

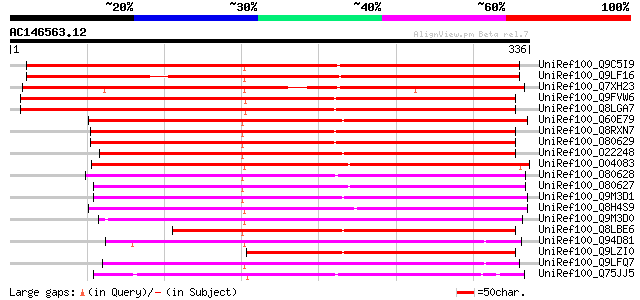
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146563.12 - phase: 0
(336 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9C5I9 Putative lipase [Arabidopsis thaliana] 424 e-117
UniRef100_Q9LF16 Lipase-like protein [Arabidopsis thaliana] 393 e-108
UniRef100_Q7XH23 Putative lipase-like protein [Oryza sativa] 376 e-103
UniRef100_Q9FVW6 Lysophospholipase isolog, putative [Arabidopsis... 352 8e-96
UniRef100_Q8LGA7 Lysophospholipase isolog, putative [Arabidopsis... 352 8e-96
UniRef100_Q60E79 Putative phospholipase [Oryza sativa] 252 1e-65
UniRef100_Q8RXN7 Putative phospholipase [Arabidopsis thaliana] 246 8e-64
UniRef100_O80629 Putative phospholipase [Arabidopsis thaliana] 246 8e-64
UniRef100_O22248 Putative phospholipase [Arabidopsis thaliana] 241 2e-62
UniRef100_O04083 Lysophospholipase isolog; 25331-24357 [Arabidop... 236 9e-61
UniRef100_O80628 Putative phospholipase; alternative splicing is... 233 6e-60
UniRef100_O80627 Putative phospholipase [Arabidopsis thaliana] 231 2e-59
UniRef100_Q9M3D1 Lipase-like protein [Arabidopsis thaliana] 223 4e-57
UniRef100_Q8H4S9 Putative lysophospholipase homolog [Oryza sativa] 212 1e-53
UniRef100_Q9M3D0 Lipase-like protein [Arabidopsis thaliana] 211 2e-53
UniRef100_Q8LBE6 Putative phospholipase [Arabidopsis thaliana] 210 5e-53
UniRef100_Q94D81 Phospholipase-like protein [Oryza sativa] 176 1e-42
UniRef100_Q9LZI0 Hypothetical protein F26K9_290 [Arabidopsis tha... 174 2e-42
UniRef100_Q9LFQ7 Lysophospholipase-like protein [Arabidopsis tha... 170 4e-41
UniRef100_Q75JJ5 Similar to Arabidopsis thaliana (Mouse-ear cres... 169 8e-41
>UniRef100_Q9C5I9 Putative lipase [Arabidopsis thaliana]
Length = 351
Score = 424 bits (1089), Expect = e-117
Identities = 198/332 (59%), Positives = 264/332 (78%), Gaps = 14/332 (4%)
Query: 12 SEELQNIYLSNMDEAPARRLAREAFKDIQLAIDHCLFQLPADGVKMEEIYEVNSRGLKVF 71
+EEL+ + N+DEAP RR R++ KDIQL +DH LF+ P +G+K +E +EVNSRG+++F
Sbjct: 10 NEELRRLREVNIDEAPGRRRVRDSLKDIQLNLDHILFKTPENGIKTKESFEVNSRGVEIF 69
Query: 72 SKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSDGLHGY 131
SKSW+PE S + +V +CHGY DTCTF+FEG+AR+LA SG+GVFA+DYPGFGLS+GLHGY
Sbjct: 70 SKSWLPEASKPRALVCFCHGYGDTCTFFFEGIARRLALSGYGVFAMDYPGFGLSEGLHGY 129
Query: 132 IPSFENLVNDVIEHFSKIK-------------GESMGGAIALNIHFKQPTAWDGAALIAP 178
IPSF+ LV DVIEH+S IK G+SMGGA++L IH KQP AW GA L+AP
Sbjct: 130 IPSFDLLVQDVIEHYSNIKANPEFSSLPSFLFGQSMGGAVSLKIHLKQPNAWAGAVLLAP 189
