

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146570.6 + phase: 0
(101 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
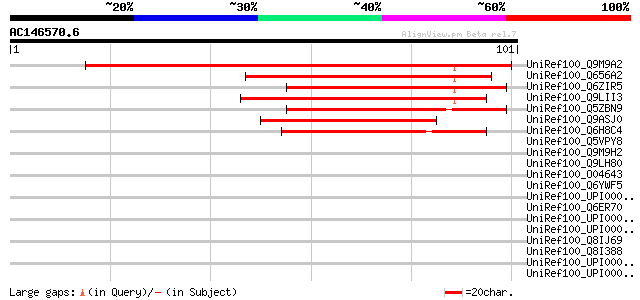
UniRef100_Q9M9A2 F27J15.21 [Arabidopsis thaliana] 65 4e-10
UniRef100_Q656A2 Hypothetical protein P0596H06.16 [Oryza sativa] 61 6e-09
UniRef100_Q6ZIR5 Hypothetical protein OJ1038_A06.25 [Oryza sativa] 60 1e-08
UniRef100_Q9LII3 Gb|AAF43227.1 [Arabidopsis thaliana] 59 3e-08
UniRef100_Q5ZBN9 Hypothetical protein P0704D04.8 [Oryza sativa] 58 6e-08
UniRef100_Q9ASJ0 Hypothetical protein P0439B06.27 [Oryza sativa] 57 1e-07
UniRef100_Q6H8C4 Hypothetical protein OJ1006_A02.44 [Oryza sativa] 44 7e-04
UniRef100_Q5VPY8 Hypothetical protein B1096D03.33 [Oryza sativa] 44 0.001
UniRef100_Q9M9H2 F14O23.12 [Arabidopsis thaliana] 39 0.023
UniRef100_Q9LH80 Gb|AAF43227.1 [Arabidopsis thaliana] 37 0.15
UniRef100_O04643 F2P16.13 protein [Arabidopsis thaliana] 36 0.19
UniRef100_Q6YWF5 Putative pattern formation protein GNOM [Oryza ... 35 0.33
UniRef100_UPI00002BD476 UPI00002BD476 UniRef100 entry 34 0.73
UniRef100_Q6ER70 Hypothetical protein OSJNBa0073A18.24 [Oryza sa... 34 0.73
UniRef100_UPI0000322900 UPI0000322900 UniRef100 entry 33 1.3
UniRef100_UPI00002FB62D UPI00002FB62D UniRef100 entry 33 1.3
UniRef100_Q8IJ69 Hypothetical protein [Plasmodium falciparum] 33 1.3
UniRef100_Q8I388 Developmental protein, putative [Plasmodium fal... 33 1.3
UniRef100_UPI0000317F2D UPI0000317F2D UniRef100 entry 33 1.6
UniRef100_UPI0000300D6D UPI0000300D6D UniRef100 entry 33 1.6
>UniRef100_Q9M9A2 F27J15.21 [Arabidopsis thaliana]
Length = 156
Score = 65.1 bits (157), Expect = 4e-10
Identities = 33/86 (38%), Positives = 52/86 (60%), Gaps = 1/86 (1%)
Query: 16 SLKTKKLEELSTSSKNIIKKIYKAIHCKILKMKKQEDDKCLWKKTILMGEKCQPLEFPGA 75
S+ + + +EL T+ N + + + K E++ LW++ ILMG KC+PL+F G
Sbjct: 67 SMSSPRPKELFTTLSNKAMTMVRRKNPPEEKATAMEEEHGLWQREILMGGKCEPLDFSGV 126
Query: 76 IFYDSEGNQLSE-PPRTPRSSPSSSF 100
I+YDS G L+E PPR+PR +P S+
Sbjct: 127 IYYDSNGRLLNEVPPRSPRGTPLPSY 152
>UniRef100_Q656A2 Hypothetical protein P0596H06.16 [Oryza sativa]
Length = 171
Score = 61.2 bits (147), Expect = 6e-09
Identities = 26/51 (50%), Positives = 38/51 (73%), Gaps = 2/51 (3%)
Query: 48 KKQEDDKCLWKKTILMGEKCQPLEFPGAIFYDSEGNQLSE--PPRTPRSSP 96
+++ D LW+K ILMGE+CQPL+F G I+YD++G +L+ PPR+P SP
Sbjct: 106 REEGDSGGLWRKEILMGERCQPLDFSGVIYYDADGRRLAHPPPPRSPMRSP 156
