

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146567.12 + phase: 0 /pseudo
(307 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
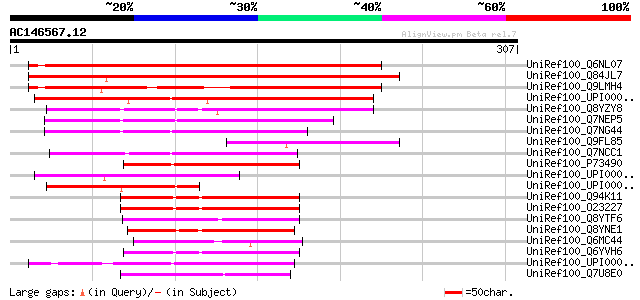
Sequences producing significant alignments: (bits) Value
UniRef100_Q6NL07 At1g13820 [Arabidopsis thaliana] 335 9e-91
UniRef100_Q84JL7 Hypothetical protein At5g39220 [Arabidopsis tha... 285 1e-75
UniRef100_Q9LMH4 F16A14.4 [Arabidopsis thaliana] 266 7e-70
UniRef100_UPI00002D2955 UPI00002D2955 UniRef100 entry 171 2e-41
UniRef100_Q8YZY8 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase [An... 127 3e-28
UniRef100_Q7NEP5 Glr3833 protein [Gloeobacter violaceus] 118 2e-25
UniRef100_Q7NG44 Glr3329 protein [Gloeobacter violaceus] 114 3e-24
UniRef100_Q9FL85 Arabidopsis thaliana genomic DNA, chromosome 5,... 113 7e-24
UniRef100_Q7NCC1 Glr3058 protein [Gloeobacter violaceus] 102 1e-20
UniRef100_P73490 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase [Sy... 91 5e-17
UniRef100_UPI00002E45B6 UPI00002E45B6 UniRef100 entry 88 3e-16
UniRef100_UPI000034CB6F UPI000034CB6F UniRef100 entry 84 4e-15
UniRef100_Q94K11 Hypothetical protein unannotated coding sequenc... 75 3e-12
UniRef100_O23227 Hypothetical protein C7A10.830 [Arabidopsis tha... 75 3e-12
UniRef100_Q8YTF6 All2761 protein [Anabaena sp.] 68 4e-10
UniRef100_Q8YNE1 Alr4625 protein [Anabaena sp.] 68 4e-10
UniRef100_Q6MC44 Hypothetical protein [Parachlamydia sp.] 68 4e-10
UniRef100_Q6YVH6 Hydrolase, alpha/beta fold family-like [Oryza s... 67 8e-10
UniRef100_UPI000034D3CA UPI000034D3CA UniRef100 entry 64 5e-09
UniRef100_Q7U8E0 Predicted alpha/beta hydrolase superfamily prot... 64 5e-09
>UniRef100_Q6NL07 At1g13820 [Arabidopsis thaliana]
Length = 339
Score = 335 bits (859), Expect = 9e-91
Identities = 159/214 (74%), Positives = 183/214 (85%), Gaps = 4/214 (1%)
Query: 12 FKVVAETDADSFPSFLPKDVNRIKDPFARTLATRIQRLPVSVKFSENPIMSSCVKPLVQN 71
FKV AE FP+FLP DV +IKD FA LA RI+RLPVSV F E+ IMSSCV PL++N
Sbjct: 25 FKVTAE----KFPTFLPTDVEKIKDTFALKLAARIERLPVSVSFREDSIMSSCVTPLMRN 80
Query: 72 KENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKREHF 131
+ P+VLLHGFDSSCLEWRYTYPLLEEAG+ETWA DILGWGFSDL+KLP CDV SKREHF
Sbjct: 81 ETTPVVLLHGFDSSCLEWRYTYPLLEEAGLETWAFDILGWGFSDLDKLPPCDVASKREHF 140
Query: 132 YQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEGTGNLATLPRAA 191
Y+FWKS+IK+P++LVGPSLG+AVAID A+N+PEAVE LVL+DASVY EGTGNLATLP+AA
Sbjct: 141 YKFWKSHIKRPVVLVGPSLGAAVAIDIAVNHPEAVESLVLMDASVYAEGTGNLATLPKAA 200
Query: 192 AYAGVYLLKSVPLRVYANYLSFTNISFSTSLDWT 225
AYAGVYLLKS+PLR+Y N++ F IS TS DWT
Sbjct: 201 AYAGVYLLKSIPLRLYVNFICFNGISLETSWDWT 234
>UniRef100_Q84JL7 Hypothetical protein At5g39220 [Arabidopsis thaliana]
Length = 330
Score = 285 bits (729), Expect = 1e-75
Identities = 137/228 (60%), Positives = 172/228 (75%), Gaps = 3/228 (1%)
