

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146570.4 - phase: 0
(338 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
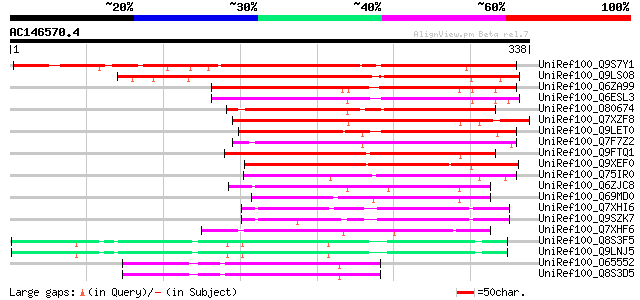
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9S7Y1 Putative DNA-binding protein; 36199-34606 [Arab... 315 1e-84
UniRef100_Q9LS08 Putative HLH DNA-binding protein [Arabidopsis t... 261 1e-68
UniRef100_Q6ZA99 DNA-binding protein-like [Oryza sativa] 209 7e-53
UniRef100_Q6ESL3 DNA binding protein-like [Oryza sativa] 208 2e-52
UniRef100_O80674 Hypothetical protein At2g41130 [Arabidopsis tha... 153 6e-36
UniRef100_Q7XZF8 Putative DNA binding protein [Oryza sativa] 152 2e-35
UniRef100_Q9LET0 Putative HLH DNA binding protein [Arabidopsis t... 149 1e-34
UniRef100_Q7F7Z2 ESTs C26093 [Oryza sativa] 137 4e-31
UniRef100_Q9FTQ1 DNA binding protein-like [Oryza sativa] 131 2e-29
UniRef100_Q9XEF0 Hypothetical protein T07M07.8 [Arabidopsis thal... 123 6e-27
UniRef100_Q75IR0 Hypothetical protein OSJNBb0099P06.13 [Oryza sa... 115 2e-24
UniRef100_Q6ZJC8 BHLH protein family-like [Oryza sativa] 111 3e-23
UniRef100_Q69MD0 BHLH-like protein [Oryza sativa] 100 4e-20
UniRef100_Q7XHI6 Putative bHLH131 transcription factor [Arabidop... 100 6e-20
UniRef100_Q9SZK7 Hypothetical protein F20D10.190 [Arabidopsis th... 91 3e-17
UniRef100_Q7XHF6 Putative bHLH transcription factor [Oryza sativa] 70 8e-11
UniRef100_Q8S3F5 Putative bHLH transcription factor [Arabidopsis... 68 4e-10
UniRef100_Q9LNJ5 F6F3.7 protein [Arabidopsis thaliana] 68 4e-10
UniRef100_O65552 Hypothetical protein F6I18.110 [Arabidopsis tha... 67 9e-10
UniRef100_Q8S3D5 Putative bHLH transcription factor [Arabidopsis... 67 9e-10
>UniRef100_Q9S7Y1 Putative DNA-binding protein; 36199-34606 [Arabidopsis thaliana]
Length = 368
Score = 315 bits (806), Expect = 1e-84
Identities = 196/358 (54%), Positives = 239/358 (66%), Gaps = 46/358 (12%)
Query: 3 QEDQGQCSSQTINNFETYQQEQFLLQQQMMRQQNSDHHIFGGGRSMFPTAGGDHN----- 57
+E++ + SS+ +NN + YQ + F Q S HH S T G N
Sbjct: 8 EEEEEEDSSEAMNNIQNYQNDLFF------HQLISHHHHHHHDPSQSETLGASGNVGSGF 61
Query: 58 ------QVSPILQQTWSMQQLHHHHHPFNPHDPFVIPQQQASPYASLFNR---RVPSLQF 108
VSPI WS+ PF+ P AS Y S FNR LQF
Sbjct: 62 TIFSQDSVSPI----WSLPPPTSIQPPFDQFPP--PSSSPASFYGSFFNRSRAHHQGLQF 115
Query: 109 AYDHHAGS--------EHLRIISDTLQH--GSGSGPFGGFQGEVGKMSAQEIMEAKALAA 158
Y+ G+ E LRI+S+ L +GSGPFG Q E+GKM+AQEIM+AKALAA
Sbjct: 116 GYEGFGGATSAAHHHHEQLRILSEALGPVVQAGSGPFG-LQAELGKMTAQEIMDAKALAA 174
Query: 159 SKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSP 218
SKSHSEAERRRRERINNHLAKLRS+LP+TTKTDKASLLAEVIQHVKELKR+TS+I+ET+
Sbjct: 175 SKSHSEAERRRRERINNHLAKLRSILPNTTKTDKASLLAEVIQHVKELKRETSVISETNL 234
Query: 219 VPTECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLKALRLRTLKADI 278
