

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146566.1 + phase: 0
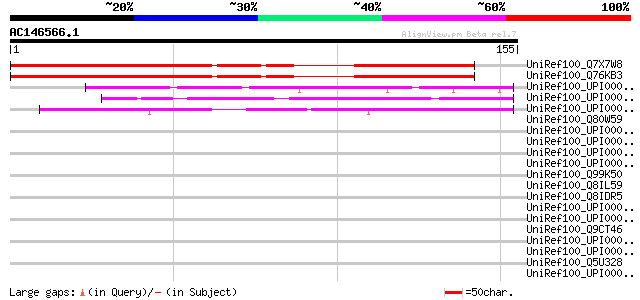
(155 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7X7W8 Makorin ring-zinc-finger protein [Pisum sativum] 167 1e-40
UniRef100_Q76KB3 Makorin ring-zinc-finger protein [Pisum sativum] 167 1e-40
UniRef100_UPI000042C34A UPI000042C34A UniRef100 entry 47 1e-04
UniRef100_UPI00003C2255 UPI00003C2255 UniRef100 entry 46 3e-04
UniRef100_UPI000045344F UPI000045344F UniRef100 entry 45 6e-04
UniRef100_Q80W59 Histidine-rich calcium binding protein [Rattus ... 44 0.002
UniRef100_UPI00004399D7 UPI00004399D7 UniRef100 entry 43 0.002
UniRef100_UPI00003189BD UPI00003189BD UniRef100 entry 41 0.011
UniRef100_UPI000045285D UPI000045285D UniRef100 entry 40 0.014
UniRef100_UPI00003C233F UPI00003C233F UniRef100 entry 40 0.014
UniRef100_Q99K50 Nucleolin [Mus musculus] 40 0.018
UniRef100_Q8IL59 Hypothetical protein [Plasmodium falciparum] 40 0.018
UniRef100_Q8IDR5 Casein kinase II regulatory subunit, putative [... 40 0.018
UniRef100_UPI000042DDF6 UPI000042DDF6 UniRef100 entry 40 0.024
UniRef100_UPI00002F1EA5 UPI00002F1EA5 UniRef100 entry 40 0.024
UniRef100_Q9CT46 Mus musculus 10 days embryo whole body cDNA, RI... 40 0.024
UniRef100_UPI00001A1BB9 UPI00001A1BB9 UniRef100 entry 39 0.031
UniRef100_UPI000021BB95 UPI000021BB95 UniRef100 entry 39 0.040
UniRef100_Q5U328 Nucleolin [Rattus norvegicus] 39 0.040
UniRef100_UPI000021BFA3 UPI000021BFA3 UniRef100 entry 39 0.052
>UniRef100_Q7X7W8 Makorin ring-zinc-finger protein [Pisum sativum]
Length = 461
Score = 167 bits (422), Expect = 1e-40
Identities = 89/142 (62%), Positives = 104/142 (72%), Gaps = 20/142 (14%)
Query: 1 MHDMLDVLGEVDLSSGEFYSIMRDSDFFEGMDPFEMMALSDSLAGGSGPCLGPFDSDDDD 60
M+DMLD+L EVDLSSGEFYSIMRDS+FF+ MDP EMMALSDSLAGGS PCLGPF SDD+
Sbjct: 339 MYDMLDMLSEVDLSSGEFYSIMRDSEFFDEMDPLEMMALSDSLAGGSVPCLGPFGSDDEG 398
Query: 61 DDGFRVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPDDFPGID 120
D+ F V RMAAM EA+ +D FGPDD G DDF PDDF +D
Sbjct: 399 DN-FNVFRMAAMSEALD-NLDDFGPDD------------------FGPDDFDPDDFDELD 438
Query: 121 PMDAALISMMMHSHMDDEEDDE 142
PM+AALISMMMHS++D++ +DE
Sbjct: 439 PMEAALISMMMHSNIDEDSEDE 460
>UniRef100_Q76KB3 Makorin ring-zinc-finger protein [Pisum sativum]
Length = 411
Score = 167 bits (422), Expect = 1e-40
Identities = 89/142 (62%), Positives = 104/142 (72%), Gaps = 20/142 (14%)
Query: 1 MHDMLDVLGEVDLSSGEFYSIMRDSDFFEGMDPFEMMALSDSLAGGSGPCLGPFDSDDDD 60
M+DMLD+L EVDLSSGEFYSIMRDS+FF+ MDP EMMALSDSLAGGS PCLGPF SDD+
