

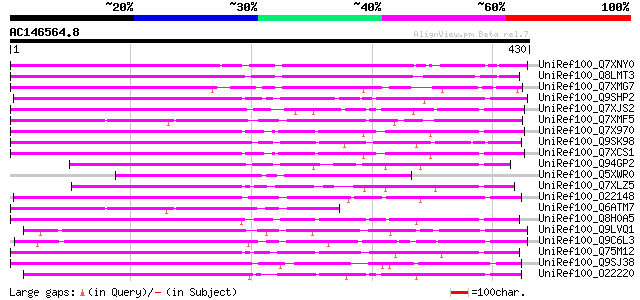
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146564.8 - phase: 0 /pseudo
(430 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XNY0 OSJNBb0015N08.11 protein [Oryza sativa] 223 8e-57
UniRef100_Q8LMT3 Putative retroelement [Oryza sativa] 194 5e-48
UniRef100_Q7XMG7 OSJNBa0028I23.15 protein [Oryza sativa] 181 4e-44
UniRef100_Q9SHP2 Putative non-LTR retroelement reverse transcrip... 170 6e-41
UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcrip... 169 2e-40
UniRef100_Q7XMF5 OSJNBa0061G20.4 protein [Oryza sativa] 164 6e-39
UniRef100_Q7X970 BZIP-like protein [Oryza sativa] 163 8e-39
UniRef100_Q9SK98 Very similar to retrotransposon reverse transcr... 160 8e-38
UniRef100_Q7XCS1 Putative reverse transcriptase [Oryza sativa] 158 3e-37
UniRef100_Q94GP2 Putative reverse transcriptase [Oryza sativa] 157 4e-37
UniRef100_Q5XWR0 Hypothetical protein [Solanum tuberosum] 156 1e-36
UniRef100_Q7XLZ5 OSJNBa0086O06.14 protein [Oryza sativa] 156 1e-36
UniRef100_O22148 Putative non-LTR retroelement reverse transcrip... 155 2e-36
UniRef100_Q6ATM7 Hypothetical protein OSJNBb0021K20.15 [Oryza sa... 152 2e-35
UniRef100_Q8H0A5 Putative reverse transcriptase [Oryza sativa] 151 4e-35
UniRef100_Q9LVQ1 Non-LTR retroelement reverse transcriptase-like... 150 5e-35
UniRef100_Q9C6L3 Hypothetical protein F2J7.11 [Arabidopsis thali... 147 4e-34
UniRef100_Q75M12 Hypothetical protein P0676G05.2 [Oryza sativa] 147 7e-34
UniRef100_Q9SJ38 Putative non-LTR retroelement reverse transcrip... 145 2e-33
UniRef100_O22220 Putative non-LTR retroelement reverse transcrip... 145 3e-33
>UniRef100_Q7XNY0 OSJNBb0015N08.11 protein [Oryza sativa]
Length = 1026
Score = 223 bits (568), Expect = 8e-57
Identities = 142/431 (32%), Positives = 226/431 (51%), Gaps = 25/431 (5%)
Query: 1 LKALLRGFEMASGLKVNFAKSCLIGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGANP 60
LK +L FE SGLK+NF KS + +E D N C+ GS P +YLG+P+
Sbjct: 506 LKYILCLFEQLSGLKINFNKSEVFCFGEAKEKQDLYSNIFTCKVGSLPLKYLGIPIDQKR 565
Query: 61 RRLSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKLPVKVWKRVV 120
W+ + ++ +L W + S+GGR++LLN+ L+++P + ++F++LP V +R+
Sbjct: 566 ILNKDWKLAENKMEHKLGCWQGRLQSIGGRLILLNSTLSSVPMYMISFYRLPKGVQERID 625
Query: 121 RIQREFLWGGVNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISLLAKWRWRLVQPEQP 180
++ FLW G +K V W VC PRD+GGLGV D++ +N ++L KW WRL + E+
Sbjct: 626 YFRKRFLWQEDQGIRKYHLVNWPLVCSPRDQGGLGVLDLEAMNKAMLGKWIWRL-ENEEG 684
Query: 181 L*KDVLRCKYGSRINFLLDPIYN-SWSSLASRWWLDLMSLEGVVGANWFNREMVRKVGNG 239
++++ KY S P+ + +S +W +M ++ + F + VGNG
Sbjct: 685 WWQEIIYAKYCSD-----KPLSGLRLKAGSSHFWQGVMEVK-----DDFFSFCTKIVGNG 734
Query: 240 ETTRFWLDHWVGNEALYLTFPRLFSISSQKEAMVGEVWVDGDWNLTWRRSLFVWEEGLIH 299
E T FW D W+G + L + FP L+ I K + ++ G + +RR L +
Sbjct: 735 EKTLFWEDSWLGGKPLAIQFPSLYGIVITKRITIADLNRKGIDCMKFRRDLHGDKLRDWR 794
Query: 300 ILLDELEGREVSEP-MDSWWWKLEEGGICSVSSSYSLHVKLQMPPEPLENTKAMVFGSVW 358
+++ EG + E D WW L + G +V S Y +KLQ P N K +W
Sbjct: 795 KIVNSWEGLNLVENCKDKLWWTLSKDGKFTVRSFYRA-LKLQQTSFP--NKK------IW 845
Query: 359 KSPTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGETAAHLFLHCDMS 418
K P K+ +F W ++I TKDNLLKR ++ +C FCD+V ET HLF C ++
Sbjct: 846 KFRVPLKIRIFIWFFTKNKILTKDNLLKRGWRKGDN--KCQFCDKV-ETVQHLFFDCPLA 902
Query: 419 FKVWSMVYCRL 429
+W+++ C L
Sbjct: 903 RLIWNIIACAL 913
>UniRef100_Q8LMT3 Putative retroelement [Oryza sativa]
Length = 764
Score = 194 bits (492), Expect = 5e-48
Identities = 122/422 (28%), Positives = 197/422 (45%), Gaps = 21/422 (4%)
Query: 1 LKALLRGFEMASGLKVNFAKSCLIGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGANP 60
+K LL +E SGLK+N+ KS ++ + + NC+ G PF YLG+P+ N
Sbjct: 237 VKFLLYCYEAMSGLKINYQKSEIMVIGGDEMETQRVADLFNCQAGKMPFTYLGIPISMNK 296
Query: 61 RRLSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKLPVKVWKRVV 120
+ + + ++KRL +W Y+S GG+ +L+N+ L++IP + + + LP V ++
Sbjct: 297 LTNADLDIPPNKIEKRLATWKCGYLSYGGKAILINSCLSSIPLYMMGVYLLPEGVHNKMD 356
Query: 121 RIQREFLWGGVNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISLLAKWRWRLVQPEQP 180
I+ F W G+ +K +KW +C P++ GGLG D + +NI+LL KW +RL ++
Sbjct: 357 SIRARFFWEGLEKKRKYHMIKWEALCRPKEFGGLGFIDTRKMNIALLCKWIYRLESGKED 416
Query: 181 L*KDVLRCKYGSRINFLLDPIYNSWSSLASRWWLDLMSLEGVVGANWFNREMVRKVGNGE 240
+LR KY + S + +S++W L ++ W + KVGNG+
Sbjct: 417 PCCVLLRNKYMKDGG----GFFQSKAEESSQFWKGLHEVK-----KWMDLGSSYKVGNGK 467