Query: 179 LCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRELAPYNVLFYKD 238
+CK A+D++P ++KQILIG+A VLPK KLVPQK ++ E +RD RKR++ PYN++ Y
Sbjct: 190 MCKIADDLVPPPVLKQILIGLANVLPKHKLVPQK-DLAEAGFRDIRKRDMTPYNMICYSG 248
Query: 239 KPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKAKVKDKKLCL 298
KPRL TA+E+L+ TQ++E++L+EVSLP+L++HGEAD +TDPS S+ LY+KAK DKK+ L
Sbjct: 249 KPRLRTAVEMLRTTQDIEKQLQEVSLPILILHGEADTVTDPSVSRELYEKAKSPDKKIVL 308
Query: 299 YKDAFHTLLEGEPDETIFHVLDDIISWLDDHS 330
Y++A+H+LLEGEPD+ I VL DIISWL+DHS
Sbjct: 309 YENAYHSLLEGEPDDMILRVLSDIISWLNDHS 340
>UniRef100_Q9LF16 Lipase-like protein [Arabidopsis thaliana]
Length = 340
Score = 393 bits (1009), Expect = e-108
Identities = 189/332 (56%), Positives = 254/332 (75%), Gaps = 25/332 (7%)
Query: 12 SEELQNIYLSNMDEAPARRLAREAFKDIQLAIDHCLFQLPADGVKMEEIYEVNSRGLKVF 71
+EEL+ + N+DEAP RR R++ KDIQL +DH LF+ P +G+K +E +EVNSRG+++F
Sbjct: 10 NEELRRLREVNIDEAPGRRRVRDSLKDIQLNLDHILFKTPENGIKTKESFEVNSRGVEIF 69
Query: 72 SKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSDGLHGY 131
SKSW+PE S + +V +CHG +AR+LA SG+GVFA+DYPGFGLS+GLHGY
Sbjct: 70 SKSWLPEASKPRALVCFCHG-----------IARRLALSGYGVFAMDYPGFGLSEGLHGY 118
Query: 132 IPSFENLVNDVIEHFSKIK-------------GESMGGAIALNIHFKQPTAWDGAALIAP 178
IPSF+ LV DVIEH+S IK G+SMGGA++L IH KQP AW GA L+AP
Sbjct: 119 IPSFDLLVQDVIEHYSNIKANPEFSSLPSFLFGQSMGGAVSLKIHLKQPNAWAGAVLLAP 178
Query: 179 LCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRELAPYNVLFYKD 238
+CK A+D++P ++KQILIG+A VLPK KLVPQK+ + E +RD RKR++ PYN++ Y
Sbjct: 179 MCKIADDLVPPPVLKQILIGLANVLPKHKLVPQKD-LAEAGFRDIRKRDMTPYNMICYSG 237
Query: 239 KPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKAKVKDKKLCL 298
KPRL TA+E+L+ TQ++E++L+EVSLP+L++HGEAD +TDPS S+ LY+KAK DKK+ L
Sbjct: 238 KPRLRTAVEMLRTTQDIEKQLQEVSLPILILHGEADTVTDPSVSRELYEKAKSPDKKIVL 297
Query: 299 YKDAFHTLLEGEPDETIFHVLDDIISWLDDHS 330
Y++A+H+LLEGEPD+ I VL DIISWL+DHS
Sbjct: 298 YENAYHSLLEGEPDDMILRVLSDIISWLNDHS 329
>UniRef100_Q7XH23 Putative lipase-like protein [Oryza sativa]
Length = 464
Score = 376 bits (965), Expect = e-103
Identities = 195/393 (49%), Positives = 255/393 (64%), Gaps = 81/393 (20%)
Query: 9 EGMSEELQNIYL-SNMDEAPARRLAREAFKDIQLAIDHCLFQLPADGVKMEEI------- 60
EG+ + L+ + L + +D+AP RR R+AFKD+QL+IDHCLF+ + +E+
Sbjct: 81 EGVDQALERMVLRACLDQAPERRRVRDAFKDVQLSIDHCLFKGQYSDIGTKEVLFLALAI 140
Query: 61 -------------------------YEVNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADT 95