VPTE DELTV A ++E+ G +F+IKASLCC+DRSDLLP++IKTLKA+RL+TLKA+I
Sbjct: 235 VPTESDELTV-AFTEEEETGD--GRFVIKASLCCEDRSDLLPDMIKTLKAMRLKTLKAEI 291
Query: 279 TTLGGRVKNVLFITGEED-----DHEYCISSIQEALKAVMEKS-VGDESASGSVKRQR 330
TT+GGRVKNVLF+TGEE + EYCI +I+EALKAVMEKS V + S+SG+ KRQR
Sbjct: 292 TTVGGRVKNVLFVTGEESSGEEVEEEYCIGTIEEALKAVMEKSNVEESSSSGNAKRQR 349
>UniRef100_Q9LS08 Putative HLH DNA-binding protein [Arabidopsis thaliana]
Length = 344
Score = 261 bits (668), Expect = 1e-68
Identities = 160/320 (50%), Positives = 199/320 (62%), Gaps = 63/320 (19%)
Query: 71 QLHHHHHP---------FNPHDPFVIPQQQA--SPYASLFNRRVPSLQFAYDHHAG---- 115
Q H HHHP DP P S FNRR S ++D++ G
Sbjct: 22 QFHLHHHPQILPWSSTSLPSFDPLHFPSNPTRYSDPVHYFNRRASSSSSSFDYNDGFVSP 81
Query: 116 -------SEHLRIISDTLQHGSGSGPFGGFQGEV-GKMSAQEIMEAKALAASKSHSEAER 167
HLRI+S+ L G GF GE+ GK+SAQE+M+AKALAASKSHSEAER
Sbjct: 82 PPSMDHPQNHLRILSEALGPIMRRGSSFGFDGEIMGKLSAQEVMDAKALAASKSHSEAER 141
Query: 168 RRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETSPVPTECDELT 227
RRRERIN HLAKLRS+LP+TTKTDKASLLAEVIQH+KELKRQTS I +T VPTECD+LT
Sbjct: 142 RRRERINTHLAKLRSILPNTTKTDKASLLAEVIQHMKELKRQTSQITDTYQVPTECDDLT 201
Query: 228 VDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLKALRLRTLKADITTLGGRVKN 287
VD++ +DE+ GN +I+AS CC DR+DL+ ++I LK+LRLRTLKA+I T+GGRVKN
Sbjct: 202 VDSSYNDEE----GN-LVIRASFCCQDRTDLMHDVINALKSLRLRTLKAEIATVGGRVKN 256
Query: 288 VLFITGEEDDHE------------------------YCISSIQEALKAVMEKSVG----- 318
+LF++ E DD E +SSI+EALKAV+EK V
Sbjct: 257 ILFLSREYDDEEDHDSYRRNFDGDDVEDYDEERMMNNRVSSIEEALKAVIEKCVHNNDES 316
Query: 319 ------DESASGSVKRQRTN 332
++S+SG +KRQRT+
Sbjct: 317 NDNNNLEKSSSGGIKRQRTS 336
>UniRef100_Q6ZA99 DNA-binding protein-like [Oryza sativa]
Length = 345
Score = 209 bits (533), Expect = 7e-53
Identities = 126/241 (52%), Positives = 158/241 (65%), Gaps = 48/241 (19%)
Query: 132 SGPFGGFQGEVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTD 191
SG G Q E+G+M+A+EIM+AKALAAS+SHSEAERRRR+RIN HLA+LRSLLP+TTKTD
Sbjct: 95 SGLLGTLQAELGRMTAKEIMDAKALAASRSHSEAERRRRQRINGHLARLRSLLPNTTKTD 154
Query: 192 KASLLAEVIQHVKELKRQTSLIAE---------TSPV---PTECDELTVDAAADDEDYGS 239
KASLLAEVI+HVKELKRQTS + E +PV PTE DEL VDAAAD+
Sbjct: 155 KASLLAEVIEHVKELKRQTSAMMEDGAAGGEAAAAPVVLLPTEDDELEVDAAADE----- 209
Query: 240 NGNKFIIKASLCCDDRSDLLPELIKTLKALRLRTLKADITTLGGRVKNVLFI-------- 291
G + + +ASLCC+DR+DL+P + + L ALRLR +A+I TLGGRV++VL I
Sbjct: 210 -GGRLVARASLCCEDRADLIPGIARALAALRLRARRAEIATLGGRVRSVLLIAAVEEEDP 268
Query: 292 --TGEEDDHEY------------CISSIQEALKAVMEK--------SVGDESASGSVKRQ 329
G +DD E+ ++SI EAL+ VM + S G GS+KRQ
Sbjct: 269 DEAGNDDDGEHGYGVAASHRRHELVASIHEALRGVMNRKAASSDTSSSGAGGGGGSIKRQ 328
Query: 330 R 330
R
Sbjct: 329 R 329