Query: 12 FKVVAETDADSFPSFLPKDVNRIKDPFARTLATRIQRLPVSVKFSE--NPIMSSCVKPLV 69
F+ + D FP+FLPK++ IKDPFAR LA RI R+PV ++ +MSSC+KPLV
Sbjct: 17 FRQRVTANGDGFPTFLPKEIQNIKDPFARALAQRIVRIPVPLQMGNFRGSVMSSCIKPLV 76
Query: 70 Q-NKENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKR 128
Q + ++P+VLLH FDSSCLEWR TYPLLE+A +ETWAID+LGWGFSDLEKLP CD SKR
Sbjct: 77 QLHDKSPVVLLHCFDSSCLEWRRTYPLLEQACLETWAIDVLGWGFSDLEKLPPCDAASKR 136
Query: 129 EHFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEGTGNLATLP 188
H ++ WK+YIK+PMILVGPSLG+ VA+DF YPEAV+KLVLI+A+ Y+EGTG L LP
Sbjct: 137 HHLFELWKTYIKRPMILVGPSLGATVAVDFTATYPEAVDKLVLINANAYSEGTGRLKELP 196
Query: 189 RAAAYAGVYLLKSVPLRVYANYLSFTNISFSTSLDWTNATMLLRR*KW 236
++ AYAGV LLKS PLR+ AN L+F + S ++DWTN L + W
Sbjct: 197 KSIAYAGVKLLKSFPLRLLANVLAFCSSPLSENIDWTNIGRLHCQMPW 244
>UniRef100_Q9LMH4 F16A14.4 [Arabidopsis thaliana]
Length = 633
Score = 266 bits (679), Expect = 7e-70
Identities = 140/221 (63%), Positives = 162/221 (72%), Gaps = 32/221 (14%)
Query: 12 FKVVAETDADSFPSFLPKDVNRIKDPFARTLATRIQRLPVSVK-------FSENPIMSSC 64
FKV AE FP+FLP DV +IKD FA LA RI+RLPVSV F E+ IMSSC
Sbjct: 333 FKVTAE----KFPTFLPTDVEKIKDTFALKLAARIERLPVSVSGLRFHVSFREDSIMSSC 388
Query: 65 VKPLVQNKENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDV 124
V PL++N+ P+VLLHGFD RYTYPLLEEAG+ETWA DILGWGFSDL
Sbjct: 389 VTPLMRNETTPVVLLHGFD------RYTYPLLEEAGLETWAFDILGWGFSDLGF------ 436
Query: 125 VSKREHFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEGTGNL 184
FWKS+IK+P++LVGPSLG+AVAID A+N+PEAVE LVL+DASVY EGTGNL
Sbjct: 437 ---------FWKSHIKRPVVLVGPSLGAAVAIDIAVNHPEAVESLVLMDASVYAEGTGNL 487
Query: 185 ATLPRAAAYAGVYLLKSVPLRVYANYLSFTNISFSTSLDWT 225
ATLP+AAAYAGVYLLKS+PLR+Y N++ F IS TS DWT
Sbjct: 488 ATLPKAAAYAGVYLLKSIPLRLYVNFICFNGISLETSWDWT 528
>UniRef100_UPI00002D2955 UPI00002D2955 UniRef100 entry
Length = 309
Score = 171 bits (433), Expect = 2e-41
Identities = 95/210 (45%), Positives = 128/210 (60%), Gaps = 6/210 (2%)
Query: 16 AETDADSFPSFLPKDVNRIKDPFARTLATRIQRLPVSVKFSENPIMSSCVKPLVQ--NKE 73
+ T FPSF P +V + +P A +A R + + V+V + +++SCV +K
Sbjct: 43 SSTSEKDFPSFFPPEVRELTEPAAIDMAKRCELVEVAVPGRADAVLTSCVGDAGPEGSKL 102
Query: 74 NPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEK--LPSCDVVSKREHF 131
P VLLHGFDSSCLE+R +P L A ETWA+D+LGWGF+D + +KR H
Sbjct: 103 APFVLLHGFDSSCLEYRRLFPKLA-AKTETWAVDLLGWGFTDARDGGVGDYSPEAKRAHL 161
Query: 132 YQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEGTGNLATLPRAA 191
Y FWK I +PM+L G SLG A AIDFA N+PEAVEKLVL+DA + EG G + LPR
Sbjct: 162 YAFWKEKIGRPMVLCGASLGGAAAIDFATNHPEAVEKLVLVDAQGFIEGLGPMGALPRPI 221
Query: 192 AYAGVYLLKSVPLRVYANYLS-FTNISFST 220
A GV +L++ LR AN ++ F +F+T
Sbjct: 222 AKVGVNVLRTTALRNAANQMAYFDKATFAT 251
>UniRef100_Q8YZY8 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase [Anabaena sp.]
