

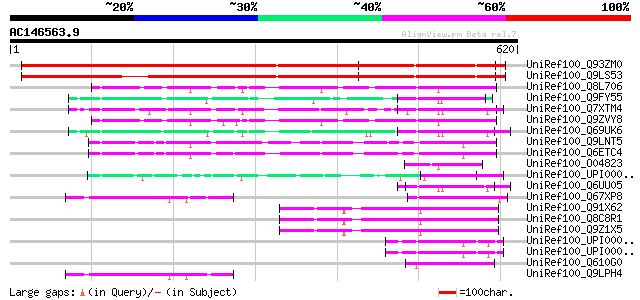
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146563.9 + phase: 0
(620 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q93ZM0 AT3g18370/MYF24_8 [Arabidopsis thaliana] 686 0.0
UniRef100_Q9LS53 Arabidopsis thaliana genomic DNA, chromosome 3,... 633 e-180
UniRef100_Q8L706 Ca2+-dependent lipid-binding protein, putative ... 91 1e-16
UniRef100_Q9FY55 CLB1-like protein [Arabidopsis thaliana] 89 4e-16
UniRef100_Q7XTM4 OSJNBa0033G05.21 protein [Oryza sativa] 89 5e-16
UniRef100_Q9ZVY8 T25N20.15 [Arabidopsis thaliana] 86 3e-15
UniRef100_Q69UK6 Putative C2 domain-containing protein [Oryza sa... 66 3e-09
UniRef100_Q9LNT5 T20H2.13 protein [Arabidopsis thaliana] 66 3e-09
UniRef100_Q6ETC4 Putative CLB1 protein [Oryza sativa] 65 8e-09
UniRef100_O04823 Hypothetical protein [Sporobolus stapfianus] 65 8e-09
UniRef100_UPI00003AE7D2 UPI00003AE7D2 UniRef100 entry 64 1e-08
UniRef100_Q6UU05 Putative Ca2+-dependent lipid-binding protein [... 64 1e-08
UniRef100_Q67XP8 Hypothetical protein At3g14590 [Arabidopsis tha... 64 1e-08
UniRef100_Q91X62 Membrane bound C2 domain containing protein [Mu... 64 2e-08
UniRef100_Q8C8R1 Mus musculus adult retina cDNA, RIKEN full-leng... 64 2e-08
UniRef100_Q9Z1X5 GLUT4 vesicle protein [Mus musculus] 64 2e-08
UniRef100_UPI0000365141 UPI0000365141 UniRef100 entry 62 4e-08
UniRef100_UPI0000365140 UPI0000365140 UniRef100 entry 62 4e-08
UniRef100_Q610G0 Hypothetical protein CBG17495 [Caenorhabditis b... 62 4e-08
UniRef100_Q9LPH4 T3F20.10 protein [Arabidopsis thaliana] 62 5e-08
>UniRef100_Q93ZM0 AT3g18370/MYF24_8 [Arabidopsis thaliana]
Length = 815
Score = 686 bits (1770), Expect = 0.0
Identities = 332/592 (56%), Positives = 444/592 (74%), Gaps = 3/592 (0%)
Query: 15 EVAVDFFNYVLQEKPKIPFFIPVILIACAVEKWVFSFSTWVPLALAVWATIQYGRYQRKL 74
E A +F N+++ E+ + +P++L A+E+WVF+FS WVPL +AVWA++QYG YQR L
Sbjct: 14 EAAREFINHLVAERHSLLLLVPLVLAFWAIERWVFAFSNWVPLVVAVWASLQYGSYQRAL 73
Query: 75 LVEDLDKKWKRIILNNSPITPLEHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRK 134
L EDL KKW++ + N S ITPLEHC+WLNKLL+EIW NY N KLS R S++VE RL+ R+
Sbjct: 74 LAEDLTKKWRQTVFNASTITPLEHCQWLNKLLSEIWLNYMNKKLSLRFSSMVEKRLRQRR 133
Query: 135 PRFLERVELQEFSLGSCPPSLALQGMRWSTIGDQRVMQLGFDWDTHEMSILLLAKLAKPL 194
R +E ++L EFSLGSCPP L L G WS G+Q++M+L F+WDT ++SILL AKL+ P
Sbjct: 134 SRLIENIQLLEFSLGSCPPLLGLHGTCWSKSGEQKIMRLDFNWDTTDLSILLQAKLSMPF 193
Query: 195 MGTARIVINSLHIKGDLIFTPILDGKALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGV 254
TARIV+NSL IKGD++ PIL+G+ALLYSFVS PEVR+GVAFG GG QSLPATE PGV
Sbjct: 194 NRTARIVVNSLCIKGDILIRPILEGRALLYSFVSNPEVRIGVAFGGGGGQSLPATELPGV 253
Query: 255 SSWLEKLFTDTLVKTMVEPRRRCFTLPAVDLRKKAVGGIIYVRVISANKLSSSSFKASRR 314
SSWL K+ T+TL K MVEPRR CF+LPA DL K A+GGIIYV V+S N L+ + S
Sbjct: 254 SSWLVKILTETLNKKMVEPRRGCFSLPATDLHKTAIGGIIYVTVVSGNNLNRRILRGSPS 313
Query: 315 QQSGSTNGSSEDVSDDKDLHTFVEVEIEELTRRTDVRLGSTPRWDAPFNMVLHDNTGTLR 374
+ S GSS + S K + TFVEVE+E+L+RRT+++ G P + + FNM+LHDNTGTL+
Sbjct: 314 KSSEIGEGSSGN-SSSKPVQTFVEVELEQLSRRTEMKSGPNPAYQSTFNMILHDNTGTLK 372
Query: 375 FNLYECIPNNVKCDYLGSCEIKLRHVEDDSTIMWAVGPDSGIIAKQAQFCGDEIEMVVPF 434
FNLYE P +V+ D L SCE+K+++V DDST+ WAVG D+G+IAK A+FCG EIEMVVPF
Sbjct: 373 FNLYENNPGSVRYDSLASCEVKMKYVGDDSTMFWAVGSDNGVIAKHAEFCGQEIEMVVPF 432
Query: 435 EGTNSGELKVSIVVKEWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKD 494
EG +SGEL V +++KEW FSDG+HSLN++ ++S SL+ SS + +TG+K+ +TV+ GK+
Sbjct: 433 EGVSSGELTVRLLLKEWHFSDGSHSLNSVNSSSLHSLDSSSALLSKTGRKIIVTVLAGKN 492
Query: 495 LAAAKEKTGKFDPYIKLQYGKVMQKTKTSHTPNPVWNQTIEFDEVGGGEYLKLKVFTEEL 554
L +K+K+GK D +KLQYGK++QKTK + VWNQ EF+E+ G EYLK+K + EE+
Sbjct: 493 L-VSKDKSGKCDASVKLQYGKIIQKTKIVNAAECVWNQKFEFEELAGEEYLKVKCYREEM 551
Query: 555 FGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIEAIKVDDQEGST 606
G +NIG+A ++L+G ++ S +W+PLE V SGEI L IEA+ + E +
Sbjct: 552 LGTDNIGTATLSLQG-INNSEMHIWVPLEDVNSGEIELLIEALDPEYSEADS 602
Score = 80.1 bits (196), Expect = 2e-13
Identities = 52/169 (30%), Positives = 88/169 (51%), Gaps = 25/169 (14%)
Query: 427 EIEMVVPFEGTNSGELKVSIVVKEWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKKLK 486
E+ + VP E NSGE+++ I + ++S+ +S + L ++
Sbjct: 571 EMHIWVPLEDVNSGEIELLIEALDPEYSEA---------DSSKGL-------------IE 608
Query: 487 ITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSH-TPNPVWNQTIEFDEVGGGEYL 545
+ +VE +DL AA + G DPY+++QYG+ Q+TK + T P WNQT+EF + G L
Sbjct: 609 LVLVEARDLVAADIR-GTSDPYVRVQYGEKKQRTKVIYKTLQPKWNQTMEFPDDGSSLEL 667
Query: 546 KLKVFTEELFGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKI 594
+K + L +IG+ V +GL D WI L+ V+ GE+ +++
Sbjct: 668 HVKDYNT-LLPTSSIGNCVVEYQGLKPNETADKWIILQGVKHGEVHVRV 715
>UniRef100_Q9LS53 Arabidopsis thaliana genomic DNA, chromosome 3, P1 clone: MYF24
[Arabidopsis thaliana]
Length = 786
Score = 633 bits (1632), Expect = e-180
Identities = 314/592 (53%), Positives = 423/592 (71%), Gaps = 33/592 (5%)
Query: 15 EVAVDFFNYVLQEKPKIPFFIPVILIACAVEKWVFSFSTWVPLALAVWATIQYGRYQRKL 74
E A +F N+++ E+ + +P++L A+E+WVF+FS WVPL +AVWA++QYG YQR L
Sbjct: 14 EAAREFINHLVAERHSLLLLVPLVLAFWAIERWVFAFSNWVPLVVAVWASLQYGSYQRAL 73
Query: 75 LVEDLDKKWKRIILNNSPITPLEHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRK 134
L EDL KKW++ + N S ITPLEHC+WLNKLL+EIW NY N KLS R S++VE RL+ R+