YE NSRG+++FSK W PE +K IV CHGY DT
Sbjct: 141 ILQFLHAKLAILMIIGCGSQGSGTSYEKNSRGVEIFSKCWYPENHRIKAIVCLCHGYGDT 200
Query: 96 CTFYFEGVARKLASSGFGVFALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKIK----- 150
CTF+ +G+ARK+AS+G+GVFALDYPGFGLS+GLHG+IPSF+ LV+DV EHF+K+K
Sbjct: 201 CTFFLDGIARKIASAGYGVFALDYPGFGLSEGLHGFIPSFDTLVDDVAEHFTKVKENPEH 260
Query: 151 --------GESMGGAIALNIHFKQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKV 202
G+SMGGA+AL IHFKQP WDGA L+AP+C KQ+LI +A++
Sbjct: 261 RGLPSFLFGQSMGGAVALKIHFKQPNEWDGAILVAPMC------------KQVLIFMARL 308
Query: 203 LPKTKLVPQKEEVKENIYRDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEE- 261
LPK KLVPQK ++ E +++ +K+E YNV+ YKDKPRL TALE+L+ T+E+E RLEE
Sbjct: 309 LPKEKLVPQK-DLAELAFKEKKKQEQCSYNVIAYKDKPRLRTALEMLRTTKEIESRLEEF 367
Query: 262 ---------------------VSLPLLVMHGEADIITDPSASKALYQKAKVKDKKLCLYK 300
VSLP++++HGE D++TDP+ SKALY KAK DK L LYK
Sbjct: 368 FTSCRIANGLLFRSITISVPLVSLPIIILHGEGDLVTDPAVSKALYDKAKSSDKTLRLYK 427
Query: 301 DAFHTLLEGEPDETIFHVLDDIISWLDDHSSTK 333
DA+H +LEGEPDE IF VLDDIISWLD HS+ K
Sbjct: 428 DAYHAILEGEPDEAIFQVLDDIISWLDQHSTKK 460
>UniRef100_Q9FVW6 Lysophospholipase isolog, putative [Arabidopsis thaliana]
Length = 382
Score = 352 bits (903), Expect = 8e-96
Identities = 167/334 (50%), Positives = 242/334 (72%), Gaps = 15/334 (4%)
Query: 8 IEGMSEELQNIYLSNMDEAPARRLAREAFKDIQLAIDHCLFQLPADGVKMEEIYEVNSRG 67
I+G+S+EL I N+D+APARRLAR AF D+QL +DHCLF+ G++ EE YE NS+G
Sbjct: 44 IDGVSDELNLIASQNLDQAPARRLARSAFVDLQLQLDHCLFKKAPSGIRTEEWYERNSKG 103
Query: 68 LKVFSKSWIPEKSP-MKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSD 126
+F KSW+P+ +K V +CHGY TCTF+F+G+A+++A G+GV+A+D+PGFGLSD
Sbjct: 104 EDIFCKSWLPKSGDEIKAAVCFCHGYGSTCTFFFDGIAKQIAGFGYGVYAIDHPGFGLSD 163
Query: 127 GLHGYIPSFENLVNDVIEHFSKIKG-------------ESMGGAIALNIHFKQPTAWDGA 173
GLHG+IPSF++L ++ IE F+K+KG +SMGGA+AL IH K+P AWDG
Sbjct: 164 GLHGHIPSFDDLADNAIEQFTKMKGRSELRNLPRFLLGQSMGGAVALKIHLKEPQAWDGL 223
Query: 174 ALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRELAPYNV 233
L+AP+CK +ED+ P LV + LI ++ + PK KL P K ++ + +RD KR+L Y+V
Sbjct: 224 ILVAPMCKISEDVKPPPLVLKTLILMSTLFPKAKLFP-KRDLSDFFFRDLSKRKLCEYDV 282
Query: 234 LFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKAKVKD 293
+ Y D+ RL TA+ELL AT+++E ++++VSLPLL++HG+ D +TDP+ SK L++ A +D
Sbjct: 283 ICYDDQTRLKTAVELLNATRDIEMQVDKVSLPLLILHGDTDKVTDPTVSKFLHKHAVSQD 342
Query: 294 KKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLD 327