Sbjct: 289 MYDMLDMLSEVDLSSGEFYSIMRDSEFFDEMDPLEMMALSDSLAGGSVPCLGPFGSDDEG 348
Query: 61 DDGFRVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPDDFPGID 120
D+ F V RMAAM EA+ +D FGPDD G DDF PDDF +D
Sbjct: 349 DN-FNVFRMAAMSEALD-NLDDFGPDD------------------FGPDDFDPDDFDELD 388
Query: 121 PMDAALISMMMHSHMDDEEDDE 142
PM+AALISMMMHS++D++ +DE
Sbjct: 389 PMEAALISMMMHSNIDEDSEDE 410
>UniRef100_UPI000042C34A UPI000042C34A UniRef100 entry
Length = 495
Score = 47.0 bits (110), Expect = 1e-04
Identities = 44/143 (30%), Positives = 64/143 (43%), Gaps = 18/143 (12%)
Query: 24 DSDFFEGMDPFEMMALSDSLAGGSGPCLGPFDSDDDDDDGFRVSRMAAMQEAMAIGMDAF 83
D +F++ DP + M + L+G S G D D+DDD + M++ E AI D
Sbjct: 191 DDEFYDAEDPVDAMMTGEDLSGASDD--GDDDDDEDDDGDDDIDDMSS--ELSAIDSDLD 246
Query: 84 GPDD-DEEEFNVFRMAAMQEAMVSGIDDFGPD--DFPGIDPMDAALISMMMHSH------ 134
G D+ D+ E + E ID+ D D ID +D L+S
Sbjct: 247 GGDNADDIEMEIEVDPYDDERGSEDIDEDASDMEDIEIIDDLD--LVSQTDGDDDGDDDD 304
Query: 135 -MDDEEDDEDYEDEE--EYTDDE 154
DDEED ++D+E EY +DE
Sbjct: 305 GRDDEEDSSSWDDDEMSEYDEDE 327
>UniRef100_UPI00003C2255 UPI00003C2255 UniRef100 entry
Length = 608
Score = 46.2 bits (108), Expect = 3e-04
Identities = 40/126 (31%), Positives = 57/126 (44%), Gaps = 11/126 (8%)
Query: 29 EGMDPFEMMALSDSLAGGSGPCLGPFDSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPDDD 88
+G+D +M AL D LAG D DDDD+D EA+ D DDD
Sbjct: 193 DGLDEVDMAAL-DELAGEEEED----DDDDDDEDDEDEENKNDSDEALEHDDD----DDD 243
Query: 89 EEEFNVFRMAAMQEAMVSGIDDFGPDDFPGIDPMDAALISMMMHSHMDDEEDDEDYEDEE 148
EE + +AA QE + + D + L+ + + DDEEDDE+ E+ E
Sbjct: 244 NEESDSEELAAEQEDEIPRVVPNANGDTLAASIARSGLVPV--EASDDDEEDDEEDEENE 301
Query: 149 EYTDDE 154
+ +DE
Sbjct: 302 QEDEDE 307
Score = 35.0 bits (79), Expect = 0.58
Identities = 26/74 (35%), Positives = 34/74 (45%), Gaps = 12/74 (16%)
Query: 86 DDDE---EEFNVFRMAAMQEAMVSGIDDFGPDDFPGIDPMDAALISMMM--HSHMDDEED 140
DDDE EE MQ M + DD G+D +D A + + DD++D
Sbjct: 166 DDDESDLEEGQDVSEKGMQRLMAALGDD-------GLDEVDMAALDELAGEEEEDDDDDD 218
Query: 141 DEDYEDEEEYTDDE 154
DED EDEE D +
Sbjct: 219 DEDDEDEENKNDSD 232
>UniRef100_UPI000045344F UPI000045344F UniRef100 entry
Length = 3919
Score = 45.1 bits (105), Expect = 6e-04
Identities = 44/154 (28%), Positives = 66/154 (42%), Gaps = 20/154 (12%)
Query: 10 EVDLSSGEFYSIMRDSDFFEGMDPFEMMALSD-----SLAGGSGPCLGPFDSDDDDDDGF 64
EVDL F D DF + +D S+ SL G G + D DDDD+D
Sbjct: 1535 EVDLDQFGFQFNSSDEDFDDDLDDSLEDDFSEEFDLTSLDGLDGEDIDDNDEDDDDND-- 1592
Query: 65 RVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGI----DDFGPDDFPGID 120