Sbjct: 74 LAEDLTKKWRQTVFNASTITPLEHCQWLNKLLSEIWLNYMNKKLSLRFSSMVEKRLRQRR 133
Query: 135 PRFLERVELQEFSLGSCPPSLALQGMRWSTIGDQRVMQLGFDWDTHEMSILLLAKLAKPL 194
R + ++M+L F+WDT ++SILL AKL+ P
Sbjct: 134 SRLI------------------------------KIMRLDFNWDTTDLSILLQAKLSMPF 163
Query: 195 MGTARIVINSLHIKGDLIFTPILDGKALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGV 254
TARIV+NSL IKGD++ PIL+G+ALLYSFVS PEVR+GVAFG GG QSLPATE PGV
Sbjct: 164 NRTARIVVNSLCIKGDILIRPILEGRALLYSFVSNPEVRIGVAFGGGGGQSLPATELPGV 223
Query: 255 SSWLEKLFTDTLVKTMVEPRRRCFTLPAVDLRKKAVGGIIYVRVISANKLSSSSFKASRR 314
SSWL K+ T+TL K MVEPRR CF+LPA DL K A+GGIIYV V+S N L+ + S
Sbjct: 224 SSWLVKILTETLNKKMVEPRRGCFSLPATDLHKTAIGGIIYVTVVSGNNLNRRILRGSPS 283
Query: 315 QQSGSTNGSSEDVSDDKDLHTFVEVEIEELTRRTDVRLGSTPRWDAPFNMVLHDNTGTLR 374
+ S GSS + S K + TFVEVE+E+L+RRT+++ G P + + FNM+LHDNTGTL+
Sbjct: 284 KSSEIGEGSSGN-SSSKPVQTFVEVELEQLSRRTEMKSGPNPAYQSTFNMILHDNTGTLK 342
Query: 375 FNLYECIPNNVKCDYLGSCEIKLRHVEDDSTIMWAVGPDSGIIAKQAQFCGDEIEMVVPF 434
FNLYE P +V+ D L SCE+K+++V DDST+ WAVG D+G+IAK A+FCG EIEMVVPF
Sbjct: 343 FNLYENNPGSVRYDSLASCEVKMKYVGDDSTMFWAVGSDNGVIAKHAEFCGQEIEMVVPF 402
Query: 435 EGTNSGELKVSIVVKEWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKD 494
EG +SGEL V +++KEW FSDG+HSLN++ ++S SL+ SS + +TG+K+ +TV+ GK+
Sbjct: 403 EGVSSGELTVRLLLKEWHFSDGSHSLNSVNSSSLHSLDSSSALLSKTGRKIIVTVLAGKN 462
Query: 495 LAAAKEKTGKFDPYIKLQYGKVMQKTKTSHTPNPVWNQTIEFDEVGGGEYLKLKVFTEEL 554
L +K+K+GK D +KLQYGK++QKTK + VWNQ EF+E+ G EYLK+K + EE+
Sbjct: 463 L-VSKDKSGKCDASVKLQYGKIIQKTKIVNAAECVWNQKFEFEELAGEEYLKVKCYREEM 521
Query: 555 FGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIEAIKVDDQEGST 606
G +NIG+A ++L+G ++ S +W+PLE V SGEI L IEA+ + E +
Sbjct: 522 LGTDNIGTATLSLQG-INNSEMHIWVPLEDVNSGEIELLIEALDPEYSEADS 572
Score = 80.1 bits (196), Expect = 2e-13
Identities = 52/169 (30%), Positives = 88/169 (51%), Gaps = 25/169 (14%)
Query: 427 EIEMVVPFEGTNSGELKVSIVVKEWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKKLK 486
E+ + VP E NSGE+++ I + ++S+ +S + L ++
Sbjct: 541 EMHIWVPLEDVNSGEIELLIEALDPEYSEA---------DSSKGL-------------IE 578
Query: 487 ITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSH-TPNPVWNQTIEFDEVGGGEYL 545
+ +VE +DL AA + G DPY+++QYG+ Q+TK + T P WNQT+EF + G L
Sbjct: 579 LVLVEARDLVAADIR-GTSDPYVRVQYGEKKQRTKVIYKTLQPKWNQTMEFPDDGSSLEL 637
Query: 546 KLKVFTEELFGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKI 594
+K + L +IG+ V +GL D WI L+ V+ GE+ +++
Sbjct: 638 HVKDYNT-LLPTSSIGNCVVEYQGLKPNETADKWIILQGVKHGEVHVRV 685
>UniRef100_Q8L706 Ca2+-dependent lipid-binding protein, putative [Arabidopsis
thaliana]
Length = 560
Score = 90.9 bits (224), Expect = 1e-16
Identities = 119/521 (22%), Positives = 223/521 (41%), Gaps = 69/521 (13%)
Query: 101 WLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRFLERVELQEFSLGSCPPSLALQGM 160
WLN LT+IWP Y + S + A VE L+ +P + + + +LG+ P +
Sbjct: 73 WLNHHLTKIWP-YVDEAASELIKASVEPVLEQYRPAIVASLTFSKLTLGTVAPQFTGVSV 131
Query: 161 RWSTIGDQR--VMQLGFDWDTHEMSILLLAKLAKPLMGTAR-IVINSLHIKG--DLIFTP 215
GD+ ++L WD + +L + K L+G + I + ++ G LIF P
Sbjct: 132 ---IDGDKNGITLELDMQWDGNPNIVLGV----KTLVGVSLPIQVKNIGFTGVFRLIFRP 184
Query: 216 ILDG----KALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGVSSWLEKLFTDTLVKTMV 271
+++ A+ S ++ + G ++P G+S +E+ D + ++
Sbjct: 185 LVEDFPCFGAVSVSLREKKKLDFTLKVVGGDISAIP-----GLSEAIEETIRDAVEDSIT 239
Query: 272 EPRRRCFTLPAV-----DLRKKAVGGIIYVRVISANKLSSSSFKASRRQQSGSTNGSSED 326
P R+ +P + DL K VG ++ V+++ A L++
Sbjct: 240 WPVRK--VIPIIPGDYSDLELKPVG-MLEVKLVQAKNLTNKDLVGK-------------- 282
Query: 327 VSDDKDLHTFVEVEIEELTRRTDVRLGSTPRWDAPFNMVLHD-NTGTLRFNLYEC----I 381
D F+ E+ R + P W+ F V+ D +T L +Y+
Sbjct: 283 --SDPFAKMFIRPLREKTKRSKTINNDLNPIWNEHFEFVVEDASTQHLVVRIYDDEGVQA 340
Query: 382 PNNVKCDYLGSCEIKLRHVEDDSTIMWAVGPDSGIIAKQAQFCGD-EIEMVVPFEGTNSG 440
+ C + CE++ V+D +W I + + G+ +E++ G+ +G
Sbjct: 341 SELIGCAQIRLCELEPGKVKD----VWLKLVKDLEIQRDTKNRGEVHLELLYIPYGSGNG 396
Query: 441 ELKVSIVVKEWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKK--LKITVVEGKDLAAA 498
+V + S T L+N++ N SS + + L +TV+ +++
Sbjct: 397 ------IVNPFVTSSMTSLERVLKNDTTDEENASSRKRKDVIVRGVLSVTVISAEEIPI- 449
Query: 499 KEKTGKFDPYIKLQYGKVMQKTKT---SHTPNPVWNQTIEFD-EVGGGEYLKLKVFTEEL 554
++ GK DPY+ L K K+KT + + NPVWNQT +F E G + L L+V+ +
Sbjct: 450 QDLMGKADPYVVLSMKKSGAKSKTRVVNDSLNPVWNQTFDFVVEDGLHDMLVLEVWDHDT 509
Query: 555 FGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIE 595
FG + IG + L ++ W PL+ ++G+++L ++
Sbjct: 510 FGKDYIGRCILTLTRVIMEEEYKDWYPLDESKTGKLQLHLK 550
>UniRef100_Q9FY55 CLB1-like protein [Arabidopsis thaliana]
Length = 574
Score = 89.0 bits (219), Expect = 4e-16
Identities = 133/557 (23%), Positives = 222/557 (38%), Gaps = 87/557 (15%)
Query: 72 RKLLVEDLDKKWKRIILNNSPITPLEHC--EWLNKLLTEIWPNYFNPKLSSRLSAIVEAR 129
RKLL D W ++ + C WLN L +IWP Y N S + + VE
Sbjct: 51 RKLLPGDFYPSW--VVFSQRQKLSYSKCLLNWLNLELEKIWP-YVNEAASELIKSSVEPV 107
Query: 130 LKLRKPRFLERVELQEFSLGSCPPSLALQGMRWSTIGDQRV-MQLGFDWDTHEMSILLLA 188
L+ P L ++ +F+LG+ P + S G + M+L WD + +L +
Sbjct: 108 LEQYTPAMLASLKFSKFTLGTVAPQFTGVSILESESGPNGITMELEMQWDGNPKIVLDV- 166
Query: 189 KLAKPLMGTA-RIVINSLHIKG--DLIFTPILDGKALLYSFVSAPEVRVGVAFG---SGG 242
K L+G + I + ++ G LIF P++D + + + G+ F GG
