjct: 408 GYKG--------------------YNNFKTNNPNLSYRSTNVANPQDQVYPPQQQQSQNK 447
Query: 298 QQVAISS*T--PGVFPGQTETNPKAHVNAISLGGNKLEDTITKAKSVKGESVKLLGEKDA 355
V + T PG+ NPK + +AI+L K T + K+V +S GE +
Sbjct: 448 PFVPYNQATKQTSQLPGKAVQNPKEYAHAITLHSGKALPTREEPKTVTEDSEDQDGEDLS 507
Query: 356 IKTLLDKNKALNPLRLT-----KLNLEAQFAKFLNILKR--------------------- 389
+K K++A PL L+ L+L+ L+ R
Sbjct: 508 LK----KDQADKPLDLSLEQPLDLSLQQSLDPPLDSFTRPTTRPVIPAASPTAPKPVAVK 563
Query: 390 ----ICIKIPFGEALSRMPLYTKFLKEIFSKKKAMDHKETIALTRESSAIVKKQ--PPKL 443
+ ++IP +AL+ +P KFLK++ ++ + + + L+ E SAI++K+ P KL
Sbjct: 564 NKEKVELRIPLVDALALIPDSHKFLKDLI-VERIQEVQGMVVLSHECSAIIQKKIIPKKL 622
Query: 444 RDPGSFVIPCVIGKETVDKALCDLGASVSLLPLSLFKRMGIGELKPTEMILKLADRSTIP 503
DPGSF +PC +G ++ LCDLGASVSL+PLS+ KR+G + K + L LADRS
Sbjct: 623 SDPGSFTLPCSLGPLAFNRCLCDLGASVSLMPLSVAKRLGFTQYKSCNISLILADRSVRI 682
Query: 504 LVGYIEDIPVKIEGIYIPTDFVVVDIEEDHDVPIILGRPFLATAGAIVDVQSGRIVFQ-A 562
G +E++P++I + IPTDFVV++++E+ P+ILGRPFLATAGA++DV+ G+I
Sbjct: 683 PHGLLENLPIRIGAVEIPTDFVVLEMDEEPKDPLILGRPFLATAGAMIDVKKGKIDLNLG 742
Query: 563 SDAMIGFELENVMK 576
D + F++++ MK
Sbjct: 743 KDFRMTFDVKDAMK 756
>UniRef100_Q9SHM3 F7F22.17 [Arabidopsis thaliana]
Length = 1799
Score = 281 bits (719), Expect = 4e-74
Identities = 221/727 (30%), Positives = 341/727 (46%), Gaps = 165/727 (22%)
Query: 2 NQFSGSPTEDPNLHISSFLRLSGTIKEN---QEAVRLHLFPFSLRDRASAWFHSLEVGSI 58
N+F G P EDP H+ F RL K N ++ +L LFPFSL D+A W +L SI
Sbjct: 35 NKFHGLPMEDPLDHLDEFDRLCNLTKINGVSEDGFKLRLFPFSLGDKAHIWEKNLPHDSI 94
Query: 59 TSWDDMRRAFLARFFPPSKTAKLRDQITRFNQKDGESLYEAWEHFKEMLRLCPHHGLEKW 118
T+WDD ++ FL++FF ++TA+LR++I+ F+QK GES EAWE FK CPHHG K
Sbjct: 95 TTWDDCKKVFLSKFFSNARTARLRNEISGFSQKTGESFCEAWERFKGYTNQCPHHGFTKA 154
Query: 119 LIVHTFYNGLSDTTKMSVDAAAGGALMNKNYTEAYALIEDMAQN--HYQWTNERAV--TT 174
++ T Y G+ +M +D A+ G NK+ E + L+E++AQ+ +Y +R V T
Sbjct: 155 SLLSTLYRGVLPRIRMLLDTASNGNFQNKDVEEGWELVENLAQSDGNYNEDCDRTVRGTA 214
Query: 175 PTPSKKEAGIHE---------VSEYNHL---------------AAKVEQLNYAQYKQGMR 210
+ K I +S++ H+ ++E+++Y QG
Sbjct: 215 DSDDKHRKEIKALNDKLDRILLSQHKHVHFLVDDEQYEVQDGEGNQLEEVSYINNNQGGY 274
Query: 211 PNQNFYK--NP------------------------------------------QGSYGQT 226
N++K NP QG+Y Q
Sbjct: 275 KGYNYFKTNNPNLSYRSTNVANPQDQVYPPQQQQSQNKPFVPYNQGFVPKQQFQGNY-QQ 333
Query: 227 APPGYTNNQR-------VAQKSSLEILLE---NCMMNQNKQLQELKNQTGSLNDSLSKLN 276
PPG+ Q K L+ LL +C M K++ EL N+ L+ S + LN
Sbjct: 334 QPPGFAPPQHQGPAAPDADMKQMLQQLLHGQASCSMEMAKKISELHNK---LDCSYNDLN 390
Query: 277 TKVDSIATHTKMLETQISQVAQQVAISS*TPGVFPGQTETNPKAHVNAISLGGNKLEDTI 336
K++++ T + LE + + S PG+ NPK + +AI+L K T
Sbjct: 391 VKMETLDTKVRYLEGHSTSSSATKQTSQ-----LPGKAVQNPKEYAHAITLRSGKALPTR 445
Query: 337 TKAKSVKGESVKLLGEKDAIKTLLDKNKALNPLRLT-----KLNLEAQFAKFLNILKRIC 391
+ K+V +S ++D L+K++A PL L+ L+L+ L+ R
Sbjct: 446 EEPKTVTEDS----EDQDGEDLSLEKDQADKPLDLSLEQPLDLSLQQSLDPPLDSFTRPT 501
Query: 392 IK--IPFGEALSRMPLYTKFLKEIF-------------SKKKAMDHK------------- 423
+ IP + P+ K +++F KKA+ K
Sbjct: 502 TRPVIPAASPTAPKPVAVKNKEKVFVPPPYKSQLPFPGRHKKALADKYRAMFAKNIKEVE 561
Query: 424 ------ETIAL-------------------------TRESSAIVKKQ--PPKLRDPGSFV 450
+ +AL + E SAI++K+ P KL DPGSF
Sbjct: 562 LRIPLVDALALIPDSHKFLKDLIVERIQEVQGMVVLSHECSAIIQKKIIPKKLSDPGSFT 621
Query: 451 IPCVIGKETVDKALCDLGASVSLLPLSLFKRMGIGELKPTEMILKLADRSTIPLVGYIED 510
+PC +G ++ LCDLGASVSL+PLS+ KR+G + K + L LADRS G +E+
Sbjct: 622 LPCSLGPLAFNRCLCDLGASVSLMPLSVAKRLGFTQYKSCNISLILADRSVRIPHGLLEN 681
Query: 511 IPVKIEGIYIPTDFVVVDIEEDHDVPIILGRPFLATAGAIVDVQSGRIVFQ-ASDAMIGF 569
+P++I + IPTDFVV++++E+ P+ILGRPFLATAGA++DV+ G+I D + F
Sbjct: 682 LPIRIGAVEIPTDFVVLEMDEEPKDPLILGRPFLATAGAMIDVKKGKIDLNLGKDFRMTF 741
Query: 570 ELENVMK 576
++++ MK
Sbjct: 742 DVKDAMK 748
>UniRef100_Q9FZN9 Retroelement pol polyprotein-like [Arabidopsis thaliana]
Length = 1864
Score = 280 bits (717), Expect = 6e-74
Identities = 211/709 (29%), Positives = 331/709 (45%), Gaps = 145/709 (20%)
Query: 2 NQFSGSPTEDPNLHISSFLRLSGTIKEN---QEAVRLHLFPFSLRDRASAWFHSLEVGSI 58
N+F G P EDP H+ F RL K N ++ +L LFPFSL D+A W SL GSI
Sbjct: 84 NKFHGLPMEDPLDHLDEFDRLCSLTKINGVSEDGFKLRLFPFSLGDKAHQWEKSLLQGSI 143
Query: 59 TSWDDMRRAFLARFFPPSKTAKLRDQITRFNQKDGESLYEAWEHFKEMLRLCPHHGLEKW 118
TSW+D ++AFLA+FF S+TA+LR+ I+ F Q + E+ EAWE FK CPHHG K
