

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146571.1 - phase: 0
(713 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
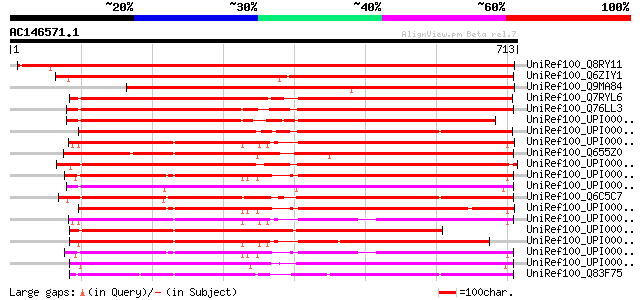
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8RY11 AT3g05350/T12H1_32 [Arabidopsis thaliana] 993 0.0
UniRef100_Q6ZIY1 Putative X-prolyl aminopeptidase [Oryza sativa] 893 0.0
UniRef100_Q9MA84 Putative aminopeptidase [Arabidopsis thaliana] 834 0.0
UniRef100_Q7RYL6 Hypothetical protein [Neurospora crassa] 568 e-160
UniRef100_Q76LL3 Aminopeptidase-P [Aspergillus oryzae] 560 e-158
UniRef100_UPI00002365AE UPI00002365AE UniRef100 entry 549 e-154
UniRef100_UPI000021B477 UPI000021B477 UniRef100 entry 549 e-154
UniRef100_UPI00003AE3CB UPI00003AE3CB UniRef100 entry 542 e-152
UniRef100_Q655Z0 Putative Xaa-Pro aminopeptidase 2 [Oryza sativa] 536 e-151
UniRef100_UPI000023DCBC UPI000023DCBC UniRef100 entry 531 e-149
UniRef100_UPI000036180F UPI000036180F UniRef100 entry 527 e-148
UniRef100_UPI000042DE85 UPI000042DE85 UniRef100 entry 512 e-143
UniRef100_Q6C5C7 Similar to tr|Q8RY11 Arabidopsis thaliana AT3g0... 511 e-143
UniRef100_UPI0000361812 UPI0000361812 UniRef100 entry 509 e-142
UniRef100_UPI00003AE3CC UPI00003AE3CC UniRef100 entry 501 e-140
UniRef100_UPI00002A6EFE UPI00002A6EFE UniRef100 entry 497 e-139
UniRef100_UPI00003AE3D1 UPI00003AE3D1 UniRef100 entry 496 e-138
UniRef100_UPI0000361810 UPI0000361810 UniRef100 entry 488 e-136
UniRef100_UPI0000430FD3 UPI0000430FD3 UniRef100 entry 457 e-127
UniRef100_Q83F75 Peptidase, M24 family protein [Coxiella burnetii] 447 e-124
>UniRef100_Q8RY11 AT3g05350/T12H1_32 [Arabidopsis thaliana]
Length = 710
Score = 993 bits (2566), Expect = 0.0
Identities = 490/705 (69%), Positives = 584/705 (82%), Gaps = 8/705 (1%)
Query: 12 SSPATVTKLALSIHSSFPTLNLNPIFFYKFKSNKHNKPPFFSIR-----NCTTSNSISAK 66
SSP+ + +L LS +L L+ K P F R + ++S+S +AK
Sbjct: 7 SSPS-LNRLVLSTSRYSHSLFLSNFNSLSLIHRKLPYKPLFGARCHASSSSSSSSSFTAK 65
Query: 67 PSSQLRKNRSSNSDSDPKLTALRRLFSKPDVSIDAYIIPSQDAHQSEFIAQSYARRKYIS 126
S ++RK ++ D KL+++RRLFS+P V IDAYIIPSQDAHQSEFIA+ YARR YIS
Sbjct: 66 SSKEIRKAQTKVV-VDEKLSSIRRLFSEPGVGIDAYIIPSQDAHQSEFIAECYARRAYIS 124
Query: 127 AFTGSNGTAVVTNDKAALWTDGRYFLQAEKQLNSNWILMRAGNPGVPTTSEWLNEVLAPG 186
FTGS GTAVVT DKAALWTDGRYFLQAEKQLNS+WILMRAGNPGVPT SEW+ +VLAPG
Sbjct: 125 GFTGSAGTAVVTKDKAALWTDGRYFLQAEKQLNSSWILMRAGNPGVPTASEWIADVLAPG 184
Query: 187 ARVGIDPFLFTSDAAEELKHVISKKNHELVYLYNSNLVDEIWKEARPEPPNKPVRVHGLK 246
RVGIDPFLF++DAAEELK VI+KKNHELVYLYN NLVDEIWK++RP+PP++ +R+H LK
Sbjct: 185 GRVGIDPFLFSADAAEELKEVIAKKNHELVYLYNVNLVDEIWKDSRPKPPSRQIRIHDLK 244
Query: 247 YAGLDVASKLSSLRSELVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEIDG 306
YAGLDVASKL SLR++++ AG+SAI+++ LDEIAW+LNLRGSD+PHSPV+YAYLIVE+D
Sbjct: 245 YAGLDVASKLLSLRNQIMDAGTSAIVISMLDEIAWVLNLRGSDVPHSPVMYAYLIVEVDQ 304
Query: 307 AKLFIDNSKVTEEVDDHLKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAY 366
A+LF+DNSKVT EV DHLK A IE+RPY+SI+ I++LAARG+ L +D S++N AI++ Y
Sbjct: 305 AQLFVDNSKVTVEVKDHLKNAGIELRPYDSILQGIDSLAARGAQLLMDPSTLNVAIISTY 364
Query: 367 KAACDRYYQNYESKHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQECH 426
K+AC+RY +N+ES+ K ++K D S + P ++ SP+S AKAIKN+ ELKGM+ H
Sbjct: 365 KSACERYSRNFESEAKVKTKFTDSSSGYTANPSGIYMQSPISWAKAIKNDAELKGMKNSH 424
Query: 427 LRDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAI 486
LRDAAALA FW WLE+E+ + LTEV+V+D+LLEFRS Q GF+DTSFDTISGSG NGAI
Sbjct: 425 LRDAAALAHFWAWLEEEVHKNANLTEVDVADRLLEFRSMQDGFMDTSFDTISGSGANGAI 484
Query: 487 IHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRTVHFGKPTTREKECFTRVLQGHIA 546
IHYKPEP SCS VD KLFLLDSGAQYVDGTTDITRTVHF +P+ REKECFTRVLQGHIA
Sbjct: 485 IHYKPEPESCSRVDPQKLFLLDSGAQYVDGTTDITRTVHFSEPSAREKECFTRVLQGHIA 544
Query: 547 LDQAVFPEDTPGFVLDAFARSFLWKVGLDYRHGTGHGVGAALNVHEGPQGISYRYGNLTP 606
LDQAVFPE TPGFVLD FARS LWK+GLDYRHGTGHGVGAALNVHEGPQ IS+RYGN+TP
Sbjct: 545 LDQAVFPEGTPGFVLDGFARSSLWKIGLDYRHGTGHGVGAALNVHEGPQSISFRYGNMTP 604
Query: 607 LVNGMIVSNEPGYYEDHAFGIRIENLLYVRNVETPNRFGGIQYLGFEKLTYVPIQIKLVD 666
L NGMIVSNEPGYYEDHAFGIRIENLL+VR+ ETPNRFGG YLGFEKLT+ PIQ K+VD
Sbjct: 605 LQNGMIVSNEPGYYEDHAFGIRIENLLHVRDAETPNRFGGATYLGFEKLTFFPIQTKMVD 664
Query: 667 VSLLSTTEIDWLNNYHSVVWEKVSPLLDGS-ARQWLWNNTQPIIR 710
VSLLS TE+DWLN+YH+ VWEKVSPLL+GS +QWLWNNT+P+ +
Sbjct: 665 VSLLSDTEVDWLNSYHAEVWEKVSPLLEGSTTQQWLWNNTRPLAK 709
>UniRef100_Q6ZIY1 Putative X-prolyl aminopeptidase [Oryza sativa]
Length = 718
Score = 893 bits (2307), Expect = 0.0
Identities = 437/662 (66%), Positives = 531/662 (80%), Gaps = 20/662 (3%)
Query: 65 AKPSSQLRKNRSSNSDS---------DPKLTALRRLFSKPDVSIDAYIIPSQDAHQSEFI 115
A PSS+LRK R S S D KL +LRRL ++PDV+IDAYI+PSQDAHQSEFI
Sbjct: 59 ALPSSELRKRRGGASSSSSAAPGGGEDEKLRSLRRLLARPDVAIDAYIVPSQDAHQSEFI 118
Query: 116 AQSYARRKYISAFTGSNGTAVVTNDKAALWTDGRYFLQAEKQLNSNWILMRAGNPGVPTT 175
A+ + RR Y++ FTGS GTAVVT DKAALWTDGRYFLQAEK+L+ +W LMR+GN GVPTT
Sbjct: 119 AECFMRRAYLTGFTGSAGTAVVTKDKAALWTDGRYFLQAEKELSHDWTLMRSGNQGVPTT 178
Query: 176 SEWLNEVLAPGARVGIDPFLFTSDAAEELKHVISKKNHELVYLYNSNLVDEIWKEARPEP 235
SEWLNEVL G RVGIDPFLF+ DAAEELK IS+KNHELV + + NLVDEIW E+RPEP
Sbjct: 179 SEWLNEVLPSGCRVGIDPFLFSFDAAEELKDAISEKNHELVLIKDLNLVDEIWGESRPEP 238
Query: 236 PNKPVRVHGLKYAGLDVASKLSSLRSELVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPV 295
P + RVHG+KYAG+DV SKLS +RS+L + G +A++++ LDE+AWLLN+RGSD+P+SPV
Sbjct: 239 PKEQTRVHGIKYAGVDVPSKLSFVRSQLAENGCNAVVISLLDEVAWLLNMRGSDVPNSPV 298
Query: 296 VYAYLIVEIDGAKLFIDNSKVTEEVDDHLKKANIEIRPYNSIVSEIENLAARGSSLWLDT 355
Y+YLIVE A LF+DN+KV+E+V +HL+KA ++++PY +I+S++E LA G+ LWLD+
Sbjct: 299 FYSYLIVEDTAATLFVDNNKVSEDVLEHLEKAGVKLKPYEAILSDVERLAENGAKLWLDS 358
Query: 356 SSVNAAIVNAYKAACDRYYQNY---------ESKHKTRSKGFDGSIANSDVPIAVHKSSP 406
SS+NAAIVN ++++C+RY + ES + G G + N V A++K SP
Sbjct: 359 SSINAAIVNVFRSSCERYVKKRGKAGRQIGKESSQGDPATGSSG-VQNGTVN-ALYKVSP 416
Query: 407 VSLAKAIKNETELKGMQECHLRDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEFRSKQ 466
+LAKA+KNE E++GM+ HLRDAAALA+FW WLE ++ LTEV+V++KLLEFR KQ
Sbjct: 417 ATLAKAVKNEAEVEGMKSSHLRDAAALAEFWCWLEGQVRESVPLTEVQVAEKLLEFRQKQ 476
Query: 467 AGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRTVHF 526
GF+DTSFDTISG G NGAIIHY+P P SCS+V ++ LFLLDSGAQY+DGTTDITRTVHF
Sbjct: 477 DGFIDTSFDTISGYGANGAIIHYRPTPESCSSVGSDNLFLLDSGAQYIDGTTDITRTVHF 536
Query: 527 GKPTTREKECFTRVLQGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLDYRHGTGHGVGA 586
G+PT R+KECFTRVLQGHIALDQAVFPE TPGFVLD ARS LWK+GLDYRHGTGHGVGA