Sbjct: 167 ---KTLLGVSLPIEVKNIGFTGVFRLIFKPLVDEFPCFGALSYSLREKKGLDFTLKVIGG 223
Query: 243 SQSLPATEWPGVSSWLEKLFTDTLVKTMVEPRRRCF-TLPA--VDLRKKAVGGIIYVRVI 299
T PG+S +E+ D + ++ P R+ LP DL K VG + V+V+
Sbjct: 224 E----LTSIPGISDAIEETIRDAIEDSITWPVRKIIPILPGDYSDLELKPVGK-LDVKVV 278
Query: 300 SANKLSSSSFKASRRQQSGSTNGSSEDVSDDKDLHTFVEVE-IEELTRRTDVRLGS-TPR 357
A L +++D+ D + V + + + T++T S P
Sbjct: 279 QAKDL------------------ANKDMIGKSDPYAIVFIRPLPDRTKKTKTISNSLNPI 320
Query: 358 WDAPFNMVLHDNT--------------------GTLRFNLYECIPNNVKCDYLGSCEIKL 397
W+ F ++ D + G + L E +P VK +L +K
Sbjct: 321 WNEHFEFIVEDVSTQHLTVRVFDDEGVGSSQLIGAAQVPLNELVPGKVKDIWLKL--VKD 378
Query: 398 RHVEDDSTIMWAVGPDSGIIAKQAQFCGDEIEMVVPFEGTNSGELKVSIVVKEWQFSDGT 457
++ D+ + G + G E + PF S + ++ E + SD T
Sbjct: 379 LEIQRDT-------KNRGQLELLYCPLGKEGGLKNPFNPDYSLTILEKVLKPESEDSDAT 431
Query: 458 HSLNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVM 517
+ L S + L +TVV +DL A + GK D ++ + K
Sbjct: 432 ---------DMKKLVTSKKKDVIVRGVLSVTVVAAEDLPAV-DFMGKADAFVVITLKKSE 481
Query: 518 QKTKTSHTP---NPVWNQTIEF-DEVGGGEYLKLKVFTEELFGDENIGSAQVNLEGLVDG 573
K+KT P NPVWNQT +F E + L L+V+ + FG + IG + L ++
Sbjct: 482 TKSKTRVVPDSLNPVWNQTFDFVVEDALHDLLTLEVWDHDKFGKDKIGRVIMTLTRVMLE 541
Query: 574 SVRDVWIPLERVRSGEI 590
W L+ +SG++
Sbjct: 542 GEFQEWFELDGAKSGKL 558
Score = 68.2 bits (165), Expect = 7e-10
Identities = 44/114 (38%), Positives = 67/114 (58%), Gaps = 7/114 (6%)
Query: 475 SNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKT----SHTPNPVW 530
S+++L+ KL + VV+ KDLA K+ GK DPY + + +TK S++ NP+W
Sbjct: 263 SDLELKPVGKLDVKVVQAKDLAN-KDMIGKSDPYAIVFIRPLPDRTKKTKTISNSLNPIW 321
Query: 531 NQTIEFD-EVGGGEYLKLKVFTEELFGDEN-IGSAQVNLEGLVDGSVRDVWIPL 582
N+ EF E ++L ++VF +E G IG+AQV L LV G V+D+W+ L
Sbjct: 322 NEHFEFIVEDVSTQHLTVRVFDDEGVGSSQLIGAAQVPLNELVPGKVKDIWLKL 375
>UniRef100_Q7XTM4 OSJNBa0033G05.21 protein [Oryza sativa]
Length = 575
Score = 88.6 bits (218), Expect = 5e-16
Identities = 130/561 (23%), Positives = 237/561 (42%), Gaps = 92/561 (16%)
Query: 72 RKLLVEDLDKKWKRIILNNSPITPLEHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLK 131
RKLL DL W ++ + + +WLN+ L +IWP + N S + VE L+
Sbjct: 51 RKLLPADLYPSW--VVFSTQ-----QKLKWLNQELIKIWP-FVNAAASELIKTSVEPVLE 102
Query: 132 LRKPRFLERVELQEFSLGSCPPSLALQGMRWSTIGDQR---VMQLGFDWDTHEMSILLLA 188
+P L ++ + +LG+ P Q S I + VM+L +WD + SI+L
Sbjct: 103 QYRPIILASLKFSKLTLGTVAP----QFTGVSIIENDESGIVMELEMNWDANP-SIILDV 157
Query: 189 KLAKPLMGTARIVINSLHIKG--DLIFTPILDG----KALLYSFVSAPEVRVGVAFGSGG 242
K L + I + + G LIF P++D A+ +S ++ + G
Sbjct: 158 KTR--LGVSLPIQVKDIGFTGVFRLIFKPLVDQLPCFGAVCFSLRKKKKLDFRLKVIGGE 215
Query: 243 SQSLPATEWPGVSSWLEKLFTDTLVKTMVEPRRRCFTLPAV-----DLRKKAVGGIIYVR 297
++ PG+S LE + + ++ P R+ +P + DL K V G + V+
Sbjct: 216 ISAI-----PGISDALEDTIKNAIEDSITWPVRK--VIPIIPGDYSDLELKPV-GTLEVK 267
Query: 298 VISANKLSSSSFKASRRQQSGSTNGSSEDVSDDKDLHTFVEVEIEELTRRTDVRLGSTPR 357
++ A L++ D +V +++ R + P
Sbjct: 268 LVQARDLTNKDLIG----------------KSDPFAIVYVRPLPDKMKRSKTINNDLNPI 311
Query: 358 WDAPFNMVLHD-NTGTLRFNLYECIPNNVKCDYLGSCEIKLRHVEDDST-IMWAV----- 410
W+ F ++ D +T T+ +Y+ + + +G ++ L+ ++ +W
Sbjct: 312 WNEHFEFIVEDADTQTVTVKIYD-DDGIQESELIGCAQVTLKDLQPGKVKDVWLKLVKDL 370
Query: 411 -----GPDSGIIAKQAQFCGDEIEMVVPFEGTNSGELKVSIVVKEWQFSDGTHSLNNLRN 465
D G + + +C +++ P N + S+ E ++ ++ N
Sbjct: 371 EIQRDRKDRGQVHLELLYCPFDMKEETP----NPFRQQFSMTSLE-------RTMTSMEN 419
Query: 466 NSQQSLNGSSNIQLRTGKK------LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQK 519
S NG + + R K+ L +TV+ G+DL A + GK DPY+ L K K
Sbjct: 420 GS--GSNGFNRLSSRKKKEIIMRGVLSVTVISGEDL-PAMDMNGKSDPYVVLSLKKSKTK 476
Query: 520 TKT---SHTPNPVWNQTIEF-DEVGGGEYLKLKVFTEELFGDENIGSAQVNL-EGLVDGS 574
KT S + NPVWNQT +F E G + L L+V+ + F + +G + L + L++
Sbjct: 477 YKTRVVSESLNPVWNQTFDFVVEDGLHDMLMLEVYDHDTFSRDYMGRCILTLTKVLIEED 536
Query: 575 VRDVWIPLERVRSGEIRLKIE 595
+D + LE +SG++ L ++
Sbjct: 537 YKDSF-KLEGAKSGKLNLHLK 556
Score = 56.6 bits (135), Expect = 2e-06
Identities = 42/143 (29%), Positives = 74/143 (51%), Gaps = 15/143 (10%)
Query: 475 SNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSHTPN----PVW 530
S+++L+ L++ +V+ +DL K+ GK DP+ + + K K S T N P+W
Sbjct: 254 SDLELKPVGTLEVKLVQARDLTN-KDLIGKSDPFAIVYVRPLPDKMKRSKTINNDLNPIW 312
Query: 531 NQTIEFD-EVGGGEYLKLKVFTEE-LFGDENIGSAQVNLEGLVDGSVRDVWIPL------ 582
N+ EF E + + +K++ ++ + E IG AQV L+ L G V+DVW+ L
Sbjct: 313 NEHFEFIVEDADTQTVTVKIYDDDGIQESELIGCAQVTLKDLQPGKVKDVWLKLVKDLEI 372
Query: 583 --ERVRSGEIRLKIEAIKVDDQE 603
+R G++ L++ D +E
Sbjct: 373 QRDRKDRGQVHLELLYCPFDMKE 395
>UniRef100_Q9ZVY8 T25N20.15 [Arabidopsis thaliana]
Length = 528
Score = 85.9 bits (211), Expect = 3e-15
Identities = 120/523 (22%), Positives = 224/523 (41%), Gaps = 70/523 (13%)
Query: 101 WLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRFLERVELQEFSLGSCPPSLALQGM 160
WLN LT+IWP Y + S + A VE L+ +P + + + +LG+ P +
Sbjct: 38 WLNHHLTKIWP-YVDEAASELIKASVEPVLEQYRPAIVASLTFSKLTLGTVAPQFTGVSV 96
Query: 161 RWSTIGDQR--VMQLGFDWDTHEMSILLLAKLAKPLMGTAR-IVINSLHIKG--DLIFTP 215
GD+ ++L WD + +L + K L+G + I + ++ G LIF P
Sbjct: 97 ---IDGDKNGITLELDMQWDGNPNIVLGV----KTLVGVSLPIQVKNIGFTGVFRLIFRP 149
Query: 216 ILDG----KALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGVSSWLEK---LFTDTLVK 268
+++ A+ S ++ + G ++P G+S +E +F L +
