

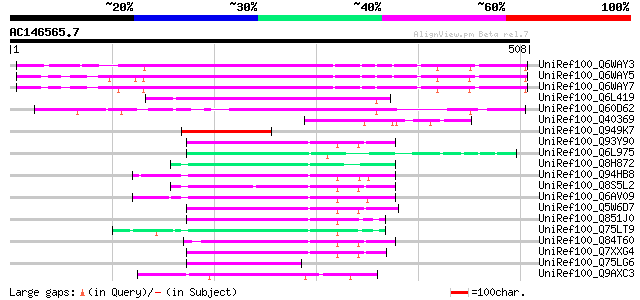
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146565.7 + phase: 0 /pseudo
(508 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum] 270 6e-71
UniRef100_Q6WAY5 Gag/pol polyprotein [Pisum sativum] 264 5e-69
UniRef100_Q6WAY7 Gag/pol polyprotein [Pisum sativum] 262 2e-68
UniRef100_Q6L419 Putative gag polyprotein [Solanum demissum] 161 4e-38
UniRef100_Q60D62 Putative gag/pol polyprotein, 3'-partial [Solan... 131 5e-29
UniRef100_Q40369 RPE15 protein [Medicago sativa] 118 5e-25
UniRef100_Q949K7 Putative gag polyprotein [Cicer arietinum] 104 5e-21
UniRef100_Q93Y90 Putative gag-pol [Oryza sativa] 72 4e-11
UniRef100_Q6L975 GAG-POL [Vitis vinifera] 72 4e-11
UniRef100_Q8H872 Putative polyprotein [Oryza sativa] 71 6e-11
UniRef100_Q94HB8 Putative retroelement [Oryza sativa] 71 6e-11
UniRef100_Q8S5L2 Hypothetical protein OSJNBa0011A24.12 [Oryza sa... 71 6e-11
UniRef100_Q6AV09 Putative polyprotein [Oryza sativa] 70 1e-10
UniRef100_Q5W6D7 Putative polyprotein [Oryza sativa] 70 1e-10
UniRef100_Q851J0 Putative GAG-POL [Oryza sativa] 70 1e-10
UniRef100_Q75LT9 Putative reverse transcriptase [Oryza sativa] 69 2e-10
UniRef100_Q84T60 Putative GAG-POL [Oryza sativa] 69 3e-10
UniRef100_Q7XXG4 OSJNBb0089K24.6 protein [Oryza sativa] 69 4e-10
UniRef100_Q75LG6 Putative gag protein [Oryza sativa] 68 5e-10
UniRef100_Q9AXC3 Hypothetical protein [Antirrhinum hispanicum] 68 5e-10
>UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum]
Length = 2262
Score = 270 bits (691), Expect = 6e-71
Identities = 182/523 (34%), Positives = 270/523 (50%), Gaps = 54/523 (10%)
Query: 7 ELESMKTNMDELSELVKVLSTAQNQPPPPPPFSTQAEASSSAIPDWTIYADTPTHSAPQR 66
EL M+ NM + +++ ++ Q + Q + + + P+ + P H P
Sbjct: 7 ELAEMRANMAQFMHMMQGVAQGQEELRA----LVQRQEAVTPPPNQALPEGNPVHDTPAA 62
Query: 67 SAPWFQPFTAGEIFRPIACEAQMPTHQYVAHVPPPPIKVPLAAMTYSAPVMHTIPQNEEP 126
+ P + GE I + Q P+A +A V+H +N P
Sbjct: 63 AIP-VNNYAVGEELMGIRVDGQ-----------------PIAPDAANARVIHAPARNRIP 104
Query: 127 IFHS--------GNMEAYDGVSDLRE-KYDELQRDVKALRGKEKFGKTAYDLCLVPNVQI 177
I E G +D R+ K D L ++A+ + G ++ LV ++I
Sbjct: 105 IVDRQEDLFTMFSEDEDIPGRNDARDRKVDALAEKIRAMECQNSLGFDVTNMGLVEGLRI 164
Query: 178 PHKFKIPNFEKYKGNSCPEEHLKMYVRRMPAYAQDDQVLIYYF*ESLAGPASKWYTNLDK 237
P+KFK P+F+KY G SCP H++ Y R++ AY D+++ +Y+F +SL+G + WY L
Sbjct: 165 PYKFKAPSFDKYNGTSCPRTHVQAYYRKISAYTDDEKMWMYFFQDSLSGASLDWYMELKS 224
Query: 238 TKIQTFRDLYEAFVEQYNYNVDMAPDRSDLQAMTQGDKETFKEYAQRWRDIAAQVSPRIE 297
I+ +RDL EAF+ QY +N+DMAP R+ LQ++ Q E+FKEYAQRWR++AA+V P +
Sbjct: 225 DSIRCWRDLGEAFLRQYKHNMDMAPSRTQLQSLCQKSNESFKEYAQRWRELAARVQPPML 284
Query: 298 EKEMTKIFLKTLNQFYYERMVGSMP-KNFAEMVGMGVQLEEGVREGRLVKDGASDSGAKK 356
E+E+T +F+ TL + +RM GS P +F+++V G + E ++ G+ ++D S S K
Sbjct: 285 ERELTDMFIGTLQGVFMDRM-GSCPFGSFSDVVICGERTESLIKTGK-IQDVGSSSSKKP 342
Query: 357 FGNNFPRKKAPEVGMVAHGRPQQNYPAYQHVAAIT-PATNAIQQPGYQPQFQQYPQQLYQ 415
F PR++ E V H R QN Y+ AA+T PA QQ Q Q Q PQQ Q
Sbjct: 343 FA-GAPRRREGETNAVQH-RRDQNRIEYRQAAAVTIPAPQPRQQ---QQQRVQQPQQQQQ 397
Query: 416 QQ--QPRPQAQRTQFDPIPMKYADFLPALLEKNLVH--TRPPPKVPERLPAWYRPDKFCA 471