Sbjct: 537 GEPTPRQKECFTRVLQGHIALDQAVFPERTPGFVLDVLARSSLWKIGLDYRHGTGHGVGA 596
Query: 587 ALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYVRNVETPNRFGG 646
ALNVHEGPQ ISYRYGNLT L GMIVSNEPGYYED++FGIRIENLL V+ V PN FGG
Sbjct: 597 ALNVHEGPQSISYRYGNLTALQKGMIVSNEPGYYEDNSFGIRIENLLLVKEVNLPNSFGG 656
Query: 647 IQYLGFEKLTYVPIQIKLVDVSLLSTTEIDWLNNYHSVVWEKVSPLLDGSARQWLWNNTQ 706
+ YLGFEKLT+VPIQ KLVD+SLLS +EI+W+N YH VWEKVSPLL G + WL NT+
Sbjct: 657 VSYLGFEKLTFVPIQSKLVDLSLLSPSEINWINEYHDEVWEKVSPLLSGHSLDWLRKNTR 716
Query: 707 PI 708
P+
Sbjct: 717 PL 718
>UniRef100_Q9MA84 Putative aminopeptidase [Arabidopsis thaliana]
Length = 569
Score = 834 bits (2154), Expect = 0.0
Identities = 402/568 (70%), Positives = 476/568 (83%), Gaps = 22/568 (3%)
Query: 165 MRAGNPGVPTTSEWLNEVLAPGARVGIDPFLFTSDAAEELKHVISKKNHELVYLYNSNLV 224
MRAGNPGVPT SEW+ +VLAPG RVGIDPFLF++DAAEELK VI+KKNHELVYLYN NLV
Sbjct: 1 MRAGNPGVPTASEWIADVLAPGGRVGIDPFLFSADAAEELKEVIAKKNHELVYLYNVNLV 60
Query: 225 DEIWKEARPEPPNKPVRVHGLKYAGLDVASKLSSLRSELVQAGSSAIIVTALDEIAWLLN 284
DEIWK++RP+PP++ +R+H LKYAGLDVASKL SLR++++ AG+SAI+++ LDEIAW+LN
Sbjct: 61 DEIWKDSRPKPPSRQIRIHDLKYAGLDVASKLLSLRNQIMDAGTSAIVISMLDEIAWVLN 120
Query: 285 LRGSDIPHSPVVYAYLIVEIDGAKLFIDNSKVTEEVDDHLKKANIEIRPYNSIVSEIENL 344
LRGSD+PHSPV+YAYLIVE+D A+LF+DNSKVT EV DHLK A IE+RPY+SI+ I++L
Sbjct: 121 LRGSDVPHSPVMYAYLIVEVDQAQLFVDNSKVTVEVKDHLKNAGIELRPYDSILQGIDSL 180
Query: 345 AARGSSLWLDTSSVNAAIVNAYKAACDRYYQNYESKHKTRSKGFDGSIANSDVPIAVHKS 404
AARG+ L +D S++N AI++ YK+AC+RY +N+ES+ K ++K D S + P ++
Sbjct: 181 AARGAQLLMDPSTLNVAIISTYKSACERYSRNFESEAKVKTKFTDSSSGYTANPSGIYMQ 240
Query: 405 SPVSLAKAIKNETELKGMQECHLRDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEFRS 464
SP+S AKAIKN+ ELKGM+ HLRDAAALA FW WLE+E+ + LTEV+V+D+LLEFRS
Sbjct: 241 SPISWAKAIKNDAELKGMKNSHLRDAAALAHFWAWLEEEVHKNANLTEVDVADRLLEFRS 300
Query: 465 KQAGFLDTSFDTISG---------------------SGPNGAIIHYKPEPGSCSTVDANK 503
Q GF+DTSFDTISG SG NGAIIHYKPEP SCS VD K
Sbjct: 301 MQDGFMDTSFDTISGMVIVTLVGGTCFLTHSQYTASSGANGAIIHYKPEPESCSRVDPQK 360
Query: 504 LFLLDSGAQYVDGTTDITRTVHFGKPTTREKECFTRVLQGHIALDQAVFPEDTPGFVLDA 563
LFLLDSGAQYVDGTTDITRTVHF +P+ REKECFTRVLQGHIALDQAVFPE TPGFVLD
Sbjct: 361 LFLLDSGAQYVDGTTDITRTVHFSEPSAREKECFTRVLQGHIALDQAVFPEGTPGFVLDG 420
Query: 564 FARSFLWKVGLDYRHGTGHGVGAALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYEDH 623
FARS LWK+GLDYRHGTGHGVGAALNVHEGPQ IS+RYGN+TPL NGMIVSNEPGYYEDH
Sbjct: 421 FARSSLWKIGLDYRHGTGHGVGAALNVHEGPQSISFRYGNMTPLQNGMIVSNEPGYYEDH 480
Query: 624 AFGIRIENLLYVRNVETPNRFGGIQYLGFEKLTYVPIQIKLVDVSLLSTTEIDWLNNYHS 683
AFGIRIENLL+VR+ ETPNRFGG YLGFEKLT+ PIQ K+VDVSLLS TE+DWLN+YH+
Sbjct: 481 AFGIRIENLLHVRDAETPNRFGGATYLGFEKLTFFPIQTKMVDVSLLSDTEVDWLNSYHA 540
Query: 684 VVWEKVSPLLDGS-ARQWLWNNTQPIIR 710
VWEKVSPLL+GS +QWLWNNT+P+ +
Sbjct: 541 EVWEKVSPLLEGSTTQQWLWNNTRPLAK 568
>UniRef100_Q7RYL6 Hypothetical protein [Neurospora crassa]
Length = 614
Score = 568 bits (1465), Expect = e-160
Identities = 306/628 (48%), Positives = 404/628 (63%), Gaps = 26/628 (4%)
Query: 84 KLTALRRLFSKPDVSIDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGTAVVTNDKAA 143
+L ALR L + +V D Y++PS+D+H SE+IA+ ARR +IS FTGS GTAVVT DKAA
Sbjct: 8 RLAALRSLMKERNV--DIYVVPSEDSHASEYIAECDARRAFISGFTGSAGTAVVTLDKAA 65
Query: 144 LWTDGRYFLQAEKQLNSNWILMRAGNPGVPTTSEWLNEVLAPGARVGIDPFLFTSDAAEE 203
L TDGRYF QA KQL+ NW L++ G VPT EW + A G VGIDP L + A++
Sbjct: 66 LATDGRYFNQASKQLDENWHLLKTGLQDVPTWQEWTADESAGGKSVGIDPTLISPAVADK 125
Query: 204 LKHVISKKNHELVYLYNSNLVDEIWKEARPEPPNKPVRVHGLKYAGLDVASKLSSLRSEL 263
L I K + N NLVD +W ++RP P++PV + G KY+G A KL++LR EL
Sbjct: 126 LDGDIKKHGGAGLKAINENLVDLVWGDSRPPRPSEPVFLLGAKYSGKGTAEKLTNLRKEL 185
Query: 264 VQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEIDGAKLFIDNSKVTEEVDDH 323
+ ++A +V+ LDE+AWL NLRG+DI ++PV ++Y IV D A L++D SK+ +EV +
Sbjct: 186 EKKKAAAFVVSMLDEVAWLFNLRGNDITYNPVFFSYAIVTKDSATLYVDESKLNDEVKQY 245
Query: 324 LKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYKAACDRYYQNYESKHKT 383
L + I+PYN + + E LA S + N KA+ KH
Sbjct: 246 LAENGTGIKPYNDLFKDTEILANAAKSTSESDKPTKYLVSN--KASWALKLALGGEKHVD 303
Query: 384 RSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQECHLRDAAALAQFWDWLEKE 443
+ SP+ AKAIKNETEL+GM+ CH+RD AAL +++ WLE +
Sbjct: 304 EVR------------------SPIGDAKAIKNETELEGMRRCHIRDGAALIKYFAWLEDQ 345
Query: 444 ITNDRI-LTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDAN 502
+ N + L EVE +D+L +FRS+QA F+ SFDTIS +GPNGAIIHYKPE G+CS +D +
Sbjct: 346 LINKKAKLDEVEAADQLEQFRSEQADFVGLSFDTISSTGPNGAIIHYKPERGACSVIDPD 405
Query: 503 KLFLLDSGAQYVDGTTDITRTVHFGKPTTREKECFTRVLQGHIALDQAVFPEDTPGFVLD 562
++L DSGAQ+ DGTTD+TRT+HFG+PT E++ +T VL+G+IALD AVFP+ T GF LD
Sbjct: 406 AIYLCDSGAQFCDGTTDVTRTLHFGQPTDAERKSYTLVLKGNIALDTAVFPKGTSGFALD 465
Query: 563 AFARSFLWKVGLDYRHGTGHGVGAALNVHEGPQGISYRYGNL-TPLVNGMIVSNEPGYYE 621
A AR FLWK GLDYRHGTGHGVG+ LNVHEGP GI R + PL G ++S EPGYYE
Sbjct: 466 ALARQFLWKYGLDYRHGTGHGVGSFLNVHEGPIGIGTRKAYIDVPLAPGNVLSIEPGYYE 525
Query: 622 DHAFGIRIENLLYVRNVETPNRFGGIQYLGFEKLTYVPIQIKLVDVSLLSTTEIDWLNNY 681
D +GIRIENL VR V+T ++FG YLGFE +T VP KL+D SLL+ E DWLN
Sbjct: 526 DGNYGIRIENLAIVREVKTEHQFGDKPYLGFEHVTMVPYCRKLIDESLLTQEEKDWLNKS 585
Query: 682 HSVVWEKVSPLLDGS--ARQWLWNNTQP 707
+ + + ++ DG +WL T P
Sbjct: 586 NEEIRKNMAGYFDGDQLTTEWLLRETSP 613
>UniRef100_Q76LL3 Aminopeptidase-P [Aspergillus oryzae]
Length = 654
Score = 560 bits (1443), Expect = e-158
Identities = 297/633 (46%), Positives = 396/633 (61%), Gaps = 29/633 (4%)
Query: 80 DSDPKLTALRRLFSKPDVSIDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGTAVVTN 139
++ +L+ LR L + V D YI+PS+D+HQSE+IA RR++IS F+GS GTA+V+
Sbjct: 45 NTSERLSRLRELMQEHKV--DVYIVPSEDSHQSEYIAPCDGRREFISGFSGSAGTAIVSL 102
Query: 140 DKAALWTDGRYFLQAEKQLNSNWILMRAGNPGVPTTSEWLNEVLAPGARVGIDPFLFTSD 199
KAAL TDGRYF QA KQL++NW L++ G G PT EW E G VG+DP L T+
Sbjct: 103 SKAALSTDGRYFNQASKQLDNNWQLLKRGVEGFPTWQEWTTEQAEGGKVVGVDPALITAS 162
Query: 200 AAEELKHVISKKNHELVYLYNSNLVDEIWKEARPEPPNKPVRVHGLKYAGLDVASKLSSL 259
A L + K LV + NLVD +W + RP PP + VRVH KYAG K+S L
Sbjct: 163 GARSLSETLKKNGSTLVGV-QQNLVDLVWGKDRPAPPREKVRVHPEKYAGKSFQEKISEL 221
Query: 260 RSELVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEIDGAKLFIDNSKVTEE 319
R EL S+ IV+ LDEIAWL NLRGSDIP++PV +++ + +L++D K+T E
Sbjct: 222 RKELESRKSAGFIVSMLDEIAWLFNLRGSDIPYNPVFFSFATITPTTTELYVDADKLTPE 281
Query: 320 VDDHLKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYKAACDRYYQNYES 379
V HL + ++ I+PY++I ++ + L+ A + A N S
