

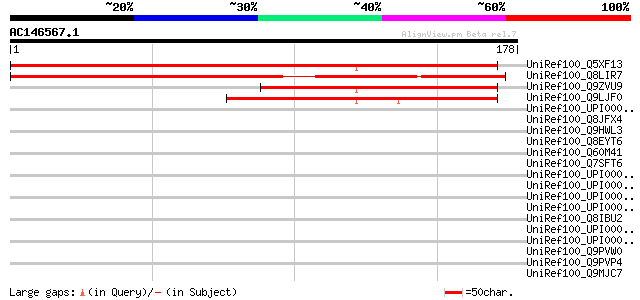
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146567.1 - phase: 0
(178 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q5XF13 At1g55535 [Arabidopsis thaliana] 255 4e-67
UniRef100_Q8LIR7 Hypothetical protein OJ1118_B03.109 [Oryza sativa] 171 8e-42
UniRef100_Q9ZVU9 T5A14.6 protein [Arabidopsis thaliana] 120 2e-26
UniRef100_Q9LJF0 Arabidopsis thaliana genomic DNA, chromosome 3,... 99 6e-20
UniRef100_UPI000032855C UPI000032855C UniRef100 entry 39 0.060
UniRef100_Q8JFX4 LBP (LPS binding protein)/BPI (Bactericidal/per... 34 1.9
UniRef100_Q9HWL3 Probable TonB-dependent receptor [Pseudomonas a... 34 1.9
UniRef100_Q8EYT6 Sensory transduction histidine kinases [Leptosp... 33 2.5
UniRef100_Q60M41 Hypothetical protein CBG23315 [Caenorhabditis b... 33 2.5
UniRef100_Q7SFT6 Hypothetical protein [Neurospora crassa] 33 2.5
UniRef100_UPI00002FEB56 UPI00002FEB56 UniRef100 entry 33 3.3
UniRef100_UPI0000332F8F UPI0000332F8F UniRef100 entry 33 4.3
UniRef100_UPI00002EC3F5 UPI00002EC3F5 UniRef100 entry 33 4.3
UniRef100_UPI00002BA675 UPI00002BA675 UniRef100 entry 33 4.3
UniRef100_Q8IBU2 Hypothetical protein PF07_0064 [Plasmodium falc... 33 4.3
UniRef100_UPI000034D04F UPI000034D04F UniRef100 entry 32 5.6
UniRef100_UPI000030A044 UPI000030A044 UniRef100 entry 32 5.6
UniRef100_Q9PVW0 Olfactory receptor 3 [Oryzias latipes] 32 5.6
UniRef100_Q9PVP4 E4 olfactory receptor [Oryzias latipes] 32 5.6
UniRef100_Q9MJC7 NADH dehydrogenase subunit 5 [Drosophila melano... 32 5.6
>UniRef100_Q5XF13 At1g55535 [Arabidopsis thaliana]
Length = 260
Score = 255 bits (651), Expect = 4e-67
Identities = 129/172 (75%), Positives = 150/172 (87%), Gaps = 1/172 (0%)
Query: 1 MISMKIGCALLGSLGASYNGVSLINLAISLFALVAIESSSQSLGRTYAFLLFCSILLDIS 60
+I +IGCAL+GSLGA YNGV LINLAI+LFALVAIES+SQSLGRTYA LLFC++LLDIS
Sbjct: 28 LIYAQIGCALIGSLGALYNGVLLINLAIALFALVAIESNSQSLGRTYAVLLFCALLLDIS 87
Query: 61 WFILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLLWIQIYRLGASYVDTA 120
WFILFT EIW+IS++ Y FFIFSVKLT+AM++IGF VRLSSSLLW QIYRLGA+ VDT+
Sbjct: 88 WFILFTEEIWSISAETYGTFFIFSVKLTMAMEMIGFFVRLSSSLLWFQIYRLGAAIVDTS 147
Query: 121 -SRAADFDLRSSFLSPVTPAVARQISNSNEILGGSIYDPAYYSSLFEDSQEN 171
R D DLR+SFL+P TPA+ARQ S + EILGGSIYDPAYY+SLFE+SQ N
Sbjct: 148 LPRETDSDLRNSFLNPPTPAIARQCSGAEEILGGSIYDPAYYTSLFEESQTN 199
>UniRef100_Q8LIR7 Hypothetical protein OJ1118_B03.109 [Oryza sativa]
Length = 247
Score = 171 bits (433), Expect = 8e-42
Identities = 94/174 (54%), Positives = 121/174 (69%), Gaps = 12/174 (6%)
Query: 1 MISMKIGCALLGSLGASYNGVSLINLAISLFALVAIESSSQSLGRTYAFLLFCSILLDIS 60