K L LY +H +LEG+ DE IF V++DI++WLD
Sbjct: 343 KTLKLYPGGYHCILEGDTDENIFTVINDIVAWLD 376
>UniRef100_Q8LGA7 Lysophospholipase isolog, putative [Arabidopsis thaliana]
Length = 382
Score = 352 bits (903), Expect = 8e-96
Identities = 167/334 (50%), Positives = 242/334 (72%), Gaps = 15/334 (4%)
Query: 8 IEGMSEELQNIYLSNMDEAPARRLAREAFKDIQLAIDHCLFQLPADGVKMEEIYEVNSRG 67
I+G+S+EL I N+D+APARRLAR AF D+QL +DHCLF+ G++ EE YE NS+G
Sbjct: 44 IDGVSDELNLIASQNLDQAPARRLARSAFVDLQLQLDHCLFKKAPSGIRTEEWYERNSKG 103
Query: 68 LKVFSKSWIPEKSP-MKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYPGFGLSD 126
+F KSW+P+ +K V +CHGY TCTF+F+G+A+++A G+GV+A+D+PGFGLSD
Sbjct: 104 EDIFCKSWLPKSGDEIKAAVCFCHGYGSTCTFFFDGIAKQIAGFGYGVYAIDHPGFGLSD 163
Query: 127 GLHGYIPSFENLVNDVIEHFSKIKG-------------ESMGGAIALNIHFKQPTAWDGA 173
GLHG+IPSF++L ++ IE F+K+KG +SMGGA+AL IH K+P AWDG
Sbjct: 164 GLHGHIPSFDDLADNAIEQFTKMKGRSELRNLPRFLLGQSMGGAVALKIHLKEPQAWDGL 223
Query: 174 ALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRELAPYNV 233
L+AP+CK +ED+ P LV + LI ++ + PK KL P K ++ + +RD KR+L Y+V
Sbjct: 224 ILVAPMCKISEDVKPPPLVLKTLILMSTLFPKAKLFP-KRDLSDFFFRDLSKRKLCEYDV 282
Query: 234 LFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQKAKVKD 293
+ Y D+ RL TA+ELL AT+++E ++++VSLPLL++HG+ D +TDP+ SK L++ A +D
Sbjct: 283 ICYDDQTRLKTAVELLNATRDIEMQVDKVSLPLLILHGDTDKVTDPTVSKFLHKHAVSQD 342
Query: 294 KKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLD 327
K L LY +H +LEG+ DE IF V++DI++WLD
Sbjct: 343 KTLKLYPGGYHCILEGDTDENIFTVINDIVAWLD 376
>UniRef100_Q60E79 Putative phospholipase [Oryza sativa]
Length = 351
Score = 252 bits (643), Expect = 1e-65
Identities = 127/297 (42%), Positives = 183/297 (60%), Gaps = 14/297 (4%)
Query: 52 ADGVKMEEIYEVNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSG 111
AD V+ E + N RGL++F+ W+P S K +++ CHGY + + + +LA++G
Sbjct: 6 ADDVEYHEEFVTNPRGLRLFTCGWLPASSSPKALIFLCHGYGMEVSGFMKACGVELATAG 65
Query: 112 FGVFALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKI-------------KGESMGGAI 158
+GVF +DY G G S G YI FE+LV+D F I GESMGGA+
Sbjct: 66 YGVFGIDYEGHGKSMGARCYIQKFEHLVDDCDRFFKSICELEEYRDKSRFLYGESMGGAV 125
Query: 159 ALNIHFKQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKEN 218
AL +H K PT WDGA L+AP+CK +E + PH LV +L V +++PK K+VP K+ + ++
Sbjct: 126 ALLLHRKDPTFWDGAVLVAPMCKISEKVKPHPLVVTLLTQVEEIIPKWKIVPTKDVI-DS 184
Query: 219 IYRDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITD 278
++D KRE N L Y+DKPRL TALELL+ + +EQ L +VS+P ++HGEAD +TD