>UniRef100_Q6ESL3 DNA binding protein-like [Oryza sativa]
Length = 363
Score = 208 bits (529), Expect = 2e-52
Identities = 127/281 (45%), Positives = 164/281 (58%), Gaps = 86/281 (30%)
Query: 132 SGPFGGFQGEVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTD 191
SG G Q E+G+++A+EIM+AKALAAS+SHSEAERRRR+RIN HLA+LRSLLP+TTKTD
Sbjct: 80 SGLLGSLQAELGRVTAREIMDAKALAASRSHSEAERRRRQRINGHLARLRSLLPNTTKTD 139
Query: 192 KASLLAEVIQHVKELKRQTSLIAETSP--------------------------VPTECDE 225
KASLLAEVI+HVKELKRQT+ IA + +PTE DE
Sbjct: 140 KASLLAEVIEHVKELKRQTTAIAAAAAAGDYHGNDEDDDDAVVGRRSAAAQQLLPTEADE 199
Query: 226 LTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLKALRLRTLKADITTLGGRV 285
L VDAA D E K +++ASLCC+DR DL+P++ + L ALRLR +A+ITTLGGRV
Sbjct: 200 LAVDAAVDAE------GKLVVRASLCCEDRPDLIPDIARALAALRLRARRAEITTLGGRV 253
Query: 286 KNVLFITGEEDDHEY----------------------------------------CISSI 305
++VL IT +E ++ CI+++
Sbjct: 254 RSVLLITADEQQQQHCDDVDDDEDGHRLLLRHGIDGAGAAADDDDECAASHRRHECIATV 313
Query: 306 QEALKAVMEK----SVGDESAS----------GSVKRQRTN 332
QEAL+ VM++ S GD S+S GS+KRQR N
Sbjct: 314 QEALRGVMDRRAAASSGDTSSSGGAVVAGGGGGSIKRQRMN 354
>UniRef100_O80674 Hypothetical protein At2g41130 [Arabidopsis thaliana]
Length = 253
Score = 153 bits (387), Expect = 6e-36
Identities = 83/178 (46%), Positives = 131/178 (72%), Gaps = 11/178 (6%)
Query: 142 VGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQ 201
+G+ AQ+ +ALAA ++H EAERRRRERIN+HL KLR++L +KTDKA+LLA+V+Q
Sbjct: 55 IGETMAQD----RALAALRNHKEAERRRRERINSHLNKLRNVLSCNSKTDKATLLAKVVQ 110
Query: 202 HVKELKRQTSLIAETSP--VPTECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLL 259
V+ELK+QT +++ +P+E DE++V DY ++G+ I KASLCC+DRSDLL
Sbjct: 111 RVRELKQQTLETSDSDQTLLPSETDEISV---LHFGDYSNDGH-IIFKASLCCEDRSDLL 166
Query: 260 PELIKTLKALRLRTLKADITTLGGRVKNVLFITGEEDDHEY-CISSIQEALKAVMEKS 316
P+L++ LK+L ++TL+A++ T+GGR ++VL + +++ H + +Q ALK+++E+S
Sbjct: 167 PDLMEILKSLNMKTLRAEMVTIGGRTRSVLVVAADKEMHGVESVHFLQNALKSLLERS 224
>UniRef100_Q7XZF8 Putative DNA binding protein [Oryza sativa]
Length = 268
Score = 152 bits (383), Expect = 2e-35
Identities = 90/213 (42%), Positives = 135/213 (63%), Gaps = 24/213 (11%)
Query: 146 SAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKE 205
+A + +ALAAS++H EAE+RRRERI +HL +LR++L K DKASLLA+ ++ V++
Sbjct: 55 AAAAAAQDRALAASRNHREAEKRRRERIKSHLDRLRAVLACDPKIDKASLLAKAVERVRD 114
Query: 206 LKRQTSLIAETSPV---PTECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPEL 262
LK++ + I E +P PTE DE+ V A+ G+ G + +AS+CCDDR DLLPEL
Sbjct: 115 LKQRMAGIGEAAPAHLFPTEHDEIVVLASGGGGVGGAGGAAAVFEASVCCDDRCDLLPEL 174
Query: 263 IKTLKALRLRTLKADITTLGGRVKNVLFIT-----------GEEDDHEYCISS------I 305
I+TL+ALRLRTL+A++ TLGGRV+NVL + G++D Y S +
Sbjct: 175 IETLRALRLRTLRAEMATLGGRVRNVLVLARDAGGAGEGGDGDDDRAGYSAVSNDGGDFL 234