Sbjct: 282 VTAHLGQ-DVVIKPYDAIYADAKALSETRKQ-------------EAGETASKFLLSNKAS 327
Query: 380 KHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQECHLRDAAALAQFWDW 439
+ S G +G + SP+ AKA+KN+ EL GM+ CH+RD AAL +++ W
Sbjct: 328 WALSLSLGGEGQVEEV--------RSPIGDAKAVKNDVELAGMRACHIRDGAALTEYFAW 379
Query: 440 LEKEITNDR-ILTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAIIHYKPEPGSCST 498
LE E+ N + L EV+ +DKL + RSK F+ SFDTIS +GPNGA+IHYKPE GSCS
Sbjct: 380 LENELVNKKSTLDEVDAADKLEQIRSKHDLFVGLSFDTISSTGPNGAVIHYKPEKGSCSI 439
Query: 499 VDANKLFLLDSGAQYVDGTTDITRTVHFGKPTTREKECFTRVLQGHIALDQAVFPEDTPG 558
+D N ++L DSGAQY+DGTTD+TRT HFG+PT EK+ FT VL+G I LD AVFP+ T G
Sbjct: 440 IDPNAIYLCDSGAQYLDGTTDVTRTFHFGQPTELEKKAFTLVLKGVIGLDTAVFPKGTSG 499
Query: 559 FVLDAFARSFLWKVGLDYRHGTGHGVGAALNVHEGPQGISYRYG-NLTPLVNGMIVSNEP 617
F LD AR +LWK GLDY HGTGHG+G+ LNVHEGP G+ R P+ G ++S+EP
Sbjct: 500 FALDVLARQYLWKEGLDYLHGTGHGIGSYLNVHEGPIGVGTRVQYTEVPIAPGNVISDEP 559
Query: 618 GYYEDHAFGIRIENLLYVRNVETPNRFGGIQYLGFEKLTYVPIQIKLVDVSLLSTTEIDW 677
G+YED FGIRIEN++ R V+T ++FG +LGFE +T PI L++ SLLS E+ W
Sbjct: 560 GFYEDGKFGIRIENVIMAREVQTTHKFGDKPWLGFEHVTMAPIGRNLIEPSLLSDAELKW 619
Query: 678 LNNYHSVVWEKVSPLL--DGSARQWLWNNTQPI 708
+N+YH +WEK D R WL TQPI
Sbjct: 620 VNDYHREIWEKTHHFFENDECTRSWLQRETQPI 652
>UniRef100_UPI00002365AE UPI00002365AE UniRef100 entry
Length = 1742
Score = 549 bits (1414), Expect = e-154
Identities = 292/606 (48%), Positives = 393/606 (64%), Gaps = 27/606 (4%)
Query: 80 DSDPKLTALRRLFSKPDVSIDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGTAVVTN 139
D+ +L++LR+L + V D YI+PS+D+HQSE+IA RR++IS F+GS GTA+++
Sbjct: 5 DTTKRLSSLRQLMREHKV--DVYIVPSEDSHQSEYIAPCDGRREFISGFSGSAGTAIISL 62
Query: 140 DKAALWTDGRYFLQAEKQLNSNWILMRAGNPGVPTTSEWLNEVLAPGARVGIDPFLFTSD 199
++AAL TDGRYF QA KQL++NW L++ G GVPT+ EW+ + G VG+DP L T
Sbjct: 63 NEAALSTDGRYFNQAAKQLDNNWTLLKRGVEGVPTSQEWITQQAEGGKVVGVDPALITGA 122
Query: 200 AAEELKHVISKKNHELVYLYNSNLVDEIWKEARPEPPNKPVRVHGLKYAGLDVASKLSSL 259
AA L + K L+ + + NLVD +W RP PP + VRVH KYAG K+S L
Sbjct: 123 AARSLSDALQKSGASLIGV-SQNLVDLVWGNDRPAPPREKVRVHPEKYAGKSFQEKVSDL 181
Query: 260 RSELVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEIDGAKLFIDNSKVTEE 319
R EL ++ +++ LDEIAWLLNLRGSDIP++PV +Y IV +L+ID+ K+T E
Sbjct: 182 RKELENKKAAGFVISMLDEIAWLLNLRGSDIPYNPVFISYCIVTPTKVELYIDDEKLTPE 241
Query: 320 VDDHLKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYKAACDRYYQNYES 379
V HL ++ I+PY+SI ++ + A+ A K D +
Sbjct: 242 VKAHLGD-DVIIKPYDSIFADAK------------------ALFEAKKKDPDAPSSKFLL 282
Query: 380 KHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQECHLRDAAALAQFWDW 439
++ S + S+ D + SP+ AKA+KNE EL GM+ CH+RD AAL +++ W
Sbjct: 283 SNRA-SWALNLSLGGEDHVEEIR--SPIGDAKAVKNEVELAGMRACHIRDGAALIEYFAW 339
Query: 440 LEKEITNDR-ILTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAIIHYKPEPGSCST 498
LE E+ N + L EV+ +DKL + RSKQ F SFDTIS +GPNGA+IHYKPE GSCS
Sbjct: 340 LENELVNKKSTLDEVDAADKLEQLRSKQELFAGLSFDTISSTGPNGAVIHYKPEKGSCSV 399
Query: 499 VDANKLFLLDSGAQYVDGTTDITRTVHFGKPTTREKECFTRVLQGHIALDQAVFPEDTPG 558
+D N ++L DSG QY+DGTTD+TRT HFG+PT EK+ FT VL+G I LD AVFP+ T G
Sbjct: 400 IDPNAIYLCDSGGQYLDGTTDVTRTFHFGQPTELEKKAFTLVLKGCIGLDSAVFPKGTSG 459
Query: 559 FVLDAFARSFLWKVGLDYRHGTGHGVGAALNVHEGPQGISYRYG-NLTPLVNGMIVSNEP 617
F LD AR LWK GLD+ HGTGHG+G+ LNVHEGP GI R PL G ++S+EP
Sbjct: 460 FALDVLARQHLWKEGLDFLHGTGHGIGSYLNVHEGPVGIGTRVQYTEVPLAPGNVISDEP 519
Query: 618 GYYEDHAFGIRIENLLYVRNVETPNRFGGIQYLGFEKLTYVPIQIKLVDVSLLSTTEIDW 677
G+YED FGIRIEN++ VR V+T ++FG +LGFE +T PI L++ SLLS +EI W
Sbjct: 520 GFYEDGKFGIRIENVIMVREVQTTHKFGERPWLGFEHVTMCPIGQNLIEPSLLSDSEIKW 579
Query: 678 LNNYHS 683
LN+YH+
Sbjct: 580 LNDYHA 585
>UniRef100_UPI000021B477 UPI000021B477 UniRef100 entry
Length = 618
Score = 549 bits (1414), Expect = e-154
Identities = 292/615 (47%), Positives = 389/615 (62%), Gaps = 23/615 (3%)
Query: 98 SIDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGTAVVTNDKAALWTDGRYFLQAEKQ 157
SID YI+P++DAH SE+IA RR++IS F+GS GTAVVTNDKAAL TDGRYF QA +
Sbjct: 21 SIDVYIVPTEDAHSSEYIAPCDGRREFISGFSGSAGTAVVTNDKAALATDGRYFNQAATE 80
Query: 158 LNSNWILMRAGNPGVPTTSEWLNEVLAPGARVGIDPFLFTSDAAEELKHVISKKNHELVY 217
L++NW L++ G P VPT EW + A G VG+DP L +S A+ L+ I K +
Sbjct: 81 LDNNWELLKQGQPDVPTWQEWTADQAAGGKTVGVDPTLLSSSEAKALQEKIKSKGGNDLV 140
Query: 218 LYNSNLVDEIWKEARPEPPNKPVRVHGLKYAGLDVASKLSSLRSELVQAGSSAIIVTALD 277
+ NLVD +W +P P+ P+ KY+G D KL LR L + +++ LD
Sbjct: 141 AISDNLVDLVWGRHKPSRPSNPIAFLPKKYSGKDTEPKLKELREVLEKKKVFGFVISTLD 200
Query: 278 EIAWLLNLRGSDIPHSPVVYAYLIVEIDGAKLFIDNSKVTEEVDDHLKKANIEIRPYNSI 337
EIAWL NLRGSDIP++PV ++Y +V D A L++D SK++EE +LK+ ++IRPY SI
Sbjct: 201 EIAWLFNLRGSDIPYNPVFFSYAVVTADNATLYVDASKLSEESHAYLKENKVDIRPYESI 260
Query: 338 VSEIENLAARGSSLWLDTSSVNAAIVNAYKAACDRYYQNYESKHKTRSKGFDGSIANSDV 397
+ E LA L + K A N S + G DG++
Sbjct: 261 FEDSEVLAKS-----LKPTEDQGEESKVKKLA----ISNKTSWALKLALGGDGAVDEI-- 309
Query: 398 PIAVHKSSPVSLAKAIKNETELKGMQECHLRDAAALAQFWDWLEKEITNDR-ILTEVEVS 456
SPV AKAIKNETEL+GM++CH+RD AAL +++ WLE ++ N + L EV+ +
Sbjct: 310 ------KSPVCDAKAIKNETELEGMRQCHIRDGAALIEYFAWLEDQVANKKATLNEVQAA 363
Query: 457 DKLLEFRSKQAGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDG 516
KL R+K F+ SF TIS G N A+IHYKPE SC+T+DA+ ++L DSGAQ++DG
Sbjct: 364 TKLENLRAKHEDFVGLSFTTISAVGANAAVIHYKPEEDSCATIDADSVYLCDSGAQFLDG 423
Query: 517 TTDITRTVHFGKPTTREKECFTRVLQGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLDY 576
TTD TRT+HFGKP+ E++ +T VL+G++ALD A+FP+ T GF LD FAR FLW+ GLDY
Sbjct: 424 TTDTTRTLHFGKPSEAERKAYTLVLKGNMALDMAIFPKGTTGFALDPFARQFLWQEGLDY 483
Query: 577 RHGTGHGVGAALNVHEGPQGISYR--YGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLY 634
RHGTGHGVG+ LNVHEGP GI R Y + PL G + S EPG+YED ++GIRIEN+
Sbjct: 484 RHGTGHGVGSYLNVHEGPIGIGTRKHYAGV-PLAPGNVTSIEPGFYEDGSYGIRIENIAM 542
Query: 635 VRNVETPNRFGGIQYLGFEKLTYVPIQIKLVDVSLLSTTEIDWLNNYHSVVWEKVSPLL- 693
+R VET + FG YLGFE +T VP +L+D SLL+ E WLN+Y+ ++ +K S
Sbjct: 543 IREVETKHMFGDKPYLGFEHVTMVPYCRRLIDESLLTPREKQWLNDYNKLILDKTSGFFK 602
Query: 694 -DGSARQWLWNNTQP 707
D WL TQP
Sbjct: 603 DDNLTMAWLERETQP 617
>UniRef100_UPI00003AE3CB UPI00003AE3CB UniRef100 entry
Length = 621
Score = 542 bits (1396), Expect = e-152
Identities = 297/654 (45%), Positives = 406/654 (61%), Gaps = 61/654 (9%)
Query: 83 PKLTA-----LRRLFSKP---DVSIDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGT 134
PK+T LR++ P + AYI+PS DAHQSE+IA RR +IS F GS GT
Sbjct: 3 PKITTELLKQLRQVMKSPRYVQEPVQAYIVPSGDAHQSEYIAPCDCRRAFISGFDGSAGT 62
