

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146564.2 - phase: 0
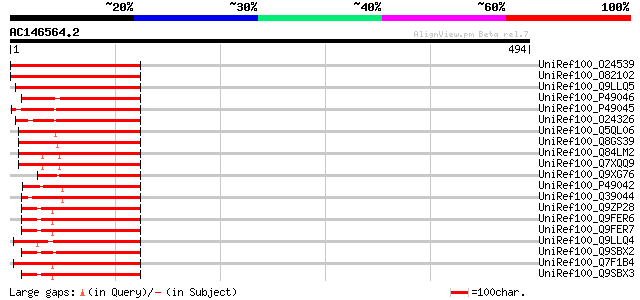
(494 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O24539 Cysteine proteinase precursor [Vicia narbonensis] 206 1e-51
UniRef100_O82102 Cysteine proteinase precursor [Vicia sativa] 205 2e-51
UniRef100_Q9LLQ5 Seed maturation protein PM40 [Glycine max] 187 5e-46
UniRef100_P49046 Legumain precursor [Canavalia ensiformis] 183 1e-44
UniRef100_P49045 Vacuolar processing enzyme precursor [Glycine max] 182 3e-44
UniRef100_O24326 Vacuolar processing enzyme precursor [Phaseolus... 174 7e-42
UniRef100_Q5QL06 Vacuolar processing enzyme 1 [Zea mays] 169 1e-40
UniRef100_Q8GS39 Asparaginyl endopeptidase REP-2 [Oryza sativa] 168 3e-40
UniRef100_Q84LM2 Vacuolar processing enzyme [Oryza sativa] 159 2e-37
UniRef100_Q7XQQ9 OSJNBa0091D06.13 protein [Oryza sativa] 159 2e-37
UniRef100_Q9XG76 Putative preprolegumain [Nicotiana tabacum] 154 4e-36
UniRef100_P49042 Vacuolar processing enzyme precursor [Ricinus c... 154 4e-36
UniRef100_Q39044 Vacuolar processing enzyme, beta-isozyme precur... 154 6e-36
UniRef100_Q9ZP28 C13 endopeptidase NP1 precursor [Zea mays] 150 1e-34
UniRef100_Q9FER6 Putative legumain precursor [Zea mays] 150 1e-34
UniRef100_Q9FER7 Putative legumain precursor [Zea mays] 149 1e-34
UniRef100_Q9LLQ4 Asparaginyl endopeptidase [Sesamum indicum] 148 3e-34
UniRef100_Q9SBX2 Legumain-like protease precursor [Zea mays] 145 3e-33
UniRef100_Q7F1B4 Asparaginyl endopeptidase [Oryza sativa] 145 3e-33
UniRef100_Q9SBX3 Legumain-like protease precursor [Zea mays] 144 6e-33
>UniRef100_O24539 Cysteine proteinase precursor [Vicia narbonensis]
Length = 488
Score = 206 bits (525), Expect = 1e-51
Identities = 103/125 (82%), Positives = 109/125 (86%), Gaps = 1/125 (0%)
Query: 1 MFSIVLSLWSWLLLLLTLDGLVA-RPNHLEWDPVIRLPGEVVDDAEVDEVGTRWAVLVAG 59
MFS VL+ W LLLL +L G VA RPN LEW+PVIRLPGE VD DE+GTRWAVLVAG
Sbjct: 1 MFSDVLASWLVLLLLSSLHGSVAARPNRLEWEPVIRLPGEPVDADVEDEMGTRWAVLVAG 60
Query: 60 SSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVY 119
S+GYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIA NE+NPRPGVIINHPQGPNVY
Sbjct: 61 SNGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIAYNEMNPRPGVIINHPQGPNVY 120
Query: 120 VGVPK 124
GVPK
Sbjct: 121 DGVPK 125
>UniRef100_O82102 Cysteine proteinase precursor [Vicia sativa]
Length = 503
Score = 205 bits (522), Expect = 2e-51