Length = 295
Score = 127 bits (320), Expect = 3e-28
Identities = 76/200 (38%), Positives = 115/200 (57%), Gaps = 7/200 (3%)
Query: 23 FPSFLPKDVNRIKDPFARTLATRIQRLPVSVKFSENPIMSSCVKPLVQNKENPIVLLHGF 82
FPSFLP V ++ + + LA I ++ S PI ++ V+ PI+L+HGF
Sbjct: 2 FPSFLPAAVGQLTESESIALAKSIHTQAIATPLSNQPITTAYVRQ--GGGGTPILLIHGF 59
Query: 83 DSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDV--VSKREHFYQFWKSYIK 140
DSS LE+R PLL + ETWA+D+LG+GF+ ++LP ++ R H + FWK+ I
Sbjct: 60 DSSVLEFRRLLPLLGKEN-ETWAVDLLGFGFT--QRLPGIKFSPIAIRTHLHSFWKTLIN 116
Query: 141 KPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEGTGNLATLPRAAAYAGVYLLK 200
+P+ILVG S+G A AIDF + YPEAV+KLVLID++ G+ + Y L+
Sbjct: 117 QPVILVGASMGGAAAIDFTLTYPEAVQKLVLIDSAGLRGGSPLSKFMFPPLDYLAAQFLR 176
Query: 201 SVPLRVYANYLSFTNISFST 220
S +R + ++ N + +T
Sbjct: 177 SPKVRDRVSRAAYKNQNLAT 196
>UniRef100_Q7NEP5 Glr3833 protein [Gloeobacter violaceus]
Length = 296
Score = 118 bits (296), Expect = 2e-25
Identities = 65/175 (37%), Positives = 105/175 (59%), Gaps = 2/175 (1%)
Query: 22 SFPSFLPKDVNRIKDPFARTLATRIQRLPVSVKFSENPIMSSCVKPLVQNKENPIVLLHG 81
+ P+FLP ++DP + L +QR PVSV F ++ + + PI+LLHG
Sbjct: 3 TLPAFLPPAAALLRDPASIGLLQTLQRDPVSVPFLPQTAETAWTFA-AGSDDPPILLLHG 61
Query: 82 FDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKREHFYQFWKSYIKK 141
FDSS LE+R+ P L + TWA+D+ G+GF++ + + + H + FW+++I++
Sbjct: 62 FDSSALEFRFVLPRLGQRH-PTWAVDLWGFGFTERQSGAAYGRAEIQTHLHAFWQAHIRR 120
Query: 142 PMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEGTGNLATLPRAAAYAGV 196
P+++VG S+G A A+DFA+N+P+AV+KLVLI ++ +T LA L A V
Sbjct: 121 PVLVVGASMGGAAAVDFAVNHPQAVQKLVLIASTGFTGPPAWLAGLAEPLREAAV 175
>UniRef100_Q7NG44 Glr3329 protein [Gloeobacter violaceus]
Length = 300
Score = 114 bits (285), Expect = 3e-24
Identities = 67/159 (42%), Positives = 93/159 (58%), Gaps = 2/159 (1%)
Query: 22 SFPSFLPKDVNRIKDPFARTLATRIQRLPVSVKFSENPIMSSCVKPLVQNKENPIVLLHG 81
S P+ D I P AR L RI+R+PV + +S V+ + P++LLHG
Sbjct: 6 SVPAPFAPDAADIDHPQARWLLERIERVPVLCPSGSQSVATSFVRA-GRADGPPVLLLHG 64
Query: 82 FDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKREHFYQFWKSYIKK 141
FDSS LE+ PLL A TWAID+LG+GF++ +C + + H + FW I +
Sbjct: 65 FDSSLLEFFRLVPLLA-AHRSTWAIDLLGFGFTERRPDLACSAAALKAHLWSFWLERIGE 123
Query: 142 PMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEG 180
P++LVG S+G A AIDFA+ +PEAV LVLID+ +T G
Sbjct: 124 PVVLVGASMGGAAAIDFALTHPEAVAGLVLIDSVGFTCG 162
>UniRef100_Q9FL85 Arabidopsis thaliana genomic DNA, chromosome 5, TAC clone:K3K3
[Arabidopsis thaliana]
Length = 239
Score = 113 bits (282), Expect = 7e-24
Identities = 62/137 (45%), Positives = 79/137 (57%), Gaps = 32/137 (23%)