Q+ QPR + +FD +PM YA+ LP LL LV T PP V LP Y + C
Sbjct: 398 QRPYQPRQRMPDRRFDSLPMSYAELLPELLRLGLVELCTMAPPTV---LPPGYDANVRCD 454
Query: 472 FHQGAPGHDTEYCYTLKAAVQKLIRDKDLSFA-------NPLP 507
FH GAPGH TE C L+ VQ LI ++FA NP+P
Sbjct: 455 FHSGAPGHHTEKCRALQHKVQDLIDANAINFAPVPNVVNNPMP 497
>UniRef100_Q6WAY5 Gag/pol polyprotein [Pisum sativum]
Length = 1105
Score = 264 bits (674), Expect = 5e-69
Identities = 183/527 (34%), Positives = 271/527 (50%), Gaps = 60/527 (11%)
Query: 7 ELESMKTNMDELSELVKVLSTAQNQPPPPPPFSTQAEASSSAIPDWTIYADTPTHSAPQR 66
EL MK NM + +++ ++ Q + + +AIP P + AP
Sbjct: 7 ELAEMKANMAQFMNMMQGVAQGQEE------LRAMVQRQEAAIP--------PVNHAPPE 52
Query: 67 SAPWFQPFTAGEIFRPIACEAQMPTHQYVA-------HVPPPPIKVPLA-AMTYSAPVMH 118
P A A +P + Y + PI A A T AP +
Sbjct: 53 GGPVNDNNVA----------AAVPINNYDVGDELGGIRINSQPIVPDAANARTIRAPARN 102
Query: 119 TIP--QNEEPIFH--SGNMEAYDGVSDLREKYDELQRDVKALRGKEKFGKTAYDLCLVPN 174
+P +E +F S + + V + K D L ++A+ + ++ LV
Sbjct: 103 PVPIVDRQEDMFTLLSEDEDVLGRVDERDRKVDALAEKIRAMECQNSLSFDVTNMGLVEG 162
Query: 175 VQIPHKFKIPNFEKYKGNSCPEEHLKMYVRRMPAYAQDDQVLIYYF*ESLAGPASKWYTN 234
++IP+KFK P+F+KY SCP H++ Y R++ AY D+++ +Y+F +SL+G + WY
Sbjct: 163 LRIPYKFKAPSFDKYNDTSCPRTHVQAYYRKISAYTDDEKMWMYFFQDSLSGASLDWYME 222
Query: 235 LDKTKIQTFRDLYEAFVEQYNYNVDMAPDRSDLQAMTQGDKETFKEYAQRWRDIAAQVSP 294
L + I+ +RDL EAF+ QY +N+DMAP R+ LQ++ Q E+FKEYAQRWR++AA+V P
Sbjct: 223 LKRDSIRCWRDLGEAFLRQYKHNMDMAPSRTQLQSLCQKSGESFKEYAQRWRELAARVQP 282
Query: 295 RIEEKEMTKIFLKTLNQFYYERMVGSMP-KNFAEMVGMGVQLEEGVREGRLVKDGASDSG 353
+ E+E+T +F+ TL + +RM GS P +F+++V G + E ++ G+ ++D S S
Sbjct: 283 PMLERELTDMFIGTLQGVFMDRM-GSCPFGSFSDVVICGERTESLIKTGK-IQDVGSSSS 340
Query: 354 AKKFGNNFPRKKAPEVGMVAHGRPQQNYPAYQHVAAIT-PATNAIQQPGYQPQFQQYPQQ 412
K F PR++ E +V H R QN Y+ AA+T PA QQ Q Q Q PQQ
Sbjct: 341 KKPFA-GAPRRREGETNVVQH-RRDQNRIEYRQAAAVTIPAPQPRQQ---QQQRVQQPQQ 395
Query: 413 LYQQQ---QPRPQAQRTQFDPIPMKYADFLPALLEKNLV--HTRPPPKVPERLPAWYRPD 467
QQQ QPR + +FD +PM YA+ LP LL +V T PP V LP Y +
Sbjct: 396 QQQQQRPYQPRQRMPDRRFDSLPMSYAELLPELLRLGMVELRTMAPPTV---LPPGYDAN 452
Query: 468 KFCAFHQGAPGHDTEYCYTLKAAVQKLIRDKDLSFA-------NPLP 507
C FH GAPGH TE C L+ VQ LI K ++FA NP+P
Sbjct: 453 VRCDFHSGAPGHHTEKCRALQHKVQDLIDAKAINFAPVPNVVNNPMP 499
>UniRef100_Q6WAY7 Gag/pol polyprotein [Pisum sativum]
Length = 1814
Score = 262 bits (669), Expect = 2e-68
Identities = 177/526 (33%), Positives = 265/526 (49%), Gaps = 58/526 (11%)
Query: 7 ELESMKTNMDELSELVKVLSTAQNQPPPPPPFSTQAEASSSAIPDWTIYADTPTHSAPQR 66
EL MK NM + +++ ++ Q + + +AIP P + AP
Sbjct: 7 ELAEMKANMAQFMNMMQGVAQGQEE------LRALVQRQEAAIP--------PVNHAPPE 52
Query: 67 SAPWFQPFTAGEIFRPIACEAQMPTHQYVAHVPPPPIKV---PLAAMTYSAPVMHTIPQN 123
P A A +P + Y I++ P+A +A + +N
Sbjct: 53 GGPVNGNNVA----------AAVPINNYAVGDELGGIRINGQPIAPDVANARAVRAPARN 102
Query: 124 EEPIFH---------SGNMEAYDGVSDLREKYDELQRDVKALRGKEKFGKTAYDLCLVPN 174
PI S + + V + K D L ++A+ + G ++ LV
Sbjct: 103 PAPIVDRQEDMFSLLSEDEDVLGRVDERDRKVDALAEKIRAMECQNSLGFDVTNMGLVEG 162
Query: 175 VQIPHKFKIPNFEKYKGNSCPEEHLKMYVRRMPAYAQDDQVLIYYF*ESLAGPASKWYTN 234