Sbjct: 150 LVEDFPCFGAVSVSLREKKKLDFTLKVVGGDISAIP-----GLSEAIEVESYVFILELAR 204
Query: 269 TMVEPRRR----CFTLPAVDLRKKAVGGIIYVRVISANKLSSSSFKASRRQQSGSTNGSS 324
+ R+ C ++P+ DL K VG ++ V+++ A L++
Sbjct: 205 QVGNLSRQLKFFCVSIPS-DLELKPVG-MLEVKLVQAKNLTNKDLVGK------------ 250
Query: 325 EDVSDDKDLHTFVEVEIEELTRRTDVRLGSTPRWDAPFNMVLHD-NTGTLRFNLYEC--- 380
D F+ E+ R + P W+ F V+ D +T L +Y+
Sbjct: 251 ----SDPFAKMFIRPLREKTKRSKTINNDLNPIWNEHFEFVVEDASTQHLVVRIYDDEGV 306
Query: 381 -IPNNVKCDYLGSCEIKLRHVEDDSTIMWAVGPDSGIIAKQAQFCGD-EIEMVVPFEGTN 438
+ C + CE++ V+D +W I + + G+ +E++ G+
Sbjct: 307 QASELIGCAQIRLCELEPGKVKD----VWLKLVKDLEIQRDTKNRGEVHLELLYIPYGSG 362
Query: 439 SGELKVSIVVKEWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKK--LKITVVEGKDLA 496
+G +V + S T L+N++ N SS + + L +TV+ +++
Sbjct: 363 NG------IVNPFVTSSMTSLERVLKNDTTDEENASSRKRKDVIVRGVLSVTVISAEEIP 416
Query: 497 AAKEKTGKFDPYIKLQYGKVMQKTKT---SHTPNPVWNQTIEFD-EVGGGEYLKLKVFTE 552
++ GK DPY+ L K K+KT + + NPVWNQT +F E G + L L+V+
Sbjct: 417 I-QDLMGKADPYVVLSMKKSGAKSKTRVVNDSLNPVWNQTFDFVVEDGLHDMLVLEVWDH 475
Query: 553 ELFGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIE 595
+ FG + IG + L ++ W PL+ ++G+++L ++
Sbjct: 476 DTFGKDYIGRCILTLTRVIMEEEYKDWYPLDESKTGKLQLHLK 518
>UniRef100_Q69UK6 Putative C2 domain-containing protein [Oryza sativa]
Length = 562
Score = 65.9 bits (159), Expect = 3e-09
Identities = 117/549 (21%), Positives = 220/549 (39%), Gaps = 76/549 (13%)
Query: 73 KLLVEDLDKKWKRIILNNSP----ITPLEHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEA 128
KL VEDL K I L P T + +WLN+ L +IWP + N S + VE
Sbjct: 44 KLTVEDLRKL---IPLELYPSWVSFTQKQKLKWLNQELVKIWP-FVNEAASELIKTSVEP 99
Query: 129 RLKLRKPRFLERVELQEFSLGSCPPSLALQGMRWSTIGDQRVMQLGFDWDTHEMSILLLA 188
+ K L + + +LG+ P + S M+L WD + +L
Sbjct: 100 IFEQYKSFILSSLHFSKLTLGTVAPQFTGVSILDSD-SSGITMELELQWDGNPNIVL--- 155
Query: 189 KLAKPLMGTARIVINSLHIKG--DLIFTPILDGKALLYSFVSAPEVRVGVAFGS---GGS 243
+ L + + + ++ G L+F P++ + + + V F GG
Sbjct: 156 DIQTTLGISLPVQVKNIGFTGVLRLVFKPLVAELPCFGAVCCSLREKSKVDFTLKVIGGE 215
Query: 244 QSLPATEWPGVSSWLEKLFTDTLVKTMVEPRRRCFTLPAV-----DLRKKAVGGIIYVRV 298
T PG+S +E DT+ + P R +P V DL K VG ++ V++
Sbjct: 216 M----TAIPGISDAIEGTIRDTIEDQLTWPNR--IVVPIVPGDYSDLELKPVG-LLEVKL 268
Query: 299 ISANKLSSSSFKASRRQQSGSTNGSSEDVSDDKDLHTFVEVEIEELTRRTDVRLGSTPRW 358
+ A L++ D ++ +++ + + P W
Sbjct: 269 VEARDLTNKDLVGK----------------SDPFAVLYIRPLQDKMKKSKTINNDLNPIW 312
Query: 359 DAPFNMVLHDNTGTLRFNLYECIPNNVKC-DYLGSCEIKLRHVEDDST--IMWAVGPDSG 415
+ + V+ D T T R + ++ + +G + L ++ + + D
Sbjct: 313 NEHYEFVVED-TSTQRLTVKIYDDEGLQASELIGCARVDLSDLQPGKVKEVWLDLVKDLE 371
Query: 416 IIAKQAQFCGDEIEMVV-PF---EGTNS---GELKVSIVVKEWQFSDGTHSLNNLRNNSQ 468
I + + +E++ PF EG ++ +++++ + K + +N +N
Sbjct: 372 IQRDKKRRGQVHLELLYYPFGKQEGVSNPFADQIQLTSLEKVLKTESNGFDVNQRKNVIM 431
Query: 469 QSLNGSSNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKT---SHT 525
+ + L +TV+ +DL + GK DP++ L K K KT + T
Sbjct: 432 RGV-------------LSVTVISAEDLPPM-DVMGKADPFVVLYLKKGETKKKTRVVTET 477
Query: 526 PNPVWNQTIEF-DEVGGGEYLKLKVFTEELFGDENIGSAQVNLEGLV-DGSVRDVWIPLE 583
NP+WNQT +F E + L ++V+ + FG + IG + L ++ +G +D ++ L+
Sbjct: 478 LNPIWNQTFDFVVEDALHDLLMVEVWDHDTFGKDYIGRCILTLTRVILEGEFQDEFV-LQ 536
Query: 584 RVRSGEIRL 592
+SG++ L
Sbjct: 537 GAKSGKLNL 545
Score = 64.3 bits (155), Expect = 1e-08
Identities = 48/152 (31%), Positives = 78/152 (50%), Gaps = 15/152 (9%)
Query: 475 SNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSHTPN----PVW 530
S+++L+ L++ +VE +DL K+ GK DP+ L + K K S T N P+W
Sbjct: 254 SDLELKPVGLLEVKLVEARDLTN-KDLVGKSDPFAVLYIRPLQDKMKKSKTINNDLNPIW 312
Query: 531 NQTIEF-DEVGGGEYLKLKVFTEE-LFGDENIGSAQVNLEGLVDGSVRDVWIPL------ 582
N+ EF E + L +K++ +E L E IG A+V+L L G V++VW+ L
Sbjct: 313 NEHYEFVVEDTSTQRLTVKIYDDEGLQASELIGCARVDLSDLQPGKVKEVWLDLVKDLEI 372
Query: 583 --ERVRSGEIRLKIEAIKVDDQEGSTVCFFSQ 612
++ R G++ L++ QEG + F Q
Sbjct: 373 QRDKKRRGQVHLELLYYPFGKQEGVSNPFADQ 404
>UniRef100_Q9LNT5 T20H2.13 protein [Arabidopsis thaliana]
Length = 535
Score = 65.9 bits (159), Expect = 3e-09
Identities = 104/514 (20%), Positives = 217/514 (41%), Gaps = 65/514 (12%)
Query: 97 EHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRF-LERVELQEFSLGSCPPSL 155
+ +WLNKL+ +WP Y + + +I + + + P + ++ VE + +LGS PPS
Sbjct: 67 DRIDWLNKLIGHMWP-YMDKAICKMAKSIAKPIIAEQIPNYKIDSVEFEMLTLGSLPPS- 124
Query: 156 ALQGMR-WSTIGDQRVMQLGFDWDTHEMSILLLAKLAKPLMGTARIVINSLHIKGDLIFT 214
QGM+ ++T + +M+L W +I+++AK A L T +++ ++ +
Sbjct: 125 -FQGMKVYATDDKEIIMELSVKW-AGNPNIIVVAK-AFGLKATVQVIDLQVYATPRITLK 181
Query: 215 PILDG----KALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGVSSWLEKLFTDTLVKTM 270
P++ + S + P+V G+ ++ PG+ +++++ D +
Sbjct: 182 PLVPSFPCFANIFVSLMDKPQVDFGLKLLGADVMAI-----PGLYRFVQEIIKDQVANMY 236
Query: 271 VEPRRRCFTLPAVDLRKKAVGGIIYVRVISANKLSSSSFKASRRQQSGSTNGSSEDVSDD 330
+ P+ + K G++ V+VI A KL GS +S D
Sbjct: 237 LWPKTLNVQIMDPSKAMKKPVGLLSVKVIKAIKLKKKDL------LGGSDPYVKLTLSGD 290
Query: 331 KDLHTFVEVEIEELTRRTDVRLGSTPRWDAPFNMVLHD-NTGTLRFNLYECIPNNVKCDY 389
K V+ L P W+ F++V+ + + L+ +Y+ K D
Sbjct: 291 KVPGKKTVVKHSNL----------NPEWNEEFDLVVKEPESQELQLIVYDW-EQVGKHDK 339
Query: 390 LGSCEIKLRHVEDDSTIMWAVGPDSGIIAKQAQFCGDEIEMVVPFEGTNSGELKVSIVVK 449
+G I+L+ + + + + + K+ P + G+L V + K
Sbjct: 340 IGMNVIQLKDLTPEEPKLMTLELLKSMEPKE------------PVSEKSRGQLVVEVEYK 387