Query: 132 YQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAV------------------------- 166
+Q WK+YIK+PMILVGPSLG+ VA+DF YPEAV
Sbjct: 17 FQLWKTYIKRPMILVGPSLGATVAVDFTATYPEAVNFQFTIIKLFWIKPILIYTSLTKFD 76
Query: 167 -------EKLVLIDASVYTEGTGNLATLPRAAAYAGVYLLKSVPLRVYANYLSFTNISFS 219
+KLVLI+A+ Y+EGTG L LP++ AYAGV LLKS PLR+ AN L+F + S
Sbjct: 77 YFRGFQVDKLVLINANAYSEGTGRLKELPKSIAYAGVKLLKSFPLRLLANVLAFCSSPLS 136
Query: 220 TSLDWTNATMLLRR*KW 236
++DWTN L + W
Sbjct: 137 ENIDWTNIGRLHCQMPW 153
>UniRef100_Q7NCC1 Glr3058 protein [Gloeobacter violaceus]
Length = 297
Score = 102 bits (254), Expect = 1e-20
Identities = 61/151 (40%), Positives = 89/151 (58%), Gaps = 4/151 (2%)
Query: 25 SFLPKDVNRIKDPFARTLATRIQRLPVSVKFSEN-PIMSSCVKPLVQNKENPIVLLHGFD 83
+FLP + +P + +A R++ + V ++ N + + CV E P +LLHGFD
Sbjct: 4 AFLPPGARILSEPGSLEMARRLELVSVRLEGRVNRTVATGCVHSGAG--EPPALLLHGFD 61
Query: 84 SSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKREHFYQFWKSYIKKPM 143
SS E+R PLL A E WA+D+LG+GF++ + D + +H FW+ YI +P
Sbjct: 62 SSVFEFRRLLPLLA-ARREVWAMDLLGFGFTERPAGIAYDPRAIGDHLASFWEQYIGRPA 120
Query: 144 ILVGPSLGSAVAIDFAINYPEAVEKLVLIDA 174
+LVG S+G A AID A+ PEAV KLVLID+
Sbjct: 121 LLVGASMGGAAAIDLALARPEAVAKLVLIDS 151
>UniRef100_P73490 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase [Synechocystis sp.]
Length = 296
Score = 90.5 bits (223), Expect = 5e-17
Identities = 44/106 (41%), Positives = 72/106 (67%), Gaps = 1/106 (0%)
Query: 70 QNKENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKRE 129
+ + P++ +HGFDSS LE+R PL+++ AID+LG+GF+ K+ + +
Sbjct: 49 EGQGEPMLFIHGFDSSVLEFRRLLPLIKK-NFRAIAIDLLGFGFTTRSKILLPTPANIKI 107
Query: 130 HFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDAS 175
H FW++ I++P+ LVG S+G AVA+DF +++PE V+KLVLID++
Sbjct: 108 HLDHFWQTIIQEPITLVGVSMGGAVALDFCLSFPERVKKLVLIDSA 153
>UniRef100_UPI00002E45B6 UPI00002E45B6 UniRef100 entry
Length = 202
Score = 88.2 bits (217), Expect = 3e-16
Identities = 47/137 (34%), Positives = 73/137 (52%), Gaps = 13/137 (9%)
Query: 16 AETDADSFPSFLPKD-VNRIKDPFARTLATRIQRLPVSVKFS-----------ENPIMSS 63
A + +FP ++P + V I++ A + +QR+ V+V F+ E+ SS
Sbjct: 37 AASSTTAFPPYVPAEAVQEIQEEAALDMLRSLQRVDVTVGFNGGVVPSSFYHKESSSSSS 96
Query: 64 CVKPLVQNKENPIV-LLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSC 122
+ + P+V LLHGFDSSCLE+R P LEE G+ +A+D+LGWGF+D
Sbjct: 97 AAAASASSSDEPVVVLLHGFDSSCLEYRRLVPCLEERGLAPYALDVLGWGFTDTRAFGDT 156
Query: 123 DVVSKREHFYQFWKSYI 139
+KR H + FW+ +
Sbjct: 157 SAAAKRAHLHAFWQQQL 173
>UniRef100_UPI000034CB6F UPI000034CB6F UniRef100 entry
Length = 152
Score = 84.3 bits (207), Expect = 4e-15
Identities = 42/96 (43%), Positives = 62/96 (63%), Gaps = 4/96 (4%)
Query: 23 FPSFLPKDVNRIKDPFARTLATRIQRLPVSVKFSENPIMSSCVK---PLVQNKENPIVLL 79