++IP+KFK P+F+KY G SCP H++ Y R++ AY D+++ +Y+F +SL+G + WY
Sbjct: 163 LRIPYKFKAPSFDKYNGTSCPRTHVQAYYRKISAYTDDEKMWMYFFQDSLSGASLDWYME 222
Query: 235 LDKTKIQTFRDLYEAFVEQYNYNVDMAPDRSDLQAMTQGDKETFKEYAQRWRDIAAQVSP 294
L + I+ ++DL EAF+ QY +N+DMAP R+ LQ++ Q E+FKEYAQRWR++AA+V P
Sbjct: 223 LKRDSIRCWKDLGEAFLRQYKHNMDMAPSRTQLQSLCQKSGESFKEYAQRWRELAARVQP 282
Query: 295 RIEEKEMTKIFLKTLNQFYYERMVGSMP-KNFAEMVGMGVQLEEGVREGRLVKDGASDSG 353
+ E+E+T +F+ TL + +RM GS P +F+++V G + E ++ G+ ++D S S
Sbjct: 283 PMLERELTDMFIGTLQGVFMDRM-GSCPFVSFSDVVICGERTESLIKTGK-IQDAGSSSS 340
Query: 354 AKKFGNNFPRKKAPEVGMVAHGRPQQNYPAYQHVAAITPATNAIQQPGYQPQFQQYPQQL 413
K F PR++ E V + R Q Q A PA QQ Q Q Q PQQ
Sbjct: 341 KKPFA-GAPRRREGETNAVQYRRDQNRSQRCQVAAVTIPAPQPRQQ---QQQRVQQPQQQ 396
Query: 414 YQQQ---QPRPQAQRTQFDPIPMKYADFLPALLEKNLV--HTRPPPKVPERLPAWYRPDK 468
QQQ QPR + +FD +PM YA+ LP LL +V T PP V LP Y +
Sbjct: 397 QQQQRPYQPRQRMPDRRFDSLPMSYAELLPELLRLGMVELRTMAPPTV---LPPGYDANV 453
Query: 469 FCAFHQGAPGHDTEYCYTLKAAVQKLIRDKDLSFA-------NPLP 507
C FH GAPGH TE C L+ VQ LI K ++FA NP+P
Sbjct: 454 RCDFHSGAPGHHTEKCRALQHKVQDLIDAKAINFAPVPNVVNNPMP 499
>UniRef100_Q6L419 Putative gag polyprotein [Solanum demissum]
Length = 421
Score = 161 bits (408), Expect = 4e-38
Identities = 85/243 (34%), Positives = 138/243 (55%), Gaps = 6/243 (2%)
Query: 134 EAYDGVSDLREKYDELQRDVKALRGKEKFGKTAYDLCLVPNVQIPHKFKIPNFEKYKGNS 193
E ++ ++ + + RD++ L G + G + DLC+ P+V +P FK PNFEKY G+
Sbjct: 152 EEHEEMTKKMKSLEHSIRDMQGLGGHK--GISFSDLCMFPHVHLPTGFKTPNFEKYDGHG 209
Query: 194 CPEEHLKMYVRRMPAYAQDDQVLIYYF*ESLAGPASKWYTNLDKTKIQTFRDLYEAFVEQ 253
P HLK Y ++ +++L+ YF ESL G ASKW+ + D T+ DL FV+Q
Sbjct: 210 DPIAHLKRYYNQLRGAEGKEELLMAYFGESLVGIASKWFIDQDIANWHTWDDLARCFVQQ 269
Query: 254 YNYNVDMAPDRSDLQAMTQGDKETFKEYAQRWRDIAAQVSPRIEEKEMTKIFLKTLNQFY 313
+ YN+D+ PDRS L M + E F+EYA RWR+ AA+V P ++E EM +FL+ Y
Sbjct: 270 FQYNIDIVPDRSSLANMRKKTTENFREYAVRWREQAARVKPPMKELEMIDVFLQAQEPDY 329
Query: 314 YERMVGSMPKNFAEMVGMGVQLEEGVREGRLVKDGASDSGAKKF----GNNFPRKKAPEV 369
+ ++ ++ K F E++ +G +E G++ G++V A + + GN +K+ +V
Sbjct: 330 FHYLLSAVGKTFTEVIKVGEMVENGIKSGKIVSQAALKATTQALPNGSGNIGGKKRREDV 389
Query: 370 GMV 372
V
Sbjct: 390 ATV 392
>UniRef100_Q60D62 Putative gag/pol polyprotein, 3'-partial [Solanum demissum]
Length = 1784
Score = 131 bits (329), Expect = 5e-29
Identities = 120/497 (24%), Positives = 202/497 (40%), Gaps = 112/497 (22%)
Query: 25 LSTAQNQPPPPPPFSTQAEASSSAIPDWTIYADTPTHSAPQ--RSAPWFQPFTAGEIFRP 82
+ + N P PP FS S+ A +TP S P +AP + +
Sbjct: 568 IQVSTNDPIYPPGFSPYTNTSNVAETSTVRPLNTPMMSNPLFVPTAPTNSTSNPTVVPKS 627
Query: 83 IACEAQMPTHQYVAHVPPPPIKVPLA---AMTYSAPV-MHTIPQNEEPIFHSGNMEAYDG 138
+ H + + P +K+P + YS+P + +NEE ++
Sbjct: 628 NNDPSFQVLHDH-GYTPEEALKIPSSYPQTHQYSSPFKIEKTVKNEE----------HEE 676
Query: 139 VSDLREKYDELQRDVKALRGKEKFGKTAYDLCLVPNVQIPHKFKIPNFEKYKGNSCPEEH 198
++ + ++ RD++ L G + G + DLC+ P+V +P G EE
Sbjct: 677 MAKKMKSLEQSIRDMQGLGGHK--GISFSDLCMFPHVHLP-----------TGAEGKEE- 722
Query: 199 LKMYVRRMPAYAQDDQVLIYYF*ESLAGPASKWYTNLDKTKIQTFRDLYEAFVEQYNYNV 258
+L+ YF ESL G AS+W+ + D T+ +L FV+Q+ YN+