Query: 450 EWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKEKTGKF--DP 507
++ D ++++ N +++ G+ + TG L + V E +DL GK+ +P
Sbjct: 388 PFKDDDIPENIDD-PNAVEKAPEGTPS----TGGLLVVIVHEAEDL------EGKYHTNP 436
Query: 508 YIKLQYGKVMQKTK-TSHTPNPVWNQTIEF--DEVGGGEYLKLKVFTEE---LFGDENIG 561
++L + +KTK P W++ +F DE + L ++V + + E +G
Sbjct: 437 SVRLLFRGEERKTKRVKKNREPRWDEDFQFPLDEPPINDKLHVEVISSSSRLIHPKETLG 496
Query: 562 SAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIE 595
+NL +V + L ++G I+++++
Sbjct: 497 YVVINLGDVVSNRRINDKYHLIDSKNGRIQIELQ 530
>UniRef100_Q6ETC4 Putative CLB1 protein [Oryza sativa]
Length = 538
Score = 64.7 bits (156), Expect = 8e-09
Identities = 100/517 (19%), Positives = 212/517 (40%), Gaps = 69/517 (13%)
Query: 97 EHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRF-LERVELQEFSLGSCPPSL 155
+ +WLNK + IWP Y + + I + + ++ ++ VE + +LGS PP+
Sbjct: 69 DRIDWLNKFVENIWP-YLDKAICKTAKEIAKPIIAENTAKYKIDSVEFETLTLGSLPPT- 126
Query: 156 ALQGMRWSTIGDQR-VMQLGFDWDTHEMSILLLAKLAKPLMGTARIVINSLHIKGDLIFT 214
QGM+ T +Q +M+ W +I ++ K A L TA+++ + +
Sbjct: 127 -FQGMKVYTTDEQELIMEPSIKW-AGNPNITVVVK-AFGLKATAQVIDLHVFALPRITLK 183
Query: 215 PILDG----KALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGVSSWLEKLFTDTLVKTM 270
P++ ++ S + P V G+ ++ PG+ +++++ +
Sbjct: 184 PLVPSFPCFAKIVVSLMEKPHVDFGLKLLGADLMAI-----PGLYVFVQEIIKTQVANMY 238
Query: 271 VEPRRRCFTLPAVDLRK--KAVGGIIYVRVISANKLSSSSFKASRRQQSGSTNGSSEDVS 328
+ P + +P +D K K GI++V ++ A KL+ F
Sbjct: 239 LWP--KVLEVPIMDPAKAQKKPVGILHVNIVRAVKLTKKDFLGK---------------- 280
Query: 329 DDKDLHTFVEVEIEEL-TRRTDV-RLGSTPRWDAPFNMVLHD-NTGTLRFNLYECIPNNV 385
D + +++ E+L +++T V R P W+ F +V+ D + L +Y+
Sbjct: 281 --SDPYVKLKLTEEKLPSKKTSVKRSNLNPEWNEDFKLVVKDPESQALELTVYDW-EQVG 337
Query: 386 KCDYLGSCEIKLRHVEDDSTIMWAVGPDSGIIAKQAQFCGDEIEMVVPFEGTNSGELKVS 445
K D +G I L+ + D + + A P G+L V
Sbjct: 338 KHDKIGMSVIPLKELIPDEAKSLTLDLHKTMDAND------------PANDKFRGQLTVD 385
Query: 446 IVVKEWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKEKTGKF 505
+ K ++ D ++ +++ +G+ G L + V E +D+ E
Sbjct: 386 VTYKPFKEGDSDVDTSDESGTIEKAPDGTP----EGGGLLVVIVHEAQDV----EGKHHT 437
Query: 506 DPYIKLQYGKVMQKTK-TSHTPNPVWNQTIEF--DEVGGGEYLKLKVFTEE----LFGDE 558
+PY+++ + +KTK +P W Q +F +E + ++++V + + E
Sbjct: 438 NPYVRIVFRGEERKTKHIKKNRDPRWEQEFQFVCEEPPINDKMQIEVISRPPSIGIHSKE 497
Query: 559 NIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIE 595
N+G ++L +++ + L ++G I+L+++
Sbjct: 498 NLGYVVISLADVINNKRINEKYHLIDSKNGRIQLELQ 534
>UniRef100_O04823 Hypothetical protein [Sporobolus stapfianus]
Length = 171
Score = 64.7 bits (156), Expect = 8e-09
Identities = 37/99 (37%), Positives = 55/99 (55%), Gaps = 6/99 (6%)
Query: 484 KLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTS---HTPNPVWNQTIEFDEVG 540
KL + VV G +LA T DPY+ L YG QK KTS PNPVWN+ ++
Sbjct: 12 KLNVRVVRGSNLAICDPLTHTSDPYVVLHYG--AQKVKTSVQKKNPNPVWNEVLQLSVTN 69
Query: 541 GGEYLKLKVFTEELF-GDENIGSAQVNLEGLVDGSVRDV 578
+ + L+VF E+ F D+++G A++NL + D + D+
Sbjct: 70 PTKPVHLEVFDEDKFTADDSMGVAEINLTDIYDAAKLDL 108
>UniRef100_UPI00003AE7D2 UPI00003AE7D2 UniRef100 entry
Length = 619
Score = 64.3 bits (155), Expect = 1e-08
Identities = 117/508 (23%), Positives = 191/508 (37%), Gaps = 101/508 (19%)
Query: 96 LEHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRFLERVELQEFSLG-SCPPS 154
+E EWLNK+L + WP YF + ++E +++ K L+ + G CP
Sbjct: 6 VERVEWLNKVLEQAWP-YFGTIMEKTFKEVLEPKIR-AKSVHLKTCTFTKIQFGEKCP-- 61
Query: 155 LALQGM--------RWSTIGDQRVMQLGFDWDTH-EMSILLLAKLAKPLMGTARIVINSL 205
+ G+ R I D ++ +G D + H ++S L L GT R+++ L
Sbjct: 62 -RINGVKVYTKEIDRRQVILDLQICYVG-DCEIHMDISKFNLGVKGVQLYGTLRVILEPL 119
Query: 206 HIKGDLIFTPILDGKALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGVSSWLEKLFTDT 265
L P + L F+ P + F G +L + PG++ + L D
Sbjct: 120 -----LTDAPFIGAVTLF--FMQKPHLE----FNWAGMSNL--LDVPGINVMSDSLIQDY 166
Query: 266 LVKTMVEPRRRCFTLP------AVDLRKKAVGGIIYVRVISANKLSSSSFKASRRQQSGS 319
+ +V P R T+P LR G+I V ++ A L Q +
Sbjct: 167 IAARLVLPNR--ITVPLKKNMSIAQLRFPVPHGVIRVHLLEAENL----------VQKDN 214
Query: 320 TNGSSEDVSDDKDLHTFVEVEIEELTRRTDVRLGSTPRWDAPFNMVLHDNTG-TLRFNLY 378
G+ SD L V+ T D+ P W+ F V+H+ G L +LY
Sbjct: 215 FLGAIRGKSDPYALLRLGTVQYRSKTISRDL----NPIWNETFEFVVHEVLGQDLEVDLY 270
Query: 379 ECIPNNVKCDYLGSCEIKLRHVEDDSTIMWAVGPDSGIIAKQAQFCGDEIEMVVPFEGTN 438
+ P+ K D++GS I L +++D T ++ P T
Sbjct: 271 DADPD--KDDFMGSLLISLLDIKNDKT----------------------VDEWFPLSKTT 306
Query: 439 SGELKVSIVVKEWQFSDGTHSLNNLRNNSQQSLNGSSN----IQLRTGKKLKITVVE--G 492
SG L + + EW SL N + + G S + L + L E
Sbjct: 307 SGHLHLKL---EWL------SLVNDQEKLHEDKKGLSTAILIVYLDSAFNLPKNHFEYSN 357
Query: 493 KDLAAAKEKTGKF--------DPYIKLQYGKVMQKTKT-SHTPNPVWNQTIEFDEVGGGE 543
+ A K K K+ ++ L G QK+KT + +P W Q F V
Sbjct: 358 GECGARKIKNNKYLKKTEREPSSFVLLTVGSKTQKSKTCNFNKDPKWGQAFTF-FVHSAH 416
Query: 544 YLKLKVFTEELFGDENIGSAQVNLEGLV 571
L + ++ D ++G++ V L L+
Sbjct: 417 SQSLHIEIKDKDQDSSLGTSVVCLSHLL 444
Score = 63.5 bits (153), Expect = 2e-08
Identities = 39/105 (37%), Positives = 61/105 (57%), Gaps = 5/105 (4%)
Query: 503 GKFDPYIKLQYGKVMQKTKT-SHTPNPVWNQTIEF--DEVGGGEYLKLKVFTEELFGDEN 559
GK DPY L+ G V ++KT S NP+WN+T EF EV G + L++ ++ + D+
Sbjct: 221 GKSDPYALLRLGTVQYRSKTISRDLNPIWNETFEFVVHEVLGQD-LEVDLYDADPDKDDF 279