+I ++IGCAL+GSLGA +NGV +INL I LFA+VAIESSSQ+LGRTYA LLF +I+LD++
Sbjct: 39 LIYLQIGCALIGSLGALFNGVLVINLVIGLFAVVAIESSSQTLGRTYAVLLFFAIVLDVA 98
Query: 61 WFILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLLWIQIYRLGASYVDTA 120
WFILF+H I + Y + F+ S+KL L MQIIGF +YRLG S
Sbjct: 99 WFILFSHAINITPEEKYGQLFVLSLKLALWMQIIGF-----------SMYRLGVSSSTPT 147
Query: 121 SRAADFDLRSSFLSPVTPAVARQISNSNEILGGSIYDPAYYSSLFEDSQENKST 174
++D R+SFLSP + +V R S +++ILGGSIYDPAYYSSLF D + N T
Sbjct: 148 YHEVNYDGRNSFLSPRSSSVRRN-SMADDILGGSIYDPAYYSSLFADVRNNTCT 200
>UniRef100_Q9ZVU9 T5A14.6 protein [Arabidopsis thaliana]
Length = 185
Score = 120 bits (300), Expect = 2e-26
Identities = 60/84 (71%), Positives = 71/84 (84%), Gaps = 1/84 (1%)
Query: 89 LAMQIIGFIVRLSSSLLWIQIYRLGASYVDTA-SRAADFDLRSSFLSPVTPAVARQISNS 147
+AM++IGF VRLSSSLLW QIYRLGA+ VDT+ R D DLR+SFL+P TPA+ARQ S +
Sbjct: 1 MAMEMIGFFVRLSSSLLWFQIYRLGAAIVDTSLPRETDSDLRNSFLNPPTPAIARQCSGA 60
Query: 148 NEILGGSIYDPAYYSSLFEDSQEN 171
EILGGSIYDPAYY+SLFE+SQ N
Sbjct: 61 EEILGGSIYDPAYYTSLFEESQTN 84
>UniRef100_Q9LJF0 Arabidopsis thaliana genomic DNA, chromosome 3, P1 clone: MRP15
[Arabidopsis thaliana]
Length = 163
Score = 98.6 bits (244), Expect = 6e-20
Identities = 58/114 (50%), Positives = 71/114 (61%), Gaps = 19/114 (16%)
Query: 77 YAKFFIFSVKLTLAMQIIGFIVRLSSSLLWIQIYRLGASYVDTA-SRAADFDLRSSFLSP 135
Y F+IFSVKLTLAM+I GF+VRLSSSLLW QIYRLGAS +D+ R +D DLR+SFL P
Sbjct: 2 YQVFYIFSVKLTLAMEIAGFVVRLSSSLLWFQIYRLGASIIDSPFPRQSDSDLRNSFLEP 61
Query: 136 ------------------VTPAVARQISNSNEILGGSIYDPAYYSSLFEDSQEN 171
PA+A+Q S S+EIL SI +PA Y+ L + N
Sbjct: 62 PLLARQRSRDPELRNSFLQAPAIAKQRSRSDEILEDSIDEPASYTPLLDGGLSN 115
>UniRef100_UPI000032855C UPI000032855C UniRef100 entry
Length = 304
Score = 38.9 bits (89), Expect = 0.060
Identities = 34/129 (26%), Positives = 58/129 (44%), Gaps = 3/129 (2%)
Query: 21 VSLINLAISLFALVAIESSSQSLGRTYAFLLFCSILL--DISWFIL-FTHEIWNISSDDY 77
+S + L I + L +S S +G ++ +L CS+LL +SW IL F ++ S
Sbjct: 156 MSYLPLIILSWVLSVWQSQSFIVGSSFVGMLICSVLLLGSLSWAILFFAGKLSQTSGSTI 215
Query: 78 AKFFIFSVKLTLAMQIIGFIVRLSSSLLWIQIYRLGASYVDTASRAADFDLRSSFLSPVT 137
+ +++ A I F+ +LL I ++ + SR ADF + S FL +
Sbjct: 216 KRMAFRNLQRNRAGAISCFLALSLGTLLINLIPQIYQGLQEEISRPADFRIPSLFLFDIQ 275
Query: 138 PAVARQISN 146
P + N
Sbjct: 276 PEQIEPLQN 284
>UniRef100_Q8JFX4 LBP (LPS binding protein)/BPI (Bactericidal/permeability-increasing
protein)-1 [Oncorhynchus mykiss]
Length = 473
Score = 33.9 bits (76), Expect = 1.9
Identities = 37/154 (24%), Positives = 64/154 (41%), Gaps = 26/154 (16%)
Query: 9 ALLGSLGASYNGVSLINLAISLFALVAIESSSQSLGRTYAF--------LLFCSILLDIS 60