Sbjct: 723 ----------------LLMAYFGESLVGIASEWFIDQDIANWHTWDNLARCFVQQFQYNI 766
Query: 259 DMAPDRSDLQAMTQGDKETFKEYAQRWRDIAAQVSPRIEEKEMTKIFLKTLNQFYYERMV 318
D+ PDRS L M + E F+EYA RWR+ AA+V ++E EM +FL+ Y+ ++
Sbjct: 767 DIVPDRSSLANMRKKTTENFREYAVRWREQAARVKLSMKESEMIDVFLQAQEPDYFHYLL 826
Query: 319 GSMPKNFAEMVGMGVQLEEGVREGRLVKDGASDSGAKKF----GNNFPRKKAPEVGMVAH 374
++ K F E++ +G +E G++ G++V A + + GN +K+ +V V +
Sbjct: 827 SAVGKTFVEVIKVGEMVENGIKSGKIVSQAALKATTQALQNGSGNIGGKKRREDVATVGN 886
Query: 375 GRPQQNYPAYQHVAAITPATNAIQQPGYQPQFQQYPQQLYQQQQPRPQAQRTQFDPIPMK 434
G + F PI
Sbjct: 887 GAKDE------------------------------------------------FTPIGES 898
Query: 435 YADFLPALLEKNLVH------TRPPPKVPERLPAWYRPDKFCAFHQGAPGHDTEYCYTLK 488
YA L N++ + PPP+ + + CA+ APGH+ E C+ LK
Sbjct: 899 YASLFQKLRTLNVLSPIERKMSNPPPRNLDY-------SQHCAYCSDAPGHNIERCWYLK 951
Query: 489 AAVQKLIRDKDLSFANP 505
A+Q LI + + +P
Sbjct: 952 KAIQDLIDTRRIIVESP 968
>UniRef100_Q40369 RPE15 protein [Medicago sativa]
Length = 219
Score = 118 bits (295), Expect = 5e-25
Identities = 82/196 (41%), Positives = 108/196 (54%), Gaps = 42/196 (21%)
Query: 289 AAQVSPRIEEKEMTKIFLKTLNQFYYERMVGSMPKNFAEMVGMGVQLEEGVREGRLV--K 346
AA+VSPR++E+E+T+ FLKTL + + ERM+ S P NF++MV MG +LEE VREG +V K
Sbjct: 14 AARVSPRLDEEELTQTFLKTLKKEHKERMIVSAPNNFSDMVTMGTRLEEAVREGIIVLEK 73
Query: 347 DGASDSGAKKFGN-NFPRKKAPEVGMVA--HGRP---QQNYPAYQHVAAITPATNAIQQP 400
S S KK+GN + RK++ +VGMV+ H P Q YQ++ Q P
Sbjct: 74 GECSASAPKKYGNGHHKRKESKKVGMVSGNHDTPVNATQLPLPYQYM-------QISQHP 126
Query: 401 GYQPQFQQYP------------------------QQLYQQQQPRPQAQRTQFDPIPMKYA 436
+ P +QQYP QQ QQQ PRP +T F PIPM+YA
Sbjct: 127 FFPPFYQQYPLPPGQPPVPVNAMAQQVQHQPPAQQQQQQQQGPRP---KTYFPPIPMRYA 183
Query: 437 DFLPALLEKNLVHTRP 452
+ LP LL K TRP
Sbjct: 184 ELLPTLLAKGHCVTRP 199
>UniRef100_Q949K7 Putative gag polyprotein [Cicer arietinum]
Length = 88
Score = 104 bits (260), Expect = 5e-21
Identities = 43/88 (48%), Positives = 68/88 (76%)
Query: 169 LCLVPNVQIPHKFKIPNFEKYKGNSCPEEHLKMYVRRMPAYAQDDQVLIYYF*ESLAGPA 228
+CLVP+V IP KFK+ FEKYKG +CP+ HL MY R+M ++A +D++LI++F +SL+G +
Sbjct: 1 MCLVPDVTIPRKFKVLEFEKYKGATCPKNHLTMYGRKMASHAHNDKLLIHFFQDSLSGAS 60
Query: 229 SKWYTNLDKTKIQTFRDLYEAFVEQYNY 256
WY +L++ +I +++DL +AF+ QY Y
Sbjct: 61 LSWYMHLERNRIHSWKDLADAFLRQYKY 88
>UniRef100_Q93Y90 Putative gag-pol [Oryza sativa]
Length = 1519
Score = 72.0 bits (175), Expect = 4e-11
Identities = 55/218 (25%), Positives = 95/218 (43%), Gaps = 16/218 (7%)
Query: 174 NVQIPHKFKIPNFEKYKGNSCPEEHLKMYVRRMPAYAQDDQVLIYYF*ESLAGPASKWYT 233
+V+ P +F+ EKY G++ PEE L++Y + A DD L Y +L G A W
Sbjct: 288 DVRWPERFRPGAIEKYDGSTNPEEFLQVYSTVLYAAGADDNALANYLSTALKGSARSWLM 347
Query: 234 NLDKTKIQTFRDLYEAFVEQYNYNVDMAPDRSDLQAMTQGDKETFKEYAQRWRDIAAQVS 293
+L I ++ DL++ FV + +L A+TQ E+ +EY +R+ + +
Sbjct: 348 HLPPYSISSWADLWQQFVANFQGTYKRHAIEDNLHALTQNSGESLREYVRRFNECRNTI- 406
Query: 294 PRIEEKEMTKIFLKTLNQFYYERMVG----SMPKNFAEMVGMGVQLEEGVR--------- 340
P I + + + F + Y + + + + E+V +++ +R
Sbjct: 407 PEITDASVIRAFKSGVRDRYTTQELATRRITTTRRLFEIVERCAHVDDALRRKNDKPKTG 466
Query: 341 -EGRLVKDGASDSGAKKFGNNFPRKKAPEVGMVAHGRP 377
E + D AS+S KK N RK EV + P