Query: 306 QEALKAVMEKSVGDESASGSVKRQRTNIISISN 338
+EAL+A++E+ A+ + +R ++S N
Sbjct: 235 KEALRALVER----PGAAAGDRPKRRRVVSDMN 263
>UniRef100_Q9LET0 Putative HLH DNA binding protein [Arabidopsis thaliana]
Length = 230
Score = 149 bits (375), Expect = 1e-34
Identities = 83/191 (43%), Positives = 128/191 (66%), Gaps = 17/191 (8%)
Query: 150 IMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQ 209
+ E KALA+ ++H EAER+RR RIN+HL KLR LL +KTDK++LLA+V+Q VKELK+Q
Sbjct: 37 VYEDKALASLRNHKEAERKRRARINSHLNKLRKLLSCNSKTDKSTLLAKVVQRVKELKQQ 96
Query: 210 TSLIAETSPVPTECDELTV----DAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKT 265
T I + + +P+E DE++V D + D+ + I K S CC+DR +LL +L++T
Sbjct: 97 TLEITDET-IPSETDEISVLNIEDCSRGDD------RRIIFKVSFCCEDRPELLKDLMET 149
Query: 266 LKALRLRTLKADITTLGGRVKNVLFITGEEDDH-EYCISSIQEALKAVMEKS-----VGD 319
LK+L++ TL AD+TT+GGR +NVL + +++ H ++ +Q ALK+++E+S VG
Sbjct: 150 LKSLQMETLFADMTTVGGRTRNVLVVAADKEHHGVQSVNFLQNALKSLLERSSKSVMVGH 209
Query: 320 ESASGSVKRQR 330
G + +R
Sbjct: 210 GGGGGEERLKR 220
>UniRef100_Q7F7Z2 ESTs C26093 [Oryza sativa]
Length = 258
Score = 137 bits (345), Expect = 4e-31
Identities = 83/195 (42%), Positives = 116/195 (58%), Gaps = 13/195 (6%)
Query: 146 SAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKE 205
S + EA+AL K HSEAERRRRERIN HL LR ++P T + DKA+LLA V+ VK+
Sbjct: 56 SGRSATEARAL---KIHSEAERRRRERINAHLTTLRRMIPDTKQMDKATLLARVVDQVKD 112
Query: 206 LKRQTSLIAETSPVPTECDELTV-----DAAADDEDYGSNGNKFIIKASLCCDDRSDLLP 260
LKR+ S I + +P+P E +E+++ DAA N IKAS+ CDDR DL+
Sbjct: 113 LKRKASEITQRTPLPPETNEVSIECFTGDAATAATTVAGNHKTLYIKASISCDDRPDLIA 172
Query: 261 ELIKTLKALRLRTLKADITTLGGRVKNVLFITGEED-DHEYCISSIQEALKAVMEKSVGD 319
+ LRLRT++A++T+LGGRV++V + EE + S++EA++ + K
Sbjct: 173 GITHAFHGLRLRTVRAEMTSLGGRVQHVFILCREEGIAGGVSLKSLKEAVRQALAKVASP 232
Query: 320 ESASGS----VKRQR 330
E GS KRQR
Sbjct: 233 ELVYGSSHFQSKRQR 247
>UniRef100_Q9FTQ1 DNA binding protein-like [Oryza sativa]
Length = 267
Score = 131 bits (330), Expect = 2e-29
Identities = 73/178 (41%), Positives = 112/178 (62%), Gaps = 4/178 (2%)
Query: 141 EVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVI 200
E+ K + E KA A KSHSEAERRRRERIN HLA LR+++P T K DKA+LLAEV+
Sbjct: 61 EMAKRNCGGGREEKAAMALKSHSEAERRRRERINAHLATLRTMVPCTDKMDKAALLAEVV 120
Query: 201 QHVKELKRQTSLIAETSPVPTECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLP 260
HVK+LK + + + VP+ DE+ VD A+ G +++A+L CDDR+DL
Sbjct: 121 GHVKKLKSAAARVGRRATVPSGADEVAVDEAS--ATGGGGEGPLLLRATLSCDDRADLFV 178
Query: 261 ELIKTLKALRLRTLKADITTLGGRVKNVLFIT--GEEDDHEYCISSIQEALKAVMEKS 316
++ + L+ L L + +++TTLGGRV+ ++ ++S++ AL++V++K+
Sbjct: 179 DVKRALQPLGLEVVGSEVTTLGGRVRLAFLVSCGSRGGAAAAAMASVRHALQSVLDKA 236