F SF P +V +++P A +A + +++ V+V + P+M+SCV P +P VL+
Sbjct: 41 FASFFPDEVTALEEPAAVEMAKKARQVSVAVPGRDKPVMTSCVSSPAPKGGEDVSPFVLI 100
Query: 80 HGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSD 115
HGFDSSCLE+R +PLL E E A+D+LGWGF+D
Sbjct: 101 HGFDSSCLEYRRLFPLLTERA-EAHAVDLLGWGFTD 135
>UniRef100_Q94K11 Hypothetical protein unannotated coding sequence from BAC CIC7A10
[Arabidopsis thaliana]
Length = 321
Score = 74.7 bits (182), Expect = 3e-12
Identities = 38/108 (35%), Positives = 65/108 (60%), Gaps = 2/108 (1%)
Query: 68 LVQNKENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSK 127
+VQ + +P+VL+HGF +S WRY P L + + +A+D+LG+G+SD + L D +
Sbjct: 37 VVQGEGSPLVLIHGFGASVFHWRYNIPELAKK-YKVYALDLLGFGWSD-KALIEYDAMVW 94
Query: 128 REHFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDAS 175
+ F K +K+P ++VG SLG A+ A+ PE V + L++++
Sbjct: 95 TDQVIDFMKEVVKEPAVVVGNSLGGFTALSVAVGLPEQVTGVALLNSA 142
>UniRef100_O23227 Hypothetical protein C7A10.830 [Arabidopsis thaliana]
Length = 378
Score = 74.7 bits (182), Expect = 3e-12
Identities = 38/108 (35%), Positives = 65/108 (60%), Gaps = 2/108 (1%)
Query: 68 LVQNKENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSK 127
+VQ + +P+VL+HGF +S WRY P L + + +A+D+LG+G+SD + L D +
Sbjct: 94 VVQGEGSPLVLIHGFGASVFHWRYNIPELAKK-YKVYALDLLGFGWSD-KALIEYDAMVW 151
Query: 128 REHFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDAS 175
+ F K +K+P ++VG SLG A+ A+ PE V + L++++
Sbjct: 152 TDQVIDFMKEVVKEPAVVVGNSLGGFTALSVAVGLPEQVTGVALLNSA 199
>UniRef100_Q8YTF6 All2761 protein [Anabaena sp.]
Length = 305
Score = 67.8 bits (164), Expect = 4e-10
Identities = 38/108 (35%), Positives = 62/108 (57%), Gaps = 3/108 (2%)
Query: 69 VQNKENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLP-SCDVVSK 127
V P+VL+HGF +S WR P+L AG + +AID+LG+G S+ + S DV
Sbjct: 37 VMGTGQPLVLVHGFGASIGHWRKNIPVLANAGYQIFAIDLLGFGGSEKAAIDYSVDVWV- 95
Query: 128 REHFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDAS 175
E FW ++I++P + VG S+G+ +++ +PE VLI+++
Sbjct: 96 -ELLKDFWTAHIQQPAVFVGNSIGALLSLIILAKHPEITSGGVLINSA 142
>UniRef100_Q8YNE1 Alr4625 protein [Anabaena sp.]
Length = 312
Score = 67.8 bits (164), Expect = 4e-10
Identities = 35/102 (34%), Positives = 66/102 (64%), Gaps = 4/102 (3%)
Query: 72 KENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPS-CDVVSKREH 130
++ P++LLHGF +S WR+ +L E+ +A+D+LG+G S EK P+ + E
Sbjct: 37 EKTPLILLHGFGASIGHWRHNLEVLGESHT-VYALDMLGFGGS--EKAPANYSIELWVEQ 93
Query: 131 FYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLI 172
Y FW+++I++P++L+G S GS +++ A +P+ V+ +V++
Sbjct: 94 VYDFWQAFIRQPVVLIGNSNGSLISLAAAAAHPDMVKGIVMM 135
>UniRef100_Q6MC44 Hypothetical protein [Parachlamydia sp.]