Sbjct: 185 AFKDPIKREKIRKNKLIYQDKPRLKTALELLRTSISVEQSLSQVSIPFFILHGEADKVTD 244
Query: 279 PSASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHSSTKNK 335
P S+ALY++A DK + LY +H L GEPD + V DI++WLD S +++
Sbjct: 245 PEVSRALYERAASADKTIKLYPGMWHGLTAGEPDHNVHLVFSDIVAWLDRRSHRQDR 301
>UniRef100_Q8RXN7 Putative phospholipase [Arabidopsis thaliana]
Length = 317
Score = 246 bits (627), Expect = 8e-64
Identities = 125/288 (43%), Positives = 176/288 (60%), Gaps = 14/288 (4%)
Query: 53 DGVKMEEIYEVNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGF 112
+ +K EE + N+RG+K+F+ W+P K K +V+ CHGYA C+ AR+L +GF
Sbjct: 6 ENIKYEESFIKNTRGMKLFTCKWVPAKQEPKALVFICHGYAMECSITMNSTARRLVKAGF 65
Query: 113 GVFALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKI-------------KGESMGGAIA 159
V+ +DY G G SDGL Y+P+F++LV+DV H++ I GESMGGA+
Sbjct: 66 AVYGIDYEGHGKSDGLSAYVPNFDHLVDDVSTHYTSICEKEENKGKMRFLLGESMGGAVL 125
Query: 160 LNIHFKQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENI 219
L +H K+P WDGA L+AP+CK AE+M P LV IL ++ V+P K++P +++ E
Sbjct: 126 LLLHRKKPQFWDGAVLVAPMCKIAEEMKPSPLVISILAKLSGVIPSWKIIP-GQDIIETA 184
Query: 220 YRDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDP 279
++ R+ N YK +PRL TA ELL+ + +LE+RL EVSLP +V+HGE D +TD
Sbjct: 185 FKQPEIRKQVRENPYCYKGRPRLKTAYELLRVSTDLEKRLNEVSLPFIVLHGEDDKVTDK 244
Query: 280 SASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLD 327
+ S+ LY+ A DK LY +H LL GE E I V DII WLD
Sbjct: 245 AVSRQLYEVASSSDKTFKLYPGMWHGLLYGETPENIETVFADIIGWLD 292
>UniRef100_O80629 Putative phospholipase [Arabidopsis thaliana]
Length = 318
Score = 246 bits (627), Expect = 8e-64
Identities = 125/288 (43%), Positives = 176/288 (60%), Gaps = 14/288 (4%)
Query: 53 DGVKMEEIYEVNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGF 112
+ +K EE + N+RG+K+F+ W+P K K +V+ CHGYA C+ AR+L +GF
Sbjct: 7 ENIKYEESFIKNTRGMKLFTCKWVPAKQEPKALVFICHGYAMECSITMNSTARRLVKAGF 66
Query: 113 GVFALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKI-------------KGESMGGAIA 159
V+ +DY G G SDGL Y+P+F++LV+DV H++ I GESMGGA+
Sbjct: 67 AVYGIDYEGHGKSDGLSAYVPNFDHLVDDVSTHYTSICEKEENKGKMRFLLGESMGGAVL 126
Query: 160 LNIHFKQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENI 219
L +H K+P WDGA L+AP+CK AE+M P LV IL ++ V+P K++P +++ E
Sbjct: 127 LLLHRKKPQFWDGAVLVAPMCKIAEEMKPSPLVISILAKLSGVIPSWKIIP-GQDIIETA 185
Query: 220 YRDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDP 279
++ R+ N YK +PRL TA ELL+ + +LE+RL EVSLP +V+HGE D +TD