Query: 560 IGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIEAIK-VDDQE 603
+GS ++L + + D W PL + SG + LK+E + V+DQE
Sbjct: 280 MGSLLISLLDIKNDKTVDEWFPLSKTTSGHLHLKLEWLSLVNDQE 324
>UniRef100_Q6UU05 Putative Ca2+-dependent lipid-binding protein [Oryza sativa]
Length = 422
Score = 64.3 bits (155), Expect = 1e-08
Identities = 48/152 (31%), Positives = 78/152 (50%), Gaps = 15/152 (9%)
Query: 475 SNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSHTPN----PVW 530
S+++L+ L++ +VE +DL K+ GK DP+ L + K K S T N P+W
Sbjct: 114 SDLELKPVGLLEVKLVEARDLTN-KDLVGKSDPFAVLYIRPLQDKMKKSKTINNDLNPIW 172
Query: 531 NQTIEF-DEVGGGEYLKLKVFTEE-LFGDENIGSAQVNLEGLVDGSVRDVWIPL------ 582
N+ EF E + L +K++ +E L E IG A+V+L L G V++VW+ L
Sbjct: 173 NEHYEFVVEDTSTQRLTVKIYDDEGLQASELIGCARVDLSDLQPGKVKEVWLDLVKDLEI 232
Query: 583 --ERVRSGEIRLKIEAIKVDDQEGSTVCFFSQ 612
++ R G++ L++ QEG + F Q
Sbjct: 233 QRDKKRRGQVHLELLYYPFGKQEGVSNPFADQ 264
Score = 52.0 bits (123), Expect = 5e-05
Identities = 36/113 (31%), Positives = 62/113 (54%), Gaps = 7/113 (6%)
Query: 485 LKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKT---SHTPNPVWNQTIEF-DEVG 540
L +TV+ +DL + GK DP++ L K K KT + T NP+WNQT +F E
Sbjct: 295 LSVTVISAEDLPPM-DVMGKADPFVVLYLKKGETKKKTRVVTETLNPIWNQTFDFVVEDA 353
Query: 541 GGEYLKLKVFTEELFGDENIGSAQVNLEGLV-DGSVRDVWIPLERVRSGEIRL 592
+ L ++V+ + FG + IG + L ++ +G +D ++ L+ +SG++ L
Sbjct: 354 LHDLLMVEVWDHDTFGKDYIGRCILTLTRVILEGEFQDEFV-LQGAKSGKLNL 405
>UniRef100_Q67XP8 Hypothetical protein At3g14590 [Arabidopsis thaliana]
Length = 692
Score = 63.9 bits (154), Expect = 1e-08
Identities = 62/218 (28%), Positives = 104/218 (47%), Gaps = 23/218 (10%)
Query: 69 RYQRKLLVEDLDKKWKRIILNNSPITPLEHCEWLNKLLTEIWPNYFNPKLSSR-LSAIVE 127
R ++KL E+ + +R +L++S E WLN + IWP S + L I+
Sbjct: 2 RLRKKLQFEERKQANQRRVLSDS-----ESVRWLNHAVERIWPICMEQIASQKILRPIIP 56
Query: 128 ARLKLRKPRFLERVELQEFSLGSCPPSLA-LQGMRWSTIGDQRVMQLGFDWDT-HEMSIL 185
L +P ++V +Q LG PP L ++ +R ST D V++LG ++ T +MS +
Sbjct: 57 WFLDKYRPWTAKKVVIQHLYLGRNPPLLTDIRVLRQSTGDDHLVLELGMNFLTADDMSAI 116
Query: 186 LLAKLAKPL---MGTARIVINSLHIKGDLIFT-------PILDGKALLYSFVSAPEVRVG 235
L KL K L M T ++ + +H++G ++ P L L F P ++
Sbjct: 117 LAVKLRKRLGFGMWT-KLHLTGMHVEGKVLIGVKFLRRWPFLG--RLRVCFAEPPYFQMT 173
Query: 236 VAFGSGGSQSLPATEWPGVSSWLEKLFTDTLVKTMVEP 273
V + + L PG++ WL+KL + +T+VEP
Sbjct: 174 VKPIT--THGLDVAVLPGIAGWLDKLLSVAFEQTLVEP 209
Score = 57.0 bits (136), Expect = 2e-06
Identities = 38/131 (29%), Positives = 63/131 (48%), Gaps = 10/131 (7%)
Query: 487 ITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTS-HTPNPVWNQTIEFDEV--GGGE 543
+ VVE D+ + + G DPY+K Q G KTK T P W + +
Sbjct: 242 VEVVEACDVKPS-DLNGLADPYVKGQLGAYRFKTKILWKTLAPKWQEEFKIPICTWDSAN 300
Query: 544 YLKLKVFTEELFGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIEAI----KV 599
L ++V ++ F D+++G VN+ G D+W+PL+ ++ G + L I + K+
Sbjct: 301 ILNIEVQDKDRFSDDSLGDCSVNIAEFRGGQRNDMWLPLQNIKMGRLHLAITVLEDEAKL 360
Query: 600 DDQ--EGSTVC 608
+D EG T+C
Sbjct: 361 NDDPFEGVTIC 371
>UniRef100_Q91X62 Membrane bound C2 domain containing protein [Mus musculus]
Length = 1092
Score = 63.5 bits (153), Expect = 2e-08
Identities = 63/283 (22%), Positives = 124/283 (43%), Gaps = 28/283 (9%)
Query: 330 DKDLHTFVEVEIEELTRRTDVRLGS-TPRWDAPFNMVLHDNTGTLRFNLYECIPNNVKCD 388
+K+ + V++ ++++TR + + +P W+ F L D L + ++ +
Sbjct: 486 NKEPNPMVQLSVQDVTRESKATYSTNSPVWEEAFRFFLQDPRSQ---ELDVQVKDDSRAL 542
Query: 389 YLGSCEIKLRHVEDDSTI---MW----AVGPDSGIIAKQAQFCGDEIEMVVPFEGTNSGE 441
LG+ + L + S + W + GP+S + K +V+ + E
Sbjct: 543 TLGALTLPLARLLTASELTLDQWFQLSSSGPNSRLYMK----------LVMRILYLDYSE 592
Query: 442 LKVSIVVKEWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKE- 500
++ V + + + + + + + N T L+I V+E +DL A
Sbjct: 593 IRFPTVPGAQDWDRESLETGSSVDAPPRPYHTTPNSHFGTENVLRIHVLEAQDLIAKDRF 652
Query: 501 ----KTGKFDPYIKLQY-GKVMQKTKTSHTPNPVWNQTIEFDEVG-GGEYLKLKVFTEEL 554
GK DPY+KL+ GK + NP WN+ E G+ L+++VF ++L
Sbjct: 653 LGGLVKGKSDPYVKLKVAGKSFRTHVVREDLNPRWNEVFEVIVTSIPGQELEIEVFDKDL 712
Query: 555 FGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIEAI 597
D+ +G +V+L +++ D W+ LE V SG + L++E +
Sbjct: 713 DKDDFLGRYKVSLTTVLNSGFLDEWLTLEDVPSGRLHLRLERL 755
Score = 47.0 bits (110), Expect = 0.002
Identities = 71/319 (22%), Positives = 126/319 (39%), Gaps = 45/319 (14%)
Query: 96 LEHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRFLERVELQEFSLGSCPPSL 155
+E EWLNK++ ++WP + + L+ V ++ P L+ LG P +
Sbjct: 126 VEKAEWLNKIVAQVWP-FLGQYMEKLLAETVAPAVRGANPH-LQTFTFTRVELGEKPLRI 183
Query: 156 ALQGMRWSTIGDQRVMQLGFDW------DTHEMSILLLAKL-AKPLMGTARIVINSLHIK 208
+ S DQ ++ L + D A + L G R+++ L
Sbjct: 184 IGVKVHPSQRKDQILLDLNVSYVGDVQIDVEVKKYFCKAGVKGMQLHGVLRVILEPL--T 241
Query: 209 GDLIFTPILDGKALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGVSSWLEKLFTDTLVK 268
GDL PI+ ++ F+ P + + G +L + PG+SS + + D++
Sbjct: 242 GDL---PIVGAVSMF--FIKRPTLDINWT----GMTNL--LDIPGLSSLSDTMIMDSIAA 290
Query: 269 TMVEPRRRCFTL-----PAVDLRKKAVGGIIYVRVISANKLSSSSFKASRRQQSGSTNGS 323
+V P R L LR GII + +++A LSS + G G
Sbjct: 291 FLVLPNRLLVPLVPDLQDVAQLRSPLPRGIIRIHLLAARGLSSKD-----KYVKGLIEGK 345
Query: 324 SEDVSDDKDLHTFVEVEIEELTRRT-DVRLGSTPRWDAPFNMVLHDNTG-TLRFNLYECI 381
S D + V V + R D L P W + +++H+ G + +++
Sbjct: 346 S-------DPYALVRVGTQTFCSRVIDEEL--NPHWGETYEVIVHEVPGQEIEVEVFDKD 396