Sbjct: 144 TSWNDCKKAFLAKFFSNSRTARLRNDISGFTQTNNETFCEAWERFKGYQTQCPHHGFSKA 203
Query: 119 LIVHTFYNGLSDTTKMSVDAAAGGALMNKNYTEAYALIEDMAQN--HYQWTNERAVTTPT 176
++ T Y G+ +M +D A+ G +NK+ + + L+E++AQ+ +Y +R+V T +
Sbjct: 204 SLLSTLYRGVLPKIRMLLDTASNGNFLNKDVEDGWELVENLAQSDGNYNEDYDRSVRTSS 263
Query: 177 PSKKEAGIHEVSEYNHLAAKV---------------------------EQLNYAQYKQGM 209
S E E+ N K+ E+++Y Q + G
Sbjct: 264 DS-DEKHRREMKAMNDKLDKLLLVQQKHIHFLGDDETFQVQDGETMQSEEVSYVQNQGGY 322
Query: 210 RPNQNFYK--------------NPQ-------------------------------GSY- 223
N +K NPQ G+Y
Sbjct: 323 NKGFNNFKQNHPNLSYRSTNVANPQDQVYPSQQQNQPRPFVPYNQGQGYVPKQQYQGNYQ 382
Query: 224 GQTAPPGYTNNQR----VAQKSSLEILLENCMMNQNKQLQELKNQTGSLND----SLSKL 275
Q PPG+T Q+ S L+ +L+ + Q +L + +++ S + +
Sbjct: 383 PQLPPPGFTQQQQQPASTTPDSDLKNMLQQILQGQATGAMDLSKRMAEIHNKVDCSYNDI 442
Query: 276 NTKVDSIATHTKMLETQISQVAQQVAISS*TPGVFPG---QTETNPKAHVNAISLGGNKL 332
N KV+++ + + +E Q A F G ++ +N + + +AI+L K
Sbjct: 443 NIKVEALTSKIRYIEGQTGSTAAP---------KFTGPSRKSMSNSEEYAHAITLRSGKE 493
Query: 333 EDTITKAKSVKGESVKLLGE-------------------------KDAIKTLLDKNKALN 367
T +SV GE A L++K A
Sbjct: 494 LPTKESPNQNTEDSVDQDGEDFCQNGNSAEKAIEEPILDQPTRLLAPAASPLVEKPAATK 553
Query: 368 -----------------PLRLTKLNLEAQFAKFLNILKRICIKIPFGEALSRMPLYTKFL 410
P R K+ ++ A LK + + +P + L+ +P K++
Sbjct: 554 TKDNVFVPPPYKPPLPFPGRFKKVMIQKYKALLEKQLKNLEVTMPLVDCLALIPDSNKYV 613
Query: 411 KEIFSKKKAMDHKETIALTRESSAIVKKQ--PPKLRDPGSFVIPCVIGKETVDKALCDLG 468
K++ + ++ + + + L+ E SAI++++ P KL DPGSF +PC +G +K LCDLG
Sbjct: 614 KDMIT-ERIKEVQGMVVLSHECSAIIQQKIIPKKLGDPGSFTLPCALGPLAFNKCLCDLG 672
Query: 469 ASVSLLPLSLFKRMGIGELKPTEMILKLADRSTIPLVGYIEDIPVKIEGIYIPTDFVVVD 528
ASVSL+PLS+ K++G + KP + L LADRS G +ED+PV I + +PTDFVV++
Sbjct: 673 ASVSLMPLSVAKKLGFNKYKPCNISLILADRSVRIPHGLLEDLPVMIGMVEVPTDFVVLE 732
Query: 529 IEEDHDVPIILGRPFLATAGAIVDVQSGRIVFQ-ASDAMIGFELENVMK 576
++E+ P+ILGRPFL T GAI+DV+ G+I D + F++ N MK
Sbjct: 733 MDEEPKDPLILGRPFLVTVGAIIDVKKGKIDLNLGRDLKMTFDITNTMK 781
>UniRef100_Q7FZN4 T17A2.3 protein [Arabidopsis thaliana]
Length = 1428
Score = 277 bits (709), Expect = 5e-73
Identities = 205/647 (31%), Positives = 311/647 (47%), Gaps = 102/647 (15%)
Query: 2 NQFSGSPTEDPNLHISSFLRLSGTIKEN---QEAVRLHLFPFSLRDRASAWFHSLEVGSI 58
N+F G P EDP H+ F RL K N ++ +L LFPFSL D+A W +L SI
Sbjct: 52 NKFHGLPMEDPLDHLDEFDRLCNLTKINGVSEDGFKLRLFPFSLGDKAHIWEKNLSHDSI 111
Query: 59 TSWDDMRRAFLARFFPPSKTAKLRDQITRFNQKDGESLYEAWEHFKEMLRLCPHHGLEKW 118
T+WDD ++AFL++FF ++TA+LR++I F+QK GES EAWE FK CPHH K
Sbjct: 112 TTWDDYKKAFLSKFFSNARTARLRNEIYGFSQKTGESFCEAWERFKGYTNQCPHHSFTKA 171
Query: 119 LIVHTFYNGLSDTTKMSVDAAAGGALMNKNYTEAYALIEDMAQN--HYQWTNERAV--TT 174
++ T Y G+ +M +D A+ G NK+ E + L+E++AQ+ +Y +R V T
Sbjct: 172 SLLSTLYRGVLPRIRMLLDTASNGNFQNKDVEEGWELVENLAQSDGNYNEDCDRTVRGTA 231
Query: 175 PTPSKKEAGIHE---------VSEYNHLAAKVEQLNY----------------------- 202
+ K I +S+ H+ V+ Y
Sbjct: 232 DSDDKHRKEIKALNDKLDRILLSQQKHVHFLVDDEQYQVQDGEGNQLEEVYLPQQKQGQN 291
Query: 203 ---AQYKQGMRPNQNFYKNPQGSYGQTAPPGYTNNQ---RVAQKSSLEILLENCMMNQNK 256
Y QG P Q F QG+Y PPG+ Q A + ++ +++ + Q
Sbjct: 292 KPFVLYNQGFVPKQQF----QGNYQPPPPPGFVTQQNQCHAAPDAKMKQMVKQLLQGQAS 347
Query: 257 QLQELKNQTGSLNDSL----SKLNTKVDSIATHTKMLETQISQVAQQVAISS*TPGV--F 310
E+ + L+ L + LN KV+++ T + LE Q + S+ +P V
Sbjct: 348 SSMEIAKKLSELHHKLDCSYNDLNAKVEALNTKVRYLEGQ--------SASTFSPKVTRL 399
Query: 311 PGQTETNPKAHVNAIS--------LGGNKLEDTITKAKSV-KGESVKLL----------- 350
PG++ NPK + A + L + D IT+ V +GE+ +
Sbjct: 400 PGKSIQNPKEYATAHAITICHDRELPTRHVLDLITRDNDVQEGEASTQVEASVVEFNHSA 459
Query: 351 ----------GEKDAIKTLLDKNKALNPLRLTKLN---LEAQFAKFLNI----LKRICIK 393
EK AI + K PL L L +A ++ ++ L I
Sbjct: 460 GSRHLTQSTSEEKAAIIERMVKRFKPTPLPLRALPWTFRKAWMERYKSVAAKQLDEIEAV 519
Query: 394 IPFGEALSRMPLYTKFLKEIFSKKKAMDHKETIALTRESSAIVKKQ--PPKLRDPGSFVI 451
+P E L+ + K ++ + ++ M H S K+ KL DPGSF +
Sbjct: 520 MPLMEVLNLILDPHKVVRNLILERIKMYHDSDDESDATPSRAADKRIVQEKLEDPGSFTL 579
Query: 452 PCVIGKETVDKALCDLGASVSLLPLSLFKRMGIGELKPTEMILKLADRSTIPLVGYIEDI 511
PC IG+ LCDLGASV+L+PLS+ +R+ + KP ++ L LADRS+ G ++D+
Sbjct: 580 PCSIGEFAFSDCLCDLGASVNLMPLSMARRLEFIQYKPCDLTLILADRSSRKHFGMLKDL 639
Query: 512 PVKIEGIYIPTDFVVVDIEEDHDVPIILGRPFLATAGAIVDVQSGRI 558
PV I G+ +PTDFVV+D+E +H P+ILGRPFLA+ GA++DV+ G+I