>UniRef100_Q9XEF0 Hypothetical protein T07M07.8 [Arabidopsis thaliana]
Length = 254
Score = 123 bits (309), Expect = 6e-27
Identities = 70/183 (38%), Positives = 115/183 (62%), Gaps = 7/183 (3%)
Query: 154 KALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLI 213
KA + S+SH AE+RRR+RIN+HL LR L+P++ K DKA+LLA VI+ VKELK++ +
Sbjct: 59 KAESLSRSHRLAEKRRRDRINSHLTALRKLVPNSDKLDKAALLATVIEQVKELKQKAAES 118
Query: 214 AETSPVPTECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLKALRLRT 273
+PTE DE+TV D+ SN N I KAS CC+D+ + + E+I+ L L+L T
Sbjct: 119 PIFQDLPTEADEVTVQPET-ISDFESNTNTIIFKASFCCEDQPEAISEIIRVLTKLQLET 177
Query: 274 LKADITTLGGRVKNVLFITGEEDDHE-----YCISSIQEALKAVMEKSVGDESASGSVKR 328
++A+I ++GGR++ + FI + + +E +++++L + + + + + SV R
Sbjct: 178 IQAEIISVGGRMR-INFILKDSNCNETTNIAASAKALKQSLCSALNRITSSSTTTSSVCR 236
Query: 329 QRT 331
R+
Sbjct: 237 IRS 239
>UniRef100_Q75IR0 Hypothetical protein OSJNBb0099P06.13 [Oryza sativa]
Length = 271
Score = 115 bits (287), Expect = 2e-24
Identities = 71/187 (37%), Positives = 105/187 (55%), Gaps = 11/187 (5%)
Query: 153 AKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELK---RQ 209
A A HSEAERRRRERIN HLA LR +LP + DKA+LLA V+ VK LK +
Sbjct: 61 AAQATAMTIHSEAERRRRERINAHLATLRRILPDAKQMDKATLLASVVNQVKHLKTRATE 120
Query: 210 TSLIAETSPVPTECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLKAL 269
+ + + +P E +E+TV A E + + ++A++ CDDR LL ++ T + L
Sbjct: 121 ATTPSTAATIPPEANEVTVQCYAGGEH--TAAARTYVRATVSCDDRPGLLADIAATFRRL 178
Query: 270 RLRTLKADITTLGGRVKNVLFITGEEDDHEYCISS---IQEALKAVMEKSVGDES---AS 323
RLR L AD++ LGGR ++ + EE++ E + ++EA++ + K E+
Sbjct: 179 RLRPLSADMSCLGGRTRHAFVLCREEEEEEDAAAEARPLKEAVRQALAKVALPETVYGGG 238
Query: 324 GSVKRQR 330
G KRQR
Sbjct: 239 GRSKRQR 245
>UniRef100_Q6ZJC8 BHLH protein family-like [Oryza sativa]
Length = 223
Score = 111 bits (277), Expect = 3e-23
Identities = 66/189 (34%), Positives = 113/189 (58%), Gaps = 19/189 (10%)
Query: 143 GKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQH 202
G+ SA + + + A +SHSEAER+RR+RIN HLA LR+L+PS ++ DKA+LL EV++H
Sbjct: 5 GECSAAAVRKGGSPAV-RSHSEAERKRRQRINAHLATLRTLVPSASRMDKAALLGEVVRH 63
Query: 203 VKELKRQTSLIAETSP--VPTECDELTVDAAADDEDYGSNGNKFI----------IKASL 250
V+EL+ + E + VP E DE+ V+ DDE G + ++A +
Sbjct: 64 VRELRCRADDATEGADVVVPGEGDEVGVEDEDDDEGERDEGCYVVGGGDRRWRRRVRAWV 123
Query: 251 CCDDRSDLLPELIKTLKALRLRTLKADITTLGGRVKNVLFI------TGEEDDHEYCISS 304
CC DR L+ +L + ++++ R ++A++ T+GGR ++VL + ++D +S+
Sbjct: 124 CCADRPGLMSDLGRAVRSVSARPVRAEVATVGGRTRSVLELDVVVASDAADNDRAVALSA 183
Query: 305 IQEALKAVM 313
++ AL+ V+
Sbjct: 184 LRAALRTVL 192
>UniRef100_Q69MD0 BHLH-like protein [Oryza sativa]
Length = 215
Score = 100 bits (250), Expect = 4e-20
Identities = 62/168 (36%), Positives = 99/168 (58%), Gaps = 14/168 (8%)