Query: 241 TTRFWLDHWVGNEALYLTFPRLFSISSQKEAMVGEVWVDGDWNLTWRRSLFVWEEGLIHI 300
T FW D W+G L +P ++ + + KE V ++ ++GDW + RRSL + +
Sbjct: 468 ATNFWSDVWIGETPLKTQYPNIYRMCADKEKTVSQMCLEGDWYIELRRSLGERDLNEWND 527
Query: 301 LLDELEGREVSEPMDSWWWKLEEGGICSVSSSYSLHVKLQMPPEPLENTKAMVFGSVWKS 360
L + + E D WKL + G + Y K V +W+S
Sbjct: 528 LHNTPREIHLKEERDCIIWKLTKNGFYKAKTLYQ--------ALSFGGVKDKVMQDLWRS 579
Query: 361 PTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGETAAHLFLHCDMSFK 420
P KV + W L RI L K + S C C Q E HL C +S
Sbjct: 580 SIPLKVKILFWLMLKGRIQAAGQLKK---MKWSGSPNCKLCGQT-EDVDHLMFRCHISQF 635
Query: 421 VW 422
+W
Sbjct: 636 MW 637
>UniRef100_Q7XMG7 OSJNBa0028I23.15 protein [Oryza sativa]
Length = 593
Score = 181 bits (459), Expect = 4e-44
Identities = 130/447 (29%), Positives = 217/447 (48%), Gaps = 66/447 (14%)
Query: 1 LKALLRGFEMASGLKVNFAKSCLIGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGANP 60
LK +L FE SGLK+NF KS L+ E C+ GS +YLGLP+
Sbjct: 150 LKLVLSAFERLSGLKINFHKSELLCFGKAIEVEREYALLFGCKTGSYTLKYLGLPMHYRK 209
Query: 61 RRLSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKLPVKVWKRVV 120
W+ + + +KRL SW K +S+GGR+VL+N+VL+++ + L+FF++P + K++
Sbjct: 210 LSNKDWKEVEERFQKRLGSWKGKLLSVGGRLVLINSVLSSLAMYMLSFFEVPKGIIKKLD 269
Query: 121 RIQREFLWGGVNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISL---LAKWRWRLVQP 177
+ F W KK +W+ +C P++ GGLG++++++ N L LAK W
Sbjct: 270 YYRSRFFWQSDEHKKKYRLARWSVLCKPKECGGLGIQNLEVQNKCLLRGLAKLVW----- 324
Query: 178 EQPL*KDVLRCKYGSRINFLLDPIYNSWSSLASRWWLDLMSLEGVVGANWFNREMVRKVG 237
+++LR KY ++ + S +W LM ++ + F+ K+
Sbjct: 325 -----QNLLRKKYLAKKTIMQVQKQRG----DSHFWTGLMGVK-----DTFSSFGSFKLQ 370
Query: 238 NGETTRFWLDHWVGNEALYLTFPRLFSISSQKEAMVGEVWVDGDWNLTWRRSLF-----V 292
+G RFW D W+GN++L +P L+++ K +V +V N+++RR++
Sbjct: 371 DGLQIRFWEDCWLGNQSLEKIYPSLYNLVRNKNVVVAKVLERVPLNVSFRRAIIGDNLKA 430
Query: 293 WEEGLIHILLDELEGREVSEPMDSWWWKLEEGGICSVSSSYSLHVKLQMPPEPLENTKAM 352
W E + +++ L G++ D + W + G +V+S Y K +
Sbjct: 431 WHEIVANVVDINLRGQK-----DRFTWDASKNGTFTVNSMY----------------KKL 469
Query: 353 VFGSV-------WKSPTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVG 405
+FG+V WK P K+ +F W I TKDNL KRR + S+RC FC +
Sbjct: 470 MFGNVIPRQSFIWKLRLPLKIKIFLWYLKEGVILTKDNLAKRR---WKGSLRCCFC-RSH 525
Query: 406 ETAAHLFLHCDMSF-------KVWSMV 425
ET +LF C M ++WS++
Sbjct: 526 ETIQYLFFDCPMVIFRGTHWARLWSLL 552
>UniRef100_Q9SHP2 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 773
Score = 170 bits (431), Expect = 6e-41
Identities = 120/432 (27%), Positives = 201/432 (45%), Gaps = 24/432 (5%)
Query: 4 LLRGFEMASGLKVNFAKSCLI-GVNVGREFMDAACNFMNCREGSPPFQYLGLPVGANPRR 62
+L+ +E ASG VNF KS ++ G + + + + +YLGLP +
Sbjct: 157 VLKLYEKASGQAVNFQKSAILFGKGLDFRTSEQLSQLLGIYKTEGFGRYLGLPEFVGRNK 216
Query: 63 LSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKLPVKVWKRVVRI 122
+ + + + +++++W NK +S G+ VL+ +++ AIP + ++ F LP+++ ++
Sbjct: 217 TNAFSFIAQTMDQKMDNWYNKLLSPAGKEVLIKSIVTAIPTYSMSCFLLPMRLIHQITSA 276
Query: 123 QREFLWGGVNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISLLAKWRWRLVQPEQPL* 182
R F W K+ WV W+ + P+ GGL +RD+K NI+LLAK WR++Q L
Sbjct: 277 MRWFWWSNTKVKHKIPWVAWSKLNDPKKMGGLAIRDLKDFNIALLAKQSWRILQQPFSLM 336
Query: 183 KDVLRCKYGSRINFLLDPIYNSWSSLASRWWLDLMSLEGVVGANWFNREMVRKVGNGETT 242
V + KY + LLD S SS A W ++ G +R + GNG
Sbjct: 337 ARVFKAKYFPK-ERLLDAKATSQSSYA---WKSILH-----GTKLISRGLKYIAGNGNNI 387
Query: 243 RFWLDHWVGNEALYLTFPRLFSISSQKEAMVGEVWVDGDWNLTWRRSLFVWEEGLIHILL 302
+ W D+W+ SI SQ + V ++ ++G WN L + + + HI
Sbjct: 388 QLWKDNWLPLNPPRPPVGTCDSIYSQLK--VSDLLIEGRWNEDLLCKL-IHQNDIPHI-- 442
Query: 303 DELEGREVSEPMDSWWWKLEEGGICSVSSSYSLHVKLQMP-----PEPLENTKAMVFGSV 357
++ D+ W G SV S Y L KL P P E + VF ++
Sbjct: 443 -RAIRPSITGANDAITWIYTHDGNYSVKSGYHLLRKLSQQQHASLPSPNEVSAQTVFTNI 501
Query: 358 WKSPTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGETAAHLFLHCDM 417
WK P K+ F W+S + +PT NL +RR++ ++ C C + E HL C +
Sbjct: 502 WKQNAPPKIKHFWWRSAHNALPTAGNLKRRRLITDDT---CQRCGEASEDVNHLLFQCRV 558
Query: 418 SFKVWSMVYCRL 429
S ++W + +L
Sbjct: 559 SKEIWEQAHIKL 570
>UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1344
Score = 169 bits (427), Expect = 2e-40
Identities = 117/439 (26%), Positives = 205/439 (46%), Gaps = 38/439 (8%)
Query: 1 LKALLRGFEMASGLKVNFAKSCL-IGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGAN 59
L L + SG +N KS + G NV + D + +YLGLP +