Identities = 102/125 (81%), Positives = 108/125 (85%), Gaps = 1/125 (0%)
Query: 1 MFSIVLSLWSWLLLLLT-LDGLVARPNHLEWDPVIRLPGEVVDDAEVDEVGTRWAVLVAG 59
MFS VL+ W LLL L+ L G VARPN LEW+PVIRLPGE VD DE+GTRWAVLVAG
Sbjct: 15 MFSDVLASWLVLLLFLSSLHGSVARPNRLEWEPVIRLPGEPVDADVEDEIGTRWAVLVAG 74
Query: 60 SSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVY 119
S+GYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIA +E NPRPGVIINHPQGPNVY
Sbjct: 75 SNGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIAYSEFNPRPGVIINHPQGPNVY 134
Query: 120 VGVPK 124
GVPK
Sbjct: 135 DGVPK 139
>UniRef100_Q9LLQ5 Seed maturation protein PM40 [Glycine max]
Length = 496
Score = 187 bits (476), Expect = 5e-46
Identities = 90/120 (75%), Positives = 99/120 (82%), Gaps = 1/120 (0%)
Query: 6 LSLWSWLLL-LLTLDGLVARPNHLEWDPVIRLPGEVVDDAEVDEVGTRWAVLVAGSSGYG 64
L LWSW+LL ++ G AR N EWD VI+LP E VD EVGTRWAVLVAGS+GYG
Sbjct: 13 LVLWSWMLLRMMMAQGAAARANRKEWDSVIKLPAEPVDADSDHEVGTRWAVLVAGSNGYG 72
Query: 65 NYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
NYRHQADVCHAYQLLIKGG+KEENIVVFMYDDIA +ELNPRPGVIINHP+G +VY GVPK
Sbjct: 73 NYRHQADVCHAYQLLIKGGLKEENIVVFMYDDIATDELNPRPGVIINHPEGQDVYAGVPK 132
>UniRef100_P49046 Legumain precursor [Canavalia ensiformis]
Length = 475
Score = 183 bits (464), Expect = 1e-44
Identities = 89/113 (78%), Positives = 97/113 (85%), Gaps = 4/113 (3%)
Query: 12 LLLLLTLDGLVARPNHLEWDPVIRLPGEVVDDAEVDEVGTRWAVLVAGSSGYGNYRHQAD 71
+L++L+L G AR N EWD VI+LP E VDD EVGTRWAVLVAGS+GYGNYRHQAD
Sbjct: 4 MLVMLSLHGTAARLNRREWDSVIQLPTEPVDD----EVGTRWAVLVAGSNGYGNYRHQAD 59
Query: 72 VCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
VCHAYQLLIKGGVKEENIVVFMYDDIA N +NPRPGVIINHPQGP+VY GVPK
Sbjct: 60 VCHAYQLLIKGGVKEENIVVFMYDDIAYNAMNPRPGVIINHPQGPDVYAGVPK 112
>UniRef100_P49045 Vacuolar processing enzyme precursor [Glycine max]
Length = 495
Score = 182 bits (461), Expect = 3e-44
Identities = 90/124 (72%), Positives = 104/124 (83%), Gaps = 6/124 (4%)
Query: 2 FSIVLSLWSWLLLLLT-LDGLVARPNHLEWDPVIRLPGEVVDDAEVDEVGTRWAVLVAGS 60
+S+VL W++++L + G ARPN EWD VI+LP E VD A+ DEVGTRWAVLVAGS
Sbjct: 14 YSVVL----WMMVVLVRVHGAAARPNRKEWDSVIKLPTEPVD-ADSDEVGTRWAVLVAGS 68
Query: 61 SGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYV 120
+GYGNYRHQADVCHAYQLLIKGG+KEENIVVFMYDDIA NELNPR GVIINHP+G ++Y
Sbjct: 69 NGYGNYRHQADVCHAYQLLIKGGLKEENIVVFMYDDIATNELNPRHGVIINHPEGEDLYA 128
Query: 121 GVPK 124
GVPK
Sbjct: 129 GVPK 132
>UniRef100_O24326 Vacuolar processing enzyme precursor [Phaseolus vulgaris]