Query: 158 ASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTSLIAETS 217
A +SHSEAER+RRERIN HL LR L+PS ++ DKA+LL EV+++V++L+ + + +
Sbjct: 28 ARRSHSEAERKRRERINAHLDTLRGLVPSASRMDKAALLGEVVRYVRKLRSEAA--GSAA 85
Query: 218 PVPTECDELTVDAAADDE-----DYGSNGNKFIIKASLCCDDRSDLLPELIKTLKALRLR 272
VP E DE+ V+ + D G +KAS+CC DR L+ EL +++ R
Sbjct: 86 VVPGEGDEVVVEEEEVEVEGCSCDAGERQAARRVKASVCCADRPGLMSELGDAERSVSAR 145
Query: 273 TLKADITTLGGRVKNVLFI-------TGEEDDHEYCISSIQEALKAVM 313
++A+I T+GGR ++ L + G + + ++Q AL+AV+
Sbjct: 146 AVRAEIATVGGRTRSDLELDVARTAAAGGGSNGASQLPALQAALRAVI 193
>UniRef100_Q7XHI6 Putative bHLH131 transcription factor [Arabidopsis thaliana]
Length = 256
Score = 100 bits (249), Expect = 6e-20
Identities = 58/174 (33%), Positives = 101/174 (57%), Gaps = 14/174 (8%)
Query: 152 EAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKELKRQTS 211
E+K +AA K HS+AERRRR RIN+ A LR++LP+ K DKAS+L E +++ ELK+
Sbjct: 87 ESKEVAAKK-HSDAERRRRLRINSQFATLRTILPNLVKQDKASVLGETVRYFNELKK--- 142
Query: 212 LIAETSPVPTECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLKALRL 271
++ + P+ D L +D N N+ + + C DR L+ E+ +++KA++
Sbjct: 143 MVQDIPTTPSLEDNLRLDHC--------NNNRDLARVVFSCSDREGLMSEVAESMKAVKA 194
Query: 272 RTLKADITTLGGRVKNVLFITGEEDDHEYCISSIQEALKAVMEKSVGDESASGS 325
+ ++A+I T+GGR K LF+ G + + ++++LK V+ E+ + +
Sbjct: 195 KAVRAEIMTVGGRTKCALFVQGVNGNEG--LVKLKKSLKLVVNGKSSSEAKNNN 246
>UniRef100_Q9SZK7 Hypothetical protein F20D10.190 [Arabidopsis thaliana]
Length = 1496
Score = 91.3 bits (225), Expect = 3e-17
Identities = 60/188 (31%), Positives = 101/188 (52%), Gaps = 28/188 (14%)
Query: 152 EAKALAASKSHSEAERRRRERINNHLAKLRSLLPS--------------TTKTDKASLLA 197
E+K +AA K HS+AERRRR RIN+ A LR++LP+ T + DKAS+L
Sbjct: 1313 ESKEVAAKK-HSDAERRRRLRINSQFATLRTILPNLVKRSNTFCIMFNETKQQDKASVLG 1371
Query: 198 EVIQHVKELKRQTSLIAETSPVPTECDELTVDAAADDEDYGSNGNKFIIKASLCCDDRSD 257
E +++ ELK+ I T P+ D L +D N N+ + + C DR
Sbjct: 1372 ETVRYFNELKKMVQDIPTT---PSLEDNLRLDHC--------NNNRDLARVVFSCSDREG 1420
Query: 258 LLPELIKTLKALRLRTLKADITTLGGRVKNVLFITGEEDDHEYCISSIQEALKAVMEKSV 317
L+ E+ +++KA++ + ++A+I T+GGR K LF+ G + + ++++LK V+
Sbjct: 1421 LMSEVAESMKAVKAKAVRAEIMTVGGRTKCALFVQGVNGNEG--LVKLKKSLKLVVNGKS 1478
Query: 318 GDESASGS 325
E+ + +
Sbjct: 1479 SSEAKNNN 1486
>UniRef100_Q7XHF6 Putative bHLH transcription factor [Oryza sativa]
Length = 361
Score = 70.1 bits (170), Expect = 8e-11
Identities = 51/203 (25%), Positives = 92/203 (45%), Gaps = 19/203 (9%)
Query: 126 LQHGSGSGPFGGFQGEVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLP 185
L+ SGP F + +M +A ++ + H AER+RRE+++ L S++P
Sbjct: 160 LEQQQDSGPMTKFCSPLSEMKRGG---RRATSSMQEHVIAERKRREKMHQQFTTLASIVP 216
Query: 186 STTKTDKASLLAEVIQHVKELKRQTSLIAET----SPVPTECDELTVDAAADDEDYGSNG 241