Query: 135 AVVTNDKAALWTDGRYFLQAEKQLNSNWILMRAGNPGVPTTSEWLNEVLAPGARVGIDPF 194
A+VT AA+WTDGRYFLQA Q+++NW LM+ G PT +WL VL G++VG+DP
Sbjct: 63 AIVTEQHAAMWTDGRYFLQAANQMDNNWTLMKMGLKDTPTQEDWLVSVLPEGSKVGVDPS 122
Query: 195 LFTSDAAEELKHVISKKNHELVYLYNSNLVDEIWKEARPEPPNKPVRVHGLKYAGLDVAS 254
+ +D + + V+ H+LV + NL+D IW + RP+ P KP+ V L Y G+
Sbjct: 123 IIPADQWKRMSKVLRSAGHDLVPV-KENLIDTIWTD-RPQRPCKPLIVLDLSYTGVSWRD 180
Query: 255 KLSSLRSELVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEIDGAKLFIDNS 314
K+ +LRS++ + +VTALDE+AWL NLRGSD+ ++PV +AY ++ ++ +LFID
Sbjct: 181 KIVALRSKMAERKVVWFVVTALDEVAWLFNLRGSDVEYNPVFFAYAVIGMNTIRLFIDGD 240
Query: 315 KVTEE-VDDHLK-------KANIEIRPYNSIVSEIENLAARGS---SLWLDTSSVNA--- 360
++ + V +HL+ + I++ PY SI++E++ + A S +WL + A
Sbjct: 241 RMMDPAVREHLQLDSTLEPEFKIQVMPYRSILTELQAVGAGLSPKEKVWLSDKASYALTE 300
Query: 361 AIVNAYKAACDRYYQNYESKHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETELK 420
AI AY RY Y +P+ +AKA+KN E +
Sbjct: 301 AIPKAY-----RYLTPY---------------------------TPICIAKAVKNALETE 328
Query: 421 GMQECHLRDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEFRSKQAGFLDTSFDTISGS 480
GM+ H++DA AL + ++WLEKE+ + TE+ +DK EFRS+Q F++ SF TIS +
Sbjct: 329 GMRRAHIKDAVALCELFNWLEKEVPKGTV-TEIIAADKAEEFRSQQKDFVELSFATISST 387
Query: 481 GPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRTVHFGKPTTREKECFTRV 540
GPNGAIIHYKP P + T+ N+++LLDSGAQY DGTTD+TRT+HFG P+ EKECFT V
Sbjct: 388 GPNGAIIHYKPVPETNRTLSVNEIYLLDSGAQYKDGTTDVTRTMHFGTPSAYEKECFTCV 447
Query: 541 LQGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLDYRHGTGHGVGAALNVHEGPQGISYR 600
L+GHIA+ A+FP T G +LD+FARS LW GLDY HGTGHGVG+ LNVHEGP GISY+
Sbjct: 448 LKGHIAVSAAIFPNGTKGHLLDSFARSALWDCGLDYLHGTGHGVGSFLNVHEGPCGISYK 507
Query: 601 YGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYVRNVETPNRFGGIQYLGFEKLTYVPI 660
PL GMIVS+EPGYYED +FGIRIEN++ V ET F L FE LT VPI
Sbjct: 508 TFADEPLEAGMIVSDEPGYYEDGSFGIRIENVVLVIPAETKYNFKNRGSLTFEPLTLVPI 567
Query: 661 QIKLVDVSLLSTTEIDWLNNYHSVVWEKVSPLLDGSAR----QWLWNNTQPIIR 710
Q K++DVSLL+ E +W+N+YH E + L+ R +WL T+P+IR
Sbjct: 568 QTKMIDVSLLTQKECNWVNDYHQKCREVIGAELERQGRHEALRWLIRETEPLIR 621
>UniRef100_Q655Z0 Putative Xaa-Pro aminopeptidase 2 [Oryza sativa]
Length = 648
Score = 536 bits (1382), Expect = e-151
Identities = 287/662 (43%), Positives = 403/662 (60%), Gaps = 51/662 (7%)
Query: 76 SSNSDSDPKLTALRRLFSKPDVSIDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGTA 135
S+ + D L LR L + S+ A ++PS+DAHQSE++++ RR+++S FTGS G A
Sbjct: 5 SAAAGRDALLDELRALMAAHSPSLHALVVPSEDAHQSEYVSERDKRRQFVSGFTGSAGLA 64
Query: 136 VVTNDKAALWTDGRYFLQAEKQLNSNWILMRAGNPGVPTTSEWLNEVLAPGARVGIDPFL 195
++T +A LWTDGRYFLQAE+QL + W LMR G P W+ + L+ A +GI+P+
Sbjct: 65 LITMKEALLWTDGRYFLQAEQQLTNRWKLMRMGED--PPVEVWIADNLSDEAVIGINPWC 122
Query: 196 FTSDAAEELKHVISKKNHELVYLYNSNLVDEIWKEARPEPPNKPVRVHGLKYAGLDVASK 255
+ D A+ +H SKK H+ ++ +S+LVDEIWK+ RP PV V ++YAG V K
Sbjct: 123 ISVDTAQRYEHAFSKK-HQTLFQLSSDLVDEIWKD-RPPVNALPVFVQPVEYAGCSVTEK 180
Query: 256 LSSLRSELVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEIDGAKLFIDNSK 315
L LR +L + II+ ALDE+AWL N+RG+D+ +SPVV++Y IV + A ++D K
Sbjct: 181 LKELREKLQHEKARGIIIAALDEVAWLYNIRGNDVHYSPVVHSYSIVTLHSAFFYVDKRK 240
Query: 316 VTEEVDDHLKKANIEIRPYNSIVSEIENLAA--------RGSSLWLDTSSVNAAIVNAYK 367
V+ EV +++ + I+I+ YN + S+ LA+ GSS + + N+ +
Sbjct: 241 VSVEVQNYMTENGIDIKDYNMVQSDASLLASGQLKGSAVNGSSHGENDMNENSKVWIDSN 300
Query: 368 AACDRYYQNYESKHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQECHL 427
+ C Y + + SP++L KA+KN EL G+++ H+
Sbjct: 301 SCCLALYSKLDQYQ------------------VLMLQSPIALPKAVKNPVELDGLRKAHI 342
Query: 428 RDAAALAQFWDWLEKEITNDR--------------------ILTEVEVSDKLLEFRSKQA 467
RD AA+ Q+ WL+K++ + LTEV VSDKL FR+ +
Sbjct: 343 RDGAAVVQYLAWLDKQMQENYGASGYFTEAKGSQKKEHMNVKLTEVSVSDKLEGFRASKE 402
Query: 468 GFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRTVHFG 527
F SF TIS GPN A+IHYKPE SC+ +DA+K++L DSGAQY+DGTTDITRTVHFG
Sbjct: 403 HFKGLSFPTISSVGPNAAVIHYKPEASSCAELDADKIYLCDSGAQYLDGTTDITRTVHFG 462
Query: 528 KPTTREKECFTRVLQGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLDYRHGTGHGVGAA 587
KP+ EK C+T VL+GHIALD AVFP T G +D AR+ LW+ GLDYRHGTGHG+G+
Sbjct: 463 KPSEHEKSCYTAVLKGHIALDTAVFPNGTTGHAIDILARTPLWRSGLDYRHGTGHGIGSY 522
Query: 588 LNVHEGPQGISYR-YGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYVRNVETPNRFGG 646
LNVHEGP IS+R PL M V++EPGYYED +FGIR+EN+L V+ T FG
Sbjct: 523 LNVHEGPHLISFRPSARNVPLQASMTVTDEPGYYEDGSFGIRLENVLIVKEANTKFNFGD 582
Query: 647 IQYLGFEKLTYVPIQIKLVDVSLLSTTEIDWLNNYHSVVWEKVSPLLDGSARQWLWNNTQ 706
YL FE +T+ P Q KL+D +LL+ EI+W+N YHS + + P L+ ++WL T+
Sbjct: 583 KGYLAFEHITWTPYQTKLIDTTLLTPAEIEWVNAYHSDCRKILQPYLNEQEKEWLRKATE 642
Query: 707 PI 708
PI
Sbjct: 643 PI 644
>UniRef100_UPI000023DCBC UPI000023DCBC UniRef100 entry
Length = 642
Score = 531 bits (1368), Expect = e-149
Identities = 301/660 (45%), Positives = 401/660 (60%), Gaps = 39/660 (5%)
Query: 67 PSSQLRKNRSSNSDSDP---------KLTALRRLFSKPDVSIDAYIIPSQDAHQSEFIAQ 117
PSSQ + + + DS P +LT LR L + +V + YIIPS+D+H SE+IA+
Sbjct: 6 PSSQNIEPPTYSIDSSPSLQFVSSLLRLTRLRGLMKERNVQV--YIIPSEDSHSSEYIAE 63
Query: 118 SYARRKYISAFTGSNGTAVVTNDKAALWTDGRYFLQAEKQLNSNWILMRAGNPGVPTTSE 177
ARR YIS FTGS G AVVT + AAL TDGRYF QA QL+ NW L++ G VPT +
Sbjct: 64 CDARRAYISGFTGSAGCAVVTLESAALATDGRYFNQAASQLDDNWTLLKQGLQDVPTWQD 123
Query: 178 WLNEVLAPGARVGIDPFLFTSDAAEELKHVISKKNHELVYLYNSNLVDEIWKEARPEPPN 237
W E + G VG+DP L + A+ L I K + + NLVD +W + RP P+
Sbjct: 124 WSAEQSSGGKNVGVDPTLISGSTAKGLAEKIRKNGGAELVAVDGNLVDLVWGDERPARPS 183
Query: 238 KPVRVHGLKYAGLDVASKLSSLRSELVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVY 297
+ V + + AG V +KL+ +R E+ + S +V+ LDEIAWL NLRGSDIP++PV +
Sbjct: 184 EKVIIQPDELAGESVLNKLNKVRQEMGKKHSPGFLVSMLDEIAWLFNLRGSDIPYNPVFF 243
Query: 298 AYLIVEIDGAKLFIDNSKVTEEVDDHLKKANIEIRPYNSIVSEIENLAARGSSLWLDTSS 357
AY V D AKL+ID+SK+ +E HL +EI+PY+++ + + L A S
Sbjct: 244 AYATVTPDAAKLYIDDSKLDDECRSHLTSNKVEIKPYDTVFEDSQALHA----------S 293
Query: 358 VNAAIVNAYKAACDRY-YQNYESKHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNE 416
V+ KA + N S R+ G D S+ S + AKAIK E
Sbjct: 294 VSEKTKTDDKAPKGNFLISNKGSWALKRAIGGDSSVDEI--------RSLIGDAKAIKTE 345
Query: 417 TELKGMQECHLRDAAALAQFWDWLEKEITNDR-ILTEVEVSDKLLEFRSKQAGFLDTSFD 475
ELKGM++CH+RD A+L Q++ WLE ++ N + L EV+ +DKL R ++ F+ SF
Sbjct: 346 AELKGMRDCHVRDGASLIQYFAWLEDQLVNKKATLDEVQAADKLEALRKEKKDFVGLSFP 405
Query: 476 TISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRTVHFGKPTTREKE 535
TIS +G N AIIHY PE G+C+T+D ++L DSGAQY DGTTD TRT+HFG PT E+E
Sbjct: 406 TISSTGANAAIIHYGPERGNCATIDPKAIYLCDSGAQYRDGTTDTTRTLHFGTPTDAERE 465
Query: 536 CFTRVLQGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLDYRHGTGHGVGAALNVHEGPQ 595
+T VL+GHI+LDQA+FP+ T GF LD AR LWK GLDYRHGTGHGVG+ LNVHEGP
Sbjct: 466 AYTLVLKGHISLDQAIFPKGTTGFALDGLARQHLWKNGLDYRHGTGHGVGSFLNVHEGPI 525
Query: 596 GISYR--YGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYVRNVETPNRFGGIQYLGFE 653
GI R Y + L G ++SNEPGYYED +GIRIEN++ V+ V+T + FG +LGFE
Sbjct: 526 GIGTRVQYAEVA-LAPGNVLSNEPGYYEDGKYGIRIENMVLVKEVKTKHSFGDKPFLGFE 584
Query: 654 KLTYVPIQIKLVDVSLLSTTEIDWLNNYHSVVWEKVSPLLDGSARQWLWNNTQPIIRETV 713
+T VP L+D +LL++ E DWLN Y+ V EK +G W + RETV
Sbjct: 585 YVTMVPYCRNLIDTTLLTSVEKDWLNTYNEKVIEKTQGYFEGDDVTTAW-----LKRETV 639
>UniRef100_UPI000036180F UPI000036180F UniRef100 entry
Length = 625
Score = 527 bits (1357), Expect = e-148
Identities = 289/658 (43%), Positives = 406/658 (60%), Gaps = 61/658 (9%)
Query: 77 SNSDSDPKLTA--LRRL---------FSKPDVSIDAYIIPSQDAHQSEFIAQSYARRKYI 125
S+S PK+T +R+L F++P I AYI+PS DAHQSE+IA RR+YI
Sbjct: 3 SDSAMSPKITGELIRKLRQAMKSCKYFAEP---IQAYIVPSGDAHQSEYIAPCDCRREYI 59
Query: 126 SAFTGSNGTAVVTNDKAALWTDGRYFLQAEKQLNSNWILMRAGNPGVPTTSEWLNEVLAP 185
F GS GTA++T AA+WTDGRYFLQA +Q+++NW LM+ G P+ +WL VL
Sbjct: 60 CGFNGSAGTAIITEQHAAMWTDGRYFLQASQQMDNNWTLMKMGLKETPSQEDWLISVLPE 119
Query: 186 GARVGIDPFLFTSDAAEELKHVISKKNHELVYLYNSNLVDEIWKEARPEPPNKPVRVHGL 245
++VG+DP++ +D + + ++ H LV + + NL+D +W + RPE P+ +R GL
Sbjct: 120 NSKVGVDPWIIAADQWKNMSKALTGAGHSLVAMQD-NLIDVVWTD-RPERPSTQLRTLGL 177
Query: 246 KYAGLDVASKLSSLRSELVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEID 305
+Y GL K+++LR+++ + S + TALDEIAWL NLRG+DI ++PV +AY IV ++
Sbjct: 178 EYTGLSWHDKVTALRAKMTERKISWFVATALDEIAWLFNLRGADIEYNPVFFAYTIVGLN 237
Query: 306 GAKLFIDNSKVTEE-VDDHLK---KANIEIR----PYNSIVSEIENLAAR---GSSLWLD 354
+LF+D ++T+ + HL+ + E+R PY S+ +E++ + A +W+
Sbjct: 238 TIRLFVDTKRLTDPALRGHLELDSPSQPELRILTFPYESVYTELQAICAALGPKDKVWIC 297
Query: 355 TSSVNAAIVNAYKAACDRYYQNYESKHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKAIK 414
+ A KA S +P +P+ L+KA+K
Sbjct: 298 DKASRALTQLLPKAN------------------------RSPIPY-----TPLCLSKAVK 328
Query: 415 NETELKGMQECHLRDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEFRSKQAGFLDTSF 474
N TE++GM+ H++DA AL + + WLEKEI + TE+ +DK E RS+Q F+ SF
Sbjct: 329 NATEIQGMKVAHIKDAVALCELFAWLEKEIPKGNV-TEISAADKAEELRSQQKDFVGLSF 387
Query: 475 DTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRTVHFGKPTTREK 534
TISG GPNGA+IHY+P P + T+ N+++L+DSGAQY+DGTTD+TRTVHFG P+ EK
Sbjct: 388 PTISGVGPNGAVIHYRPLPETNRTLSMNEVYLIDSGAQYIDGTTDVTRTVHFGTPSAFEK 447
Query: 535 ECFTRVLQGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLDYRHGTGHGVGAALNVHEGP 594
ECFT VL+GHIA+ AVFP T G +LD+FAR+ LW GLDY HGTGHGVG LNVHEGP
Sbjct: 448 ECFTYVLKGHIAVSAAVFPNGTKGHLLDSFARAALWDSGLDYLHGTGHGVGCFLNVHEGP 507
Query: 595 QGISYRYGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYVRNVETPNRFGGIQYLGFEK 654
GISY+ PL GMIVS+EPGYYED AFGIRIEN++ V + + L FE
Sbjct: 508 CGISYKTFADEPLEAGMIVSDEPGYYEDGAFGIRIENVVLVVPAQPKYNYRNRGSLTFEP 567
Query: 655 LTYVPIQIKLVDVSLLSTTEIDWLNNYHSVVWEKVSPLLDGSAR----QWLWNNTQPI 708
LT VPIQ+K+++ +LL+ E DWLN YH E + L+ R +WL TQP+
Sbjct: 568 LTLVPIQVKMINTALLTQKERDWLNEYHRTCREVIGAELERQGRKEALEWLVRETQPV 625
>UniRef100_UPI000042DE85 UPI000042DE85 UniRef100 entry
Length = 647
Score = 512 bits (1318), Expect = e-143
Identities = 280/646 (43%), Positives = 388/646 (59%), Gaps = 20/646 (3%)
Query: 81 SDPKLTALRRLFSKPDVSIDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGTAVVTND 140
S KL LR+L + V DAY++PS+DAH SE++A ARR YI+ FTGS G AV+T+D
Sbjct: 4 SSSKLADLRQLMKEQGV--DAYVVPSEDAHASEYLAPCDARRAYITGFTGSAGCAVITHD 61
Query: 141 KAALWTDGRYFLQAEKQLNSNWILMRAGNPGVPTTSEWLNEVLAPGARVGIDPFLFT-SD 199
KA WTDGRY+LQAEKQL W LM++G P VPT ++WL+ ++P + +GIDP + S+
Sbjct: 62 KALCWTDGRYWLQAEKQLGEGWALMKSGLPEVPTWAQWLSTEVSPNSLIGIDPTVIPYSE 121
Query: 200 AAEELKHVISKKNHELV-----YLYNSNLVDEIW-KEARPEPPNKPVRVHGLKYAGLDVA 253
A L + S + NL+D +W +RP P++P+ +Y G V+
Sbjct: 122 ALSLLSSLPSLSPAPSAASPSRLIATPNLIDSLWVPPSRPLRPSQPIFHLADRYTGEPVS 181
Query: 254 SKLSSLRSELVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEIDGAKLFIDN 313
SKL LR +L++ GS +V +LDEIAW+ NLRG+DIP++PV +AY I+ D LF+
Sbjct: 182 SKLRRLRDKLIRIGSPGTVVASLDEIAWVFNLRGADIPYNPVFFAYTIITPDDCTLFVSP 241
Query: 314 SKVTEEVDDHLKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYKAACDRY 373
S +T EV +L I + Y+ + + +E R + S V + +
Sbjct: 242 SSLTIEVRSYLHSNGIAVLDYSHVWTSLEAWKKRVKFDQENKSREQRDGVKRARLEEEAK 301
Query: 374 YQNYESKHKTRSKGFDGSIANSDVPIAVH------KSSPVSLAKAIKNETELKGMQECHL 427
+ + K K G+ + V AV + S + KA KN TE++G ++CH+
Sbjct: 302 KEEEGERLKKTDKILIGNKTSWAVAKAVGEDNVEVRRSLIEEMKAKKNATEIEGFRQCHI 361
Query: 428 RDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAII 487
RD AAL ++ WLE+ + N TE + + KL +FR + F+ SF+TIS +G N A+I
Sbjct: 362 RDGAALVRYLAWLEEALENGESWTEYDAATKLEDFRKENKLFMGLSFETISSTGANAAVI 421
Query: 488 HYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRTVHFGKPTTREKECFTRVLQGHIAL 547
HY P ++ +++L DSGAQY+DGTTD+TRT+HFG P +K FTRVLQGHI+L
Sbjct: 422 HYSPPAEGSKMIEKKQMYLCDSGAQYLDGTTDVTRTLHFGTPNEDQKRAFTRVLQGHISL 481
Query: 548 DQAVFPEDTPGFVLDAFARSFLWKVGLDYRHGTGHGVGAALNVHEGPQGISYRYG-NLTP 606
D VFP+ T G++LD AR LW GLDYRH T HG+G+ LNVHEGPQGI R N P
Sbjct: 482 DTIVFPQGTTGYILDVLARRALWSEGLDYRHSTSHGIGSFLNVHEGPQGIGQRPAYNEVP 541
Query: 607 LVNGMIVSNEPGYYEDHAFGIRIENLLYVRNVETPNRFGGIQYLGFEKLTYVPIQIKLVD 666
L GM++SNEPGYY+D +GIRIE + + ET FGG +LGFE++T PIQ KLVD
Sbjct: 542 LQEGMVISNEPGYYKDGEWGIRIEGVDVIERRETRENFGGKGWLGFERITMCPIQTKLVD 601
Query: 667 VSLLSTTEIDWLNNYHSVVWEKVSPLL----DGSARQWLWNNTQPI 708
SLL+ E DWLN YH+ V K++P+L D A +WL QP+
Sbjct: 602 SSLLTIEEKDWLNEYHAEVLAKLAPVLKEMGDERAGKWLERECQPL 647
>UniRef100_Q6C5C7 Similar to tr|Q8RY11 Arabidopsis thaliana AT3g05350/T12H1_32
[Yarrowia lipolytica]
Length = 651
Score = 511 bits (1317), Expect = e-143
Identities = 292/670 (43%), Positives = 416/670 (61%), Gaps = 59/670 (8%)
Query: 69 SQLRKNRSSNSD------SDPKLTALRRLFSKPDVSIDAYIIPSQDAHQSEFIAQSYARR 122
S L + RS ++D S KL LR+L + + + YI+PS+DAHQSE+ + RR
Sbjct: 10 SFLTRTRSFHTDFTMTASSGEKLALLRQLMASKGLGV--YIVPSEDAHQSEYTSVCDQRR 67
Query: 123 KYISAFTGSNGTAVVTNDKAALWTDGRYFLQAEKQLNSN-WILMRAGNPGVPTTSEW-LN 180
YIS FTGS GTAV+T+D AAL TDGRYFLQA++QL+ W L++ G GVPT E+ ++
Sbjct: 68 AYISGFTGSAGTAVITSDTAALATDGRYFLQADEQLDKKYWNLLKQGVKGVPTWQEYAID 127