Length = 493
Score = 174 bits (440), Expect = 7e-42
Identities = 86/119 (72%), Positives = 99/119 (82%), Gaps = 4/119 (3%)
Query: 6 LSLWSWLLLLLTLDGLVARPNHLEWDPVIRLPGEVVDDAEVDEVGTRWAVLVAGSSGYGN 65
++ W W+L+++ + N E D VI+LP + VD AE DEVGTRWAVLVAGS+GYGN
Sbjct: 16 VAFWWWMLVMVMR---IQGTNGKEQDSVIKLPTQEVD-AESDEVGTRWAVLVAGSNGYGN 71
Query: 66 YRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
YRHQADVCHAYQLLIKGGVKEENIVVFMYDDIA +ELNPRPGVIIN+PQGP+VY GVPK
Sbjct: 72 YRHQADVCHAYQLLIKGGVKEENIVVFMYDDIATHELNPRPGVIINNPQGPDVYAGVPK 130
>UniRef100_Q5QL06 Vacuolar processing enzyme 1 [Zea mays]
Length = 494
Score = 169 bits (429), Expect = 1e-40
Identities = 86/128 (67%), Positives = 95/128 (74%), Gaps = 12/128 (9%)
Query: 9 WSWLLLLLTLDGLVARPNHLEWDPVIRLPGEVVD-----------DAEVDE-VGTRWAVL 56
W LLL+L L A EWDPVIR+PGE D EVD+ VGTRWAVL
Sbjct: 5 WCLLLLVLVLAAAAACAEKGEWDPVIRMPGEKEPAGSHSHSGEGFDGEVDDAVGTRWAVL 64
Query: 57 VAGSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGP 116
VAGSSGYGNYRHQAD+CHAYQ+L KGG+KEENIVVFMYDDIAN+ LNPR GVIINHP+G
Sbjct: 65 VAGSSGYGNYRHQADICHAYQILQKGGIKEENIVVFMYDDIANSALNPRQGVIINHPEGE 124
Query: 117 NVYVGVPK 124
+VY GVPK
Sbjct: 125 DVYAGVPK 132
>UniRef100_Q8GS39 Asparaginyl endopeptidase REP-2 [Oryza sativa]
Length = 496
Score = 168 bits (426), Expect = 3e-40
Identities = 84/130 (64%), Positives = 93/130 (70%), Gaps = 14/130 (10%)
Query: 9 WSWLLLLLTLDGLVARPNHLEWDPVIRLPGEVVDDA--------------EVDEVGTRWA 54
W + LLL W+PVIR+PGEVV++ E D VGTRWA
Sbjct: 5 WCFALLLALSAAAAGAGAKRTWEPVIRMPGEVVEEEVATVPRGSEGTEEEEKDGVGTRWA 64
Query: 55 VLVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQ 114
VLVAGSSGYGNYRHQADVCHAYQ+L KGG+KEENIVVFMYDDIANN LNPRPGVI+NHPQ
Sbjct: 65 VLVAGSSGYGNYRHQADVCHAYQILRKGGLKEENIVVFMYDDIANNILNPRPGVIVNHPQ 124
Query: 115 GPNVYVGVPK 124
G +VY GVPK
Sbjct: 125 GEDVYAGVPK 134
>UniRef100_Q84LM2 Vacuolar processing enzyme [Oryza sativa]
Length = 497
Score = 159 bits (402), Expect = 2e-37
Identities = 82/129 (63%), Positives = 91/129 (69%), Gaps = 13/129 (10%)
Query: 9 WSWLLLLLTLDGLVARPNHLE---WDPVIRLPGEVVDDAEV----------DEVGTRWAV 55
W W ++ L A E W+P+IR+P E DDAE D GTRWAV
Sbjct: 6 WVWGFVVALLAVAAAADGEEEEGKWEPLIRMPTEEGDDAEAAAPAPAPAAADYGGTRWAV 65
Query: 56 LVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQG 115
LVAGSSGYGNYRHQADVCHAYQ+L KGGVKEENIVVFMYDDIA+N LNPRPG IINHP+G
Sbjct: 66 LVAGSSGYGNYRHQADVCHAYQILQKGGVKEENIVVFMYDDIAHNILNPRPGTIINHPKG 125
Query: 116 PNVYVGVPK 124
+VY GVPK