TKTDK S+L I++V L+ + ++ + S P D + + DDED N
Sbjct: 217 EITKTDKVSVLGSTIEYVHHLRERVKILQDIQSMGSTQPPISDARSRAGSGDDEDDDGNN 276
Query: 242 NKFIIKAS-----------LCCDDRSDLLPELIKTLKALRLRTLKADITTLGGRVKNVLF 290
N+ IK + C ++ +L +L+ L+ L L T+ ++ N+
Sbjct: 277 NEVEIKVEANLQGTTVLLRVVCPEKKGVLIKLLTELEKLGLSTMNTNVVPFADSSLNIT- 335
Query: 291 ITGEEDDHEYCISSIQEALKAVM 313
IT + D+ + + LK+ +
Sbjct: 336 ITAQIDNASCTTVELVKNLKSTL 358
>UniRef100_Q8S3F5 Putative bHLH transcription factor [Arabidopsis thaliana]
Length = 590
Score = 67.8 bits (164), Expect = 4e-10
Identities = 79/357 (22%), Positives = 134/357 (37%), Gaps = 57/357 (15%)
Query: 2 IQEDQGQCSSQTINNFETYQQEQFLLQQQMMRQQNSDHHIF------------------- 42
I +++ + + ++N Q Q QQQ Q H F
Sbjct: 256 IDDNRTKIFGKDLHNSGFLQHHQHHQQQQQQPPQQQQHRQFREKLTVRKMDDRAPKRLDA 315
Query: 43 ---GGGRSMFPTAGGDHNQVSPILQQTWSMQQLHHHHHPFNPHDPFVIPQQQASPYASLF 99
G R MF G ++N +L TW Q ++ P N + + + P
Sbjct: 316 YPNNGNRFMFSNPGTNNNT---LLSPTWV--QPENYTRPINVKEVPSTDEFKFLPLQQSS 370
Query: 100 NRRVPSLQFAYDHHAGSEHLRIISDTLQHGSGSGPFGGFQG--EVGKMSAQEI--MEAKA 155
R +P Q D A S S+ G G G + G E G ++ A
Sbjct: 371 QRLLPPAQMQIDFSAASSRA---SENNSDGEGGGEWADAVGADESGNNRPRKRGRRPANG 427
Query: 156 LAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKEL-------KR 208
A + +H EAER+RRE++N LRS++P+ +K DKASLL + + ++ EL +
Sbjct: 428 RAEALNHVEAERQRREKLNQRFYALRSVVPNISKMDKASLLGDAVSYINELHAKLKVMEA 487
Query: 209 QTSLIAETSPVPTECD-ELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLK 267
+ + +S P D ++ V + +D + + C S + +
Sbjct: 488 ERERLGYSSNPPISLDSDINVQTSGED-----------VTVRINCPLESHPASRIFHAFE 536
Query: 268 ALRLRTLKADITTLGGRVKNVLFITGEEDDHEYCISSIQEALKAVMEKSVGDESASG 324
++ + +++ V + + EE E IS AL SV ++SG
Sbjct: 537 ESKVEVINSNLEVSQDTVLHTFVVKSEELTKEKLIS----ALSREQTNSVQSRTSSG 589
>UniRef100_Q9LNJ5 F6F3.7 protein [Arabidopsis thaliana]
Length = 590
Score = 67.8 bits (164), Expect = 4e-10
Identities = 79/357 (22%), Positives = 134/357 (37%), Gaps = 57/357 (15%)
Query: 2 IQEDQGQCSSQTINNFETYQQEQFLLQQQMMRQQNSDHHIF------------------- 42
I +++ + + ++N Q Q QQQ Q H F
Sbjct: 256 IDDNRTKIFGKDLHNSGFLQHHQHHQQQQQQPPQQQQHRQFREKLTVRKMDDRAPKRLDA 315
Query: 43 ---GGGRSMFPTAGGDHNQVSPILQQTWSMQQLHHHHHPFNPHDPFVIPQQQASPYASLF 99
G R MF G ++N +L TW Q ++ P N + + + P
Sbjct: 316 YPNNGNRFMFSNPGTNNNT---LLSPTWV--QPENYTRPINVKEVPSTDEFKFLPLQQSS 370
Query: 100 NRRVPSLQFAYDHHAGSEHLRIISDTLQHGSGSGPFGGFQG--EVGKMSAQEI--MEAKA 155
R +P Q D A S S+ G G G + G E G ++ A
Sbjct: 371 QRLLPPAQMQIDFSAASSRA---SENNSDGEGGGEWADAVGADESGNNRPRKRGRRPANG 427
Query: 156 LAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDKASLLAEVIQHVKEL-------KR 208
A + +H EAER+RRE++N LRS++P+ +K DKASLL + + ++ EL +
Sbjct: 428 RAEALNHVEAERQRREKLNQRFYALRSVVPNISKMDKASLLGDAVSYINELHAKLKVMEA 487