E M ++ DDDE + ++F M V I DD DD G D
Sbjct: 1593 --------VEDMDDDVEDDVEDDDEND-DLFLDETMINDGVIDIEGEDDDLSNDDIVGDD 1643
Query: 121 PMDAALISMMMHSHMDDEEDDEDYEDEEEYTDDE 154
+ + + + DD++DD+D +D+++ DD+
Sbjct: 1644 DLTSDVDDVDDDDDDDDDDDDDDDDDDDDDDDDD 1677
>UniRef100_Q80W59 Histidine-rich calcium binding protein [Rattus norvegicus]
Length = 755
Score = 43.5 bits (101), Expect = 0.002
Identities = 31/103 (30%), Positives = 45/103 (43%), Gaps = 10/103 (9%)
Query: 52 GPFDSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDF 111
G D DDDD+DG VS Q D DD +EE + R+ ++ D+
Sbjct: 215 GHHDEDDDDEDG--VSTEGEHQAHRYQDHDEEDDDDSDEESHAHRLQGQED---ENDDED 269
Query: 112 GPDDFPGIDPMDAALISMMMHSHMDDEEDDEDYEDEEEYTDDE 154
G G D H DD++D+E+ E+EEE ++E
Sbjct: 270 GDSSENGHQTQDH-----QGHKEQDDDDDEEEEEEEEEEEEEE 307
>UniRef100_UPI00004399D7 UPI00004399D7 UniRef100 entry
Length = 240
Score = 43.1 bits (100), Expect = 0.002
Identities = 32/105 (30%), Positives = 47/105 (44%), Gaps = 22/105 (20%)
Query: 59 DDDDGFRVSRMAAMQEAMAIGMD----AFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPD 114
+DDDG + + M D A G DDD++E + DD G +
Sbjct: 148 EDDDGVDQLEVKMATKIMIDEEDDDDVAGGEDDDDDEGD-------------DEDDAGGE 194
Query: 115 DFPGIDPMDAALISMMMHSHMDDE-----EDDEDYEDEEEYTDDE 154
D G+D ++ + + +M DD+ EDDED DEEE DD+
Sbjct: 195 DDDGVDQLEVKMTTKIMIDEEDDDDVAAGEDDEDVNDEEEDDDDD 239
>UniRef100_UPI00003189BD UPI00003189BD UniRef100 entry
Length = 665
Score = 40.8 bits (94), Expect = 0.011
Identities = 45/145 (31%), Positives = 63/145 (43%), Gaps = 15/145 (10%)
Query: 1 MHDMLDVLGEVDLSSGEFYSIMRDSDFFEGMDPFEMMALSDSLAGGSGPCLGPFDSDDDD 60
M DM ++ G S G+ + R+S G D + S S A GSG G DD+
Sbjct: 1 MADMDELFG----SDGDSDNDQRESGSGSGSDSDQERPPSVSNASGSGS--GSERDQDDE 54
Query: 61 DDGFRVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSG--IDDFGPDDFPG 118
D+ + A+M + + FG DD E+E + + Q + SG D D
Sbjct: 55 DEEDHEAGNASMNKEL------FG-DDSEDEQHSQHSGSDQHSDRSGNQSDASMHSDQGD 107
Query: 119 IDPMDAALISMMMHSHMDDEEDDED 143
DP DA S H D++EDDED
Sbjct: 108 NDPSDAEQHSGSERGHQDEDEDDED 132
>UniRef100_UPI000045285D UPI000045285D UniRef100 entry
Length = 1090
Score = 40.4 bits (93), Expect = 0.014
Identities = 25/74 (33%), Positives = 36/74 (47%), Gaps = 5/74 (6%)
Query: 80 MDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPDDFPGIDPMDAALISMMMHSHMDDEE 139
+D FG D D + ++ + + + +DDF DDF D +D DD E
Sbjct: 967 VDNFGEDLDNNDDDIDLLDGVN---MDDMDDF--DDFEDFDDLDGESDFAGEEGEEDDSE 1021
Query: 140 DDEDYEDEEEYTDD 153
D+ED EDEE+ DD
Sbjct: 1022 DEEDEEDEEDDEDD 1035
>UniRef100_UPI00003C233F UPI00003C233F UniRef100 entry