>UniRef100_Q6ZIR5 Hypothetical protein OJ1038_A06.25 [Oryza sativa]
Length = 138
Score = 60.1 bits (144), Expect = 1e-08
Identities = 26/46 (56%), Positives = 35/46 (75%), Gaps = 2/46 (4%)
Query: 56 LWKKTILMGEKCQPLEFPGAIFYDSEGNQLSE--PPRTPRSSPSSS 99
+W+K ILMGE+CQPL+F G I+YD+EG +L + PPR+P SP S
Sbjct: 79 VWRKEILMGERCQPLDFSGVIYYDAEGRRLEQPPPPRSPLRSPLPS 124
>UniRef100_Q9LII3 Gb|AAF43227.1 [Arabidopsis thaliana]
Length = 195
Score = 58.9 bits (141), Expect = 3e-08
Identities = 24/50 (48%), Positives = 39/50 (78%), Gaps = 1/50 (2%)
Query: 47 MKKQEDDKCLWKKTILMGEKCQPLEFPGAIFYDSEGNQLSE-PPRTPRSS 95
M++ E++ +W++ ILMG KC+PL++ G I+YD G+QL + PPR+PR+S
Sbjct: 124 MEEDEEEYGVWQREILMGGKCEPLDYSGVIYYDCSGHQLKQVPPRSPRAS 173
>UniRef100_Q5ZBN9 Hypothetical protein P0704D04.8 [Oryza sativa]
Length = 505
Score = 57.8 bits (138), Expect = 6e-08
Identities = 24/44 (54%), Positives = 35/44 (79%), Gaps = 1/44 (2%)
Query: 56 LWKKTILMGEKCQPLEFPGAIFYDSEGNQLSEPPRTPRSSPSSS 99
LW++ ILMGE+C+PL FPGAI YDS G +LS+ PR ++ P+++
Sbjct: 428 LWRRAILMGERCEPLSFPGAIHYDSRGRRLSQ-PRRAKAKPAAA 470
>UniRef100_Q9ASJ0 Hypothetical protein P0439B06.27 [Oryza sativa]
Length = 175
Score = 56.6 bits (135), Expect = 1e-07
Identities = 22/35 (62%), Positives = 31/35 (87%)
Query: 51 EDDKCLWKKTILMGEKCQPLEFPGAIFYDSEGNQL 85
E+++ LW+KTI+MG+KC+PL+F G I YDS+GNQL
Sbjct: 121 EEEEALWRKTIMMGDKCRPLQFSGHIAYDSDGNQL 155
>UniRef100_Q6H8C4 Hypothetical protein OJ1006_A02.44 [Oryza sativa]
Length = 299
Score = 44.3 bits (103), Expect = 7e-04
Identities = 21/41 (51%), Positives = 27/41 (65%), Gaps = 1/41 (2%)
Query: 55 CLWKKTILMGEKCQPLEFPGAIFYDSEGNQLSEPPRTPRSS 95
C+W+K ILMG KCQ EF G I YD+ GN + P PR++
Sbjct: 97 CVWQKNILMGGKCQLPEFSGVINYDAAGN-IVAPSGRPRAA 136
>UniRef100_Q5VPY8 Hypothetical protein B1096D03.33 [Oryza sativa]
Length = 138
Score = 43.5 bits (101), Expect = 0.001
Identities = 20/39 (51%), Positives = 26/39 (66%), Gaps = 2/39 (5%)
Query: 51 EDDKCLWKKTILMGEKCQPLEFPGAIFYDSEGNQLSEPP 89
ED C +K ILM E+CQ L+F G I+YD G +L +PP
Sbjct: 78 EDGVC--RKEILMEERCQSLDFSGMIYYDVAGRRLEQPP 114
>UniRef100_Q9M9H2 F14O23.12 [Arabidopsis thaliana]
Length = 130
Score = 39.3 bits (90), Expect = 0.023
Identities = 16/27 (59%), Positives = 20/27 (73%)
Query: 56 LWKKTILMGEKCQPLEFPGAIFYDSEG 82
LW+K ILMG KCQ +F G I YD++G
Sbjct: 88 LWQKNILMGGKCQLPDFSGVILYDADG 114
>UniRef100_Q9LH80 Gb|AAF43227.1 [Arabidopsis thaliana]
Length = 168