Sbjct: 186 FKQPEIRKQVRENPYCYKGRPRLKTAYELLRVSTDLEKRLNEVSLPFIVLHGEDDKVTDK 245
Query: 280 SASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLD 327
+ S+ LY+ A DK LY +H LL GE E I V DII WLD
Sbjct: 246 AVSRQLYEVASSSDKTFKLYPGMWHGLLYGETPENIETVFADIIGWLD 293
>UniRef100_O22248 Putative phospholipase [Arabidopsis thaliana]
Length = 351
Score = 241 bits (616), Expect = 2e-62
Identities = 125/283 (44%), Positives = 174/283 (61%), Gaps = 15/283 (5%)
Query: 59 EIYEVNSRGLKVFSKSWIPEKSPM-KGIVYYCHGYADTCTFYFEGVARKLASSGFGVFAL 117
E Y NSRG+++F+ WIP S K +V+ CHGY C+ + +LAS+G+ VF +
Sbjct: 10 EEYVRNSRGVELFACRWIPSSSSSPKALVFLCHGYGMECSDSMKECGIRLASAGYAVFGM 69
Query: 118 DYPGFGLSDGLHGYIPSFENLVNDVIEHFSKI-------------KGESMGGAIALNIHF 164
DY G G S G YI F N+VND ++++ I GESMGGA+ L +H
Sbjct: 70 DYEGHGRSMGSRCYIKKFANVVNDCYDYYTSICAQEEYMDKGRFLYGESMGGAVTLLLHK 129
Query: 165 KQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDAR 224
K P W+GA L+AP+CK +E + PH +V +L V +++PK K+VP K+ + + ++D
Sbjct: 130 KDPLFWNGAILVAPMCKISEKVKPHPIVINLLTRVEEIIPKWKIVPTKDVI-DAAFKDLV 188
Query: 225 KRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKA 284
KRE N L Y+DKPRL TALE+L+ + LE L E+++P V+HGEAD +TDP SKA
Sbjct: 189 KREEVRNNKLIYQDKPRLKTALEMLRTSMNLEDTLHEITMPFFVLHGEADTVTDPEVSKA 248
Query: 285 LYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLD 327
LY+KA +DK L LY +H L GEPD + V DII+WLD
Sbjct: 249 LYEKASTRDKTLKLYPGMWHALTSGEPDCNVDLVFADIINWLD 291
>UniRef100_O04083 Lysophospholipase isolog; 25331-24357 [Arabidopsis thaliana]
Length = 324
Score = 236 bits (601), Expect = 9e-61
Identities = 121/300 (40%), Positives = 185/300 (61%), Gaps = 18/300 (6%)
Query: 54 GVKMEEIYEVNSRGLKVFSKSWIPEKS-PMKGIVYYCHGYADTCTFYFEGVARKLASSGF 112
G+ + + + RGL +F++SW+P S P +G+++ HGY + ++ F+ LA GF
Sbjct: 26 GIIGSKSFFTSPRGLNLFTRSWLPSSSSPPRGLIFMVHGYGNDVSWTFQSTPIFLAQMGF 85
Query: 113 GVFALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKIK-------------GESMGGAIA 159
FALD G G SDG+ Y+PS + +V+D+I F+ IK GESMGGAI
Sbjct: 86 ACFALDIEGHGRSDGVRAYVPSVDLVVDDIISFFNSIKQNPKFQGLPRFLFGESMGGAIC 145
Query: 160 LNIHFKQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENI 219
L I F P +DGA L+AP+CK ++ + P W V Q LI +++ LP +VP ++ ++++I
Sbjct: 146 LLIQFADPLGFDGAVLVAPMCKISDKVRPKWPVDQFLIMISRFLPTWAIVPTEDLLEKSI 205
Query: 220 YRDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDP 279
+ K+ +A N + Y +KPRLGT +ELL+ T L ++L++VS+P +++HG AD +TDP