Sbjct: 677 LMQCLSQYGQLSGQMINLNKSAITFGKNVDIQIKDWIKSRSGISLEGGTGKYLGLPECLS 736
Query: 60 PRRLSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKLPVKVWKRV 119
+ + + + L+ RL W K +S GG+ VLL ++ A+P + ++ FKLP + +++
Sbjct: 737 GSKRDLFGFIKEKLQSRLTGWYAKTLSQGGKEVLLKSIALALPVYVMSCFKLPKNLCQKL 796
Query: 120 VRIQREFLWGGVNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISLLAKWRWRLVQPEQ 179
+ +F W + +K+ W+ W + LP+D+GG G +D++ N +LLAK WR++Q +
Sbjct: 797 TTVMMDFWWNSMQQKRKIHWLSWQRLTLPKDQGGFGFKDLQCFNQALLAKQAWRVLQEKG 856
Query: 180 PL*KDVLRCKYGSRINFLLDPIYNSWSSLASRWWLDLMSLEGVVGANWFNREMVRK---- 235
L V + +Y S +FL + S S W ++ F RE++ +
Sbjct: 857 SLFSRVFQSRYFSNSDFL----SATRGSRPSYAWRSIL----------FGRELLMQGLRT 902
Query: 236 -VGNGETTRFWLDHWV--GNEALYLTFPRLFSISSQKEAMVGEVWVDGDWNLTWRRSLFV 292
+GNG+ T W D W+ G+ L R ++ + ++ +WNL R LF
Sbjct: 903 VIGNGQKTFVWTDKWLHDGSNRRPLNRRRFINVDLKVSQLIDP--TSRNWNLNMLRDLFP 960
Query: 293 WEEGLIHILLDELEGREVSEPMDSWWWKLEEGGICSVSSSY-----SLHVKLQMPPEPLE 347
W++ + I+L + R + DS+ W G+ SV + Y +H +L +
Sbjct: 961 WKD--VEIILKQ---RPLFFKEDSFCWLHSHNGLYSVKTGYEFLSKQVHHRLYQEAKVKP 1015
Query: 348 NTKAMVFGSVWKSPTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGET 407
+ ++ F +W T K+ +F W++L IP +D L R I S C+ CD ET
Sbjct: 1016 SVNSL-FDKIWNLHTAPKIRIFLWKALHGAIPVEDRLRTRGI---RSDDGCLMCDTENET 1071
Query: 408 AAHLFLHCDMSFKVWSMVY 426
H+ C ++ +VW++ +
Sbjct: 1072 INHILFECPLARQVWAITH 1090
>UniRef100_Q7XMF5 OSJNBa0061G20.4 protein [Oryza sativa]
Length = 632
Score = 164 bits (414), Expect = 6e-39
Identities = 129/434 (29%), Positives = 200/434 (45%), Gaps = 38/434 (8%)
Query: 1 LKALLRGFEMASGLKVNFAKSCLIGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGANP 60
LKALL+ F++ + LKVNF KSCLI +NV C+ GS PF YLGLP+G
Sbjct: 165 LKALLQTFQLGTSLKVNFNKSCLIPINVDEGKAQNLAAVFGCQLGSLPFTYLGLPLGTIK 224
Query: 61 RRLSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKLPVKVWKRVV 120
++ + PL+D ++RL S ++S G R+ L+N+V +++P +Y+ LP V +
Sbjct: 225 PKVIDFAPLVDRTERRLTS-NAYFLSYGVRLTLVNSVFSSLPTYYMCTLMLPKTVIDSID 283
Query: 121 RIQREFLWGG--VNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISLLAKWRWRLVQP- 177
R +R LW G VN KK W VC P+ KGGLGV ++ + N +LL K+ + +
Sbjct: 284 RARRHCLWRGSEVNSNKK-SLAAWHKVCKPKRKGGLGVLNLSIQNQALLIKFLDKFSKKN 342
Query: 178 EQPL*KDVLRCKYGSRINFLLDPIYNSWSSLASRWWLDLMSLEGVVGANWFNREMVR-KV 236
+ P V YG+ + L + + WW D+ L V R + R +
Sbjct: 343 DLPWVNLVWSSYYGNYVPHLT-------NLKGTFWWRDICKLMDVF------RGIARCHL 389
Query: 237 GNGETTRFWLDHWVGNEALYLTFPRLFSISSQKEAMVGE-VWVDGDWNLTWRRSLFVWEE 295
+G T W W L+ T RL S + + A V + + ++ + N T S+ ++E
Sbjct: 390 KSGNTCSIWYYVWADQPPLFHTMSRLCSFAQNQNASVSDFLSIEAEENCTLPLSVQAFQE 449
Query: 296 GLIHILLDELEGREVSEPMDSW---WWKLEEGGICSVSSSYSLHVKLQMPPEPLENTKAM 352
L L E+ D+W W ++ S Y+L+ + PP +
Sbjct: 450 --FQELTSSLVDIELVNENDAWSYIWGNMK----YSSQKFYALNFRAIKPP--------V 495
Query: 353 VFGSVWKSPTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVG-ETAAHL 411
F +WK V VF W +++R TK L + P + + CV C V E HL
Sbjct: 496 HFLWLWKCNAVSNVKVFGWLLMMNRFNTKFMLDYKNCAPPDCDLSCVLCQSVAIEDPMHL 555
Query: 412 FLHCDMSFKVWSMV 425
C + WS++
Sbjct: 556 IFLCPFAQNCWSLL 569
>UniRef100_Q7X970 BZIP-like protein [Oryza sativa]
Length = 2367
Score = 163 bits (413), Expect = 8e-39
Identities = 120/432 (27%), Positives = 198/432 (45%), Gaps = 26/432 (6%)
Query: 1 LKALLRGFEMASGLKVNFAK-SCLIGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGAN 59
+K +L + M +G +N AK S L+G +A + +YLG P
Sbjct: 1433 VKEVLSSYAMGTGQLINPAKCSILMGGASTPAVSEAISEILQVERDRFEDRYLGFPTPEG 1492
Query: 60 PRRLSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKLPVKVWKRV 119
++ L + KR+ WG ++S GG+ VL+ AV+ AIP + + FKLP V +
Sbjct: 1493 RMHKGRFQSLQAKIWKRVIQWGENHLSTGGKEVLIKAVIQAIPVYVMGIFKLPESVIDDL 1552
Query: 120 VRIQREFLWGGVNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISLLAKWRWRLVQPEQ 179
++ + F W +NG +K W W ++ P+ GGLG RD +L N +LLA+ WRL+
Sbjct: 1553 TKLTKNFWWDSMNGQRKTHWKAWDSLTKPKSLGGLGFRDYRLFNQALLARQAWRLITYPD 1612
Query: 180 PL*KDVLRCKYGSRINFLLDPIYNSWSSLASRWWLDLMSLEGVVGANWFNREMVRKVGNG 239
L VL+ KY L+D + S SS A R S+E G + + ++ +VGNG
Sbjct: 1613 SLCARVLKAKYFPH-GSLIDTSFGSNSSPAWR------SIE--YGLDLLKKGIIWRVGNG 1663
Query: 240 ETTRFWLDHWVGNEALYLTFPRLFSISSQKEAMVGEVWVDGDWNLTWRRSLF--VWEEGL 297
+ R W D W+ + + P + + + + + DG W++ F + E +
Sbjct: 1664 NSIRIWRDSWLPRD--HSRRPITGKANCRLKWVSDLITEDGSWDVPKIHQYFHNLDAEVI 1721