Sbjct: 640 PVMINGVEVPTDFVVLDMEVEHKDPLILGRPFLASVGAVIDVKEGKI 686
>UniRef100_Q7G6J5 Putative polyprotein [Oryza sativa]
Length = 689
Score = 260 bits (664), Expect = 9e-68
Identities = 188/605 (31%), Positives = 292/605 (48%), Gaps = 79/605 (13%)
Query: 4 FSGSPTEDPNLHISSFLRLSGTIK---ENQEAVRLHLFPFSLRDRASAWFHSLEVGSITS 60
F G P ED N H+ FL + T + + V+L LFPFSL RA WF++ ++ +
Sbjct: 45 FCGKPNEDANAHLQQFLEICSTYTIKGVSPDVVKLRLFPFSLLGRAKQWFYANRT-AVNT 103
Query: 61 WDDMRRAFLARFFPPSKTAKLRDQITRFNQKDGESLYEAWEHFKEMLRLCPHHGLEKWLI 120
WD AFL++FFP EAWE +E + CPH G++ WLI
Sbjct: 104 WDKCSTAFLSKFFP----------------------MEAWERLQEYVAACPHLGMDDWLI 141
Query: 121 VHTFYNGLSDTTKMSVDAAAGGALMNKNYTEAYALIEDMAQNHYQWTNERAVTTPTPSKK 180
+ FYNGL+ ++ +DAAA GA +K A LIE M N W+ ER T +
Sbjct: 142 LQNFYNGLTPMSRDHLDAAARGAFFSKTVQGAVELIEKMVSN-MGWSEERLQT------R 194
Query: 181 EAGIHEVSEYNHLAAKVEQLNYAQYKQGMRPNQNFYKNPQ------GSYGQTAPPGYTNN 234
+ G+H V E LAAK++ L M+ + K PQ S+ G T +
Sbjct: 195 QRGMHTVKETELLAAKLDLL--------MKRLDDHEKGPQRTVKALDSHVMCEVCGNTGH 246
Query: 235 QRVAQKSSLEILLENCMMNQNK----------QLQELKNQTGSLNDSLSKLNTKVDSIAT 284
+ LE E M N Q G+ N + + D +
Sbjct: 247 ---SGNDCLETCEEAMYMGNNNGYCPQGGQGWNQPRPYYQGGNNNSNFPNQPSLKDLVFA 303
Query: 285 HTKMLETQISQVAQQVAISS*TP----GVFPGQTETNPKAHVNAISLGGNKLE------- 333
K + ++A ++S P G PGQ + + + +V AI++ K
Sbjct: 304 QAKTTDALSKKLAANDKLASLVPANETGRIPGQPDPSIE-NVKAITMRRGKSTRDPPYPN 362
Query: 334 ----DTITKAKSVKGESVKLLGEKDAIKTLLDKNKALN-PLRLTKLNLEAQFAKFLNILK 388
+ I+K + K + ++ + + L+ P R+ K +++ QFA+F+ +++
Sbjct: 363 PVGTNEISKGAPSNDSADKEVQPENTVPQEYCDTRLLSFPQRMRKPSVDEQFARFVEVIQ 422
Query: 389 RICIKIPFGEALSRMPLYTKFLKEIFSKKKAMDHKETIALTRE-SSAIVKKQPPKLRDPG 447
+I I +P +A+ ++P Y ++LK+I + K+ + E + LT + S+ I+ K P K +DPG
Sbjct: 423 KIHINVPLLDAM-QVPTYARYLKDILNNKRPLPTTEVVKLTEQCSNLILHKLPEKKKDPG 481
Query: 448 SFVIPCVIGKETVDKALCDLGASVSLLPLSLFKRMGIGELKPTEMILKLADRSTIPLVGY 507
I C IG + D+ C+LGASVS++ +F ++ L PT M L+LAD S G
Sbjct: 482 CPTITCSIGAQQFDQVSCNLGASVSVVLKDVFDKLNFTVLAPTPMRLQLADSSVRYPAGI 541
Query: 508 IEDIPVKIEGIYIPTDFVVVDIEEDHDVPIILGRPFLATAGAIVDVQSGRIVFQASDAMI 567
ED+PVKI +IP DFVV+D++ + P+ILGRPFL+TAGA +DV +G I F +
Sbjct: 542 AEDVPVKIRDFFIPVDFVVLDMDTGKETPLILGRPFLSTAGANIDVGTGSIRFHINGKKE 601
Query: 568 GFELE 572
FE +
Sbjct: 602 KFEFQ 606
>UniRef100_Q9LPB1 T32E20.9 [Arabidopsis thaliana]
Length = 1586
Score = 253 bits (645), Expect = 1e-65
Identities = 202/695 (29%), Positives = 318/695 (45%), Gaps = 136/695 (19%)
Query: 2 NQFSGSPTEDPNLHISSFLRLSGTIKEN---QEAVRLHLFPFSLRDRASAWFHSLEVGSI 58
N+F G P EDP H+ F RL K N ++ +L LFPFSL D+A W SL GSI
Sbjct: 84 NKFHGLPMEDPLDHLDEFDRLCSLTKINRVSEDGFKLRLFPFSLGDKAHQWEKSLPQGSI 143
Query: 59 TSWDDMRRAFLARFFPPSKTAKLRDQITRFNQKDGESLYEAWEHFKEMLRLCPHHGLEKW 118
TSW+D ++AFLA+FF S+TA+LR+ I+ F Q + E+ YEAWE FK CPHH +
Sbjct: 144 TSWNDCKKAFLAKFFSNSRTARLRNDISGFTQTNNETFYEAWERFKGYQTQCPHHEM--- 200
Query: 119 LIVHTFYNG------LSDTTKMSVDAAAGGALMNKNYTEAYALIED-------------- 158
++ T NG + D ++ + A N++Y + D
Sbjct: 201 -LLDTASNGNFLNKDVEDGWEVVENLAQSDGNYNEDYDRSIRTSSDSDEKHRREMKAMND 259
Query: 159 -------MAQNHYQWTNERAVTTPTPSKKEAGIHEVS---------------EYNH---- 192
M Q H + + T + + EVS + NH
Sbjct: 260 KLDKLLLMQQKHIHFLGDDE-TLQVQDGETLQLEEVSYVQNQGGYNKGFNNFKQNHPNLS 318
Query: 193 -----LAAKVEQLNYAQYK------------QGMRPNQNFYKNPQGSYGQTAPP-GYTNN 234
+A + +Q+ +Q + QG P Q + QG+Y Q PP G+T
Sbjct: 319 YRSTNVANRQDQVYPSQQQNQPKPFVPYNQGQGYVPKQQY----QGNYQQQLPPPGFTQQ 374
Query: 235 QR----VAQKSSLEILLENCMMNQNKQLQELKNQTGSLND----SLSKLNTKVDSIATHT 286
Q+ S L+ +L+ + Q +L + +++ S + +N KV+++ +
Sbjct: 375 QQQPALTTPDSDLKNMLQQILQGQATGAMDLSKRMAEIHNKVDCSYNDINIKVEALTSKI 434
Query: 287 KMLETQISQVAQQVAISS*TPGVFPGQTETNPKAHVNAISLGGNKLEDTITKAKSVKGES 346
+ +E Q A G++ +N K + +AI+L K T +S
Sbjct: 435 RYIEGQTGSTAAPKFTGP------SGKSMSNSKEYAHAITLRSGKELPTKESPNQNTEDS 488
Query: 347 VKLLGEK-------------------------DAIKTLLDKNKALN-------------- 367
+ GE A L++K A
Sbjct: 489 LDQDGEDFCQNGNSAEKAIEEPILHQPTRPLAPAASPLVEKPAAAKTKENVFIPPPYKPP 548
Query: 368 ---PLRLTKLNLEAQFAKFLNILKRICIKIPFGEALSRMPLYTKFLKEIFSKKKAMDHKE 424
P R K+ ++ A LK + + +P + L+ +P K++K++ +++ + +
Sbjct: 549 LPFPGRFKKVMIQKYKALLEKQLKNLEVTMPLVDCLALIPDSNKYVKDMITER-IKEVQG 607
Query: 425 TIALTRESSAIVKKQ--PPKLRDPGSFVIPCVIGKETVDKALCDLGASVSLLPLSLFKRM 482