Sbjct: 126 GDVYAGVPK 134
>UniRef100_Q7XQQ9 OSJNBa0091D06.13 protein [Oryza sativa]
Length = 517
Score = 159 bits (402), Expect = 2e-37
Identities = 82/129 (63%), Positives = 91/129 (69%), Gaps = 13/129 (10%)
Query: 9 WSWLLLLLTLDGLVARPNHLE---WDPVIRLPGEVVDDAEV----------DEVGTRWAV 55
W W ++ L A E W+P+IR+P E DDAE D GTRWAV
Sbjct: 6 WVWGFVVALLAVAAAADGEEEEGKWEPLIRMPTEEGDDAEAAAPAPAPAAADYGGTRWAV 65
Query: 56 LVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQG 115
LVAGSSGYGNYRHQADVCHAYQ+L KGGVKEENIVVFMYDDIA+N LNPRPG IINHP+G
Sbjct: 66 LVAGSSGYGNYRHQADVCHAYQILQKGGVKEENIVVFMYDDIAHNILNPRPGTIINHPKG 125
Query: 116 PNVYVGVPK 124
+VY GVPK
Sbjct: 126 GDVYAGVPK 134
>UniRef100_Q9XG76 Putative preprolegumain [Nicotiana tabacum]
Length = 494
Score = 154 bits (390), Expect = 4e-36
Identities = 72/98 (73%), Positives = 84/98 (85%), Gaps = 1/98 (1%)
Query: 27 HLEWDPVIRLPGEVVDDAEVDEVGTRWAVLVAGSSGYGNYRHQADVCHAYQLLIKGGVKE 86
H WDP+IR P + D++E D+ G RWAVLVAGS+GYGNYRHQADVCHAYQ+L +GG+K+
Sbjct: 36 HRWWDPLIRSPVDRDDESE-DKDGVRWAVLVAGSNGYGNYRHQADVCHAYQILKRGGLKD 94
Query: 87 ENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
ENIVVFMYDDIA +ELNPRPGVIINHP G +VY GVPK
Sbjct: 95 ENIVVFMYDDIAKSELNPRPGVIINHPNGSDVYAGVPK 132
>UniRef100_P49042 Vacuolar processing enzyme precursor [Ricinus communis]
Length = 497
Score = 154 bits (390), Expect = 4e-36
Identities = 80/116 (68%), Positives = 91/116 (77%), Gaps = 6/116 (5%)
Query: 13 LLLLTLDGLVA-RPNHLEWDPVIRLPGEVVDDAEVDE---VGTRWAVLVAGSSGYGNYRH 68
L L + GL+A R N E P I +P E + +VD+ +GTRWAVLVAGS G+GNYRH
Sbjct: 21 LSFLPIPGLLASRLNPFE--PGILMPTEEAEPVQVDDDDQLGTRWAVLVAGSMGFGNYRH 78
Query: 69 QADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
QADVCHAYQLL KGG+KEENI+VFMYDDIA NELNPRPGVIINHPQG +VY GVPK
Sbjct: 79 QADVCHAYQLLRKGGLKEENIIVFMYDDIAKNELNPRPGVIINHPQGEDVYAGVPK 134
>UniRef100_Q39044 Vacuolar processing enzyme, beta-isozyme precursor [Arabidopsis
thaliana]
Length = 486
Score = 154 bits (389), Expect = 6e-36
Identities = 79/115 (68%), Positives = 90/115 (77%), Gaps = 4/115 (3%)
Query: 12 LLLLLTLDGLVARPNHLEWDPVIRLPGEVVDDAEVDE--VGTRWAVLVAGSSGYGNYRHQ 69
LLLLL L LV + ++P I +P E + A+ DE VGTRWAVLVAGSSGYGNYRHQ
Sbjct: 11 LLLLLVL--LVHAESRGRFEPKILMPTEEANPADQDEDGVGTRWAVLVAGSSGYGNYRHQ 68
Query: 70 ADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
ADVCHAYQ+L KGG+KEENIVV MYDDIAN+ LNPRPG +INHP G +VY GVPK
Sbjct: 69 ADVCHAYQILRKGGLKEENIVVLMYDDIANHPLNPRPGTLINHPDGDDVYAGVPK 123