Query: 181 EVLAPGARVGIDPFLFTSDAAEELKHVISKKNHE--------------LVYLYNSNLVDE 226
+ G +G+D L ++ AE++ ++ K E LV L++ NLVD
Sbjct: 128 YAIKHGVDIGVDSRLVSAVEAEDITKKLALKIEEAGVQADEKNASSVKLVGLHD-NLVDA 186
Query: 227 IWKEARPEP--PNKPVRVHGLKYAGLDVASKLSSLRSELVQAGSSAIIVTALDEIAWLLN 284
+W + +P P P +KY G KL LR ++ ++G SAII++ALDEIAWLLN
Sbjct: 187 VWSKLDTQPCRPGDPAFPLDVKYTGKPFDLKLEELRVKMRESGGSAIIISALDEIAWLLN 246
Query: 285 LRGSDIPHSPVVYAYLIVEIDGAKLFIDNSKVTEEVDDHLKKANIEIRPYNSIVSEIENL 344
LRGSDIP++PV + Y+IV + L+ D+ K+TE + HL I++RPY+ + ++ + L
Sbjct: 247 LRGSDIPYNPVFFGYVIVTPNYTTLYCDSKKITEACEKHLDGL-IDLRPYDDVFADFKKL 305
Query: 345 --AARGSSLWLDTSSVNAAIVNAYKAACDRYYQNYESKHKTRSKGFDGSIANSDVPIAVH 402
AA+ L + + A+V +++++KT ++
Sbjct: 306 GEAAQHDKLVFVPKNSSWALVECLGG--------FKNENKTYTQ---------------- 341
Query: 403 KSSPVSLAKAIKNETELKGMQECHLRDAAALAQFWDWLEK--EITNDRILTEVEVSDKLL 460
+SPV AKA+KN+TE +G + HL+D AAL +F+ WLE + N L EV+ + KL+
Sbjct: 342 ITSPVLKAKAVKNKTEQEGARAAHLKDGAALCEFFCWLEGVYDAGNPDKLDEVDAASKLV 401
Query: 461 EFRSKQAGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDI 520
EFR KQ F+ SF++IS GPN AIIHY PE + +D +K++L D+G+Q+++GTTD
Sbjct: 402 EFREKQPNFVGLSFESISSVGPNAAIIHYAPEKPKAAILDPSKVYLSDTGSQFLEGTTDT 461
Query: 521 TRTVHFGKPTTREKECFTRVLQGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLDYRHGT 580
TRT HFG P+ E+ T VL+GHIAL ++VFPE T GF LD AR FLWK GLDYRHGT
Sbjct: 462 TRTWHFGSPSDEERTSNTLVLKGHIALAESVFPEGTTGFALDILARQFLWKYGLDYRHGT 521
Query: 581 GHGVGAALNVHEGPQGISYR--YGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYVRNV 638
GHG+GA LNVHEGP GI +R Y + P+ G +VSNEPGYY+D +GIRIE++L +
Sbjct: 522 GHGIGAFLNVHEGPFGIGFRPAYRDF-PMEIGNVVSNEPGYYKDGEYGIRIESVLICKEK 580
Query: 639 ETPNRFGGIQYLGFEKLTYVPIQIKLVDVSLLSTTEIDWLNNYHSVVWEKVSPLLDGSAR 698
+T FGG +YLGFE +T VP+ KL+DVS+L +E W+N+YH VV +V PL++G +
Sbjct: 581 KTQENFGGKKYLGFETITRVPLCHKLIDVSMLEDSEKKWVNHYHQVVRNEVGPLVEGEVK 640
Query: 699 QWLWNNTQPI 708
+WL T P+
Sbjct: 641 EWLLKETAPL 650
>UniRef100_UPI0000361812 UPI0000361812 UniRef100 entry
Length = 604
Score = 509 bits (1310), Expect = e-142
Identities = 279/631 (44%), Positives = 390/631 (61%), Gaps = 54/631 (8%)
Query: 97 VSIDAYIIP-SQDAH--QSEFIAQSYARRKYISAFTGSNGTAVVTNDKAALWTDGRYFLQ 153
+S+ Y+ P S H QSE+IA RR+YI F GS GTA++T AA+WTDGRYFLQ
Sbjct: 10 LSLPLYVSPLSSPPHVFQSEYIAPCDCRREYICGFNGSAGTAIITEQHAAMWTDGRYFLQ 69
Query: 154 AEKQLNSNWILMRAGNPGVPTTSEWLNEVLAPGARVGIDPFLFTSDAAEELKHVISKKNH 213
A +Q+++NW LM+ G P+ +WL VL ++VG+DP++ +D + + ++ H
Sbjct: 70 ASQQMDNNWTLMKMGLKETPSQEDWLISVLPENSKVGVDPWIIAADQWKNMSKALTGAGH 129
Query: 214 ELVYLYNSNLVDEIWKEARPEPPNKPVRVHGLKYAGLDVASKLSSLRSELVQAGSSAIIV 273
LV + + NL+D +W + RPE P+ +R GL+Y GL K+++LR+++ + S +
Sbjct: 130 SLVAMQD-NLIDVVWTD-RPERPSTQLRTLGLEYTGLSWHDKVTALRAKMTERKISWFVA 187
Query: 274 TALDEIAWLLNLRGSDIPHSPVVYAYLIVEIDGAKLFIDNSKVTEE-VDDHLK---KANI 329
TALDEIAWL NLRG+DI ++PV +AY IV ++ +LF+D ++T+ + HL+ +
Sbjct: 188 TALDEIAWLFNLRGADIEYNPVFFAYTIVGLNTIRLFVDTKRLTDPALRGHLELDSPSQP 247
Query: 330 EIR----PYNSIVSEIENLAAR---GSSLWLDTSSVNAAIVNAYKAACDRYYQNYESKHK 382
E+R PY S+ +E++ + A +W+ + A KA
Sbjct: 248 ELRILTFPYESVYTELQAICAALGPKDKVWICDKASRALTQLLPKAN------------- 294
Query: 383 TRSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQECHLRDAAALAQFWDWLEK 442
S +P +P+ L+KA+KN TE++GM+ H++DA AL + + WLEK
Sbjct: 295 -----------RSPIPY-----TPLCLSKAVKNATEIQGMKVAHIKDAVALCELFAWLEK 338
Query: 443 EITNDRILTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDAN 502
EI + TE+ +DK E RS+Q F+ SF TISG GPNGA+IHY+P P + T+ N
Sbjct: 339 EIPKGNV-TEISAADKAEELRSQQKDFVGLSFPTISGVGPNGAVIHYRPLPETNRTLSMN 397
Query: 503 KLFLLDSGAQYVDGTTDITRTVHFGKPTTREKECFTRVLQGHIALDQAVFPEDTPGFVLD 562
+++L+DSGAQY+DGTTD+TRTVHFG P+ EKECFT VL+GHIA+ AVFP T G +LD
Sbjct: 398 EVYLIDSGAQYIDGTTDVTRTVHFGTPSAFEKECFTYVLKGHIAVSAAVFPNGTKGHLLD 457
Query: 563 AFARSFLWKVGLDYRHGTGHGVGAALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYED 622
+FAR+ LW GLDY HGTGHGVG LNVHEGP GISY+ PL GMIVS+EPGYYED
Sbjct: 458 SFARAALWDSGLDYLHGTGHGVGCFLNVHEGPCGISYKTFADEPLEAGMIVSDEPGYYED 517
Query: 623 HAFGIRIENLLYVRNVETPNRFGGIQYLGFEKLTYVPIQIKLVDVSLLSTTEIDWLNNYH 682
AFGIRIEN++ V NR L FE LT VPIQ+K+++ +LL+ E DWLN YH
Sbjct: 518 GAFGIRIENVVLVVPASQKNRGS----LTFEPLTLVPIQVKMINTALLTQKERDWLNEYH 573
Query: 683 SVVWEKVSPLLDGSAR----QWLWNNTQPII 709
E + L+ R +WL TQP+I
Sbjct: 574 RTCREVIGAELERQGRKEALEWLVRETQPVI 604
>UniRef100_UPI00003AE3CC UPI00003AE3CC UniRef100 entry
Length = 597
Score = 501 bits (1289), Expect = e-140
Identities = 283/652 (43%), Positives = 387/652 (58%), Gaps = 85/652 (13%)
Query: 83 PKLTA-----LRRLFSKP---DVSIDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGT 134
PK+T LR++ P + AYI+PS DAHQSE+IA RR +IS F GS GT
Sbjct: 5 PKITTELLKQLRQVMKSPRYVQEPVQAYIVPSGDAHQSEYIAPCDCRRAFISGFDGSAGT 64
Query: 135 AVVTNDKAALWTDGRYFLQAEKQLNSNWILMRAGNPGVPTTSEWLNEVLAPGARVGIDPF 194
A+VT AA+WTDGRYFLQA Q+++NW LM+ G PT +WL VL G++VG+DP
Sbjct: 65 AIVTEQHAAMWTDGRYFLQAANQMDNNWTLMKMGLKDTPTQEDWLVSVLPEGSKVGVDPS 124
Query: 195 LFTSDAAEELKHVISKKNHELVYLYNSNLVDEIWKEARPEPPNKPVRVHGLKYAGLDVAS 254
+ +D + + V+ H+LV + NL+D IW + RP+ P KP+ V L Y G+
Sbjct: 125 IIPADQWKRMSKVLRSAGHDLVPV-KENLIDTIWTD-RPQRPCKPLIVLDLSYTGVSWRD 182
Query: 255 KLSSLRSELVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEIDGAKLFIDNS 314
K+ +LRS++ + +VTALDE+AWL NLRGSD+ ++PV +AY ++ ++ +LFID
Sbjct: 183 KIVALRSKMAERKVVWFVVTALDEVAWLFNLRGSDVEYNPVFFAYAVIGMNTIRLFIDGD 242
Query: 315 KVTEE-VDDHLK-------KANIEIRPYNSIVSEIENLAARGS---SLWLDTSSVNA--- 360
++ + V +HL+ + I++ PY SI++E++ + A S +WL + A
Sbjct: 243 RMMDPAVREHLQLDSTLEPEFKIQVMPYRSILTELQAVGAGLSPKEKVWLSDKASYALTE 302
Query: 361 AIVNAYKAACDRYYQNYESKHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETELK 420
AI AY RY Y +P+ +AKA+KN E +
Sbjct: 303 AIPKAY-----RYLTPY---------------------------TPICIAKAVKNALETE 330
Query: 421 GMQECHLRDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEFRSKQAGFLDTSFDTISGS 480
GM+ H++DA AL + ++WLEKE+ + TE+ +DK EFRS+Q F++ SF TIS +
Sbjct: 331 GMRRAHIKDAVALCELFNWLEKEVPKGTV-TEIIAADKAEEFRSQQKDFVELSFATISST 389
Query: 481 GPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRTVHFGKPTTREKECFTRV 540
GPNGAIIHYK DGTTD+TRT+HFG P+ EKECFT V
Sbjct: 390 GPNGAIIHYK------------------------DGTTDVTRTMHFGTPSAYEKECFTCV 425
Query: 541 LQGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLDYRHGTGHGVGAALNVHEGPQGISYR 600
L+GHIA+ A+FP T G +LD+FARS LW GLDY HGTGHGVG+ LNVHEGP GISY+
Sbjct: 426 LKGHIAVSAAIFPNGTKGHLLDSFARSALWDCGLDYLHGTGHGVGSFLNVHEGPCGISYK 485