+ L+ E SAI++++ P KL DPGSF +PC +G K LCDLGASVSL+PL + K++
Sbjct: 608 MVVLSHECSAIIQQKIIPKKLGDPGSFTLPCALGPLAFSKCLCDLGASVSLMPLPVAKKL 667
Query: 483 GIGELKPTEMILKLADRSTIPLVGYIEDIPVKIEGIYIPTDFVVVDIEEDHDVPIILGRP 542
G + KP + L LADRS G +ED+PV I + +PTDFVV++++E+ P+ILGRP
Sbjct: 668 GFNKYKPCNISLILADRSVRISHGLLEDLPVMIGVVEVPTDFVVLEMDEEPKDPLILGRP 727
Query: 543 FLATAGAIVDVQSGRIVFQ-ASDAMIGFELENVMK 576
FLA A AI+DV+ G+I D + F++ N MK
Sbjct: 728 FLARARAIIDVKKGKIDLNLGRDLKMTFDITNTMK 762
>UniRef100_Q7Y1N4 Putative retrotransposon gag protein [Oryza sativa]
Length = 697
Score = 246 bits (628), Expect = 1e-63
Identities = 194/610 (31%), Positives = 285/610 (45%), Gaps = 102/610 (16%)
Query: 4 FSGSPTEDPNLHISSFLRLSG--TIKE-NQEAVRLHLFPFSLRDRASAWFHSLEVGSITS 60
F G P ED N H+ FL + TIK + VRL LFPFSL R WF++ + +
Sbjct: 45 FCGKPNEDANAHLQQFLEICSMYTIKGVSPNTVRLRLFPFSLLRRVKQWFYA-NCAAANT 103
Query: 61 WDDMRRAFLARFFPPSKTAKLRDQITRFNQKDGESLYEAWEHFKEMLRLCPHHGLEKWLI 120
WD AFL++FFP KT LR +I+ F Q ES+ EAWE +E + CPHHG++ WLI
Sbjct: 104 WDKCSMAFLSKFFPMGKTNALRGRISSFQQTMDESIPEAWERLQEYVAACPHHGMDDWLI 163
Query: 121 VHTFYNGLSDTTKMSVDAAAGGALMNKNYTEAYALIEDMAQNHYQWTNERAVTTPTPSKK 180
+ FYNGL+ ++ +DAAAGGA +K A IE M N W+ ER T +
Sbjct: 164 LQNFYNGLTPMSRDHLDAAAGGAFFSKTVQGAVEFIEKMVSN-MGWSEERLQT------R 216
Query: 181 EAGIHEVSEYNHLAAKVEQLNYAQYKQGMRPNQNFYKNPQGSY------------GQTAP 228
+ G+H V E L AK++ L M+ + K PQG+ G T
Sbjct: 217 QRGMHTVKETELLPAKLDLL--------MKRLDDHEKMPQGTVKALDSHVTCEVCGDTGH 268
Query: 229 PGYTNNQRVAQKSSLEILLENCMMNQNKQLQELKNQTGSLNDSLS-------KLNTKVDS 281
PG NN ++ ++ + N Q + T SL+ L+ N K+D
Sbjct: 269 PG--NNCPETREEAMYMGNNNGYRRQGDLVFAQAKTTDSLSKKLAANDKIVENRNVKLDG 326
Query: 282 IAT-------HTKMLETQISQVAQQVAISS*TPGVFPGQTETNPKAHVNAISLGGNKL-- 332
A+ KM+ETQ++Q+A V + G PGQ +++ + +V AI+ G K
Sbjct: 327 FASAFQNQLCFNKMMETQLAQLASLVPANE--TGRIPGQPDSSIE-NVKAITTRGGKSTR 383
Query: 333 -----EDTITKAKSVKGESVKLLG-----EKDAIKTLLDKNKALNPLRLTKLNLEAQFAK 382
T T S + S L EK + D P R+ K +++ QFA+
Sbjct: 384 DPPYPNPTRTNGLSKEAPSNGLADKEVQPEKTMPQEYCDIRLLPFPQRMRKPSVDEQFAR 443
Query: 383 FLNILKRICIKIPFGEALSRMPLYTKFLKEIFSKKKAMDHKETIALTRESSAIVKKQPPK 442
F+ ++++I I +P +A+ ++P Y + LK+I + K+ + E + LT +QP
Sbjct: 444 FVEVIQKIHINMPLLDAM-QVPTYARCLKDILNNKRPLPTTEVVKLTG-----TMQQPDT 497
Query: 443 LRDPGSFVIPCVIGKETVDKALCDLGASVSLLPLSLFKRMGIGELKPTEMILKLADRSTI 502
+ PG +R G PT M L+L D S
Sbjct: 498 PQAPGE------------------------------EERSG----APTPMRLQLVDSSIR 523
Query: 503 PLVGYIEDIPVKIEGIYIPTDFVVVDIEEDHDVPIILGRPFLATAGAIVDVQSGRIVFQA 562
+D+PVKI +IP DFVV+D++ + P ILGRPFL+TAGA +DV +G I F
Sbjct: 524 YTARIAKDVPVKIRYFFIPVDFVVLDMDTGKETPFILGRPFLSTAGANIDVGTGSIYFHI 583
Query: 563 SDAMIGFELE 572
+ FE +
Sbjct: 584 NGKEEKFEFQ 593
>UniRef100_Q7XLB2 OSJNBa0011K22.17 protein [Oryza sativa]
Length = 551
Score = 230 bits (587), Expect = 7e-59
Identities = 163/501 (32%), Positives = 257/501 (50%), Gaps = 51/501 (10%)
Query: 77 KTAKLRDQITRFNQKDGESLYEAWEHFKEMLRLCPHHGLEKWLIVHTFYNGLSDTTKMSV 136
KT LR +I+ F Q ES+ EAWE +E + CPHHG++ WLI+ FYNGL+ + +
Sbjct: 3 KTNALRGRISSFQQTRDESIPEAWERLQEYMAACPHHGMDDWLILQNFYNGLTPMSHDHL 62
Query: 137 DAAAGGALMNKNYTEAYALIEDMAQNHYQWTNERAVTTPTPSKKEAGIHEVSEYNHLAAK 196
DAAA GA +K A LIE M + ++ KK G +V + +H+ +
Sbjct: 63 DAAARGAFFSKTVQGAVDLIEKMLDLLMKRLDDH-------DKKPQGTVKVLD-SHVTCE 114
Query: 197 V------EQLNYAQYKQGMRPNQNFYKNPQGSYGQTAPPGYTNNQRVAQKSSLEIL---L 247
V + ++ M N PQ G P Y + + A + L + L
Sbjct: 115 VCGNTGHSGNDCLDTREAMYMGNNNGYCPQRGQGWNQPCPYVDRKHEAWEICLTPVQPSL 174
Query: 248 ENCMMNQNKQLQELKNQTGSLNDSLSKLNTKVDSIAT-------HTKMLETQISQVAQQV 300
++ + Q K + L + + + L +N K+D A+ KMLETQ++Q A V
Sbjct: 175 KDLVFAQAKTIDALSKKLATNDKILENINVKLDGFASAFQNQLRFNKMLETQLAQFASLV 234
Query: 301 AISS*TPGVFPGQTETNPKAHVNAISLGGNKLEDTITKAKSVKGESVKLLGEKDAIKTLL 360
+ +T TN A ++ + + K V+ E K++ ++ LL
Sbjct: 235 PAN---------ETGTNGIA---------KEVPSSDSADKEVQLE--KIVPQEYCDTWLL 274
Query: 361 DKNKALNPLRLTKLNLEAQFAKFLNILKRICIKIPFGEALSRMPLYTKFLKEIFSKKKAM 420
++ R K +++ QFA+F ++++I I +P +A+ ++P Y ++LK++ + K+ +
Sbjct: 275 SFHQ-----RSRKPSVDEQFARFAEVIQKIHINVPLLDAM-QVPTYARYLKDMLNNKRLL 328
Query: 421 DHKETIALTRE-SSAIVKKQPPKLRDPGSFVIPCVIGKETVDKALCDLGASVSLLPLSLF 479
E + LT + SS I+ K P K +DPG I C IG + D+A CDLGAS+S++P +