Query: 209 QTSLIAETSPVPTECD-ELTVDAAADDEDYGSNGNKFIIKASLCCDDRSDLLPELIKTLK 267
+ + +S P D ++ V + +D + + C S + +
Sbjct: 488 ERERLGYSSNPPISLDSDINVQTSGED-----------VTVRINCPLESHPASRIFHAFE 536
Query: 268 ALRLRTLKADITTLGGRVKNVLFITGEEDDHEYCISSIQEALKAVMEKSVGDESASG 324
++ + +++ V + + EE E IS AL SV ++SG
Sbjct: 537 ESKVEVINSNLEVSQDTVLHTFVVKSEELTKEKLIS----ALSREQTNSVQSRTSSG 589
>UniRef100_O65552 Hypothetical protein F6I18.110 [Arabidopsis thaliana]
Length = 367
Score = 66.6 bits (161), Expect = 9e-10
Identities = 50/171 (29%), Positives = 80/171 (46%), Gaps = 11/171 (6%)
Query: 74 HHHHPFNPHDPFVIPQQQASPYASLFNR-RVPSLQFAYDHHAGSEHLRIISDTLQHGSGS 132
HHH + + +IP P +LFN SL F +G + + T + S
Sbjct: 117 HHHDADSRNQITMIPLSHNHPNDALFNGFSTGSLPFHLPQGSGGQ-----TQTQSQATAS 171
Query: 133 GPFGGFQGEVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDK 192
GG + + + A+ A+ HS AER RRERI + L+ L+P+ KTDK
Sbjct: 172 ATTGG---ATAQPQTKPKVRARRGQATDPHSIAERLRRERIAERMKSLQELVPNGNKTDK 228
Query: 193 ASLLAEVIQHVKELKRQTSLI--AETSPVPTECDELTVDAAADDEDYGSNG 241
AS+L E+I +VK L+ Q ++ + + +++ DA E+ S+G
Sbjct: 229 ASMLDEIIDYVKFLQLQVKVLSMSRLGGAASASSQISEDAGGSHENTSSSG 279
>UniRef100_Q8S3D5 Putative bHLH transcription factor [Arabidopsis thaliana]
Length = 310
Score = 66.6 bits (161), Expect = 9e-10
Identities = 50/171 (29%), Positives = 80/171 (46%), Gaps = 11/171 (6%)
Query: 74 HHHHPFNPHDPFVIPQQQASPYASLFNR-RVPSLQFAYDHHAGSEHLRIISDTLQHGSGS 132
HHH + + +IP P +LFN SL F +G + + T + S
Sbjct: 60 HHHDADSRNQITMIPLSHNHPNDALFNGFSTGSLPFHLPQGSGGQ-----TQTQSQATAS 114
Query: 133 GPFGGFQGEVGKMSAQEIMEAKALAASKSHSEAERRRRERINNHLAKLRSLLPSTTKTDK 192
GG + + + A+ A+ HS AER RRERI + L+ L+P+ KTDK
Sbjct: 115 ATTGG---ATAQPQTKPKVRARRGQATDPHSIAERLRRERIAERMKSLQELVPNGNKTDK 171
Query: 193 ASLLAEVIQHVKELKRQTSLI--AETSPVPTECDELTVDAAADDEDYGSNG 241
AS+L E+I +VK L+ Q ++ + + +++ DA E+ S+G
Sbjct: 172 ASMLDEIIDYVKFLQLQVKVLSMSRLGGAASASSQISEDAGGSHENTSSSG 222
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.315 0.130 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 533,375,520
Number of Sequences: 2790947
Number of extensions: 21934751
Number of successful extensions: 90223
Number of sequences better than 10.0: 1223
Number of HSP's better than 10.0 without gapping: 394
Number of HSP's successfully gapped in prelim test: 837
Number of HSP's that attempted gapping in prelim test: 88633
Number of HSP's gapped (non-prelim): 1724
length of query: 338
length of database: 848,049,833
effective HSP length: 128
effective length of query: 210
effective length of database: 490,808,617
effective search space: 103069809570
effective search space used: 103069809570
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 75 (33.5 bits)
Medicago: description of AC146570.4