Query: 601 YGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYVRNVETPNRFGGIQYLGFEKLTYVPI 660
PL GMIVS+EPGYYED +FGIRIEN++ V ET F L FE LT VPI
Sbjct: 486 TFADEPLEAGMIVSDEPGYYEDGSFGIRIENVVLVIPAETKYNFKNRGSLTFEPLTLVPI 545
Query: 661 QIKLVDVSLLSTTEIDWLNNYHSVVWEKVSPLLDGSAR----QWLWNNTQPI 708
Q K++DVSLL+ E +W+N+YH E + L+ R +WL T+P+
Sbjct: 546 QTKMIDVSLLTQKECNWVNDYHQKCREVIGAELERQGRHEALRWLIRETEPL 597
>UniRef100_UPI00002A6EFE UPI00002A6EFE UniRef100 entry
Length = 529
Score = 497 bits (1279), Expect = e-139
Identities = 248/525 (47%), Positives = 342/525 (64%), Gaps = 7/525 (1%)
Query: 84 KLTALRRLFSKPDVSIDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGTAVVTNDKAA 143
KL LR+ + V A I+PSQD H SE++A + RR++IS FTGS GT VVT+D+A
Sbjct: 11 KLAELRKAMASRGVR--AIIVPSQDPHFSEYVAACFERRRWISEFTGSAGTVVVTDDEAL 68
Query: 144 LWTDGRYFLQAEKQLNSNWILMRAGNPGVPTTSEWLNEVLAPGARVGIDPFLFTSDAAEE 203
LWTDGRYF+QAE++L+ +W LMR+G G P WL +A G+ VGIDP + + A E
Sbjct: 69 LWTDGRYFVQAEEELSEDWTLMRSGTKGTPDAKTWLKTTMAAGSAVGIDPNVHSVSEARE 128
Query: 204 LKHVISKKNHELVYLYNSNLVDEIWKEARPEPPNKPVRVHGLKYAGLDVASKLSSLRSEL 263
L+ + + E+V L NLVD IW + RP P +RVH ++AG V KL+ +R ++
Sbjct: 129 LRKALKESECEVVAL-EENLVDSIWSD-RPAFPKTKLRVHPAEFAGQTVEEKLTHIREKM 186
Query: 264 VQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEIDGAKLFIDNSKVTEEVDDH 323
+ G+ ++V++LD++ WL N+RG D P +PV +Y++V A L++D K T+ V H
Sbjct: 187 KENGTQRLVVSSLDDVMWLCNVRGGDAPCNPVTLSYVLVGETEASLYVDVDKATDAVRAH 246
Query: 324 LKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYKAACDRYYQNYESKHKT 383
L +AN+ I+PY + ++ A G +LW+D V+ A++ +A ++ +S
Sbjct: 247 LAEANVTIKPYEDMSKDVYASAHAGETLWMDVDRVSIAMLEEAEAGAVAAPRHAKSARSG 306
Query: 384 RSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQECHLRDAAALAQFWDWLEKE 443
S S + S V + P+++AKA+KN+ E+ GM E HLRD AA+A+FW W+EKE
Sbjct: 307 ASGDVSSSASASPVK---EGTCPIAIAKAVKNDAEMAGMVEAHLRDGAAMAEFWCWMEKE 363
Query: 444 ITNDRILTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDANK 503
I + R + E E ++++ R KQ F++ SF TI+G GP+GA++HY+ S +
Sbjct: 364 IASGRTVDEYEAGERVMACRKKQGDFIEESFPTIAGEGPHGAVVHYRATRESARVIGGES 423
Query: 504 LFLLDSGAQYVDGTTDITRTVHFGKPTTREKECFTRVLQGHIALDQAVFPEDTPGFVLDA 563
L L DSG QYV GTTD+TRT HFG PT +KEC+TRVLQGHIALDQ VFP T GFVLDA
Sbjct: 424 LLLCDSGGQYVCGTTDVTRTAHFGTPTDHQKECYTRVLQGHIALDQMVFPTGTKGFVLDA 483
Query: 564 FARSFLWKVGLDYRHGTGHGVGAALNVHEGPQGISYRYGNLTPLV 608
FARS LW GLDYRHGTGHGVGAALNVHEGPQGIS R+GN+T L+
Sbjct: 484 FARSHLWASGLDYRHGTGHGVGAALNVHEGPQGISPRFGNMTALL 528
>UniRef100_UPI00003AE3D1 UPI00003AE3D1 UniRef100 entry
Length = 579
Score = 496 bits (1276), Expect = e-138
Identities = 277/608 (45%), Positives = 375/608 (61%), Gaps = 53/608 (8%)
Query: 85 LTALRRLFSKP---DVSIDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGTAVVTNDK 141
L LR++ P + AYI+PS DAHQSE+IA RR +IS F GS GTA+VT
Sbjct: 7 LKQLRQVMKSPRYVQEPVQAYIVPSGDAHQSEYIAPCDCRRAFISGFDGSAGTAIVTEQH 66
Query: 142 AALWTDGRYFLQAEKQLNSNWILMRAGNPGVPTTSEWLNEVLAPGARVGIDPFLFTSDAA 201
AA+WTDGRYFLQA Q+++NW LM+ G PT +WL VL G++VG+DP + +D
Sbjct: 67 AAMWTDGRYFLQAANQMDNNWTLMKMGLKDTPTQEDWLVSVLPEGSKVGVDPSIIPADQW 126
Query: 202 EELKHVISKKNHELVYLYNSNLVDEIWKEARPEPPNKPVRVHGLKYAGLDVASKLSSLRS 261
+ + V+ H+LV + NL+D IW + RP+ P KP+ V L Y G+ K+ +LRS
Sbjct: 127 KRMSKVLRSAGHDLVPV-KENLIDTIWTD-RPQRPCKPLIVLDLSYTGVSWRDKIVALRS 184
Query: 262 ELVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEIDGAKLFIDNSKVTEE-V 320
++ + +VTALDE+AWL NLRGSD+ ++PV +AY ++ ++ +LFID ++ + V
Sbjct: 185 KMAERKVVWFVVTALDEVAWLFNLRGSDVEYNPVFFAYAVIGMNTIRLFIDGDRMMDPAV 244
Query: 321 DDHLK-------KANIEIRPYNSIVSEIENLAARGS---SLWLDTSSVNA---AIVNAYK 367
+HL+ + I++ PY SI++E++ + A S +WL + A AI AY
Sbjct: 245 REHLQLDSTLEPEFKIQVMPYRSILTELQAVGAGLSPKEKVWLSDKASYALTEAIPKAY- 303
Query: 368 AACDRYYQNYESKHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQECHL 427
RY Y +P+ +AKA+KN E +GM+ H+
Sbjct: 304 ----RYLTPY---------------------------TPICIAKAVKNALETEGMRRAHI 332
Query: 428 RDAAALAQFWDWLEKEITNDRILT-EVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAI 486
+DA AL + ++WLEKE + V + F S+Q F++ SF TIS +GPNGAI
Sbjct: 333 KDAVALCELFNWLEKEFHFIFCIPLACAVVITIFSF-SQQKDFVELSFATISSTGPNGAI 391
Query: 487 IHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRTVHFGKPTTREKECFTRVLQGHIA 546
IHYKP P + T+ N+++LLDSGAQY DGTTD+TRT+HFG P+ EKECFT VL+GHIA
Sbjct: 392 IHYKPVPETNRTLSVNEIYLLDSGAQYKDGTTDVTRTMHFGTPSAYEKECFTCVLKGHIA 451
Query: 547 LDQAVFPEDTPGFVLDAFARSFLWKVGLDYRHGTGHGVGAALNVHEGPQGISYRYGNLTP 606
+ A+FP T G +LD+FARS LW GLDY HGTGHGVG+ LNVHEGP GISY+ P
Sbjct: 452 VSAAIFPNGTKGHLLDSFARSALWDCGLDYLHGTGHGVGSFLNVHEGPCGISYKTFADEP 511
Query: 607 LVNGMIVSNEPGYYEDHAFGIRIENLLYVRNVETPNRFGGIQYLGFEKLTYVPIQIKLVD 666
L GMIVS+EPGYYED +FGIRIEN++ V ET F L FE LT VPIQ K++D
Sbjct: 512 LEAGMIVSDEPGYYEDGSFGIRIENVVLVIPAETKYNFKNRGSLTFEPLTLVPIQTKMID 571
Query: 667 VSLLSTTE 674
VSLL+ E
Sbjct: 572 VSLLTQKE 579
>UniRef100_UPI0000361810 UPI0000361810 UniRef100 entry
Length = 599
Score = 488 bits (1257), Expect = e-136
Identities = 278/658 (42%), Positives = 387/658 (58%), Gaps = 85/658 (12%)
Query: 77 SNSDSDPKLTA--LRRL---------FSKPDVSIDAYIIPSQDAHQSEFIAQSYARRKYI 125
S+S PK+T +R+L F++P I AYI+PS DAHQSE+IA RR+YI
Sbjct: 1 SDSAMSPKITGELIRKLRQAMKSCKYFAEP---IQAYIVPSGDAHQSEYIAPCDCRREYI 57
Query: 126 SAFTGSNGTAVVTNDKAALWTDGRYFLQAEKQLNSNWILMRAGNPGVPTTSEWLNEVLAP 185
F GS GTA++T AA+WTDGRYFLQA +Q+++NW LM+ G P+ +WL VL
Sbjct: 58 CGFNGSAGTAIITEQHAAMWTDGRYFLQASQQMDNNWTLMKMGLKETPSQEDWLISVLPE 117
Query: 186 GARVGIDPFLFTSDAAEELKHVISKKNHELVYLYNSNLVDEIWKEARPEPPNKPVRVHGL 245
++VG+DP++ +D + + ++ H LV + + NL+D +W + RPE P+ +R GL
Sbjct: 118 NSKVGVDPWIIAADQWKNMSKALTGAGHSLVAMQD-NLIDVVWTD-RPERPSTQLRTLGL 175
Query: 246 KYAGLDVASKLSSLRSELVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEID 305
+Y GL K+++LR+++ + S + TALDEIAWL NLRG+DI ++PV +AY IV ++
Sbjct: 176 EYTGLSWHDKVTALRAKMTERKISWFVATALDEIAWLFNLRGADIEYNPVFFAYTIVGLN 235
Query: 306 GAKLFIDNSKVTEE-VDDHLK---KANIEIR----PYNSIVSEIENLAAR---GSSLWLD 354
+LF+D ++T+ + HL+ + E+R PY S+ +E++ + A +W+
Sbjct: 236 TIRLFVDTKRLTDPALRGHLELDSPSQPELRILTFPYESVYTELQAICAALGPKDKVWIC 295
Query: 355 TSSVNAAIVNAYKAACDRYYQNYESKHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKAIK 414
+ A KA S +P +P+ L+KA+K
Sbjct: 296 DKASRALTQLLPKAN------------------------RSPIPY-----TPLCLSKAVK 326
Query: 415 NETELKGMQECHLRDAAALAQFWDWLEKEITNDRILTEVEVSDKLLEFRSKQAGFLDTSF 474
N TE++GM+ H++DA AL + + WLEKEI + TE+ +DK E RS+Q F+ SF
Sbjct: 327 NATEIQGMKVAHIKDAVALCELFAWLEKEIPKGNV-TEISAADKAEELRSQQKDFVGLSF 385