Query: 298 IHILLDELEGREVSEPMDSWWWKLEEGGICSVSSSYSLHVKLQMPPEPLE---NTKAMVF 354
++I + D W ++ G+ SV S+Y L +L E N +
Sbjct: 1722 LNICISS------RSEEDFIAWHPDKNGMFSVRSAYRLAAQLVNIEESSSSGTNNINKAW 1775
Query: 355 GSVWKSPTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGETAAHLFLH 414
+WK P KV +F+W+ + + T N KR++ E S C CD+ E AH
Sbjct: 1776 EMIWKCKVPQKVKIFAWRVASNCLATMVNKKKRKL---EQSDMCQICDRENEDDAHALCR 1832
Query: 415 CDMSFKVWSMVY 426
C + ++WS ++
Sbjct: 1833 CIQASQLWSCMH 1844
>UniRef100_Q9SK98 Very similar to retrotransposon reverse transcriptase [Arabidopsis
thaliana]
Length = 1231
Score = 160 bits (404), Expect = 8e-38
Identities = 124/434 (28%), Positives = 201/434 (45%), Gaps = 32/434 (7%)
Query: 1 LKALLRGFEMASGLKVNFAKSCL-IGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGAN 59
L+++L + A+G +N AKS + G V + + YLGLP +
Sbjct: 547 LQSILGAYGAATGQIINLAKSSITFGSKVETVKKAEIQTCLGIFKEGDASTYLGLPECFS 606
Query: 60 PRRLSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKLPVKVWKRV 119
++ + + LK +L+SW +K +S GG+ ++L AV A+P F ++ FKLP +
Sbjct: 607 GSKIEMLDYIKVRLKSKLSSWFSKSLSQGGKEIMLKAVAMAMPVFAMSCFKLPKTTCTNL 666
Query: 120 VRIQREFLWGGVNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISLLAKWRWRLVQPEQ 179
+F W + +K+ W+ W +CLP++ GGLG RD+ L N +LLAK WRL+Q
Sbjct: 667 ASAMADFWWSANDHQRKIHWLSWEKLCLPKEHGGLGFRDIGLFNQALLAKQAWRLLQFPD 726
Query: 180 PL*KDVLRCKYGSRINFLLDPIYNSWSSLASRWWLDLMSLEGVVGANWFNREMVRKVGNG 239
L +++ +Y FL + S S W ++ G + R +V++VGNG
Sbjct: 727 CLFARLIKSRYFPVGEFLDSDV----GSRPSFGWRSILH-----GRDLLCRGLVKRVGNG 777
Query: 240 ETTRFWLDHWVGNEALYLTFPRLFSISSQKEAMVGEV--WVDGDWNLTWRRSLFVWEEGL 297
++ R W+D+W+ + L P + + + +V ++ + DW L F ++
Sbjct: 778 KSIRVWIDYWLDDNG--LRAPWIKNPIINVDLLVSDLIDYEKRDWRLDKLEEQFFPDD-- 833
Query: 298 IHILLDELEGREVSEPMDSWWWKLEEGGICSVSSSYSL-------HVKLQMPPEPLENTK 350
++ E R V D W WK + G SV Y L V ++ +P N
Sbjct: 834 ---VVKIRENRPVVSLDDFWIWKHNKSGDYSVKLGYWLASNQNLGQVAIEAMMQPSLND- 889
Query: 351 AMVFGSVWKSPTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGETAAH 410
+ VWK T K+ VF W+ L IP D LL R + +S RC C GE+ H
Sbjct: 890 --LKTQVWKLQTEPKIKVFLWKVLSGAIPVVD-LLSYRGMKLDS--RCQTCGCEGESIQH 944
Query: 411 LFLHCDMSFKVWSM 424
+ C +VW+M
Sbjct: 945 VLFSCSFPRQVWAM 958
>UniRef100_Q7XCS1 Putative reverse transcriptase [Oryza sativa]
Length = 791
Score = 158 bits (399), Expect = 3e-37
Identities = 119/432 (27%), Positives = 196/432 (44%), Gaps = 26/432 (6%)
Query: 1 LKALLRGFEMASGLKVNFAK-SCLIGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGAN 59
+K +L + M +G +N AK S L+G +A + +YL P
Sbjct: 109 VKEVLSSYAMGTGQLINPAKCSILMGGASTPAVSEAISEILQVERDRFEDRYLEFPTPEG 168
Query: 60 PRRLSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKLPVKVWKRV 119
++ L + KR+ WG ++S GG+ VL+ AV+ AIP + + FKLP V +
Sbjct: 169 RMHKGRFQSLQAKIWKRVIQWGENHLSTGGKEVLIKAVIQAIPVYVMGIFKLPESVIDDL 228
Query: 120 VRIQREFLWGGVNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISLLAKWRWRLVQPEQ 179
++ + F W +NG +K W W ++ P+ GGLG RD L N +LLA+ WRL+
Sbjct: 229 TKLTKNFWWDSMNGQRKTHWKAWDSLTKPKSLGGLGFRDYWLFNQALLARQAWRLITYPD 288
Query: 180 PL*KDVLRCKYGSRINFLLDPIYNSWSSLASRWWLDLMSLEGVVGANWFNREMVRKVGNG 239
L VL+ KY L+D + S SS A R S+E G + + ++ +VGNG
Sbjct: 289 SLCARVLKAKYFPH-GSLIDTSFGSNSSPAWR------SIE--YGLDLLKKGIIWRVGNG 339
Query: 240 ETTRFWLDHWVGNEALYLTFPRLFSISSQKEAMVGEVWVDGDWNLTWRRSLF--VWEEGL 297
+ R W D W+ + + P + + + + + DG W++ F + E +
Sbjct: 340 NSIRIWRDPWLPRD--HSRRPITGKANCRLKWVSDLITEDGSWDVPKIHQYFHNLDAEVI 397
Query: 298 IHILLDELEGREVSEPMDSWWWKLEEGGICSVSSSYSLHVKLQMPPEPLE---NTKAMVF 354
++I + D W ++ G+ SV S+Y L +L E N +
Sbjct: 398 LNICISS------RSEEDFIAWHPDKNGMFSVRSAYRLAAQLVNIEESSSSGTNNINKAW 451
Query: 355 GSVWKSPTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGETAAHLFLH 414
+WK P KV +F+W+ + + T N KR++ E S C CD+ E AH
Sbjct: 452 EMIWKCKVPQKVKIFAWRVASNCLATMVNKKKRKL---EQSDMCQICDRENEDDAHALCR 508
Query: 415 CDMSFKVWSMVY 426
C + ++WS ++
Sbjct: 509 CIQASQLWSCMH 520
>UniRef100_Q94GP2 Putative reverse transcriptase [Oryza sativa]
Length = 1833
Score = 157 bits (398), Expect = 4e-37
Identities = 108/381 (28%), Positives = 173/381 (45%), Gaps = 41/381 (10%)
Query: 50 QYLGLPVGANPRRLSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFF 109
+YLGLP + ++P+ D L KR++ W KY+S G + VL+ +V A+P + + F
Sbjct: 661 KYLGLPTPDGRMKAEQFQPINDKLSKRMSDWNEKYMSSGAKEVLIKSVAQALPTYIMGVF 720
Query: 110 KLPVKVWKRVVRIQREFLWGGVNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISLLAK 169