A +G + S G++++NL + AL + + SL T AF + + S + D
Sbjct: 62 APIGKVKYSLTGITIVNLGLPYSALALVPDTGISLSITNAFISLHGNWKIRYLSFIKDSG 121
Query: 61 WF------ILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLLWIQIYRLGA 114
F + T I I SD+ + + SV + ++ W +Y L +
Sbjct: 122 SFDLEVDGLTVTDSI-TIKSDETGRPTVSSV--NCVANVGSASIKFHGGASW--LYNLFS 176
Query: 115 SYVDTASRAADFDLRSSFLSPVTPAVARQISNSN 148
+Y+D A LRS+ + P VA I++ N
Sbjct: 177 AYIDKA-------LRSALQKQICPLVADTITDMN 203
>UniRef100_Q9HWL3 Probable TonB-dependent receptor [Pseudomonas aeruginosa]
Length = 802
Score = 33.9 bits (76), Expect = 1.9
Identities = 23/71 (32%), Positives = 34/71 (47%), Gaps = 4/71 (5%)
Query: 108 QIYRLGASYVDTASRAADFDLRSSFLSPVTPAVARQISNSNEILGGSIYDPAYYSSLFED 167
Q Y + ASY D ++FD L P+T +I E GG++ +LF+
Sbjct: 570 QTYSVYASYTDIFKPQSNFDAGGGLLDPIT-GKNYEIGLKGEHFGGALNSQI---ALFQI 625
Query: 168 SQENKSTCGVG 178
QEN++T VG
Sbjct: 626 DQENRATEDVG 636
>UniRef100_Q8EYT6 Sensory transduction histidine kinases [Leptospira interrogans]
Length = 376
Score = 33.5 bits (75), Expect = 2.5
Identities = 22/81 (27%), Positives = 45/81 (55%), Gaps = 3/81 (3%)
Query: 25 NLAISLFALVAIESSSQSLGRTYAFLLFCSILLDISWFILFTHEIWNISSDDYAKFFIFS 84
N+ +S+F ++ + + + +T +L+C IL+ I F L H++ ++D FF+FS
Sbjct: 109 NIILSIFIMIFLFFTD--VKKTIFIMLYCLILIFIDEFYLKEHDV-VYTADRIGLFFMFS 165
Query: 85 VKLTLAMQIIGFIVRLSSSLL 105
V L L ++ + V+ + L+
Sbjct: 166 VILILTVRALYGSVQEKNELI 186
>UniRef100_Q60M41 Hypothetical protein CBG23315 [Caenorhabditis briggsae]
Length = 315
Score = 33.5 bits (75), Expect = 2.5
Identities = 23/100 (23%), Positives = 45/100 (45%), Gaps = 19/100 (19%)
Query: 19 NGVSLINLAISLFALVAIESSSQ-----------SLGRTYAFLLFCSILLDISWFILFTH 67
N V L+ I F + ++ SS S Y + +C +L+ ++ F
Sbjct: 60 NSVILVWGFIGTFVPITLKESSSFSNVYETIVIISCNNIYIAVQYCGLLIALNRFCAMFF 119
Query: 68 EIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLLWI 107
+W YAKFF S+K+T + + F ++L+ ++ ++
Sbjct: 120 PLW------YAKFF--SIKITFVIVVFHFALKLADNIYYL 151
>UniRef100_Q7SFT6 Hypothetical protein [Neurospora crassa]
Length = 671
Score = 33.5 bits (75), Expect = 2.5
Identities = 19/32 (59%), Positives = 22/32 (68%), Gaps = 2/32 (6%)
Query: 118 DTASRAADFDLRSSFLSPVTPAVARQISNSNE 149
DTASRAA F LRS L P++ AVA + N NE
Sbjct: 408 DTASRAAPFLLRS--LRPISTAVASMLENPNE 437
>UniRef100_UPI00002FEB56 UPI00002FEB56 UniRef100 entry
Length = 213
Score = 33.1 bits (74), Expect = 3.3
Identities = 29/78 (37%), Positives = 41/78 (52%), Gaps = 14/78 (17%)
Query: 47 YAFLLFCSILLDISWFILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLLW 106
Y FL F I L I++F+ F +N SS +FS LT +M IGF ++ S S LW