Sbjct: 329 PTMEVVKLTEQCSSVILHKLPEKKKDPGCPTITCSIGAQQFDQAFCDLGASISVMPKDVP 388
Query: 480 KRMGIGELKPTEMILKLADRSTIPLVGYIEDIPVKIEGIYIPTDFVVVDIEEDHDVPIIL 539
++ L PT M L+LAD S G ED+PVKI +IP DFVV+D++ + P+IL
Sbjct: 389 DKLNFTVLTPTLMCLQLADLSVHYPTGIAEDVPVKIRDFFIPVDFVVLDMDMGKETPLIL 448
Query: 540 GRPFLATAGAIVDVQSGRIVF 560
GRPFL+TAGA +DV G I F
Sbjct: 449 GRPFLSTAGANIDVGMGSIRF 469
>UniRef100_Q9MA70 F2J6.11 protein [Arabidopsis thaliana]
Length = 823
Score = 227 bits (578), Expect = 8e-58
Identities = 189/642 (29%), Positives = 296/642 (45%), Gaps = 134/642 (20%)
Query: 2 NQFSGSPTEDPNLHISSFLRLSGTIKEN---QEAVRLHLFPFSLRDRASAWFHSLEVGSI 58
N+F G P EDP H+ +F RL K N +++ +L LFPFSL D+A W +L V S+
Sbjct: 88 NKFHGLPMEDPLDHLDNFDRLCSLTKINGVSEDSFKLRLFPFSLGDKAHLWEKTLPVDSV 147
Query: 59 TSWDDMRRAFLARFFPPSKTAKLRDQITRFNQKDGESLYEAWEHFKEMLRLCPHHGLEKW 118
+ DD ++AFLA+FF S+TA+LR++I+ FNQK+ ES EAWE FK CPHHG +K
Sbjct: 148 DTLDDCKKAFLAKFFSNSRTARLRNEISGFNQKNSESFAEAWERFKGYSTQCPHHGFKKA 207
Query: 119 LIVHTFYNGLSDTTKMSVDAAAGGALMNKNYTEAYALIEDMAQN------HYQWTN---- 168
++ T Y G +M +D + G +NK+ E + L+E++AQ+ +Y TN
Sbjct: 208 SLLSTLYRGALPKIRMLLDTTSNGNFLNKDVAEGWELVENLAQSDGNYNENYDRTNRGSS 267
Query: 169 -----------------------ERAVTTPTPSK-----KEAGIHEVSEYNHLAAKVEQL 200
R ++T K K+ I + N++A +Q+
Sbjct: 268 DSEDRHKRRTKLLMIRLTNWCLLNREMSTTLQKKSLHNSKKGRILLLRRSNNVANPQDQV 327
Query: 201 -----------NYAQYKQGMRPNQNFYKNPQGSYGQTAPPGYTN--NQRVAQKSSLEILL 247
+ Y QG QNF PPG+T Q AQ S ++ LL
Sbjct: 328 YPPQNQPPQAKPFIPYNQGYNQKQNF-----------GPPGFTQQPQQTSAQDSEMKTLL 376
Query: 248 E-------NCMMNQNKQLQELKNQTGSLNDSLSKLNTKVDSIATHTKMLETQISQVAQQV 300
+C M +K+L EL T ++ S + LN K+D++ T K +E I+ +
Sbjct: 377 HQLVQGQASCSMTMDKKLAEL---TTRIDCSYNDLNIKIDALNTRVKTMEGHIASTS--- 430
Query: 301 AISS*TPGVFPGQTETNPKAHVNAISLGGNK--LEDTITKAKSVKGESVKLLGEKDAIKT 358
+ PG P + NPK + +AI+ + I + + ++ S + + E D
Sbjct: 431 --APKHPGQLPRNSVQNPKEYAHAITTVSTSATADSGIQEGEVLRPRSRQEI-ELDFFAW 487
Query: 359 LLDK-NKALNPLRLTKLNLEAQF--AKFLNILKRICIKIPFGEALSRMPLYTKFLKEIFS 415
L+++ NP+R+ + + + RI K ++ K +KE+
Sbjct: 488 LVERAYDPSNPIRIPPAYEPKPYFPERIAQVNARIFQK--------HKMMFIKCIKELEE 539
Query: 416 KKKAMDHKETIALTRESSAIVKKQPPKLRDPGSFVIPCVIGKETVDKALCDLGASVSLLP 475
K +D + + + R A +Q +L SF +I ++ + K
Sbjct: 540 KIPLVDTPKEVIMERPQEA---QQIVEL----SFECSAIIQRKVIPK------------- 579
Query: 476 LSLFKRMGIGELKPTEMILKLADRSTIPLVGYIEDIPVKIEGIYIPTDFVVVDIEEDHDV 535
KLADRS G +ED+PVKI I IPTDFVV++++E+
Sbjct: 580 -------------------KLADRSVRVPHGMLEDLPVKIGSIEIPTDFVVLEMDEEPKD 620
Query: 536 PIILGRPFLATAGAIVDVQSGRIVFQ-ASDAMIGFELENVMK 576
P+ILGRPFLATAGA++DVQ G+I + + F++ MK
Sbjct: 621 PLILGRPFLATAGALIDVQMGKIDLNLGKNLHMSFDIAKKMK 662
>UniRef100_Q6F2G0 Hypothetical protein OSJNBa0083K01.26 [Oryza sativa]
Length = 585
Score = 225 bits (574), Expect = 2e-57
Identities = 161/509 (31%), Positives = 251/509 (48%), Gaps = 38/509 (7%)
Query: 77 KTAKLRDQITRFNQKDGESLYEAWEHFKEMLRLCPHHGLEKWLIVHTFYNGLSDTTKMSV 136
KT L +I+ F Q ES+ EAWE +E + CPHHG++ WLI+ FYN L+ ++ +
Sbjct: 3 KTDALCGRISSFQQTRDESIPEAWELLQEYVAACPHHGMDDWLILQNFYNRLTPMSRDHL 62
Query: 137 DAAAGGALMNKNYTEAYALIEDMAQNHYQWTNERAVTTPTPSKKEAGIHEVSEYNHLAAK 196
DAAAGGA +K LIE M N W+ ER T ++ G+H V E LAAK
Sbjct: 63 DAAAGGAFFSKTVQGTVELIEKMVSN-MGWSKERLQT------RQRGMHTVKETELLAAK 115
Query: 197 VEQLNYAQYKQGMRPNQNFYKNPQGSYGQTAPPGYTNNQRVAQKSSLEILLENCMMNQNK 256
++ L M+ N K PQG+ ++ + +C + +
Sbjct: 116 LDLL--------MKRLDNHEKRPQGTV-----KALDSHVTCEVCGGTDHSGNDCPETREE 162
Query: 257 QLQELKNQTGSLNDSLSKLNTKVDSIATHTKMLETQISQVAQQVAISS*TPGVFPGQTET 316
+ N G + N + S ETQ++Q+A V + G PGQ +
Sbjct: 163 AMYMGNNNNGYRPQGVQGWN-QPRSYYQGGNNNETQLAQLAYLVPANE--TGRIPGQPYS 219
Query: 317 NPKAHVNAISLGGNK------------LEDTITKAKSVKGESVKLLGEKDAIKTLLDKNK 364
+ + +V AI+ G K + S ++ EK + D
Sbjct: 220 SIE-NVKAITTRGGKSTRDPPYPNPAGTNGMSKETPSNDSADKEIQPEKTVPQEYCDTRL 278
Query: 365 ALNPLRLTKLNLEAQFAKFLNILKRICIKIPFGEALSRMPLYTKFLKEIFSKKKAMDHKE 424
P K +++ FA+F+ +++RI I + +A+ ++P+Y ++LK+I + K+ + E
Sbjct: 279 LPFPQWSRKTSVDELFARFVEVIQRIHINVLLLDAM-QVPIYARYLKDILNNKRPLPTTE 337
Query: 425 TIALTRE-SSAIVKKQPPKLRDPGSFVIPCVIGKETVDKALCDLGASVSLLPLSLFKRMG 483
+ LT S+ I+ K P K + I C IG + D+ALCDLGASVS++P +F R+
Sbjct: 338 VVKLTEHCSNVILHKLPEKKKYSECPTITCSIGAQQFDQALCDLGASVSVMPKDVFDRLN 397