Length = 322
Score = 67.8 bits (164), Expect = 4e-10
Identities = 36/106 (33%), Positives = 57/106 (52%), Gaps = 8/106 (7%)
Query: 76 IVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKREHFYQFW 135
++L+HGF + WRY L +AG W ID++G+G SD + D + F +
Sbjct: 69 LLLIHGFRAHSFTWRYLIEPLTQAGYHVWTIDLIGYGLSDKPLNAAYDA----DFFIEQL 124
Query: 136 KSYIKKPMI----LVGPSLGSAVAIDFAINYPEAVEKLVLIDASVY 177
KS++ I L+G S+G +A++ ++YPE V L LI+A Y
Sbjct: 125 KSFMDAKQISSAHLIGSSMGGGLALNLTLDYPEKVSSLTLINALGY 170
>UniRef100_Q6YVH6 Hydrolase, alpha/beta fold family-like [Oryza sativa]
Length = 410
Score = 66.6 bits (161), Expect = 8e-10
Identities = 37/106 (34%), Positives = 58/106 (53%), Gaps = 2/106 (1%)
Query: 70 QNKENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKRE 129
Q PIVL+HGF +S WRY P L + + +AID+LG+G+S+ + L + E
Sbjct: 120 QGAGQPIVLIHGFGASAFHWRYNIPELAKK-YKVYAIDLLGFGWSE-KALVEYEATIWME 177
Query: 130 HFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDAS 175
F + +K P ++VG SLG + A PE V +VL++++
Sbjct: 178 QVRDFLRDVVKDPAVIVGNSLGGFTTLFAATEVPELVRGVVLLNSA 223
>UniRef100_UPI000034D3CA UPI000034D3CA UniRef100 entry
Length = 262
Score = 63.9 bits (154), Expect = 5e-09
Identities = 47/163 (28%), Positives = 83/163 (50%), Gaps = 13/163 (7%)
Query: 12 FKVVAETDADSFPSFLPKDVNRIKDPFARTLATRIQRLPVSVKFSENPIMSSCVKPLV-Q 70
F+ +++ + D F S N + DP +A ++ + ++ K++ S P+V +
Sbjct: 6 FEKLSDLNVDFFES----TKNSLLDPLGLQMANDVKWIKLNEKWN------SLKFPVVME 55
Query: 71 NKENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKREH 130
K PI+LLHGFDSS LE+R Y LL+ + D+LG+GFS + +
Sbjct: 56 GKGQPILLLHGFDSSFLEFRRIYQLLKR-NFQVIVPDLLGFGFSPRCATNDYNPSKIISY 114
Query: 131 FYQFWKS-YIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLI 172
K+ I K + ++G S+G + A+ A PE++ K++L+
Sbjct: 115 LIDLLKTLQITKNLQIIGASMGGSTALKLAFEMPESINKIILL 157
>UniRef100_Q7U8E0 Predicted alpha/beta hydrolase superfamily protein [Synechococcus
sp.]
Length = 303
Score = 63.9 bits (154), Expect = 5e-09
Identities = 35/103 (33%), Positives = 60/103 (57%), Gaps = 1/103 (0%)
Query: 68 LVQNKENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSK 127
L Q+ + IVLLHGF ++ WR+T P L G +++D+LG+G SD +P + V
Sbjct: 29 LGQDNDPAIVLLHGFGAASGHWRHTAPRLASQGWRVFSLDLLGFGASDQPAIPLDNRVWG 88
Query: 128 REHFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLV 170
++ F + +++P +L+G SLG+ A+ A+ PE + +V
Sbjct: 89 QQ-VNAFVEQVVQRPAVLLGNSLGALTALTAAVLKPEQIRAVV 130
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.335 0.145 0.460
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 477,998,703
Number of Sequences: 2790947
Number of extensions: 19344989
Number of successful extensions: 70977
Number of sequences better than 10.0: 1313
Number of HSP's better than 10.0 without gapping: 404
Number of HSP's successfully gapped in prelim test: 910
Number of HSP's that attempted gapping in prelim test: 69905
Number of HSP's gapped (non-prelim): 1340
length of query: 307
length of database: 848,049,833
effective HSP length: 127
effective length of query: 180
effective length of database: 493,599,564
effective search space: 88847921520
effective search space used: 88847921520
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 74 (33.1 bits)
Medicago: description of AC146567.12