Sbjct: 467 GEKKSATD-ASESSKKKNRKNGKRKAQAEVLAAEYANP 503
>UniRef100_Q6L975 GAG-POL [Vitis vinifera]
Length = 549
Score = 72.0 bits (175), Expect = 4e-11
Identities = 74/325 (22%), Positives = 126/325 (38%), Gaps = 47/325 (14%)
Query: 174 NVQIPHKFKIPNFEKYKGNSCPEEHLKMYVRRMPAYAQDDQVLIYYF*ESLAGPASKWYT 233
N + P F +P F Y G S P +H+ Y + M +D +L F SL G A W+
Sbjct: 12 NYEPPRGFLVPKFSTYDGTSDPFDHIMHYRQLMTLDIGNDALLCKVFPASLQGQALSWFH 71
Query: 234 NLDKTKIQTFRDLYEAFVEQYNYNVDMAPDRSDLQAMTQGDKETFKEYAQRWRDIAAQVS 293
L + FRDL EAFV QY + + S LQ + D E+ +E+ +R+ QV
Sbjct: 72 RLPPNSVGNFRDLSEAFVGQYLCSARHKQNISTLQNIKMQDNESLREFVKRFGQAVLQVE 131
Query: 294 PRIEEKEMTKIFLKTL--NQFYYERMVGSMPKNFAEMVGMGVQLEEGVREGRLVKDGASD 351
+ +IF +++ ++E + P ++
Sbjct: 132 -ACSMDAVLQIFKRSICPGTPFFESLAKKPPTTMDDLF---------------------- 168
Query: 352 SGAKKFGNNFPRKKAPEVGMVAHGRPQQNYPAYQHVAAITPATNAIQQPGYQPQFQQYPQ 411
A K+ +A ++ GRP + + + +P + P
Sbjct: 169 RRANKYSMLEDDVRAATQQVLVAGRP----------------SGSNTERNTKPPDRPKPS 212
Query: 412 QLYQQQQPRPQAQRTQFDPIPMKYADFLPALLEKNLVHTRPPPKVPERLPAWYRPDKFCA 471
Q+ RP + P+ + Y LP + + L + P + P+ + CA
Sbjct: 213 DRRQEGPSRP--KMPPLTPLSISYEKLLPMI--QGLSDFKWPRPIATD-PSTRDRSRRCA 267
Query: 472 FHQGAPGHDTEYCYTLKAAVQKLIR 496
FH+ GH TE C + + V++LI+
Sbjct: 268 FHKD-HGHTTETCRSFQYLVERLIK 291
>UniRef100_Q8H872 Putative polyprotein [Oryza sativa]
Length = 1469
Score = 71.2 bits (173), Expect = 6e-11
Identities = 57/221 (25%), Positives = 90/221 (39%), Gaps = 23/221 (10%)
Query: 158 GKEKFGKTAYDLCLVPNVQIPHKFKIPNFEKYKGNSCPEEHLKMYVRRMPAYAQDDQVLI 217
G FG++ D V+ P +F+ +KY G++ PEE L++Y + A DD L
Sbjct: 325 GCRAFGRSLRD------VRWPERFRPRAIKKYDGSTDPEEFLQVYSTVLYAAGADDNALA 378
Query: 218 YYF*ESLAGPASKWYTNLDKTKIQTFRDLYEAFVEQYNYNVDMAPDRSDLQAMTQGDKET 277
Y +L G A W +L I ++ DL++ FV + DL A+TQ E+
Sbjct: 379 NYLPTALKGSARSWLMHLPPYSISSWADLWQQFVANFQGTYKHHAIEDDLHALTQNSGES 438
Query: 278 FKEYAQRWRDIAAQVSPRIEEKEMTKIFLKTLNQFY-YERMVGSMPKNFAEMVGMGVQLE 336
+EY +R+ + + P I + + + F + Y + PK E
Sbjct: 439 LREYVRRFNECRNTI-PEITDSSVIRTFKSGVRDRYTTQERKNDKPKTGGE--------- 488
Query: 337 EGVREGRLVKDGASDSGAKKFGNNFPRKKAPEVGMVAHGRP 377
+ AS+S KK N RK EV + P
Sbjct: 489 ------KKPAADASESSKKKNHKNRKRKAQAEVLAAEYANP 523
>UniRef100_Q94HB8 Putative retroelement [Oryza sativa]
Length = 2079
Score = 71.2 bits (173), Expect = 6e-11
Identities = 66/271 (24%), Positives = 111/271 (40%), Gaps = 22/271 (8%)
Query: 121 PQNEEPIFHSGNMEAYDGVSDLREKYDELQRDVKA-LRGKEKFGKTAYDLCLVPNVQIPH 179
P+ +P S + A +SD + R A G FG++ D V+ P
Sbjct: 524 PEPSDPS-SSSSSSASSSLSDRHPRRSRDHRQPTAPSAGCRAFGRSLRD------VRWPE 576
Query: 180 KFKIPNFEKYKGNSCPEEHLKMYVRRMPAYAQDDQVLIYYF*ESLAGPASKWYTNLDKTK 239
+F+ EKY G+ PEE L++Y + A DD + Y +L G A W +L
Sbjct: 577 RFRPGAIEKYDGSIDPEEFLQVYSTVLYAAGADDNAVANYLPTALKGSARSWLMHLPPYS 636
Query: 240 IQTFRDLYEAFVEQYNYNVDMAPDRSDLQAMTQGDKETFKEYAQRWRDIAAQVSPRIEEK 299
I ++ DL++ FV + DL A+TQ E+ +EY +R+ + + P I +
Sbjct: 637 ISSWADLWQQFVANFQGTYKRHAIEDDLHALTQNSGESLREYVRRFNECRNTI-PEITDA 695
Query: 300 EMTKIFLKTLNQFYYERMVG----SMPKNFAEMVGMGVQLEEGVRE-------GRLVKDG 348
+ + F + Y + + + + ++V +++ +R G K