Query: 475 DTISGSGPNGAIIHYKPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRTVHFGKPTTREK 534
TISG GPNGA+IHY DGTTD+TRTVHFG P+ EK
Sbjct: 386 PTISGVGPNGAVIHYS------------------------DGTTDVTRTVHFGTPSAFEK 421
Query: 535 ECFTRVLQGHIALDQAVFPEDTPGFVLDAFARSFLWKVGLDYRHGTGHGVGAALNVHEGP 594
ECFT VL+GHIA+ AVFP T G +LD+FAR+ LW GLDY HGTGHGVG LNVHEGP
Sbjct: 422 ECFTYVLKGHIAVSAAVFPNGTKGHLLDSFARAALWDSGLDYLHGTGHGVGCFLNVHEGP 481
Query: 595 QGISYRYGNLTPLVNGMIVSNEPGYYEDHAFGIRIENLLYVRNVETPNRFGGIQYLGFEK 654
GISY+ PL GMIVS+EPGYYED AFGIRIEN++ V + + L FE
Sbjct: 482 CGISYKTFADEPLEAGMIVSDEPGYYEDGAFGIRIENVVLVVPAQPKYNYRNRGSLTFEP 541
Query: 655 LTYVPIQIKLVDVSLLSTTEIDWLNNYHSVVWEKVSPLLDGSAR----QWLWNNTQPI 708
LT VPIQ+K+++ +LL+ E DWLN YH E + L+ R +WL TQP+
Sbjct: 542 LTLVPIQVKMINTALLTQKERDWLNEYHRTCREVIGAELERQGRKEALEWLVRETQPV 599
>UniRef100_UPI0000430FD3 UPI0000430FD3 UniRef100 entry
Length = 613
Score = 457 bits (1175), Expect = e-127
Identities = 263/644 (40%), Positives = 366/644 (55%), Gaps = 50/644 (7%)
Query: 84 KLTALRRLFSKPDVS------IDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGTAVV 137
KL LR L + I A I+ S DAHQSE++ + R +IS FTGS GTA++
Sbjct: 1 KLAKLRELMKAVQIGGLKEKGIQALIVSSDDAHQSEYLREHDKRICFISGFTGSFGTAII 60
Query: 138 TNDKAALWTDGRYFLQAEKQLN--SNWILMRAGNPGVPTTSEWLNEVLAPGARVGIDPFL 195
T +KA LWTDGRY++QA + + W LM+ G PT + WL L P + VG D L
Sbjct: 61 TQNKALLWTDGRYYMQALAEFDPPEEWTLMKEGLLDTPTRAAWLTCNLPPKSTVGADSNL 120
Query: 196 FTSDAAEELKHVISKKNHELVYLYNSNLVDEIWKEARPEPPNKPVRVHGLKYAGLDVASK 255
+ L ++ H L+ L NL+D++W + +P P + L+++G K
Sbjct: 121 ISYTEWVVLHTSLTAAGHCLMPL-EENLIDKVWGDEQPVPTANIIVPQPLQFSGCTAGKK 179
Query: 256 LSSLRSELVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEIDGAKLFIDNSK 315
+ R + + A+++TALDEIA++LNLRGSDIP++PV +AY+I+ +D LFID +K
Sbjct: 180 VKLCREVMSKNKVKALVITALDEIAYILNLRGSDIPYNPVFFAYIILTLDHLHLFIDINK 239
Query: 316 VTEEVDDHL--KKANIEIRPYNSI---VSEIENLAARGSSLWLDTSSVNAAIVNAYKAAC 370
+TEE L ++ N+ PY I + +I NL +WL SS A + +A
Sbjct: 240 LTEEARQQLIIEEVNLIYHPYEDIHNYLKKIANLCINDDKIWLSNSSSYALHADCGEA-- 297
Query: 371 DRYYQNYESKHKTRSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQECHLRDA 430
KH K +P+S+ KA+KN TE+ GM+ H+RD+
Sbjct: 298 --------KKH--------------------IKITPISVMKAVKNNTEIAGMKAAHIRDS 329
Query: 431 AALAQFWDWLEKEITNDR-ILTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAIIHY 489
AL +++ WLE +I N + +TE+ + +L +FR +Q F+ SF TIS GP+ AIIHY
Sbjct: 330 VALIKYFAWLEDQIKNKKNTVTEISGATQLEKFRQEQEHFIGLSFPTISSVGPHAAIIHY 389
Query: 490 KPEPGSCSTVDANKLFLLDSGAQYVDGTTDITRTVHFGKPTTREKECFTRVLQGHIALDQ 549
P + + +++L DSGAQY DGTTD+TRT+HFG PT E+ECFTRV +G L
Sbjct: 390 LPTLKTDVPITDKEIYLCDSGAQYQDGTTDVTRTLHFGNPTNFERECFTRVFKGQCRLSS 449
Query: 550 AVFPEDTPGFVLDAFARSFLWKVGLDYRHGTGHGVGAALNVHEGPQGISYR-YGNLTPLV 608
+FP G LD AR LW VGL+Y HGTGHG+G+ LNVHEGP IS+R Y + L
Sbjct: 450 TIFPLMIQGNYLDTLARENLWNVGLNYLHGTGHGIGSYLNVHEGPISISWRPYPDDPGLQ 509
Query: 609 NGMIVSNEPGYYEDHAFGIRIENLLYVRNVETPNRFGGIQYLGFEKLTYVPIQIKLVDVS 668
GM +SNEPGYYED FGIR+EN+ + T +L FE +T VPIQ KL+DVS
Sbjct: 510 PGMFLSNEPGYYEDEKFGIRLENIELIVKANTHYNHKNRGFLTFETVTLVPIQTKLLDVS 569
Query: 669 LLSTTEIDWLNNYHSVVWEKVSPLLDG----SARQWLWNNTQPI 708
LL+ EI +LNNYH+ + PLL G A +WL T+P+
Sbjct: 570 LLTDVEIQYLNNYHAKCLNTIKPLLQGPENVQALEWLERETRPL 613
>UniRef100_Q83F75 Peptidase, M24 family protein [Coxiella burnetii]
Length = 597
Score = 447 bits (1151), Expect = e-124
Identities = 255/629 (40%), Positives = 368/629 (57%), Gaps = 43/629 (6%)
Query: 84 KLTALRRLFSKPDVSIDAYIIPSQDAHQSEFIAQSYARRKYISAFTGSNGTAVVTNDKAA 143
+L ALRRL ++ +D Y +PS D H++E++ + RR +IS FTGS G VV DKA
Sbjct: 8 RLAALRRLMH--EIGVDYYYVPSSDPHKNEYVPSCWQRRAWISGFTGSAGDVVVGIDKAF 65
Query: 144 LWTDGRYFLQAEKQLNSN-WILMRAGNPGVPTTSEWLNEVLAPGARVGIDPFLFTSDAAE 202
LWTD RYFLQAE+QL+ + + LM+ G P +WL + G +DP L +E
Sbjct: 66 LWTDPRYFLQAEQQLDDSLYHLMKMGQGETPAIDQWLTQQ-RNGIVFAVDPRLINLQQSE 124
Query: 203 ELKHVISKKNHELVYLYNSNLVDEIWKEARPEPPNKPVRVHGLKYAGLDVASKLSSLRSE 262
+++ + K+N +L+ L + NL+D +WK+ +P P +++ L+YAGL KL++LR
Sbjct: 125 KIQRALEKQNGKLLAL-DENLIDRVWKD-QPPLPQSAIQLQPLQYAGLSAEDKLAALRQT 182
Query: 263 LVQAGSSAIIVTALDEIAWLLNLRGSDIPHSPVVYAYLIVEIDGAKLFIDNSKVTEEVDD 322
L + + AI++ LD IAWL N+RG+D+ ++P+V +Y ++ + A LF+D K+TE
Sbjct: 183 LQKESADAIVLNTLDAIAWLFNIRGNDVAYNPLVISYAVITQNEASLFVDPHKITEGDRS 242
Query: 323 HLKKANIEIRPYNSIVSEIENLAARGSSLWLDTSSVNAAIVNAYKAACDRYYQNYESKHK 382
+ KK + I PY I +E+L+ S+WLD + N + + K
Sbjct: 243 YFKKIPVHIEPYEGIGKLLESLS---GSVWLDPGATNLWLRDQLK--------------- 284
Query: 383 TRSKGFDGSIANSDVPIAVHKSSPVSLAKAIKNETELKGMQECHLRDAAALAQFWDWLEK 442
+ + K SP++LAKA+KN E KG +E H+ DA A+ QF WLE
Sbjct: 285 -------------NTASLILKPSPITLAKALKNPVEQKGAREAHIIDAIAMIQFLHWLEN 331
Query: 443 EITNDRILTEVEVSDKLLEFRSKQAGFLDTSFDTISGSGPNGAIIHYKPEPGSCSTVDAN 502
+ ++E+ ++KL FR + LD SF +ISG GP+GAI+HY + +T++ +
Sbjct: 332 HWQSG--VSEISAAEKLEFFRRGDSRCLDLSFPSISGFGPHGAIVHYSATTDTDATINDS 389
Query: 503 KLFLLDSGAQYVDGTTDITRTVHFGKPTTREKECFTRVLQGHIALDQAVFPEDTPGFVLD 562
+L+DSG QY GTTDITRT+H G PT EK +T VL+GH+A+ QAVFP+ T G L+
Sbjct: 390 APYLIDSGGQYHYGTTDITRTIHLGTPTEEEKRLYTLVLKGHLAIRQAVFPKGTCGEHLN 449
Query: 563 AFARSFLWKVGLDYRHGTGHGVGAALNVHEGPQGISYRYGNLTPLVNGMIVSNEPGYYED 622
A A FLW+ LDY HGTGHGVG+ L VHEGPQ I+ RY + PL GMIVSNEPG Y
Sbjct: 450 ALAHQFLWREALDYGHGTGHGVGSYLCVHEGPQAITSRYTGI-PLQPGMIVSNEPGVYLT 508
Query: 623 HAFGIRIENLLYVRNVET--PNRFGGIQYLGFEKLTYVPIQIKLVDVSLLSTTEIDWLNN 680
H +GIRIENL V T + G + FE LT VP KL++ +LL++ EI +N+
Sbjct: 509 HKYGIRIENLCLVTEKFTVDDSLTGDGPFYSFEDLTLVPYCRKLINPNLLTSEEIQQIND 568
Query: 681 YHSVVWEKVSPLLDGS-ARQWLWNNTQPI 708
YH V + + LL + WL T P+
Sbjct: 569 YHQRVDQTLRDLLPANELNDWLHEATAPL 597
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.133 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,237,844,598
Number of Sequences: 2790947
Number of extensions: 53862982
Number of successful extensions: 149529
Number of sequences better than 10.0: 1188
Number of HSP's better than 10.0 without gapping: 761
Number of HSP's successfully gapped in prelim test: 428
Number of HSP's that attempted gapping in prelim test: 146033
Number of HSP's gapped (non-prelim): 1604
length of query: 713
length of database: 848,049,833
effective HSP length: 135
effective length of query: 578
effective length of database: 471,271,988
effective search space: 272395209064
effective search space used: 272395209064
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 79 (35.0 bits)
Medicago: description of AC146571.1