>UniRef100_Q9ZP28 C13 endopeptidase NP1 precursor [Zea mays]
Length = 485
Score = 150 bits (378), Expect = 1e-34
Identities = 75/115 (65%), Positives = 88/115 (76%), Gaps = 5/115 (4%)
Query: 12 LLLLLTLDGLVARPNHLEWDPVIRLPGE--VVDDAEVDEVGTRWAVLVAGSSGYGNYRHQ 69
LLL L ARP +P IRLP E D+ + D VGTRWAVL+AGS+GY NYRHQ
Sbjct: 10 LLLSACLCSAWARPR---LEPTIRLPSERAAADETDDDAVGTRWAVLIAGSNGYYNYRHQ 66
Query: 70 ADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
AD+CHAYQ++ KGG+K+ENIVVFMYDDIA++ NPRPGVIINHPQG +VY GVPK
Sbjct: 67 ADICHAYQIMKKGGLKDENIVVFMYDDIAHSPENPRPGVIINHPQGGDVYAGVPK 121
>UniRef100_Q9FER6 Putative legumain precursor [Zea mays]
Length = 485
Score = 150 bits (378), Expect = 1e-34
Identities = 75/115 (65%), Positives = 88/115 (76%), Gaps = 5/115 (4%)
Query: 12 LLLLLTLDGLVARPNHLEWDPVIRLPGE--VVDDAEVDEVGTRWAVLVAGSSGYGNYRHQ 69
LLL L ARP +P IRLP E D+ + D VGTRWAVL+AGS+GY NYRHQ
Sbjct: 10 LLLSACLCSAWARPR---LEPTIRLPSERAAADETDDDAVGTRWAVLIAGSNGYYNYRHQ 66
Query: 70 ADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
AD+CHAYQ++ KGG+K+ENIVVFMYDDIA++ NPRPGVIINHPQG +VY GVPK
Sbjct: 67 ADICHAYQIMKKGGLKDENIVVFMYDDIAHSPENPRPGVIINHPQGGDVYAGVPK 121
>UniRef100_Q9FER7 Putative legumain precursor [Zea mays]
Length = 486
Score = 149 bits (377), Expect = 1e-34
Identities = 73/116 (62%), Positives = 89/116 (75%), Gaps = 6/116 (5%)
Query: 12 LLLLLTLDGLVARPNHLEWDPVIRLPGE---VVDDAEVDEVGTRWAVLVAGSSGYGNYRH 68
LLL + L ARP +P IRLP + D+ + +VGTRWAVL+AGSSGY NYRH
Sbjct: 10 LLLSVCLSSAWARPR---LEPAIRLPSQRAAAADETDDGDVGTRWAVLIAGSSGYYNYRH 66
Query: 69 QADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
QAD+CHAYQ++ KGG+K+ENI+VFMYDDIA++ NPRPGVIINHPQG +VY GVPK
Sbjct: 67 QADICHAYQIMKKGGLKDENIIVFMYDDIAHSPENPRPGVIINHPQGGDVYAGVPK 122
>UniRef100_Q9LLQ4 Asparaginyl endopeptidase [Sesamum indicum]
Length = 489
Score = 148 bits (374), Expect = 3e-34
Identities = 75/129 (58%), Positives = 93/129 (71%), Gaps = 11/129 (8%)
Query: 4 IVLSLWSWLLLLLTLDGLVARP--------NHLEWDPVIRLPGEVVDDAEVDEVGTRWAV 55
+ S ++ L++L L +VA P WDP+IR P +D E ++ TRWAV
Sbjct: 1 MAFSCFAGRLMILVLCVVVALPFAAAGGGRRSGPWDPIIRWP---LDRRETEDNATRWAV 57
Query: 56 LVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQG 115
LVAGS+G+GNYRHQADVCHAYQ+L KGG+++ENI+VFMYDDIA NELNPR GVIINHP G
Sbjct: 58 LVAGSNGFGNYRHQADVCHAYQILKKGGLRDENIIVFMYDDIAMNELNPRKGVIINHPTG 117
Query: 116 PNVYVGVPK 124
+VY GVPK
Sbjct: 118 GDVYAGVPK 126
>UniRef100_Q9SBX2 Legumain-like protease precursor [Zea mays]
Length = 481
Score = 145 bits (365), Expect = 3e-33