K+ K R+ R F WG NG +KV W+ W + P+ GGLG RDMK N +LLA+
Sbjct: 721 KMTDKFCDDYTRMVRNFWWGHDNGERKVHWIAWEKILYPKSHGGLGFRDMKCFNQALLAR 780
Query: 170 WRWRLVQPEQPL*KDVLRCKYGSRINFLLDPIYNSWSSLASRWWLDLMSLEGVVGANWFN 229
WRL+ L V + KY N + ++ +++S W + G +
Sbjct: 781 QAWRLLTSPDSLCARVFKAKYYPNGNL----VDTAFPTVSSSVWKGIQH-----GLDLLK 831
Query: 230 REMVRKVGNGETTRFWLDHWV--GNEALYLTFPRLFSISSQKEAMVGEVWVDGDWNLTWR 287
+ + +VGNG + W WV G L +K+ ++V+ N R
Sbjct: 832 QGTIWRVGNGHNIKIWRHKWVPHGEHINVL----------EKKGRNRLIYVNELINEENR 881
Query: 288 RSLFVWEEGLIHILLDELEGREV------SEPMDSW-WWKLEEGGICSVSSSYSLHVKL- 339
W E L+ +L E + EV + MD + W E+ G+ +V S+Y L L
Sbjct: 882 ----CWNETLVRHVLKEEDANEVLKIRLPNHQMDDFPAWHHEKSGLFTVKSAYKLAWNLS 937
Query: 340 -----QMPPEPLENTKAMVFGSVWKSPTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTES 394
Q + + ++ VW + KV +F W+ D++PT +N +R+I E
Sbjct: 938 GKGVVQSSSSTATSGERKIWSRVWNAKVQAKVKIFIWKLAQDKLPTWENKRRRKI---EM 994
Query: 395 SVRCVFCDQVGETAAHLFLHC 415
+ C C GE + H + C
Sbjct: 995 NGTCPVCGTKGENSYHATVEC 1015
>UniRef100_Q5XWR0 Hypothetical protein [Solanum tuberosum]
Length = 327
Score = 156 bits (394), Expect = 1e-36
Identities = 86/247 (34%), Positives = 137/247 (54%), Gaps = 9/247 (3%)
Query: 88 GGRVVLLNAVLNAIPNFYLAFFKLPVKVWKRVVRIQREFLWGGVNGGKKVCWVKWATVCL 147
GGR+ L+N+VL++IP ++++ +P KV +R+ +I+R+FLW G N K+ VKWA V L
Sbjct: 72 GGRLTLINSVLDSIPTYFMSLLLMPAKVKQRLDKIRRDFLWEGNNKDHKIHLVKWAKVTL 131
Query: 148 PRDKGGLGVRDMKLVNISLLAKWRWRLVQPEQPL*KDVLRCKYGSRINFLLDPIYNSWSS 207
P+ GGLG++D+ L N LL KW WR Q L K+V++ KYGS ++ + + S+ +
Sbjct: 132 PKQYGGLGIKDLALHNKCLLLKWHWRYNQEAAGLWKEVIQAKYGSDSHWCSNKVSTSYGT 191
Query: 208 LASRWWLDLMSLEGVVGANWFNREMVRKVGNGETTRFWLDHWVGNEALYLTFPRLFSISS 267
W D+ L + F + VGNGE +FW D W+G+ L FP +F I+S
Sbjct: 192 GV---WRDIRKLWEI-----FFGDASLMVGNGEHIQFWKDIWLGDTPLMNVFPSIFLIAS 243
Query: 268 QKEAMVGEVWVDGDWNLTWRRSLFVWEEGLIHILLDELEGREV-SEPMDSWWWKLEEGGI 326
++ + + D WNL RR+L WE + L+ ++ + S+ D W+ G
Sbjct: 244 HPDSTISQYREDNVWNLILRRNLNDWELEEVFSLMATIQTSAINSQRRDKLIWRHSRDGS 303
Query: 327 CSVSSSY 333
+V + Y
Sbjct: 304 YTVKACY 310
>UniRef100_Q7XLZ5 OSJNBa0086O06.14 protein [Oryza sativa]
Length = 1205
Score = 156 bits (394), Expect = 1e-36
Identities = 109/391 (27%), Positives = 178/391 (44%), Gaps = 59/391 (15%)
Query: 52 LGLPVGANPRRLSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKL 111
LGLP + ++P+ + +KRL W +++SL G+ L+ +V A+P + + FK+
Sbjct: 739 LGLPTPDGRMKAEQFQPIKERFEKRLTDWSERFLSLAGKEALIKSVAQALPTYTMGVFKM 798
Query: 112 PVKVWKRVVRIQREFLWGGVNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISLLAKWR 171
P + + ++ R F WG G KKV W+ W + P+ GGLG RD++ N +LLA+
Sbjct: 799 PERFCEEYEQLVRNFWWGHEKGEKKVHWIAWEKLTSPKLLGGLGFRDIRCFNQALLARQA 858
Query: 172 WRLVQPEQPL*KDVLRCKYGSRINFLLDPIYNSWSSLASRWWLDLMSLEGVV-GANWFNR 230
WRL++ L VL+ KY + D ++ S++S W +G+V G +
Sbjct: 859 WRLIESPDSLCARVLKAKYYPN-GTITD---TAFPSVSSPTW------KGIVHGLELLKK 908
Query: 231 EMVRKVGNGETTRFWLDHWVGNEALYLTFPRLFSISSQKEAMVGEVWVDGDWNLTWRRSL 290
++ ++G+G T+ W +HWV + I +K TW R +
Sbjct: 909 GLIWRIGDGSKTKIWRNHWVAH-------GENLKILEKK---------------TWNRVI 946
Query: 291 FV----------WEEGLIHILLDELEGREV-------SEPMDSWWWKLEEGGICSVSSSY 333
+V W E LI ++ E + E+ E D W E+ GI SV S Y
Sbjct: 947 YVRELIVTDTKTWNEPLIRHIIREEDADEILKIRIPQREEEDFPAWHYEKTGIFSVRSVY 1006
Query: 334 SLHVKLQMPPEPLENTKA------MVFGSVWKSPTPFKVVVFSWQSLLDRIPTKDNLLKR 387
L L ++ + ++ +VWK+ KV VF+W+ DR+ T +N KR
Sbjct: 1007 RLAWNLARKTSEQASSSSGGADGRKIWDNVWKANVQPKVRVFAWKLAQDRLATWENKKKR 1066
Query: 388 RILPTESSVRCVFCDQVGETAAHLFLHCDMS 418
+I E C C Q ET H + C ++
Sbjct: 1067 KI---EMFGTCPICGQKEETGFHATVECTLA 1094
>UniRef100_O22148 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1374
Score = 155 bits (392), Expect = 2e-36
Identities = 114/434 (26%), Positives = 194/434 (44%), Gaps = 30/434 (6%)
Query: 4 LLRGFEMASGLKVNFAKSCL-IGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGANPRR 62
++ + +ASG +VN+ KS + G ++ E + YLGLP +
Sbjct: 702 IIEEYSLASGQRVNYLKSSIYFGKHISEERRCLVKRKLGIEREGGEGVYLGLPESFQGSK 761
Query: 63 LSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKLPVKVWKRVVRI 122
++T L D L K++ W + ++S GG+ +LL AV A+P + ++ FK+P + +++ +
Sbjct: 762 VATLSYLKDRLGKKVLGWQSNFLSPGGKEILLKAVAMALPTYTMSCFKIPKTICQQIESV 821
Query: 123 QREFLWGGVNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISLLAKWRWRLVQPEQPL* 182
EF W G+ + W W + P+ GGLG ++++ NI+LL K WR++ + L