Query: 382 PNNVKCDYLGSCEIKLRHV 400
P+ K D+LG ++ + V
Sbjct: 397 PD--KDDFLGRMKLDVGKV 413
Score = 39.3 bits (90), Expect = 0.35
Identities = 27/96 (28%), Positives = 50/96 (51%), Gaps = 5/96 (5%)
Query: 503 GKFDPYIKLQYG-KVMQKTKTSHTPNPVWNQTIEF--DEVGGGEYLKLKVFTEELFGDEN 559
GK DPY ++ G + NP W +T E EV G E ++++VF ++ D+
Sbjct: 344 GKSDPYALVRVGTQTFCSRVIDEELNPHWGETYEVIVHEVPGQE-IEVEVFDKDPDKDDF 402
Query: 560 IGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIE 595
+G ++++ ++ V D W PL+ G++ L++E
Sbjct: 403 LGRMKLDVGKVLQAGVLDNWYPLQG-GQGQVHLRLE 437
>UniRef100_Q8C8R1 Mus musculus adult retina cDNA, RIKEN full-length enriched library,
clone:A930030A18 product:membrane bound C2 domain
containing protein, full insert sequence [Mus musculus]
Length = 893
Score = 63.5 bits (153), Expect = 2e-08
Identities = 63/283 (22%), Positives = 124/283 (43%), Gaps = 28/283 (9%)
Query: 330 DKDLHTFVEVEIEELTRRTDVRLGS-TPRWDAPFNMVLHDNTGTLRFNLYECIPNNVKCD 388
+K+ + V++ ++++TR + + +P W+ F L D L + ++ +
Sbjct: 486 NKEPNPMVQLSVQDVTRESKATYSTNSPVWEEAFRFFLQDPRSQ---ELDVQVKDDSRAL 542
Query: 389 YLGSCEIKLRHVEDDSTI---MW----AVGPDSGIIAKQAQFCGDEIEMVVPFEGTNSGE 441
LG+ + L + S + W + GP+S + K +V+ + E
Sbjct: 543 TLGALTLPLARLLTASELTLDQWFQLSSSGPNSRLYMK----------LVMRILYLDYSE 592
Query: 442 LKVSIVVKEWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKE- 500
++ V + + + + + + + N T L+I V+E +DL A
Sbjct: 593 IRFPTVPGAQDWDRESLETGSSVDAPPRPYHTTPNSHFGTENVLRIHVLEAQDLIAKDRF 652
Query: 501 ----KTGKFDPYIKLQY-GKVMQKTKTSHTPNPVWNQTIEFDEVG-GGEYLKLKVFTEEL 554
GK DPY+KL+ GK + NP WN+ E G+ L+++VF ++L
Sbjct: 653 LGGLVKGKSDPYVKLKVAGKSFRTHVVREDLNPRWNEVFEVIVTSIPGQELEIEVFDKDL 712
Query: 555 FGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIEAI 597
D+ +G +V+L +++ D W+ LE V SG + L++E +
Sbjct: 713 DKDDFLGRYKVSLTTVLNSGFLDEWLTLEDVPSGRLHLRLERL 755
Score = 44.3 bits (103), Expect = 0.011
Identities = 70/319 (21%), Positives = 125/319 (38%), Gaps = 45/319 (14%)
Query: 96 LEHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRFLERVELQEFSLGSCPPSL 155
+E EWLNK++ ++WP + + L+ V ++ P L+ LG P +
Sbjct: 126 VEKAEWLNKIVAQVWP-FLGQYMEKLLAETVAPAVRGANPH-LQTFTFTRVELGEKPLRI 183
Query: 156 ALQGMRWSTIGDQRVMQLGFDW------DTHEMSILLLAKL-AKPLMGTARIVINSLHIK 208
+ S DQ ++ L + D A + L G R+++ L
Sbjct: 184 IGVKVHPSQRKDQILLDLNVSYVGDVQIDVEVKKYFCKAGVKGMQLHGVLRVILEPL--T 241
Query: 209 GDLIFTPILDGKALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGVSSWLEKLFTDTLVK 268
GDL PI+ ++ F+ P + + G +L + PG+SS + + D++
Sbjct: 242 GDL---PIVGAVSMF--FIKRPTLDINWT----GMTNL--LDIPGLSSLSDTMIMDSIAA 290
Query: 269 TMVEPRRRCFTL-----PAVDLRKKAVGGIIYVRVISANKLSSSSFKASRRQQSGSTNGS 323
+V P R L LR GII + +++A LSS + G G
Sbjct: 291 FLVLPNRLLVPLVPDLQDVAQLRSPLPRGIIRIHLLAARGLSSKD-----KYVKGLIEGK 345
Query: 324 SEDVSDDKDLHTFVEVEIEELTRRT-DVRLGSTPRWDAPFNMVLHD-NTGTLRFNLYECI 381
S D + V V + R D L P W + +++H+ + +++
Sbjct: 346 S-------DPYALVRVGTQTFCSRVIDEEL--NPHWGETYEVIVHEVPRQEIEVEVFDKD 396
Query: 382 PNNVKCDYLGSCEIKLRHV 400
P+ K D+LG ++ + V
Sbjct: 397 PD--KDDFLGRMKLDVGKV 413
Score = 36.2 bits (82), Expect = 2.9
Identities = 26/96 (27%), Positives = 49/96 (50%), Gaps = 5/96 (5%)
Query: 503 GKFDPYIKLQYG-KVMQKTKTSHTPNPVWNQTIEF--DEVGGGEYLKLKVFTEELFGDEN 559
GK DPY ++ G + NP W +T E EV E ++++VF ++ D+
Sbjct: 344 GKSDPYALVRVGTQTFCSRVIDEELNPHWGETYEVIVHEVPRQE-IEVEVFDKDPDKDDF 402
Query: 560 IGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIE 595
+G ++++ ++ V D W PL+ G++ L++E
Sbjct: 403 LGRMKLDVGKVLQAGVLDNWYPLQG-GQGQVHLRLE 437
>UniRef100_Q9Z1X5 GLUT4 vesicle protein [Mus musculus]
Length = 701
Score = 63.5 bits (153), Expect = 2e-08
Identities = 63/283 (22%), Positives = 124/283 (43%), Gaps = 28/283 (9%)
Query: 330 DKDLHTFVEVEIEELTRRTDVRLGS-TPRWDAPFNMVLHDNTGTLRFNLYECIPNNVKCD 388
+K+ + V++ ++++TR + + +P W+ F L D L + ++ +
Sbjct: 95 NKEPNPMVQLSVQDVTRESKATYSTNSPVWEEAFRFFLQDPRSQ---ELDVQVKDDSRAL 151
Query: 389 YLGSCEIKLRHVEDDSTI---MW----AVGPDSGIIAKQAQFCGDEIEMVVPFEGTNSGE 441
LG+ + L + S + W + GP+S + K +V+ + E
Sbjct: 152 TLGALTLPLARLLTASELTLDQWFQLSSSGPNSRLYMK----------LVMRILYLDYSE 201
Query: 442 LKVSIVVKEWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKE- 500
++ V + + + + + + + N T L+I V+E +DL A
Sbjct: 202 IRFPTVPGAQDWDRESLETGSSVDAPPRPYHTTPNSHFGTENVLRIHVLEAQDLIAKDRF 261
Query: 501 ----KTGKFDPYIKLQY-GKVMQKTKTSHTPNPVWNQTIEFDEVG-GGEYLKLKVFTEEL 554
GK DPY+KL+ GK + NP WN+ E G+ L+++VF ++L
Sbjct: 262 LGGLVKGKSDPYVKLKVAGKSFRTHVVREDLNPRWNEVFEVIVTSIPGQELEIEVFDKDL 321
Query: 555 FGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLKIEAI 597
D+ +G +V+L +++ D W+ LE V SG + L++E +
Sbjct: 322 DKDDFLGRYKVSLTTVLNSGFLDEWLTLEDVPSGRLHLRLERL 364
>UniRef100_UPI0000365141 UPI0000365141 UniRef100 entry
Length = 1576
Score = 62.4 bits (150), Expect = 4e-08
Identities = 47/162 (29%), Positives = 83/162 (51%), Gaps = 26/162 (16%)
Query: 460 LNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQK 519
+ ++++ Q L+GSS + K+ I VV + L A K++TG DPY+ +Q GK ++
Sbjct: 560 VQHMKSIKQNVLDGSS----KWSAKIAINVVSAQGLQA-KDRTGSSDPYVTIQVGKTKKR 614
Query: 520 TKTSH-TPNPVWNQTIEFDEVGGGEYLKLKVFTEE------------LFGDENIGSAQVN 566
TKT + NPVW + F+ + +KL+V+ E+ D+ +G + +
Sbjct: 615 TKTIYGNLNPVWEEKFSFECHNSSDRIKLRVWDEDDDIKSRVKQRLKRESDDFLGQSIIE 674
Query: 567 LEGLVDGSVRDVWIPLER-----VRSGEIRLKIEAIKVDDQE 603