Query: 484 IGELKPTEMILKLADRSTIPLVGYIEDIPVKIEGIYIPTDFVVVDIEEDHDVPIILGRPF 543
L PT M L++AD S G ED+P+KI +IP DFVV+D++ + P+ILGRPF
Sbjct: 398 FTVLAPTPMRLQMADSSVRYQAGIAEDVPIKIWDFFIPVDFVVLDMDTRKETPLILGRPF 457
Query: 544 LATAGAIVDVQSGRIVFQASDAMIGFELE 572
L+TAGA +DV++ I F ++ FE +
Sbjct: 458 LSTAGANIDVETSSICFHINEKEEKFEFQ 486
>UniRef100_Q9MA68 F2J6.13 protein [Arabidopsis thaliana]
Length = 780
Score = 223 bits (568), Expect = 1e-56
Identities = 189/658 (28%), Positives = 298/658 (44%), Gaps = 139/658 (21%)
Query: 2 NQFSGSPTEDPNLHISSFLRLSGTIKEN---QEAVRLHLFPFSLRDRASAWFHSLEVGSI 58
N+F G P EDP H+ +F R K N +++ +L LFPFSL D A W +L V S+
Sbjct: 52 NKFHGLPMEDPLDHLDNFDRFCSLTKINGVSEDSFKLRLFPFSLGDEAHLWEKTLLVDSV 111
Query: 59 TSWDDMRRAFLARFFPPSKTAKLRDQITRFNQKDGESLYEAWEHFKEMLRLCPHHGLEKW 118
+WDD ++AFLA+FF S+TA+LR++I+ FNQK+ ES EAWE FK CPHHG +K
Sbjct: 112 DTWDDCKKAFLAKFFSNSRTARLRNEISGFNQKNSESFAEAWERFKRYSTQCPHHGFKKA 171
Query: 119 LIVHTFYNGLSDTTKMSVDAAAGGALMNKNYTEAYALIEDMAQN------HYQWTNERAV 172
++ T Y G +M +D + G +NK+ E + L+E++AQ+ Y TN +
Sbjct: 172 SLLSTLYRGALPKIRMLLDTTSNGNFLNKDVAEGWELVENLAQSDGNYNEDYDRTNRGSS 231
Query: 173 TTPTPSKKE----------------AGIHEVSEYNHLAAK------VEQLNYAQYKQGMR 210
+ KKE +H ++E + VE+++Y Q + G
Sbjct: 232 DSEDRHKKEIKALNDKIDKLVLAQQRNVHYITEEELTQLQEGENLTVEEVSYLQNQGGYN 291
Query: 211 PNQNFYK--------------NPQG--------------------SYGQT---APPGYTN 233
N YK NPQ Y Q PP +T
Sbjct: 292 KGFNNYKPPHPNLSYRSNNVANPQDQVYPPQNQPPQAKPFIPYNQGYNQKQNFRPPSFTQ 351
Query: 234 --NQRVAQKSSLEILLE-------NCMMNQNKQLQELKNQTGSLNDSLSKLNTKVDSIAT 284
Q AQ S ++ LL +C + +K+L EL T ++ S + LN K+D++ T
Sbjct: 352 QPQQTSAQDSEMKTLLHQLVQGQASCSITMDKKLAEL---TTRIDCSYNDLNIKIDALNT 408
Query: 285 HTKMLETQISQVAQQVAISS*TPGVFPGQTETNPKAHVNAISLGGNK--LEDTITKAKSV 342
K +E I+ + + PG ++ NPK + +AI+ + I + + +
Sbjct: 409 RVKTMEGHIASTS-----APKHPGQLSRKSVQNPKEYAHAITTVSTSATADSGIQEGEVL 463
Query: 343 KGESVKLLGEKDAIKTLLDK-NKALNPLRLTKLNLEAQF--AKFLNILKRICIKIPFGEA 399
+ S + + E D L+++ NP+R+ + + + RI K
Sbjct: 464 RPRSRQEI-ELDFFALLVERAYDPSNPIRIPPAYEPKPYFPERIAQVNARIFQK------ 516
Query: 400 LSRMPLYTKFLKEIFSKKKAMDHKETIALTRESSAIVKKQPPKLRDPGSFVIPCVIGKET 459
++ K +KE+ K +D + + + R A +Q +L SF +I ++
Sbjct: 517 --HKMMFIKCIKELEEKIPLVDTPKEVIMERPQEA---QQIIEL----SFECSAIIQRKV 567
Query: 460 VDKALCDLGASVSLLPLSLFKRMGIGELKPTEMILKLADRSTIPLVGYIEDIPVKIEGIY 519
+ K KLADRS G +ED+PVKI I
Sbjct: 568 IPK--------------------------------KLADRSVRVPHGMLEDLPVKIGSIE 595
Query: 520 IPTDFVVVDIEEDHDVPIILGRPFLATAGAIVDVQSGRIVFQ-ASDAMIGFELENVMK 576
IPTDF+V++++E+ P+ILGRPFLATAGA++DVQ G+I + + F++ MK
Sbjct: 596 IPTDFMVLEMDEEPKDPLILGRPFLATAGALIDVQMGKIDLNLGKNLHMSFDIAKKMK 653
>UniRef100_Q9SHR2 F28L22.6 protein [Arabidopsis thaliana]
Length = 719
Score = 167 bits (424), Expect = 6e-40
Identities = 167/668 (25%), Positives = 286/668 (42%), Gaps = 149/668 (22%)
Query: 46 ASAWFHSL-EVGSITSWDDMRRAFLARFFPPSKTAKLRDQITRFNQKDGESLYEAWE--H 102
+S FHSL E I+ + FL PP TA+ ++ F+Q G Y
Sbjct: 4 SSDCFHSLLETKHISG----KSHFLKALSPPGMTARKPSWLS-FSQTPGLRDYGMTYLVS 58
Query: 103 FKEMLRLCPHHGLEKWLI-------VHTFYNGLSDTTKMSVDAAAGGALMNKNYTEAYAL 155
+ ++RL HG +I V L+ +T+M +D A+ G +NK+ + + +
Sbjct: 59 LRRIMRLSMKHGSASRVIRRNVLIMVSPKLRFLALSTEMLLDTASNGNFLNKDVEDGWEV 118
Query: 156 IEDMAQN--HYQWTNERAVTTPTPS--------------------KKEAGIHEVSEYNHL 193
+E++AQ+ +Y +R++ T + S ++ IH + + L
Sbjct: 119 VENLAQSDGNYNEDYDRSIRTSSDSDEKHRREMKAMNDKLDKLLLMQQKHIHFLGDDETL 178
Query: 194 ------AAKVEQLNYAQYKQGMRPNQNFYK--NPQGSYGQT------------------- 226
++E+++Y Q + G N +K +P SY T
Sbjct: 179 QVQDGETLQLEEVSYVQNQGGYNKGFNNFKQNHPNLSYRSTNVANRQDQVYPSQQQNQPK 238
Query: 227 -------------------------APPGYTNNQR----VAQKSSLEILLENCMMNQNKQ 257
PPG+T Q+ S L+ +L+ + Q
Sbjct: 239 PFVPYNQGQGYVPKQQYQGNYQQQLPPPGFTQQQQQPALTTPDSDLKNMLQQILQGQATG 298
Query: 258 LQELKNQTGSLND----SLSKLNTKVDSIATHTKMLETQISQVAQQVAISS*TPGVFPGQ 313
+L + +++ S + +N KV+++ + + +E Q A G+
Sbjct: 299 AMDLSKRMAEIHNKVDCSYNDINIKVEALTSKIRYIEGQTGSTAAPKFTGP------SGK 352
Query: 314 TETNPKAHVNAISLGGNKLEDTITKAKSVKGESVKLLGE--------------------- 352
+ +N K + +AI+L K T +S+ GE
Sbjct: 353 SMSNSKEYAHAITLRSGKELPTKESPNQNTEDSLDQDGEDFCQNGNSAEKAIEEPILHQP 412
Query: 353 ----KDAIKTLLDKNKALN-----------------PLRLTKLNLEAQFAKFLNILKRIC 391
A L++K A P R K+ ++ A LK +