Sbjct: 822 MAEFWWKNKKEGRGLHWKAWCHLSRPKAVGGLGFKEIEAFNIALLGKQLWRMITEKDSLM 881
Query: 183 KDVLRCKYGSRINFLLDPIYNSWSSLASRWWLDLMSLEGVVGANWFNREMVRKV-GNGET 241
V + +Y S+ DP+ S S W + + ++ ++ +R V GNGET
Sbjct: 882 AKVFKSRYFSK----SDPLNAPLGSRPSFAWKSIYEAQVLI------KQGIRAVIGNGET 931
Query: 242 TRFWLDHWVG-NEALYLTFPRLFSISSQKEAMVGEVWVD------GDWNLTWRRSLFVWE 294
W D W+G A + + SQ A V D DWN W ++
Sbjct: 932 INVWTDPWIGAKPAKAAQAVKRSHLVSQYAANSIHVVKDLLLPDGRDWN--WNLVSLLFP 989
Query: 295 EGLIHILLDELEGREVSEPMDSWWWKLEEGGICSVSSSYSLHVKL----QMPPEPLENTK 350
+ +L G + E D + W+ G SV S Y + ++ P E L+ +
Sbjct: 990 DNTQENILALRPGGK--ETRDRFTWEYSRSGHYSVKSGYWVMTEIINQRNNPQEVLQPSL 1047
Query: 351 AMVFGSVWKSPTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGETAAH 410
+F +WK P K+ F W+ + + + NL R + +S CV C GET H
Sbjct: 1048 DPIFQQIWKLDVPPKIHHFLWRCVNNCLSVASNLAYRHLAREKS---CVRCPSHGETVNH 1104
Query: 411 LFLHCDMSFKVWSM 424
L C + W++
Sbjct: 1105 LLFKCPFARLTWAI 1118
>UniRef100_Q6ATM7 Hypothetical protein OSJNBb0021K20.15 [Oryza sativa]
Length = 352
Score = 152 bits (383), Expect = 2e-35
Identities = 97/276 (35%), Positives = 143/276 (51%), Gaps = 17/276 (6%)
Query: 1 LKALLRGFEMASGLKVNFAKSCLIGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGANP 60
LKALL+ F +GLKVNF KSCLI +NV + + R GS PF YLGLP+G
Sbjct: 17 LKALLQSFAQGTGLKVNFHKSCLIPINVSEDKAEMLAGVFGYRTGSMPFTYLGLPLGTTK 76
Query: 61 RRLSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKLPVKVWKRVV 120
R+ + PL+D +++RL++ + ++S G R+ L+N+V +++P +Y+ +LP V + +
Sbjct: 77 PRVIDFAPLVDRIERRLSA-NSAFLSNGDRLTLMNSVFSSLPTYYMCILQLPKSVIENID 135
Query: 121 RIQREFLW--GGVNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISLLAKWRWRLVQPE 178
R +R LW G +N KK V W VC P+ KGGLGV ++ + N +LL K+ +
Sbjct: 136 RARRHCLWRGGDINSNKK-SLVAWDKVCKPKVKGGLGVINLSIQNQALLLKFLDKFYNRR 194
Query: 179 QPL*KDVLRCKYGSRINFLLDPIYNSWSSLASRWWLDLMSLEGVVGANWFNREMVRKV-G 237
Q ++ Y L PI S+ WW D+ L + R + R + G
Sbjct: 195 QVPWVSLIWSSYYENKVPHLAPIKGSF------WWKDICKLMDIY------RGISRCILG 242
Query: 238 NGETTRFWLDHWVGNEALYLTFPRLFSISSQKEAMV 273
NG + FW D W L L PRL S + K V
Sbjct: 243 NGSSCSFWDDIWSDGPPLSLHMPRLHSFAINKNVSV 278
>UniRef100_Q8H0A5 Putative reverse transcriptase [Oryza sativa]
Length = 1557
Score = 151 bits (381), Expect = 4e-35
Identities = 114/430 (26%), Positives = 193/430 (44%), Gaps = 30/430 (6%)
Query: 1 LKALLRGFEMASGLKVNFAK-SCLIGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGAN 59
+K + + ++G +N K S L G ++ DA N + +YLGLP
Sbjct: 1068 IKEVFGSYATSTGQLINPTKCSILFGNSLPIASRDAITNCLQIASTEFEDKYLGLPTPGG 1127
Query: 60 PRRLSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKLPVKVWKRV 119
++ L + + K++ WG Y+S GG+ VL+ AV+ AIP + + FKLP V + +
Sbjct: 1128 RMHKGRFQSLRERIWKKILQWGENYLSSGGKEVLIKAVIQAIPVYVMGIFKLPESVCEDL 1187
Query: 120 VRIQREFLWGGVNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISLLAKWRWRLVQPEQ 179
+ R F WG G +K W W ++ + GGLG +D++L N +LLA+ WRL+
Sbjct: 1188 TSLTRNFWWGAEKGKRKTHWKAWKSLTKSKSLGGLGFKDIRLFNQALLARQAWRLIDNPD 1247
Query: 180 PL*KDVLRCKY---GSRINFLLDPIYNSWSSLASRWWLDLMSLEGVVGANWFNREMVRKV 236
L VL+ KY GS ++ S+ AS W + G + ++ ++
Sbjct: 1248 SLCARVLKAKYYPNGSIVD-------TSFGGNASPGWQAIEH-----GLELVKKGIIWRI 1295
Query: 237 GNGETTRFWLDHWVGNEALYLTFPRLFSISSQKEAMVGEVWVD-GDWNLTWRRSLFVWEE 295
GNG + R W D W+ + L+ + ++ + V ++ +D G W+ +F+ +
Sbjct: 1296 GNGRSVRVWQDPWLPRD---LSRRPITPKNNCRIKWVADLMLDNGMWDANKINQIFLPVD 1352
Query: 296 GLIHILLDELEGREVSEPMDSWWWKLEEGGICSVSSSYSL---HVKLQMPPEPLENTKAM 352
+ I+L E D W ++ G SV ++Y L K + +
Sbjct: 1353 --VEIILKLRTSSRDEE--DFIAWHPDKLGNFSVRTAYRLAENWAKEEASSSSSDVNIRK 1408
Query: 353 VFGSVWKSPTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGETAAHLF 412
+ +WK P KV +F+W++ + +PT DN KR + E S CV C E H
Sbjct: 1409 AWELLWKCNVPSKVKIFTWRATSNCLPTWDNKKKRNL---EISDTCVICGMEKEDTMHAL 1465
Query: 413 LHCDMSFKVW 422
C + +W
Sbjct: 1466 CRCPQAKHLW 1475
>UniRef100_Q9LVQ1 Non-LTR retroelement reverse transcriptase-like [Arabidopsis
thaliana]
Length = 1072
Score = 150 bits (380), Expect = 5e-35
Identities = 129/429 (30%), Positives = 189/429 (43%), Gaps = 42/429 (9%)
Query: 12 SGLKVNFAKSCLI--GVNVGREFMDAACNFMNCREGSPPFQYLGLPVGANPRRLSTWEPL 69
SGLKVN KS L G+++ AA F G+ P +YLGLP+ R++ + PL
Sbjct: 586 SGLKVNKDKSQLFQAGLDLSERITSAAYGFP---AGTFPIRYLGLPLMCRKLRIADYGPL 642
Query: 70 LDVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKLPVKVWKRVVRIQREFLWG 129