Sbjct: 696 SVIRAFKSGVRDRYTTQELATRHITTTRRLFQIVERCAHVDDALRRKNDKPKTGGEKKSA 755
Query: 349 A--SDSGAKKFGNNFPRKKAPEVGMVAHGRP 377
A S+S KK N RK EV + P
Sbjct: 756 ADTSESSKKKNRKNGKRKAQAEVLAAEYANP 786
>UniRef100_Q8S5L2 Hypothetical protein OSJNBa0011A24.12 [Oryza sativa]
Length = 1001
Score = 71.2 bits (173), Expect = 6e-11
Identities = 60/234 (25%), Positives = 98/234 (41%), Gaps = 24/234 (10%)
Query: 158 GKEKFGKTAYDLCLVPNVQIPHKFKIPNFEKYKGNSCPEEHLKMYVRRMPAYAQDDQVLI 217
G FG++ D V+ P +F+ EKY G++ PEE L++Y + A DD L
Sbjct: 505 GCRAFGRSLRD------VRWPERFRPGAIEKYDGSADPEEFLQVYSTVLYATGADDNALA 558
Query: 218 YYF*ESLAGPASKWYTNLDKTKIQTFRDLYEAFVEQYNYNVDMAPDRSDLQAMTQGDKET 277
Y +L G A W +L T Q DL++ FV + + DL A+TQ E+
Sbjct: 559 NYLPAALKGSARSWLMHLPPT--QFLHDLWQQFVANFQGTYKRHAIKDDLHALTQNSGES 616
Query: 278 FKEYAQRWRDIAAQVSPRIEEKEMTKIFLKTLNQFYYERMVG----SMPKNFAEMVGMGV 333
+EY R+ + + P I + + F + Y + + + + E++
Sbjct: 617 LREYVSRFNECRNTI-PEITDASVIHAFKSGVRDRYTTQELATRRITTTRRLFEIIERCA 675
Query: 334 QLEEGVR----------EGRLVKDGASDSGAKKFGNNFPRKKAPEVGMVAHGRP 377
+++ +R E +L D S+S KK N RK EV + P
Sbjct: 676 HVDDALRRKNDKPKIGGEKKLAAD-TSESNKKKNRKNGKRKAHAEVLAAGYANP 728
>UniRef100_Q6AV09 Putative polyprotein [Oryza sativa]
Length = 1551
Score = 70.5 bits (171), Expect = 1e-10
Identities = 64/270 (23%), Positives = 110/270 (40%), Gaps = 21/270 (7%)
Query: 121 PQNEEPIFHSGNMEAYDGVSDLREKYDELQRDVKALRGKEKFGKTAYDLCLVPNVQIPHK 180
P+ +P S ++ + R D Q + G FG++ D V+ P +
Sbjct: 339 PEPTDPSSSSSSVSSSSSDRHPRRSRDRRQPTAPSA-GCRAFGRSLRD------VRWPER 391
Query: 181 FKIPNFEKYKGNSCPEEHLKMYVRRMPAYAQDDQVLIYYF*ESLAGPASKWYTNLDKTKI 240
+ EKY G++ PEE L++Y + A DD L Y +L G A W +L I
Sbjct: 392 LRPGAIEKYDGSTDPEEFLQVYSTVLYAAGADDNALANYLPTALKGSARSWLMHLPPYSI 451
Query: 241 QTFRDLYEAFVEQYNYNVDMAPDRSDLQAMTQGDKETFKEYAQRWRDIAAQVSPRIEEKE 300
++ DL++ FV + DL +TQ E+ +EY +R+ + + P I +
Sbjct: 452 SSWIDLWQQFVANFQGTYKRHAIEDDLHTLTQNSGESLREYVRRFNECRNTI-PEITDAF 510
Query: 301 MTKIFLKTLNQFYYERMVG----SMPKNFAEMVGMGVQLEEGV-REGRLVKDG------- 348
+ + F + Y + + + + E+V +++ + R+ K G
Sbjct: 511 VIRAFKSGVRDRYTTQELATRRITTTRRLFEIVERCAHVDDALRRKNNKPKTGGEKKPAV 570
Query: 349 -ASDSGAKKFGNNFPRKKAPEVGMVAHGRP 377
AS+S KK N RK EV + P
Sbjct: 571 DASESSKKKNRKNGKRKAQAEVLAAEYANP 600
>UniRef100_Q5W6D7 Putative polyprotein [Oryza sativa]
Length = 1185
Score = 70.5 bits (171), Expect = 1e-10
Identities = 56/221 (25%), Positives = 95/221 (42%), Gaps = 16/221 (7%)
Query: 174 NVQIPHKFKIPNFEKYKGNSCPEEHLKMYVRRMPAYAQDDQVLIYYF*ESLAGPASKWYT 233
+V+ P +F+ EKY G++ PEE L++Y A DD L Y +L G A W
Sbjct: 316 DVRWPERFRPGAIEKYDGSTDPEEFLQVYSTVPYAAGADDNALANYLPTALKGSARSWLM 375
Query: 234 NLDKTKIQTFRDLYEAFVEQYNYNVDMAPDRSDLQAMTQGDKETFKEYAQRWRDIAAQVS 293
+L I ++ DL++ FV + DL A+TQ E+ ++Y +R+ D +
Sbjct: 376 HLPPYSISSWADLWQQFVGNFQGTYKRHMIEDDLHALTQNPGESLRDYVRRFNDCRNTI- 434
Query: 294 PRIEEKEMTKIFLKTLNQFYYERMVG----SMPKNFAEMVGMGVQLEEGVR--------- 340
P I + + + F + Y + + + E+V ++ +R
Sbjct: 435 PEITDASVIRAFKTGVRDRYTTLELATRQITTTRRLFEVVDRCAHADDALRRKNDKPKAG 494
Query: 341 -EGRLVKDGASDSGAKKFGNNFPRKKAPEVGMVAHGRPQQN 380
E + KD A + KK G + RK EV + RP ++