Length = 543
Score = 40.4 bits (93), Expect = 0.014
Identities = 35/125 (28%), Positives = 54/125 (43%), Gaps = 23/125 (18%)
Query: 53 PFDSDDDDDDGFRVS---RMAAMQEAMAIGMDAFGPDDDEEEFNVFR---------MAAM 100
PF+ D+DD DG + R A ++ A G DDDE+ F+V A+
Sbjct: 89 PFNEDEDDSDGEADAGSHRRRADKKRRGDAAGAAGDDDDEDMFDVENDDEKQTSSASKAI 148
Query: 101 QEAMVSGI----------DDFGPDDFPGIDPMDAALISMMMHSHMDDEEDDEDYEDEE-E 149
++A G DFG ++ +D D ++ DD++DD+D D E E
Sbjct: 149 RKAQTRGKYLKLGELEEGQDFGSNNRSTLDAADDEDAKLLRGKGKDDDDDDDDDLDPELE 208
Query: 150 YTDDE 154
DD+
Sbjct: 209 LEDDD 213
>UniRef100_Q99K50 Nucleolin [Mus musculus]
Length = 707
Score = 40.0 bits (92), Expect = 0.018
Identities = 27/77 (35%), Positives = 36/77 (46%), Gaps = 1/77 (1%)
Query: 79 GMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPDDFPGIDPMDAALISMMMHSHMDDE 138
G +A D DE+E + ++ D+F P G+ P AA + DDE
Sbjct: 137 GKNAKKEDSDEDEDEEDEEDSDEDEDDEEEDEFEPPIVKGVKPAKAAPAAPASEDEEDDE 196
Query: 139 -EDDEDYEDEEEYTDDE 154
EDDE+ EDEEE D E
Sbjct: 197 DEDDEEEEDEEEEDDSE 213
Score = 35.4 bits (80), Expect = 0.44
Identities = 29/108 (26%), Positives = 47/108 (42%), Gaps = 8/108 (7%)
Query: 55 DSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPD-------DDEEEFNVFRMAAMQEAMVSG 107
D DD+++D F + ++ A A D DDEEE + +E ++
Sbjct: 160 DEDDEEEDEFEPPIVKGVKPAKAAPAAPASEDEEDDEDEDDEEEEDEEEEDDSEEEVMEI 219
Query: 108 IDDFGPDDFPGIDPMDAALISMMMHSHMDDEED-DEDYEDEEEYTDDE 154
G + PM A ++ +DE+D DED E+E++ DDE
Sbjct: 220 TTAKGKKTPAKVVPMKAKSVAEEEDEEEEDEDDEDEDDEEEDDEDDDE 267
>UniRef100_Q8IL59 Hypothetical protein [Plasmodium falciparum]
Length = 227
Score = 40.0 bits (92), Expect = 0.018
Identities = 31/100 (31%), Positives = 38/100 (38%), Gaps = 40/100 (40%)
Query: 55 DSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPD 114
D DDDDDD D DDDE++ DD D
Sbjct: 119 DEDDDDDD------------------DDDEDDDDEDD--------------EDDDDDDED 146
Query: 115 DFPGIDPMDAALISMMMHSHMDDEEDDEDYEDEEEYTDDE 154
D D MD DD++DDED +DEE+Y DD+
Sbjct: 147 DDDDFDDMD--------EDDDDDDDDDEDDDDEEDYDDDD 178
Score = 37.0 bits (84), Expect = 0.15
Identities = 30/100 (30%), Positives = 38/100 (38%), Gaps = 27/100 (27%)
Query: 55 DSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPD 114
D DDDDDD M E D DDDEE+++ DD D
Sbjct: 137 DEDDDDDDEDDDDDFDDMDEDDDDDDDDDEDDDDEEDYD---------------DDDDDD 181
Query: 115 DFPGIDPMDAALISMMMHSHMDDEEDDEDYEDEEEYTDDE 154
D D + DD+EDD+D D+ EY D+
Sbjct: 182 DDDDEDDDE------------DDDEDDDDENDQNEYAGDD 209
Score = 33.5 bits (75), Expect = 1.7
Identities = 29/101 (28%), Positives = 37/101 (35%), Gaps = 35/101 (34%)