Sbjct: 12 YLFLYF--IFLTITFFVFFNFN-YNFSS------ILFS--LTSSMSNIGFSLKNSPSNLW 60
Query: 107 ---IQIYRLGASYVDTAS 121
+ I +G S+ T+S
Sbjct: 61 FIFLLIVVIGGSFFSTSS 78
>UniRef100_UPI0000332F8F UPI0000332F8F UniRef100 entry
Length = 280
Score = 32.7 bits (73), Expect = 4.3
Identities = 19/69 (27%), Positives = 38/69 (54%), Gaps = 1/69 (1%)
Query: 50 LLFCSILLDISWFI-LFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLLWIQ 108
+ F SIL I+ FI L+++ + + ++ FF+ S+KL+ + G ++ L + L I+
Sbjct: 158 IFFLSILFTIAVFIYLYSNISFTVKENNLLIFFLTSLKLSFCFLVSGILLVLMTKLKIIE 217
Query: 109 IYRLGASYV 117
+ SY+
Sbjct: 218 KKMIAESYI 226
>UniRef100_UPI00002EC3F5 UPI00002EC3F5 UniRef100 entry
Length = 151
Score = 32.7 bits (73), Expect = 4.3
Identities = 18/66 (27%), Positives = 32/66 (48%), Gaps = 4/66 (6%)
Query: 40 SQSLGRTYAFLLFCSILLDISWFILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVR 99
S LG LLF ++ W++ F + N Y FF FS+++ L + + + +
Sbjct: 66 SHRLGALIVLLLFLALF----WYLFFIQKYTNQKKIGYVVFFFFSLQILLGISNVVYSLP 121
Query: 100 LSSSLL 105
LS ++L
Sbjct: 122 LSVAVL 127
>UniRef100_UPI00002BA675 UPI00002BA675 UniRef100 entry
Length = 321
Score = 32.7 bits (73), Expect = 4.3
Identities = 18/66 (27%), Positives = 32/66 (48%), Gaps = 4/66 (6%)
Query: 40 SQSLGRTYAFLLFCSILLDISWFILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVR 99
S LG LLF ++ W++ F + N Y FF FS+++ L + + + +
Sbjct: 236 SHRLGALIVLLLFLALF----WYLFFIQKYTNQKKIGYVVFFFFSLQILLGISNVVYSLP 291
Query: 100 LSSSLL 105
LS ++L
Sbjct: 292 LSVAVL 297
>UniRef100_Q8IBU2 Hypothetical protein PF07_0064 [Plasmodium falciparum]
Length = 434
Score = 32.7 bits (73), Expect = 4.3
Identities = 20/77 (25%), Positives = 39/77 (49%), Gaps = 6/77 (7%)
Query: 19 NGVSLINLAISLF-ALVAIESSSQSLGRTYAFLLFCSILLDISWFILFTHEIWNISSDDY 77
+ V++I I +F AL++ + + AF +F S + + T EIWN+ + D
Sbjct: 227 HNVAIIQPTIPIFTALISYYLKIEKMNYITAFSIFLSF-----FGLAITAEIWNLKTFDL 281
Query: 78 AKFFIFSVKLTLAMQII 94
+ + +V +T +Q+I
Sbjct: 282 GFYLLLTVPITKGLQVI 298
>UniRef100_UPI000034D04F UPI000034D04F UniRef100 entry
Length = 194
Score = 32.3 bits (72), Expect = 5.6
Identities = 29/101 (28%), Positives = 44/101 (42%), Gaps = 11/101 (10%)
Query: 54 SILLDISWFILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLLWIQIYRLG 113
SILL + +L T + W+ISS++Y +F L A+ +I+ + S IY
Sbjct: 24 SILLGLLGLVLITGQSWDISSNNYKLGILFG--LLTAISYAAYIISMKS------IYHNA 75
Query: 114 ASYVDTASRAADFDLRSS---FLSPVTPAVARQISNSNEIL 151
AS D L S+ FL + +I S EI+
Sbjct: 76 ASKSDPIFNLLFVSLISAALLFLFSIIEQAPLEIKTSGEIV 116
>UniRef100_UPI000030A044 UPI000030A044 UniRef100 entry
Length = 290
Score = 32.3 bits (72), Expect = 5.6