L+ L RL SW +K +S GR L+++V+ + NF+++ F LP K++ + +FLW
Sbjct: 643 LEKLSARLRSWVSKALSFAGRTQLISSVIFGLINFWMSTFLLPKGCIKKIESLCSKFLWA 702
Query: 130 GVNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISLLAKWRWRLVQPEQPL*KDVLRCK 189
G G+K V W CLP+ +GGLG R N +LL + W L + L R
Sbjct: 703 GSIDGRKSSKVSWVDCCLPKSEGGLGFRSFGEWNKTLLLRLIWVLFDRDTSLWAQWQRHH 762
Query: 190 YGSRINFLLDPIYNSWSSLASR---W-WLDLMSLEGVVGANWFNREMVRKVGNGETTRFW 245
+F W A + W W L++L + + + KVGNG T FW
Sbjct: 763 RLGHASF--------WQVNALQTDPWTWKMLLNLRPLA-----EKFIKAKVGNGGTVSFW 809
Query: 246 LDHW--VGNEALYLTFPRLFSISSQKEAMVGEVWVDGDWNLTWRRSLFVWEEGLIHILLD 303
D W +G YL + A V + W L RSL +L
Sbjct: 810 FDCWTSLGPLIKYLGDVGSRPLRIPFSAKVADAIDGSGWRLPLSRSL------TADSILS 863
Query: 304 ELEGREVSEPM---DSWWWKLEEGGICSVSSSYSLHVKLQMPPEPLENTKAMVFGSVWKS 360
L P+ DS+ W +++ S++ + V P P++ SVW
Sbjct: 864 HLASLPPPSPLMVSDSYSWCVDDVDCQGFSAAKTWEV--LRPRRPVKRWAK----SVWFK 917
Query: 361 PTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGETAAHLFLHCDMSFK 420
K W + L+R+PT+ L+ ++ SS C C ET HL L CD S +
Sbjct: 918 GAVPKHAFNFWTAQLNRLPTRQRLVSWGLV---SSAECCLCSFDTETRDHLLLLCDFSSQ 974
Query: 421 VWSMVYCRL 429
VW MV+ RL
Sbjct: 975 VWRMVFLRL 983
>UniRef100_Q9C6L3 Hypothetical protein F2J7.11 [Arabidopsis thaliana]
Length = 1213
Score = 147 bits (372), Expect = 4e-34
Identities = 127/431 (29%), Positives = 195/431 (44%), Gaps = 31/431 (7%)
Query: 5 LRGFEMASGLKVNFAKS--CLIGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGANPRR 62
L F SGLKVN KS L G+N +AA F G+ P +YLGLP+ R
Sbjct: 719 LDDFASWSGLKVNKDKSHLYLAGLNQLESNANAAYGFPI---GTLPIRYLGLPLMNRKLR 775
Query: 63 LSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKLPVKVWKRVVRI 122
++ +EPLL+ + R SW NK +S GR+ L+++V+ NF+++ F LP KR+ +
Sbjct: 776 IAEYEPLLEKITARFRSWVNKCLSFAGRIQLISSVIFGSINFWMSTFLLPKGCIKRIESL 835
Query: 123 QREFLWGGVNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISLLAKWRWRLVQPEQPL* 182
FLW G K V WA +CLP+ +GGLG+R + N +L + WRL + L
Sbjct: 836 CSRFLWSGNIEQAKGIKVSWAALCLPKSEGGLGLRRLLEWNKTLSMRLIWRLFVAKDSLW 895
Query: 183 KDVLRCKYGSRINF--LLDPIYNSWSSLASRWWLDLMSLEGVVGANWFNREMVRKVGNGE 240
D + SR +F + +SW+ W L+SL + ++ +V KVGNG
Sbjct: 896 ADWQHLHHLSRGSFWAVEGGQSDSWT------WKRLLSLRPLA-----HQFLVCKVGNGL 944
Query: 241 TTRFWLDHWVGNEALYLTFPRL--FSISSQKEAMVGEVWVDGDWNLTWRRSLFVWEEGLI 298
+W D+W L+ + S+ A V + + W L RS +G+
Sbjct: 945 KADYWYDNWTSLGPLFRIIGDIGPSSLRVPLLAKVASAFSEDGWRLPVSRS--APAKGIH 1002
Query: 299 HILLDELEGREVSEPMDSWWWKLEEGGICSVSSSYSLHVKLQMPPEPLENTKAMVFGSVW 358
L E +D + W + G +C S+ K P K+ S+W
Sbjct: 1003 DHLCTVPVPSTAQEDVDRYEWSV-NGFLCQGFSA----AKTWEAIRPKATVKSWA-SSIW 1056
Query: 359 KSPTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGETAAHLFLHCDMS 418
K W S L+R+ T+ L + +++ CV C E+ HL L C+ S
Sbjct: 1057 FKGAVPKYAFNMWVSHLNRLLTRQRLASWGHIQSDA---CVLCSFASESRDHLLLICEFS 1113
Query: 419 FKVWSMVYCRL 429
+VW +V+ R+
Sbjct: 1114 AQVWRLVFRRI 1124
>UniRef100_Q75M12 Hypothetical protein P0676G05.2 [Oryza sativa]
Length = 1936
Score = 147 bits (370), Expect = 7e-34
Identities = 112/429 (26%), Positives = 195/429 (45%), Gaps = 28/429 (6%)
Query: 1 LKALLRGFEMASGLKVNFAK-SCLIGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGAN 59
+K +L + +G +N AK S L G + N + + +YLG P
Sbjct: 1339 VKEVLTNYAQGTGQLINPAKCSILFGEASPSSVSEDIRNTLQVERDNFEDRYLGFPTPEG 1398
Query: 60 PRRLSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKLPVKVWKRV 119
++ L + KR+ WG ++S GG+ +L+ AV+ AIP + + FK P V+ +
Sbjct: 1399 RMHKGRFQSLQAKIAKRVIQWGENFLSSGGKEILIKAVIQAIPVYVMGLFKFPDSVYDEL 1458
Query: 120 VRIQREFLWGGVNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISLLAKWRWRLVQPEQ 179
++ R F WG NG ++ W W ++ + GGLG RD KL N +LL + WRL++
Sbjct: 1459 TKMTRNFWWGADNGRRRTHWRAWDSLTKAKINGGLGFRDYKLFNQALLTRQAWRLIEFPN 1518
Query: 180 PL*KDVLRCKYGSRINFLLDPIYNSWSSLASRWWLDLMSLEGVVGANWFNREMVRKVGNG 239
L VL+ KY L D ++S+ AS W + G + + ++ ++GNG
Sbjct: 1519 SLCAQVLKAKYFPH-GSLTD---TTFSANASPTWHGI-----EYGLDLLKKGIIWRIGNG 1569
Query: 240 ETTRFWLDHWVGNEALYLTFPRLFSISSQKEAMVGE-VWVDGDWNLTWRRSLFVWEEGLI 298
+ R W D W+ + L+ + S ++ + V + + DG W+ F +
Sbjct: 1570 NSVRIWRDPWIPRD---LSRRPVSSKANCRLKWVSDLIAEDGTWDSAKINQYF------L 1620
Query: 299 HILLDELEGREVSEPMDSWW--WKLEEGGICSVSSSYSLHVKLQMPPEPLENTKAMVFGS 356
I D ++ +S ++ + W ++ G SV S+Y L ++L ++ + + S
Sbjct: 1621 KIDADIIQKICISARLEEDFIAWHPDKTGRFSVRSAYKLALQLADMNNCSSSSSSRLNKS 1680
Query: 357 ---VWKSPTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGETAAHLFL 413