Query: 55 DSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPD 114
D DDDDDD D DDDE++ DD D
Sbjct: 86 DDDDDDDD------------------DDDEDDDDEDD-----------------DDDDDD 110
Query: 115 DFPGIDPMDAALISMMMHSHMDDEEDDEDYEDEEEYTDDEY 155
D D D DD+EDDED +D++E DD++
Sbjct: 111 DDDDDDDDDEDDDDDDDDDEDDDDEDDEDDDDDDEDDDDDF 151
>UniRef100_Q8IDR5 Casein kinase II regulatory subunit, putative [Plasmodium
falciparum]
Length = 385
Score = 40.0 bits (92), Expect = 0.018
Identities = 34/149 (22%), Positives = 66/149 (43%), Gaps = 17/149 (11%)
Query: 9 GEVDLSSGEFY--SIMRDSDFFEGMDPFEMMALSDSLAGGSGPCLGPFDSDDDDDDGFRV 66
GEV+L+ +FY +++ D E ++ E A +D + + ++ DD+D+D
Sbjct: 21 GEVELTDADFYDLTVINDKIDEEIIEDDEEEADNDDQENDNVQEV--YNIDDEDNDIHND 78
Query: 67 SRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPDDFPGIDPMDAAL 126
+ Q + + +++EEE +E D+ DD D D
Sbjct: 79 KLLLDQQRDNDVNEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDEDDDDDDDDDDDDD--- 135
Query: 127 ISMMMHSHMDDEEDDEDYEDEEEYTDDEY 155
D++DD+DY+D++EY +D++
Sbjct: 136 ----------DDDDDDDYDDDDEYDEDDF 154
Score = 33.9 bits (76), Expect = 1.3
Identities = 31/111 (27%), Positives = 47/111 (41%), Gaps = 34/111 (30%)
Query: 55 DSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPDD--DEEEFNV--------FRMAAMQEAM 104
D DDDDDD D + DD DE++FN F +
Sbjct: 133 DDDDDDDD------------------DDYDDDDEYDEDDFNEATVSWIEWFCQLKQNLFL 174
Query: 105 VSGIDDFGPDDFPGID-----PMDAALISMMM-HSHMDDEEDDEDYEDEEE 149
V +DF D+F I P L+ +++ DD++DD+DY+DE++
Sbjct: 175 VEVDEDFIRDEFNLIGLQTKVPHFKKLLKIILDEDDDDDDDDDDDYDDEDD 225
>UniRef100_UPI000042DDF6 UPI000042DDF6 UniRef100 entry
Length = 777
Score = 39.7 bits (91), Expect = 0.024
Identities = 29/93 (31%), Positives = 42/93 (44%), Gaps = 9/93 (9%)
Query: 5 LDVLGEVDLSSGEFYSIMRDSDFFEGMDPFEMMALSDSLAGGSGPCLGPFDSDDDDDDGF 64
L+ E+ SGE M + + E MD FE DS GSG D DDDD++G
Sbjct: 203 LEEFREMLKESGEDVDDMTEEELMERMDEFEGDESDDSEESGSG---SDEDEDDDDEEGD 259
Query: 65 RVSRMAAM------QEAMAIGMDAFGPDDDEEE 91
+ A E +++G D ++DE+E
Sbjct: 260 EDDELLASASEGENDEEVSVGDDEDEDEEDEDE 292
>UniRef100_UPI00002F1EA5 UPI00002F1EA5 UniRef100 entry
Length = 223
Score = 39.7 bits (91), Expect = 0.024
Identities = 35/139 (25%), Positives = 50/139 (35%), Gaps = 38/139 (27%)
Query: 16 GEFYSIMRDSDFFEGMDPFEMMALSDSLAGGSGPCLGPFDSDDDDDDGFRVSRMAAMQEA 75
G+ + + R + P LS + G D DDDDDD
Sbjct: 86 GKGHPLGRTTPRGSAQSPLSQNGLSQNGYGDDDDSYDDDDDDDDDDD------------- 132
Query: 76 MAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPDDFPGIDPMDAALISMMMHSHM 135
D DDD+++ + DD DD D D H H