Score = 36.6 bits (83), Expect = 0.15
Identities = 17/43 (39%), Positives = 23/43 (52%)
Query: 56 LWKKTILMGEKCQPLEFPGAIFYDSEGNQLSEPPRTPRSSPSS 98
+W++ ILMGEKC+ F G I YD G+ P + SS
Sbjct: 116 VWQRPILMGEKCELPRFSGLILYDECGHPRHHPQLQEKVKQSS 158
>UniRef100_O04643 F2P16.13 protein [Arabidopsis thaliana]
Length = 164
Score = 36.2 bits (82), Expect = 0.19
Identities = 18/39 (46%), Positives = 26/39 (66%), Gaps = 4/39 (10%)
Query: 50 QEDDKCLWKKTILMGEKCQPLEFPGAIFYDSEGNQLSEP 88
+E++ C KK ILMGE+C+P+ G + YD +G L EP
Sbjct: 130 EEEELC--KKRILMGERCKPMN--GVLQYDGDGILLPEP 164
>UniRef100_Q6YWF5 Putative pattern formation protein GNOM [Oryza sativa]
Length = 1424
Score = 35.4 bits (80), Expect = 0.33
Identities = 25/98 (25%), Positives = 46/98 (46%), Gaps = 3/98 (3%)
Query: 2 IINKINKAIISYIISLKTKKLEELSTSSKNIIKKIYKAIHCKILKMKKQEDDKCLWKKTI 61
+++ I K + + +T+KL+E+ I + KA ++L ++ LW+ T
Sbjct: 1325 VLDMIEKLMKVKVRGRRTEKLQEVIPELLKNILLVLKAN--RVLSKTSTSEENSLWEATW 1382
Query: 62 LMGEKCQPLEFPGAIFYDSEGNQLSEPPRTPRSSPSSS 99
L K P P +F DSEG+ ++ + SP+ S
Sbjct: 1383 LQVNKIAPSLQP-EVFPDSEGDVATQSAKNKSDSPAQS 1419
>UniRef100_UPI00002BD476 UPI00002BD476 UniRef100 entry
Length = 189
Score = 34.3 bits (77), Expect = 0.73
Identities = 22/62 (35%), Positives = 33/62 (52%), Gaps = 7/62 (11%)
Query: 2 IINKINKAIISYIISLKTKKLEELST--SSKNIIKKIYKAIHCKILK-----MKKQEDDK 54
I K+NKA+ YI KK E + T + KN ++KI K IH ++ K +KK + K
Sbjct: 42 IFIKLNKALPQYITQFPIKKQEIIKTILAKKNNLRKIIKIIHPEVRKKMNIFLKKNKKSK 101
Query: 55 CL 56
+
Sbjct: 102 II 103
>UniRef100_Q6ER70 Hypothetical protein OSJNBa0073A18.24 [Oryza sativa]
Length = 143
Score = 34.3 bits (77), Expect = 0.73
Identities = 13/27 (48%), Positives = 17/27 (62%)
Query: 56 LWKKTILMGEKCQPLEFPGAIFYDSEG 82
+W++ +LMG KCQ F G I YD G
Sbjct: 89 IWQRKVLMGVKCQLPRFSGMILYDERG 115
>UniRef100_UPI0000322900 UPI0000322900 UniRef100 entry
Length = 100
Score = 33.5 bits (75), Expect = 1.3
Identities = 20/47 (42%), Positives = 27/47 (56%), Gaps = 2/47 (4%)
Query: 2 IINKINKAIISYIISLKTKKLEELST--SSKNIIKKIYKAIHCKILK 46
I K+NK + YI KK E +T ++KN +KKI K IH +I K
Sbjct: 42 IFIKLNKTLPKYITQFPIKKQEVTNTILANKNNLKKIIKIIHPEIRK 88
>UniRef100_UPI00002FB62D UPI00002FB62D UniRef100 entry
Length = 302
Score = 33.5 bits (75), Expect = 1.3
Identities = 23/61 (37%), Positives = 36/61 (58%), Gaps = 2/61 (3%)
Query: 4 NKINKAIISYIISLKTKKLEELSTSSKNIIKKIY--KAIHCKILKMKKQEDDKCLWKKTI 61
N I+KAI +I +L K + S ++K+IIKK+ K + KI+K+K+ D L + I