Sbjct: 495 GEKKPAKD-APEPSKKKSGKSGKRKAQAEVFASEYERPPKH 534
>UniRef100_Q851J0 Putative GAG-POL [Oryza sativa]
Length = 831
Score = 70.5 bits (171), Expect = 1e-10
Identities = 51/199 (25%), Positives = 85/199 (42%), Gaps = 13/199 (6%)
Query: 174 NVQIPHKFKIPNFEKYKGNSCPEEHLKMYVRRMPAYAQDDQVLIYYF*ESLAGPASKWYT 233
+V+ P +F+ EKY G++ PEE L++Y + A DD L+ Y +L G A W
Sbjct: 343 DVRWPERFRPGAIEKYDGSTNPEEFLQVYSTVLYAAGADDSALVNYLPTALKGSARSWLM 402
Query: 234 NLDKTKIQTFRDLYEAFVEQYNYNVDMAPDRSDLQAMTQGDKETFKEYAQRWRDIAAQVS 293
+L I + DL++ FV + DL A+TQ E+ +Y R+ + +
Sbjct: 403 HLPPYSISLWADLWQQFVANFQGTYKRHAIEDDLHALTQNPGESLSDYVWRFNECRNTI- 461
Query: 294 PRIEEKEMTKIFLKTLNQFYYERMVG----SMPKNFAEMVGMGVQLEEGVREGRLVKDGA 349
P I + + + F + Y + + + + E+V ++ +R D
Sbjct: 462 PEITDASVIRAFKSGVRDRYTTQELATRRVTTTRRLFEIVDRCAHTDDALRRN---NDKP 518
Query: 350 SDSGAKKFGNNFPRKKAPE 368
G KK P K APE
Sbjct: 519 KTGGEKK-----PAKDAPE 532
>UniRef100_Q75LT9 Putative reverse transcriptase [Oryza sativa]
Length = 1445
Score = 69.3 bits (168), Expect = 2e-10
Identities = 67/276 (24%), Positives = 109/276 (39%), Gaps = 26/276 (9%)
Query: 101 PPIKVPLAAMTYSAPVMHTIPQNEEPIFHSGNMEAYDGVSDL----REKYDELQRDVKAL 156
PP + + T S P P + N E S++ R Y+ Q +
Sbjct: 108 PPAEAEIGTSTTSEPEKD--PSEAKSCLSDKNHEPTRMTSEVTRHPRRAYNRRQPTAPSA 165
Query: 157 RGKEKFGKTAYDLCLVPNVQIPHKFKIPNFEKYKGNSCPEEHLKMYVRRMPAYAQDDQVL 216
R + FG++ D V+ P +F+ EKY G++ PEE L++Y + A DD L
Sbjct: 166 RCRA-FGRSLRD------VRWPKRFRSGAIEKYDGSTDPEEFLQVYSTVLYAAGADDNAL 218
Query: 217 IYYF*ESLAGPASKWYTNLDKTKIQTFRDLYEAFVEQYNYNVDMAPDRSDLQAMTQGDKE 276
Y +L G A W +L I + DL++ FV + DL A TQ E
Sbjct: 219 ANYLPTALKGSARSWLMHLPPYSISLWADLWQQFVANFQGTYKRHAIEDDLHASTQNPGE 278
Query: 277 TFKEYAQRWRDIAAQVSPRIEEKEMTKIFLKTLNQFYYERMVG----SMPKNFAEMVGMG 332
+ ++Y +R+ + + P I + + F + Y + + + + E+V
Sbjct: 279 SLRDYVRRFNECRNTI-PEITDASVICAFKSGVRDRYTTQELATRRVTTTRRLFEIVDRC 337
Query: 333 VQLEEGVREGRLVKDGASDSGAKKFGNNFPRKKAPE 368
++ VR D G KK P K AP+
Sbjct: 338 AHADDAVRR---KNDKPKTGGEKK-----PAKDAPK 365
>UniRef100_Q84T60 Putative GAG-POL [Oryza sativa]
Length = 1380
Score = 68.9 bits (167), Expect = 3e-10
Identities = 57/221 (25%), Positives = 93/221 (41%), Gaps = 23/221 (10%)
Query: 171 LVPNVQIPHKFKIPNFEKYKGNSCPEEHLKMYVRRMPAYAQDDQVLIYYF*ESLAGPASK 230
+VP+V H EKY G++ PEE L++Y + A DD L Y +L G A
Sbjct: 299 IVPDVMSLH-------EKYDGSTDPEEFLQVYSTVLYAAGADDNALANYLPTALKGSARS 351
Query: 231 WYTNLDKTKIQTFRDLYEAFVEQYNYNVDMAPDRSDLQAMTQGDKETFKEYAQRWRDIAA 290
W +L I ++ DL++ FV + DL A+TQ E+ +EY QR+ +
Sbjct: 352 WLMHLPPYSISSWADLWQQFVANFQGTYKRHAIEDDLHALTQNPGESLREYVQRFNECRN 411
Query: 291 QVSPRIEEKEMTKIFLKTLNQFYYERMVG----SMPKNFAEMVGMGVQLEEGVR------ 340
+ P+I + + + F + Y + + + + E+V ++ +R
Sbjct: 412 TI-PKITDASVIRAFKSGVRDRYTTQELATRRITTNRRLFEIVERCAHADDALRRKNDKP 470
Query: 341 ----EGRLVKDGASDSGAKKFGNNFPRKKAPEVGMVAHGRP 377
E + D A +S KK N RK EV + P
Sbjct: 471 KTGGEKKSATD-APESSKKKNRKNGKRKAQAEVLAAEYANP 510
>UniRef100_Q7XXG4 OSJNBb0089K24.6 protein [Oryza sativa]
Length = 1400
Score = 68.6 bits (166), Expect = 4e-10
Identities = 54/210 (25%), Positives = 91/210 (42%), Gaps = 16/210 (7%)