Sbjct: 133 -----DDDDDDDDDDDDD------------DDDDDDDDDDDDNDDDDD--------HDHD 167
Query: 136 DDEEDDEDYEDEEEYTDDE 154
DD++DDED +D++E DD+
Sbjct: 168 DDDDDDEDDDDDDENDDDD 186
>UniRef100_Q9CT46 Mus musculus 10 days embryo whole body cDNA, RIKEN full-length
enriched library, clone:2600013M03 product:nucleolin,
full insert sequence [Mus musculus]
Length = 444
Score = 39.7 bits (91), Expect = 0.024
Identities = 22/75 (29%), Positives = 37/75 (49%)
Query: 79 GMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPDDFPGIDPMDAALISMMMHSHMDDE 138
G +A + DE+E + ++ G D+F P G+ P AA + DDE
Sbjct: 137 GKNAKKEESDEDEDEEDEDDSDEDEDDEGEDEFEPPIVKGVKPAKAAPAAPASEDEGDDE 196
Query: 139 EDDEDYEDEEEYTDD 153
++D++ +D+EE DD
Sbjct: 197 DEDDEEDDDEEEEDD 211
Score = 34.7 bits (78), Expect = 0.76
Identities = 28/107 (26%), Positives = 46/107 (42%), Gaps = 11/107 (10%)
Query: 55 DSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPD-------DDEEEFNVFRMAAMQEAMVSG 107
D DD+ +D F + ++ A A D DDEE+ + +E ++
Sbjct: 160 DEDDEGEDEFEPPIVKGVKPAKAAPAAPASEDEGDDEDEDDEEDDDEEEEDDSEEEVMEI 219
Query: 108 IDDFGPDDFPGIDPMDAALISMMMHSHMDDEEDDEDYEDEEEYTDDE 154
G + PM A ++ DDEE+DED EDE++ +D+
Sbjct: 220 TTAKGKKTPAKVVPMKAKSVA----EEEDDEEEDEDDEDEDDEEEDD 262
>UniRef100_UPI00001A1BB9 UPI00001A1BB9 UniRef100 entry
Length = 225
Score = 39.3 bits (90), Expect = 0.031
Identities = 19/26 (73%), Positives = 22/26 (84%), Gaps = 1/26 (3%)
Query: 130 MMHSHMDDEEDDEDYEDEEEYTDDEY 155
MM S+MDDEEDDE+ E+EEE DDEY
Sbjct: 197 MMKSNMDDEEDDEEDEEEEE-DDDEY 221
>UniRef100_UPI000021BB95 UPI000021BB95 UniRef100 entry
Length = 755
Score = 38.9 bits (89), Expect = 0.040
Identities = 29/93 (31%), Positives = 44/93 (47%), Gaps = 18/93 (19%)
Query: 61 DDGFRVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPDDFPGID 120
+DGF V + +E+ A G A G DDDE VSG+ + + GI
Sbjct: 164 EDGFEVGSDDSEEESDASG-SASGSDDDE---------------VSGVSEDESELENGIQ 207
Query: 121 PMDAALISMMMHSHMDDEEDDEDYEDEEEYTDD 153
D+ + M+D D++DYE+EE+Y +D
Sbjct: 208 GSDSE--QNVTDGDMEDMGDEDDYEEEEDYEED 238
>UniRef100_Q5U328 Nucleolin [Rattus norvegicus]
Length = 714
Score = 38.9 bits (89), Expect = 0.040
Identities = 30/105 (28%), Positives = 48/105 (45%), Gaps = 6/105 (5%)
Query: 55 DSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPD 114
D D+D++D F + ++ A A D+DEE+ + + DD +
Sbjct: 158 DEDEDEEDEFEPPVVKGVKPAKAAPAAPASEDEDEEDDDDEEDDDDDDEEEEEEDD-SEE 216
Query: 115 DFPGIDPMD-----AALISMMMHSHMDDEEDDEDYEDEEEYTDDE 154
+ I P A ++ + S ++EEDDED EDEEE D+E
Sbjct: 217 EVMEITPAKGKKTPAKVVPVKAKSVAEEEEDDEDDEDEEEDEDEE 261