Identities = 74/113 (65%), Positives = 86/113 (75%), Gaps = 5/113 (4%)
Query: 12 LLLLLTLDGLVARPNHLEWDPVIRLPGEVVDDAEVDEVGTRWAVLVAGSSGYGNYRHQAD 71
LLL L ARP +P IRLP + A D VGTRWAVL+AGS+GY NYRHQAD
Sbjct: 10 LLLSACLCSAWARPR---LEPTIRLPSDRA--AADDAVGTRWAVLIAGSNGYYNYRHQAD 64
Query: 72 VCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
+CHAYQ++ KGG+K+ENIVVFMYDDIA++ NPRPGVIINHPQG +VY GVPK
Sbjct: 65 ICHAYQIMKKGGLKDENIVVFMYDDIAHSPENPRPGVIINHPQGGDVYAGVPK 117
>UniRef100_Q7F1B4 Asparaginyl endopeptidase [Oryza sativa]
Length = 501
Score = 145 bits (365), Expect = 3e-33
Identities = 77/129 (59%), Positives = 91/129 (69%), Gaps = 8/129 (6%)
Query: 4 IVLSLWSWLLLLLTLDGL-VARPNHLEWDPVIRLPGE-------VVDDAEVDEVGTRWAV 55
+VL + LLL L + VARP E +RLP E DDA GTRWAV
Sbjct: 9 LVLPPLAALLLFAHLAAVAVARPRWEEEGSNLRLPSERAVAAGAAADDAAEAAEGTRWAV 68
Query: 56 LVAGSSGYGNYRHQADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQG 115
L+AGS+GY NYRHQADVCHAYQ++ +GG+K+ENI+VFMYDDIA+N NPRPGVIINHPQG
Sbjct: 69 LIAGSNGYYNYRHQADVCHAYQIMKRGGLKDENIIVFMYDDIAHNPENPRPGVIINHPQG 128
Query: 116 PNVYVGVPK 124
+VY GVPK
Sbjct: 129 GDVYAGVPK 137
>UniRef100_Q9SBX3 Legumain-like protease precursor [Zea mays]
Length = 486
Score = 144 bits (363), Expect = 6e-33
Identities = 72/116 (62%), Positives = 87/116 (74%), Gaps = 6/116 (5%)
Query: 12 LLLLLTLDGLVARPNHLEWDPVIRLPGE---VVDDAEVDEVGTRWAVLVAGSSGYGNYRH 68
LLL + L ARP + IRLP + D+ + VGTRWAVL+AGSSGY NYRH
Sbjct: 10 LLLSVCLCSAWARPR---LETAIRLPSQRAAAADETDDGAVGTRWAVLIAGSSGYYNYRH 66
Query: 69 QADVCHAYQLLIKGGVKEENIVVFMYDDIANNELNPRPGVIINHPQGPNVYVGVPK 124
QAD+CHAYQ++ KGG+K+ENI+VFMYDDIA++ NPRPGVIINHPQG +VY GVPK
Sbjct: 67 QADICHAYQIMKKGGLKDENIIVFMYDDIAHSPENPRPGVIINHPQGGDVYAGVPK 122
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.348 0.155 0.614
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 831,768,131
Number of Sequences: 2790947
Number of extensions: 34086798
Number of successful extensions: 92000
Number of sequences better than 10.0: 132
Number of HSP's better than 10.0 without gapping: 129
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 91854
Number of HSP's gapped (non-prelim): 133
length of query: 494
length of database: 848,049,833
effective HSP length: 131
effective length of query: 363
effective length of database: 482,435,776
effective search space: 175124186688
effective search space used: 175124186688
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC146564.2