+ L DVW LE+ SG IRL+I +++++ +E
Sbjct: 675 VRTL--SGEMDVWYNLEKRTDKSAVSGAIRLQI-SVEIEGEE 713
>UniRef100_UPI0000365140 UPI0000365140 UniRef100 entry
Length = 1550
Score = 62.4 bits (150), Expect = 4e-08
Identities = 47/162 (29%), Positives = 83/162 (51%), Gaps = 26/162 (16%)
Query: 460 LNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQK 519
+ ++++ Q L+GSS + K+ I VV + L A K++TG DPY+ +Q GK ++
Sbjct: 553 VQHMKSIKQNVLDGSS----KWSAKIAINVVSAQGLQA-KDRTGSSDPYVTIQVGKTKKR 607
Query: 520 TKTSH-TPNPVWNQTIEFDEVGGGEYLKLKVFTEE------------LFGDENIGSAQVN 566
TKT + NPVW + F+ + +KL+V+ E+ D+ +G + +
Sbjct: 608 TKTIYGNLNPVWEEKFSFECHNSSDRIKLRVWDEDDDIKSRVKQRLKRESDDFLGQSIIE 667
Query: 567 LEGLVDGSVRDVWIPLER-----VRSGEIRLKIEAIKVDDQE 603
+ L DVW LE+ SG IRL+I +++++ +E
Sbjct: 668 VRTL--SGEMDVWYNLEKRTDKSAVSGAIRLQI-SVEIEGEE 706
>UniRef100_Q610G0 Hypothetical protein CBG17495 [Caenorhabditis briggsae]
Length = 713
Score = 62.4 bits (150), Expect = 4e-08
Identities = 37/114 (32%), Positives = 62/114 (53%), Gaps = 5/114 (4%)
Query: 485 LKITVVEGKDLA---AAKEKTGKFDPYIKLQYGKVMQKTKTSHTP-NPVWNQTIE-FDEV 539
+++ V+E K+L + K GK DPY ++Q G KT+T NP+WN+ E +
Sbjct: 278 VRLKVIEAKNLENRDISFIKKGKSDPYAEIQVGSQFFKTRTIDDDLNPIWNEYFEAVVDQ 337
Query: 540 GGGEYLKLKVFTEELFGDENIGSAQVNLEGLVDGSVRDVWIPLERVRSGEIRLK 593
G+ L++++F E+ DE +G V+L+ + D W PLE + G++ LK
Sbjct: 338 ADGQKLRIELFDEDQGKDEELGRLSVDLKMVQAKGTVDKWYPLEGCKHGDLHLK 391
Score = 38.5 bits (88), Expect = 0.59
Identities = 82/453 (18%), Positives = 169/453 (37%), Gaps = 74/453 (16%)
Query: 97 EHCEWLNKLLTEIWPNYFNPKLSSRLSAIVEARLKLRKPRFLERVELQEFSLGSCPPSLA 156
E EW+NK++ ++WP Y + ++ + ++K + P + + + +G P +
Sbjct: 85 ERVEWMNKVIHQLWP-YVGEYTKTFMNDFIIPQVKAQMPGMFKNFKFTKMDMGDIPCRVG 143
Query: 157 LQGMRWSTIGDQRV---MQLGFDWDTHEMSILLLAKLAK----PLMGTARIVINSLHIKG 209
+ + +G R+ M + + D + ++ G R ++ L
Sbjct: 144 GIKVYTTNVGRDRIIVDMDVAYAGDA-DFTVSCCGFTGGMNNIQFSGKLRAILKPL---- 198
Query: 210 DLIFTPILDGKALLYSFVSAPEVRVGVAFGSGGSQSLPATEWPGVSSWLEKLFTDTLVKT 269
L + P++ G + +F+ P++ + G G E PG+ + + +
Sbjct: 199 -LPYPPMVGG--VSGTFLEMPKMDFNLT-GMG-----EMVELPGLIDAIRSVINSQIAAL 249
Query: 270 MVEPRRRCFTL-PAVDLRK---KAVGGIIYVRVISANKLSSSSFKASRRQQSGSTNGSSE 325
V P L P VD+ K G++ ++VI A L + ++ +S
Sbjct: 250 CVLPNEIVVPLAPDVDVTKLYFPEPDGVVRLKVIEAKNLENRDISFIKKGKS-------- 301
Query: 326 DVSDDKDLHTFVEVEIEELTRRTDVRLGSTPRWDAPFNMVLHDNTG-TLRFNLYECIPNN 384
D + ++V + RT + P W+ F V+ G LR L++ +
Sbjct: 302 ------DPYAEIQVGSQFFKTRT-IDDDLNPIWNEYFEAVVDQADGQKLRIELFD--EDQ 352
Query: 385 VKCDYLGSCEIKLRHVEDDSTIMWAVGPDSGIIAKQAQFCGDEIEMVVPFEGTNSGELKV 444
K + LG + L+ V+ T ++ P EG G+L +
Sbjct: 353 GKDEELGRLSVDLKMVQAKGT----------------------VDKWYPLEGCKHGDLHL 390
Query: 445 SIVVKEWQFSDGTHSLNNLRNNSQQSLNGSSNIQLRTGKKLKITVVEGKDLAAAKEKTGK 504
+ + L +L ++ G ++ + + L + + DL K K +
Sbjct: 391 KAT-----WMSLSTDLKHLERQEWEAEWGQADKPIHSA-LLMVYIDSVADLPYPKSKL-E 443
Query: 505 FDPYIKLQYGKVMQKTKTS-HTPNPVWNQTIEF 536
P++++ GK Q+T T NP++ F
Sbjct: 444 PSPFVEVSLGKETQRTPVKVKTVNPLFQSKFMF 476
>UniRef100_Q9LPH4 T3F20.10 protein [Arabidopsis thaliana]
Length = 768
Score = 62.0 bits (149), Expect = 5e-08
Identities = 57/218 (26%), Positives = 104/218 (47%), Gaps = 23/218 (10%)
Query: 69 RYQRKLLVEDLDKKWKRIILNNSPITPLEHCEWLNKLLTEIWPNYFNPKLSSR-LSAIVE 127
R +RKL E+ + +R +L++S E W+N + +IWP S + L I+
Sbjct: 47 RLKRKLQFEERKQANQRRVLSDS-----ESVRWMNYAVEKIWPICMEQIASQKILGPIIP 101
Query: 128 ARLKLRKPRFLERVELQEFSLGSCPPSLA-LQGMRWSTIGDQRVMQLGFDW-DTHEMSIL 185
L+ +P ++ +Q +G PP L ++ +R ST D V++LG ++ +MS +
Sbjct: 102 WFLEKYRPWTAKKAVIQHLYMGRNPPLLTDIRVLRQSTGDDHLVLELGMNFLAADDMSAI 161
Query: 186 LLAKLAKPL---MGTARIVINSLHIKGDLIFT-------PILDGKALLYSFVSAPEVRVG 235
L KL K L M T ++ + +H++G ++ P L + ++ ++ V
Sbjct: 162 LAVKLRKRLGFGMWT-KLHLTGMHVEGKVLIGVKFLRRWPFLGRLRVCFAEPPYFQMTVK 220
Query: 236 VAFGSGGSQSLPATEWPGVSSWLEKLFTDTLVKTMVEP 273
F G L PG++ WL+KL + +T+V+P
Sbjct: 221 PIFTHG----LDVAVLPGIAGWLDKLLSIAFEQTLVQP 254
Score = 42.0 bits (97), Expect = 0.053
Identities = 33/132 (25%), Positives = 56/132 (42%), Gaps = 21/132 (15%)
Query: 487 ITVVEGKDLAAAKEKTGKFDPYIKLQYGKVMQKTKTSH-TPNPVWNQTIEFDEV--GGGE 543
+ V E DL + + G DPY+K + G KTK T +P W++ +
Sbjct: 286 VEVFEASDLKPS-DLNGLADPYVKGKLGAYRFKTKIQKKTLSPKWHEEFKIPIFTWDSPS 344
Query: 544 YLKLKVFTEELFGDENIGSA-----------------QVNLEGLVDGSVRDVWIPLERVR 586
L ++V ++ F D+ +G A VN+E G D+W+ L+ ++
Sbjct: 345 ILNIEVGDKDRFVDDTLGFAPEPQFQYSKLVEYQNECSVNIEEFRGGQRNDMWLSLQNIK 404
Query: 587 SGEIRLKIEAIK 598
G + L I I+
Sbjct: 405 MGRLHLAITVIE 416
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,058,239,255
Number of Sequences: 2790947
Number of extensions: 45413457
Number of successful extensions: 114037
Number of sequences better than 10.0: 664
Number of HSP's better than 10.0 without gapping: 94
Number of HSP's successfully gapped in prelim test: 572
Number of HSP's that attempted gapping in prelim test: 113207
Number of HSP's gapped (non-prelim): 1164
length of query: 620
length of database: 848,049,833
effective HSP length: 134
effective length of query: 486
effective length of database: 474,062,935
effective search space: 230394586410
effective search space used: 230394586410
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 78 (34.7 bits)
Medicago: description of AC146563.9