Sbjct: 413 TRPLAPAASPLVEKPAAAKTKENVFIPPPYKPPLPFPGRFKKVMIQKYKALLEKQLKNLE 472
Query: 392 IKIPFGEALSRMPLYTKFLKEIFSKKKAMDHKETIALTRESSAIVKKQ--PPKLRDPGSF 449
+ +P + L+ +P K++K++ + ++ + + + L+ E SAI++++ P KL DPGSF
Sbjct: 473 VTMPLVDCLALIPDSNKYVKDMIT-ERIKEVQGMVVLSHECSAIIQQKIIPKKLGDPGSF 531
Query: 450 VIPCVIGKETVDKALCDLGASVSLLPLSLFKRMGIGELKPTEMILKLADRSTIPLVGYIE 509
+PC +G K LCDLGASVSL+PL + K++G + KP + L LADRS G +E
Sbjct: 532 TLPCALGPLAFSKCLCDLGASVSLMPLPVAKKLGFNKYKPCNISLILADRSVRISHGLLE 591
Query: 510 DIPVKIEGIYIPTDFVVVDIEEDHDVPIILGRPFLATAGAIVDVQSGRIVFQ-ASDAMIG 568
D+PV I + +PTDFVV++++E+ P+ILGRPFLA A AI+DV+ G+I D +
Sbjct: 592 DLPVMIGVVEVPTDFVVLEMDEEPKDPLILGRPFLARARAIIDVKKGKIDLNLGRDLKMT 651
Query: 569 FELENVMK 576
F++ N MK
Sbjct: 652 FDITNTMK 659
>UniRef100_Q96467 Hypothetical protein [Hordeum vulgare]
Length = 337
Score = 162 bits (410), Expect = 2e-38
Identities = 82/203 (40%), Positives = 125/203 (61%), Gaps = 10/203 (4%)
Query: 1 QNQFSGSPTEDPNLHISSFLRLSGTIKE---NQEAVRLHLFPFSLRDRASAWFHSLEVGS 57
+ QFSG P+ED H+++F+ L K+ + + ++L LFPFSLRDRA WF SL S
Sbjct: 44 KEQFSGLPSEDVASHLNTFIELCDMQKKKDVDNDVIKLKLFPFSLRDRAKTWFSSLPKSS 103
Query: 58 ITSWDDMRRAFLARFFPPSKTAKLRDQITRFNQKDGESLYEAWEHFKEMLRLCPHHGLEK 117
I SWD + A+++++FPP+K LR+ I F Q D E + +AWE K M+R CP +GL
Sbjct: 104 IDSWDKCKDAYISKYFPPAKIISLRNDIMNFKQLDHEHVAQAWERMKLMIRNCPANGLSL 163
Query: 118 WLIVHTFYNGLSDTTKMSVDAAAGGALMNKNYTEAYALIEDMAQNHYQWTNERAVTTPTP 177
W+I+ FY GL+ ++ +D+A GG M EA L++++ N+ QW ER+ T
Sbjct: 164 WMIIQIFYAGLNFASRNILDSATGGTFMEITLGEATKLLDNIMTNYSQWHTERSPT---- 219
Query: 178 SKKEAGIHEVSEYNHLAAKVEQL 200
SKK +H + E N L++K+++L
Sbjct: 220 SKK---VHVIEEINSLSSKMDEL 239
>UniRef100_Q8S6J2 Hypothetical protein OSJNBa0019N10.34 [Oryza sativa]
Length = 237
Score = 153 bits (387), Expect = 1e-35
Identities = 84/195 (43%), Positives = 124/195 (63%), Gaps = 2/195 (1%)
Query: 368 PLRLTKLNLEAQFAKFLNILKRICIKIPFGEALSRMPLYTKFLKEIFSKKKAMDHKETIA 427
P R K +++ QFA F+ ++++I I +P +A+ ++P Y +LK+I + K+ + E +
Sbjct: 36 PQRSRKPSVDEQFAHFVEVIQKIHIDVPLLDAM-QVPTYACYLKDILNNKRPLPTTEVVK 94
Query: 428 LTRE-SSAIVKKQPPKLRDPGSFVIPCVIGKETVDKALCDLGASVSLLPLSLFKRMGIGE 486
LT + S+ I+ K P K +DPG I C IG + D+ALCDLGASVS++P +F ++
Sbjct: 95 LTLQCSNVILHKLPEKKKDPGCPTITCSIGAQQFDQALCDLGASVSVMPKYVFDKLNFTV 154
Query: 487 LKPTEMILKLADRSTIPLVGYIEDIPVKIEGIYIPTDFVVVDIEEDHDVPIILGRPFLAT 546
L PT M L+LAD S G ED+PVKI +I DFVV+D++ ++ IILGRPFL T
Sbjct: 155 LAPTPMRLQLADSSVRYPAGIAEDVPVKIRDFFILVDFVVLDMDTGKEMSIILGRPFLGT 214
Query: 547 AGAIVDVQSGRIVFQ 561
AGA + V +G I FQ
Sbjct: 215 AGANIHVGTGSIRFQ 229
>UniRef100_Q9LKS8 Hypothetical protein T15F17.m [Arabidopsis thaliana]
Length = 346
Score = 150 bits (380), Expect = 7e-35
Identities = 86/240 (35%), Positives = 132/240 (54%), Gaps = 10/240 (4%)
Query: 2 NQFSGSPTEDPNLHISSFLRLSGTIKEN---QEAVRLHLFPFSLRDRASAWFHSLEVGSI 58
N+F G P EDP H+ F RL K N ++ +L LFPFSL D+A W +L GSI
Sbjct: 52 NKFHGLPMEDPLDHLDEFERLCDLTKINGVSEDGFKLRLFPFSLGDKAHLWEKTLPQGSI 111
Query: 59 TSWDDMRRAFLARFFPPSKTAKLRDQITRFNQKDGESLYEAWEHFKEMLRLCPHHGLEKW 118
T+WDD ++ FL +FF S+TA+L ++I+ F QK ES EAWE FK CPHHG ++
Sbjct: 112 TTWDDCKKVFLEKFFSNSRTARLWNEISGFTQKQNESFCEAWERFKGYQTKCPHHGFKEA 171
Query: 119 LIVHTFYNGLSDTTKMSVDAAAGGALMNKNYTEAYALIEDMAQN--HYQWTNERAVTTPT 176
++ T Y G+ +M +D A+ G +NK+ + + L+E++AQ+ +Y +R++ T T
Sbjct: 172 SLLSTLYRGVLPKIRMLLDTASNGNFLNKDVEDGWELVENLAQSDGNYNEDYDRSIPTST 231
Query: 177 PSKKEAGIHEVSEYNHLAAKVEQLNYAQYKQGMRPNQN-FYKNPQGSYGQTAPPGYTNNQ 235
S + E L K+++L Q K +++ ++ +G Q A Y NQ
Sbjct: 232 ESDDK----HRREIKALHEKIDKLLQVQQKHVHSASEDEVFQIQEGENDQDAEISYVQNQ 287
>UniRef100_Q9C8Q7 Athila ORF 1, putative; 43045-40843 [Arabidopsis thaliana]
Length = 530
Score = 150 bits (380), Expect = 7e-35
Identities = 82/218 (37%), Positives = 137/218 (62%), Gaps = 5/218 (2%)
Query: 363 NKALNPLRLTKLNLEAQFAKFLNILKRICIK-IPFGEALSRMPLYTKFLKEIFSKKKAMD 421
N+ + L + F + +N IK +P + L+ +P K++K++ +++ +
Sbjct: 89 NRLFITTKAKSLFQSSSFKEDINNSSHHLIKDMPLVDCLALIPNEHKYVKDLITER-IKE 147
Query: 422 HKETIALTRESSAIVKKQ--PPKLRDPGSFVIPCVIGKETVDKALCDLGASVSLLPLSLF 479
+ + L+ E SAI++++ KL DPGSF +PC I + T +LCDLGASVS++PLS+
Sbjct: 148 VQGMVVLSHECSAIIQQKIVQEKLEDPGSFTLPCSIRQLTFSNSLCDLGASVSIMPLSMA 207
Query: 480 KRMGIGELKPTEMILKLADRSTIPLVGYIEDIPVKIEGIYIPTDFVVVDIEEDHDVPIIL 539