Query: 174 NVQIPHKFKIPNFEKYKGNSCPEEHLKMYVRRMPAYAQDDQVLIYYF*ESLAGPASKWYT 233
+V+ P +F+ EKY G++ PEE L++Y + A DD L Y +L G A W
Sbjct: 435 DVRWPERFRPRAIEKYDGSTDPEEFLQVYSTVLYAAGADDNALANYLPTALKGYARSWLM 494
Query: 234 NLDKTKIQTFRDLYEAFVEQYNYNVDMAPDRSDLQAMTQGDKETFKEYAQRWRDIAAQVS 293
+L I ++ DL++ FV + DL A+TQ E+ +EY +R+ + +
Sbjct: 495 HLPPYSISSWADLWQQFVANFQGTYKRHAIEDDLHALTQNSGESLREYVRRFNECRNTI- 553
Query: 294 PRIEEKEMTKIFLKTLNQFYYERMVG----SMPKNFAEMVGMGVQLEEGVR--------- 340
P I + + + + Y + + + + E+V ++ +R
Sbjct: 554 PEITDASVIRALKSGVRDRYTTQELATRRITTTRRLFEIVERCAHADDALRRKNDKPKTG 613
Query: 341 -EGRLVKDGASDSGAKKFGNNFPRKKAPEV 369
E + D AS+S KK N RK EV
Sbjct: 614 GEKKPATD-ASESSKKKNRKNGKRKAQAEV 642
>UniRef100_Q75LG6 Putative gag protein [Oryza sativa]
Length = 641
Score = 68.2 bits (165), Expect = 5e-10
Identities = 35/112 (31%), Positives = 58/112 (51%)
Query: 174 NVQIPHKFKIPNFEKYKGNSCPEEHLKMYVRRMPAYAQDDQVLIYYF*ESLAGPASKWYT 233
NV+ P +F+ EKY G+ P E L++Y + A DD+V+ +F +L G A W
Sbjct: 230 NVRWPPRFRPTITEKYDGSVNPTEFLQVYTTGIEAAGGDDRVMANFFPMALKGQARGWLV 289
Query: 234 NLDKTKIQTFRDLYEAFVEQYNYNVDMAPDRSDLQAMTQGDKETFKEYAQRW 285
NL + ++ DL + F + + +DL A+ +GD E+ + Y QR+
Sbjct: 290 NLPPASVHSWEDLCQQFTMNFQGTYPRPGEEADLHAVQRGDDESLRSYIQRF 341
>UniRef100_Q9AXC3 Hypothetical protein [Antirrhinum hispanicum]
Length = 1455
Score = 68.2 bits (165), Expect = 5e-10
Identities = 55/243 (22%), Positives = 104/243 (42%), Gaps = 13/243 (5%)
Query: 126 PIFHSGNMEAYDGVSDLREKYDELQRDVKALRGKEKFGKTAYDLCLVPNVQIPHKFKIPN 185
P + N + + V++L+ + L RD +++ G L P +IP ++ N
Sbjct: 117 PGMDNNNQQFINMVTELKREVQALNRDRDGTAEEDEMGSPFAPHILAP--EIPVSMRLSN 174
Query: 186 FEKYKGNSC---PEEHLKMYVRRMPAYAQDDQVLIYYF*ESLAGPASKWYTNLDKTKIQT 242
Y G+ P +H+ ++ M + D L + F +L G A W++ L + I++
Sbjct: 175 PVVYTGDRNARDPRDHINQFLAVMDILSTPDHALCWIFRSTLTGRAQAWFSQLPRGSIRS 234
Query: 243 FRDLYEAFVEQYNYNVDMAPDRSDLQAMTQGDKETFKEYAQRWRDI---AAQVSPRIEEK 299
F +L E FV Q+ N L M + + E+ +EY +R+ + V P I +
Sbjct: 235 FDELRELFVRQFACNRRYPKTIGHLMTMVEAEGESLREYMRRFANTVIDVRHVGPHILSE 294
Query: 300 EMTKIFLKTLNQFYYERMVGSMPKNFAEMVGMG--VQLEEGVREGRLVKDGASDSGAKKF 357
+ + T + + +VG ++ E++ L+E R +L G D +
Sbjct: 295 FVAQ---GTRSVEFANSLVGRPARSLEELMHRAERYMLQEDTRLAKLGIQGHEDRSRNRR 351
Query: 358 GNN 360
G+N
Sbjct: 352 GDN 354
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.134 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 930,092,533
Number of Sequences: 2790947
Number of extensions: 43115484
Number of successful extensions: 208864
Number of sequences better than 10.0: 1116
Number of HSP's better than 10.0 without gapping: 416
Number of HSP's successfully gapped in prelim test: 746
Number of HSP's that attempted gapping in prelim test: 201482
Number of HSP's gapped (non-prelim): 4687
length of query: 508
length of database: 848,049,833
effective HSP length: 132
effective length of query: 376
effective length of database: 479,644,829
effective search space: 180346455704
effective search space used: 180346455704
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC146565.7