+WK P KV +F+W+ + + T +N KR + E C CD+ E A H
Sbjct: 1681 WELIWKCNVPQKVRIFAWRVASNSLATMENKKKRNL---ERFDVCGICDREKEDAGHALC 1737
Query: 414 HCDMSFKVW 422
C + +W
Sbjct: 1738 RCVHANSLW 1746
>UniRef100_Q9SJ38 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1229
Score = 145 bits (367), Expect = 2e-33
Identities = 108/435 (24%), Positives = 196/435 (44%), Gaps = 42/435 (9%)
Query: 1 LKALLRGFEMASGLKVNFAKSCL-IGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGAN 59
L +L + ASG +NF KS + R + R+ +YLGLP
Sbjct: 594 LSEILSRYGKASGQSINFHKSSVTFSSKTPRSVKGQVKRILKIRKEGGTGKYLGLPEHFG 653
Query: 60 PRRLSTWEPLLDVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKLPVKVWKRV 119
R+ + ++D ++++ +SW ++++S G+ V+L AVL ++P + ++ FKLP + +++
Sbjct: 654 RRKRDIFGAIIDKIRQKSHSWASRFLSQAGKQVMLKAVLASMPLYSMSCFKLPSALCRKI 713
Query: 120 VRIQREFLWGGVNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISLLAKWRWRLVQPEQ 179
+ F W +K WV W+ + P++ GGLG RD++ N SLLAK WRL+ +
Sbjct: 714 QSLLTRFWWDTKPDVRKTSWVAWSKLTNPKNAGGLGFRDIERCNDSLLAKLGWRLLNSPE 773
Query: 180 PL*KDVLRCKYGSRINFLLDPIYNSWSSLASRWWLDLMSLEGVV--GANWFNREMVRKVG 237
L +L KY +F+ + S S W +++ ++ G W +
Sbjct: 774 SLLSRILLGKYCHSSSFMECKL----PSQPSHGWRSIIAGREILKEGLGWL-------IT 822
Query: 238 NGETTRFWLDHWVG-NEALYLTFPRLFSISSQK-EAMVGEVWVDGDWNLTWRRSLFVWEE 295
NGE W D W+ ++ L P L + A++ + + DWN
Sbjct: 823 NGEKVSIWNDPWLSISKPLVPIGPALREHQDLRVSALINQNTLQWDWN------------ 870
Query: 296 GLIHILLDELEG--REVSEP----MDSWWWKLEEGGICSVSSSYSLHVKLQMPPEPLENT 349
I ++L E +++ P +D W + G + S Y + + P+ T
Sbjct: 871 -KIAVILPNYENLIKQLPAPSSRGVDKLAWLPVKSGQYTSRSGYGI---ASVASIPIPQT 926
Query: 350 KAMVFGSVWKSPTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGETAA 409
+ ++WK T K+ W++ ++ +P L++R I P+ + RC E+
Sbjct: 927 QFNWQSNLWKLQTLPKIKHLMWKAAMEALPVGIQLVRRHISPSAACHRC----GAPESTT 982
Query: 410 HLFLHCDMSFKVWSM 424
HLF HC+ + +VW +
Sbjct: 983 HLFFHCEFAAQVWEL 997
>UniRef100_O22220 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1094
Score = 145 bits (365), Expect = 3e-33
Identities = 112/425 (26%), Positives = 186/425 (43%), Gaps = 36/425 (8%)
Query: 12 SGLKVNFAKSCL-IGVNVGREFMDAACNFMNCREGSPPFQYLGLPVGANPRRLSTWEPLL 70
SG +N KS + G V E N + +YLGLP + + +
Sbjct: 439 SGQMINSEKSAITFGAKVNEETKQWIMNRSGIQTEGGTGKYLGLPECFQGSKQVLFGFIK 498
Query: 71 DVLKKRLNSWGNKYVSLGGRVVLLNAVLNAIPNFYLAFFKLPVKVWKRVVRIQREFLWGG 130
+ L+ RL+ W K +S GG+ +LL ++ A P + + F+L + ++ + +F W
Sbjct: 499 EKLQSRLSGWYAKTLSQGGKDILLKSIAMAFPVYAMTCFRLSKTLCTKLTSVMMDFWWNS 558
Query: 131 VNGGKKVCWVKWATVCLPRDKGGLGVRDMKLVNISLLAKWRWRLVQPEQPL*KDVLRCKY 190
V KK+ W+ + LP+ GG G +D++ N +LLAK RL L +L+ +Y
Sbjct: 559 VQDKKKIHWIGAQKLMLPKFLGGFGFKDLQCFNQALLAKQASRLHTDSDSLLSQILKSRY 618
Query: 191 GSRINFL-----LDPIYNSWSSLASRWWLDLMSLEGVVGANWFNREMVRKVGNGETTRFW 245
+FL P Y +W S+ + G + + +GNGE T W
Sbjct: 619 YMNSDFLSATKGTRPSY-AWQSI-------------LYGRELLVSGLKKIIGNGENTYVW 664
Query: 246 LDHWVGNEALYLTFPRLFSISSQKEAMVGEVW--VDGDWNLTWRRSLFVWEEGLIHILLD 303
+D+W+ ++ P I + V ++ +WNL R LF W+E I I+
Sbjct: 665 MDNWIFDDK--PRRPESLQIMVDIQLKVSQLIDPFSRNWNLNMLRDLFPWKE--IQIICQ 720
Query: 304 ELEGREVSEPMDSWWWKLEEGGICSVSSSYSL---HVKLQMPPEPLENTKA-MVFGSVWK 359
+ R ++ DS+ W G+ +V S Y L V QM E E +FG +W
Sbjct: 721 Q---RPMASRQDSFCWFGTNHGLYTVKSEYDLCSRQVHKQMFKEAEEQPSLNPLFGKIWN 777
Query: 360 SPTPFKVVVFSWQSLLDRIPTKDNLLKRRILPTESSVRCVFCDQVGETAAHLFLHCDMSF 419
+ K+ VF W+ L + +D L R +L + C C + ET H+ C ++
Sbjct: 778 LNSAPKIKVFLWKVLKGAVAVEDRLRTRGVLIEDG---CSMCPEKNETLNHILFQCPLAR 834
Query: 420 KVWSM 424
+VW++
Sbjct: 835 QVWAL 839
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.325 0.141 0.475
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 798,973,072
Number of Sequences: 2790947
Number of extensions: 35630965
Number of successful extensions: 83914
Number of sequences better than 10.0: 433
Number of HSP's better than 10.0 without gapping: 240
Number of HSP's successfully gapped in prelim test: 194
Number of HSP's that attempted gapping in prelim test: 82959
Number of HSP's gapped (non-prelim): 712
length of query: 430
length of database: 848,049,833
effective HSP length: 130
effective length of query: 300
effective length of database: 485,226,723
effective search space: 145568016900
effective search space used: 145568016900
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 76 (33.9 bits)
Medicago: description of AC146564.8