Sbjct: 241 NNISKAIDLFIKNLNQFKKDINSKNNKSIIKKLIQTKKVRSKIIKLKQDIDKPDLEEINI 300
Query: 62 L 62
L
Sbjct: 301 L 301
>UniRef100_Q8IJ69 Hypothetical protein [Plasmodium falciparum]
Length = 682
Score = 33.5 bits (75), Expect = 1.3
Identities = 15/46 (32%), Positives = 30/46 (64%)
Query: 2 IINKINKAIISYIISLKTKKLEELSTSSKNIIKKIYKAIHCKILKM 47
+INKI + +IS++++L + +S++S N K I + +H KI ++
Sbjct: 200 VINKITEGLISFLVTLGVVPIIRVSSNSSNPSKMIAQKLHEKIYEL 245
>UniRef100_Q8I388 Developmental protein, putative [Plasmodium falciparum]
Length = 195
Score = 33.5 bits (75), Expect = 1.3
Identities = 21/60 (35%), Positives = 34/60 (56%), Gaps = 4/60 (6%)
Query: 4 NKINKAIISYIISLKTKKLEELSTSS----KNIIKKIYKAIHCKILKMKKQEDDKCLWKK 59
NKI+ + + LKTK+LE+LS S K +I + KAI +++ + +KC+ KK
Sbjct: 3 NKISTEDHIFRLKLKTKELEKLSQRSELEEKKLIGDVKKAIQAGKIELARLYAEKCIRKK 62
>UniRef100_UPI0000317F2D UPI0000317F2D UniRef100 entry
Length = 146
Score = 33.1 bits (74), Expect = 1.6
Identities = 18/39 (46%), Positives = 25/39 (63%), Gaps = 2/39 (5%)
Query: 5 KINKAIISYIIS--LKTKKLEELSTSSKNIIKKIYKAIH 41
K+NK +I+S +K KKL L S+KN +KKI K +H
Sbjct: 45 KLNKLFPKHILSFPIKKKKLVNLILSNKNNLKKINKIVH 83
>UniRef100_UPI0000300D6D UPI0000300D6D UniRef100 entry
Length = 158
Score = 33.1 bits (74), Expect = 1.6
Identities = 25/99 (25%), Positives = 44/99 (44%), Gaps = 4/99 (4%)
Query: 7 NKAIISYI--ISLKTKKLEELSTSSKNIIKKIYKAIHCKILKMKKQE--DDKCLWKKTIL 62
NK I I I++ T K ++ T + NI+ K ++ + ++ +K+ D + +I
Sbjct: 37 NKTIKDSIKTIAISTTKYKKFGTLTGNIVTKHFEPLFVRLFSFEKENIFHDAIVNSSSIF 96
Query: 63 MGEKCQPLEFPGAIFYDSEGNQLSEPPRTPRSSPSSSFE 101
K ++ +FYD +GN SPS FE
Sbjct: 97 KISKVPEGKYYLMVFYDKDGNTKYSQGHLNPYSPSEWFE 135
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.315 0.132 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 172,127,023
Number of Sequences: 2790947
Number of extensions: 6508102
Number of successful extensions: 17292
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 49
Number of HSP's that attempted gapping in prelim test: 17263
Number of HSP's gapped (non-prelim): 71
length of query: 101
length of database: 848,049,833
effective HSP length: 77
effective length of query: 24
effective length of database: 633,146,914
effective search space: 15195525936
effective search space used: 15195525936
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 68 (30.8 bits)
Medicago: description of AC146570.6