Score = 31.6 bits (70), Expect = 6.4
Identities = 29/133 (21%), Positives = 51/133 (37%), Gaps = 24/133 (18%)
Query: 24 DSDFFEGMDPFEMMALSDSLAGGSGPCLGPFDSDDDDDDGFRVSRMAAMQEAMAIGMDAF 83
D D + +P + + + A + P D +DDDD+
Sbjct: 160 DEDEEDEFEPPVVKGVKPAKAAPAAPASEDEDEEDDDDE--------------------- 198
Query: 84 GPDDDEEEFNVFRMAAMQEAMVSGIDDFGPDDFPGIDPMDAALISMMMHSHMDDE--EDD 141
DDD+++ +E ++ G + P+ A ++ DDE E+D
Sbjct: 199 -EDDDDDDEEEEEEDDSEEEVMEITPAKGKKTPAKVVPVKAKSVAEEEEDDEDDEDEEED 257
Query: 142 EDYEDEEEYTDDE 154
ED EDEE+ D++
Sbjct: 258 EDEEDEEDDEDED 270
>UniRef100_UPI000021BFA3 UPI000021BFA3 UniRef100 entry
Length = 4048
Score = 38.5 bits (88), Expect = 0.052
Identities = 34/102 (33%), Positives = 49/102 (47%), Gaps = 25/102 (24%)
Query: 55 DSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDF--G 112
DSDDDD+D + D PD+ E + + M+ E VS DD G
Sbjct: 2412 DSDDDDEDMY----------------DDGYPDEMEYDED---MSQDDEENVSDDDDEIDG 2452
Query: 113 PDDFPGIDPMDAALISMMMHSHMDDEEDDEDYEDEEEYTDDE 154
G+ P DA ++ ++M DD++DDE +D+EE T DE
Sbjct: 2453 MGHVEGL-PGDAGVVEVIMG---DDDDDDESMDDDEEDTSDE 2490
Score = 36.2 bits (82), Expect = 0.26
Identities = 34/146 (23%), Positives = 59/146 (40%), Gaps = 27/146 (18%)
Query: 12 DLSSGEFYSIMRDSDFFEGMDPFEMMALSDSLAGGSGPCLGPFDSDDDDDDGFRVSRMAA 71
D+S + ++ D D +GM E L G +G DDDDD+ +
Sbjct: 2434 DMSQDDEENVSDDDDEIDGMGHVE------GLPGDAGVVEVIMGDDDDDDE--------S 2479
Query: 72 MQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSG----IDDFGPDDFPGIDPMDAALI 127
M + D DD+ + ++ + + E +V+G ++D G ++ D
Sbjct: 2480 MDDDEEDTSDEDDEDDEMDSGDIEELEEVVEEIVNGEGSPVEDDGASEWESESEND---- 2535
Query: 128 SMMMHSHMDDEEDDEDYEDEEEYTDD 153
DDE+D+ DYED + D+
Sbjct: 2536 -----DEDDDEQDEIDYEDGAQDIDE 2556
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.138 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 290,976,572
Number of Sequences: 2790947
Number of extensions: 13029367
Number of successful extensions: 143155
Number of sequences better than 10.0: 702
Number of HSP's better than 10.0 without gapping: 298
Number of HSP's successfully gapped in prelim test: 426
Number of HSP's that attempted gapping in prelim test: 127513
Number of HSP's gapped (non-prelim): 5934
length of query: 155
length of database: 848,049,833
effective HSP length: 116
effective length of query: 39
effective length of database: 524,299,981
effective search space: 20447699259
effective search space used: 20447699259
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 69 (31.2 bits)
Medicago: description of AC146566.1