+++G + KP ++ L LADR++ G +ED+PV I G+ +P DFVV++++E+ P+IL
Sbjct: 208 RKLGFVQYKPCDLTLILADRTSRRPFGLLEDVPVMINGVEVPIDFVVLEMDEESKDPLIL 267
Query: 540 GRPFLATAGAIVDVQSGRIVFQ-ASDAMIGFELENVMK 576
GRPFLA+AGA++DV+ G+I D + FE+ N MK
Sbjct: 268 GRPFLASAGAVIDVKQGKINLNLGEDFKMKFEIRNTMK 305
>UniRef100_O22104 Vicia faba mRNA expressed from retrotransposon-like gene [Vicia
faba]
Length = 238
Score = 150 bits (378), Expect = 1e-34
Identities = 79/202 (39%), Positives = 121/202 (59%), Gaps = 3/202 (1%)
Query: 377 EAQFAKFLNILKRICIKIPFGEALSRMPLYTKFLKEIFSKKKAMDHKETIALTRESSAIV 436
E F FL + K++ + I EAL +MP Y KF+K+I SKK+ +D + I T S+I+
Sbjct: 11 EKNFEIFLEMFKKLELNILVLEALEKMPTYAKFMKDIISKKRTID-TDPIIHTETCSSIL 69
Query: 437 K--KQPPKLRDPGSFVIPCVIGKETVDKALCDLGASVSLLPLSLFKRMGIGELKPTEMIL 494
+ K P K +D G PC IG + K +LGASVSLLP ++K++GI ++ T M L
Sbjct: 70 QGMKIPVKKKDRGFVTTPCTIGDRSFKKYFINLGASVSLLPFYIYKKLGISNVQNTRMTL 129
Query: 495 KLADRSTIPLVGYIEDIPVKIEGIYIPTDFVVVDIEEDHDVPIILGRPFLATAGAIVDVQ 554
+ AD S G +ED+ VKI+ P DFVV+++ ED ++ +ILGRPFL T ++D++
Sbjct: 130 RFADHSVKIPYGIVEDVLVKIDKFVFPVDFVVLEMPEDEEITLILGRPFLETRRCLIDIE 189
Query: 555 SGRIVFQASDAMIGFELENVMK 576
G + + D + +++N MK
Sbjct: 190 EGTVTLKVYDEELKIDVQNTMK 211
>UniRef100_Q7XF80 Hypothetical protein [Oryza sativa]
Length = 725
Score = 149 bits (375), Expect = 3e-34
Identities = 83/204 (40%), Positives = 128/204 (62%), Gaps = 3/204 (1%)
Query: 368 PLRLTKLNLEAQFAKFLNILKRICIKIPFGEALSRMPLYTKFLKEIFSKKKAMDHKETIA 427
P R K +++ QFA F+ ++++I I +P +A+ ++P Y +LK+I + K+ + E +
Sbjct: 67 PQRSRKPSVDEQFAHFVEVIQKIHIDVPLLDAM-QVPTYACYLKDILNNKRPLPTTEVVK 125
Query: 428 LTRE-SSAIVKKQPPKLRDPGSFVIPCVIGKETVDKALCDLGASVSLLPLSLFKRMGIGE 486
LT + S+ I+ K P K +DPG I C IG + D+ALCDLGASVS++P +F ++
Sbjct: 126 LTLQCSNVILHKLPEKKKDPGCPTITCSIGAQQFDQALCDLGASVSVMPKYVFDKLNFTV 185
Query: 487 LKPTEMILKLADRSTIPLVGYIEDIPVKIEGIYIPTDFVVVDIEEDHDVPIILGRPFLAT 546
L PT M L+LAD S G ED+PVKI +I DFVV+D++ ++ IILGRPFL T
Sbjct: 186 LAPTPMRLQLADSSVRYPAGIAEDVPVKIRDFFILVDFVVLDMDTGKEMSIILGRPFLGT 245
Query: 547 AGA-IVDVQSGRIVFQASDAMIGF 569
AGA I ++Q + +D+++ F
Sbjct: 246 AGANIHNIQEVEVEPPKTDSLVKF 269
>UniRef100_Q8LM79 Hypothetical protein OSJNAa0082N11.11 [Oryza sativa]
Length = 322
Score = 148 bits (374), Expect = 4e-34
Identities = 101/304 (33%), Positives = 158/304 (51%), Gaps = 26/304 (8%)
Query: 276 NTKVDSIAT-------HTKMLETQISQVAQQVAISS*TPGVFPGQTETNPKAHVNAISLG 328
N K+D A+ KM+ETQ++Q+A V + PGQ +++ + + G
Sbjct: 27 NVKLDGFASAFQNQLSFNKMIETQLAQLASLVPANE--SERIPGQPDSSIENIKAITTRG 84
Query: 329 GNKLEDTITKAKSVKGESVKLLGEKDAIKTLLDKNKALNPLRLTKLNLEAQFAKFLNILK 388
G T + + + ++ EK + D N QFA+ + +++
Sbjct: 85 GTNGMSKETPSTDLADKEIQ--PEKTVPQEYCDTRGVGN---------RQQFARSVEVIQ 133
Query: 389 RICIKIPFGEALSRMPLYTKFLKEIFSKKKAMDHKETIALTRESSAIVKKQPPKLRDPGS 448
+I I +P +A+ ++P Y ++LK+I + K+ + E S+ I+ K P K +DP
Sbjct: 134 KIHINVPLLDAM-QVPTYARYLKDILNNKRPLPTTEHC-----SNVILHKLPEKKKDPRC 187
Query: 449 FVIPCVIGKETVDKALCDLGASVSLLPLSLFKRMGIGELKPTEMILKLADRSTIPLVGYI 508
I C IG + D+ALCDLGASVS++P +F ++ L PT M L+LAD S VG
Sbjct: 188 PTITCSIGAQQFDQALCDLGASVSVMPKDVFDKLNFTVLAPTPMRLQLADSSVHCPVGIE 247
Query: 509 EDIPVKIEGIYIPTDFVVVDIEEDHDVPIILGRPFLATAGAIVDVQSGRIVFQASDAMIG 568
ED+PVKI +IP DFVV+D++ + +ILG PFL+TAGA +DV G I F +
Sbjct: 248 EDVPVKIWDFFIPVDFVVLDMDTGKETSLILGHPFLSTAGANIDVGMGSIRFHINGKEEK 307
Query: 569 FELE 572
FE +
Sbjct: 308 FEFQ 311
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.134 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 940,113,874
Number of Sequences: 2790947
Number of extensions: 38871564
Number of successful extensions: 118365
Number of sequences better than 10.0: 739
Number of HSP's better than 10.0 without gapping: 331
Number of HSP's successfully gapped in prelim test: 408
Number of HSP's that attempted gapping in prelim test: 117369
Number of HSP's gapped (non-prelim): 1164
length of query: 577
length of database: 848,049,833
effective HSP length: 133
effective length of query: 444
effective length of database: 476,853,882
effective search space: 211723123608
effective search space used: 211723123608
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 78 (34.7 bits)
Medicago: description of AC146583.2