Identities = 28/86 (32%), Positives = 43/86 (49%), Gaps = 11/86 (12%)
Query: 28 ISLFALVAIESSSQ---SLGRTYAFLLFCSILLDISWFILFTHEIWNISSDDYAKFFIFS 84
ISLFA+ + E +L Y F++F I L IS+F + E W S+ Y +F+
Sbjct: 9 ISLFAMYSTEQGKLGYFTLSHLYRFIIFFIIFLTISFFRI---EFWFRST--YIFYFLI- 62
Query: 85 VKLTLAMQIIGFIVRLSSSLLWIQIY 110
+ L A++ G V S S WI ++
Sbjct: 63 LLLLFAVEFYG--VTASGSKRWINLF 86
>UniRef100_Q9PVW0 Olfactory receptor 3 [Oryzias latipes]
Length = 248
Score = 32.3 bits (72), Expect = 5.6
Identities = 26/80 (32%), Positives = 45/80 (55%), Gaps = 7/80 (8%)
Query: 24 INLAISLFALVAIESSSQSLGRTYAFLLFCSILLDISWFILFTH-EIWNISSDDYAKFFI 82
I LA LF ++A+ +++ L R L FC+ ++ S+F H +I+ I+ +DY+ +I
Sbjct: 151 IILAFWLFVIIAVSTAAGLLTR----LSFCNSVVINSYFC--DHGQIFGIACNDYSPSYI 204
Query: 83 FSVKLTLAMQIIGFIVRLSS 102
FS L + I I+ +SS
Sbjct: 205 FSDVLIAIILWIPLIIVISS 224
>UniRef100_Q9PVP4 E4 olfactory receptor [Oryzias latipes]
Length = 321
Score = 32.3 bits (72), Expect = 5.6
Identities = 26/80 (32%), Positives = 45/80 (55%), Gaps = 7/80 (8%)
Query: 24 INLAISLFALVAIESSSQSLGRTYAFLLFCSILLDISWFILFTH-EIWNISSDDYAKFFI 82
I LA LF ++A+ +++ L R L FC+ ++ S+F H +I+ I+ +DY+ +I
Sbjct: 151 IILAFWLFVIIAVSTAAGLLTR----LSFCNSVVINSYFC--DHGQIFGIACNDYSPSYI 204
Query: 83 FSVKLTLAMQIIGFIVRLSS 102
FS L + I I+ +SS
Sbjct: 205 FSDVLIAIILWIPLIIVISS 224
>UniRef100_Q9MJC7 NADH dehydrogenase subunit 5 [Drosophila melanogaster]
Length = 577
Score = 32.3 bits (72), Expect = 5.6
Identities = 28/109 (25%), Positives = 49/109 (44%), Gaps = 11/109 (10%)
Query: 1 MISMKIGCALLGSLGASYNGVSLINLAISLFALVAIESSSQSLGRTYAF----LLFCSIL 56
+ISM + C LL L+N I + +S S+ T+ F LLF S +
Sbjct: 14 LISMSLSCFLLSLY-------FLLNDMIYFIEWELVSLNSMSIVMTFLFDWMSLLFMSFV 66
Query: 57 LDISWFILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLL 105
L IS ++F + + ++ + +F + + L+M ++ L S LL
Sbjct: 67 LMISSLVIFYSKEYMMNDNHINRFIMLVLMFVLSMMLLIISPNLISILL 115
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.324 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 262,686,187
Number of Sequences: 2790947
Number of extensions: 9184953
Number of successful extensions: 32876
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 24
Number of HSP's that attempted gapping in prelim test: 32863
Number of HSP's gapped (non-prelim): 30
length of query: 178
length of database: 848,049,833
effective HSP length: 119
effective length of query: 59
effective length of database: 515,927,140
effective search space: 30439701260
effective search space used: 30439701260
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 70 (31.6 bits)
Medicago: description of AC146567.1