Sbjct: 206 -KVEEKKPIAKRNPMRYNEKPRLGTVMELLRVTDYLGKKLKDVSIPFIIVHGSADAVTDP 264
Query: 280 SASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDH---SSTKNKV 336
S+ LY+ AK KDK L +Y H++L GEPD+ I V DI+SWL+D TK +V
Sbjct: 265 EVSRELYEHAKSKDKTLKIYDGMMHSMLFGEPDDNIEIVRKDIVSWLNDRCGGDKTKTQV 324
>UniRef100_O80628 Putative phospholipase; alternative splicing isoform [Arabidopsis
thaliana]
Length = 317
Score = 233 bits (594), Expect = 6e-60
Identities = 123/298 (41%), Positives = 175/298 (58%), Gaps = 14/298 (4%)
Query: 50 LPADGVKMEEIYEVNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLAS 109
+ + +K EE + N+RG K+F+ W+P + +V+ CHGY C+ AR+L
Sbjct: 3 IETEDIKYEESFIKNTRGFKLFTCRWLPTNREPRALVFLCHGYGMECSITMNSTARRLVK 62
Query: 110 SGFGVFALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKI-------------KGESMGG 156
+GF V+ +DY G G SDGL YI +F+ LV+DV H++ I GESMGG
Sbjct: 63 AGFAVYGMDYEGHGKSDGLSAYISNFDRLVDDVSTHYTAICEREENKWKMRFMLGESMGG 122
Query: 157 AIALNIHFKQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVK 216
A+ L + K P WDGA L+AP+CK AE+M P V IL + ++PK K++P +++
Sbjct: 123 AVVLLLGRKNPDFWDGAILVAPMCKIAEEMKPSPFVISILTKLISIIPKWKIIPS-QDII 181
Query: 217 ENIYRDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADII 276
E Y++ R+ N L K +PRL TA ELL+ + +LE+RL+EVSLP LV+HG+ D +
Sbjct: 182 EISYKEPEIRKQVRENPLCSKGRPRLKTAYELLRISNDLEKRLQEVSLPFLVLHGDDDKV 241
Query: 277 TDPSASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHSSTKN 334
TD + S+ LY+ A DK L LY +H LL GE E I V D+ISWL+ S N
Sbjct: 242 TDKAVSQELYKVALSADKTLKLYPGMWHGLLTGETPENIEIVFADVISWLEKRSDYGN 299
>UniRef100_O80627 Putative phospholipase [Arabidopsis thaliana]
Length = 311
Score = 231 bits (590), Expect = 2e-59
Identities = 123/293 (41%), Positives = 177/293 (59%), Gaps = 14/293 (4%)
Query: 55 VKMEEIYEVNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGV 114
V EE + +NSRG+K+F+ W P K K +++ CHGYA + A +LA++GF V
Sbjct: 2 VMYEEDFVLNSRGMKLFTCVWKPVKQEPKALLFLCHGYAMESSITMNSAATRLANAGFAV 61
Query: 115 FALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKI-------------KGESMGGAIALN 161
+ +DY G G S+GL+GYI +F++LV+DV H+S I GESMGGA+ L
Sbjct: 62 YGMDYEGHGKSEGLNGYISNFDDLVDDVSNHYSTICEREENKGKMRFLLGESMGGAVVLL 121
Query: 162 IHFKQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYR 221
+ K+P WDGA L+AP+CK A+++ PH +V ILI +AK +P K+VP + + I +
Sbjct: 122 LARKKPDFWDGAVLVAPMCKLADEIKPHPVVISILIKLAKFIPTWKIVPGNDIIDIAI-K 180
Query: 222 DARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSA 281
+ R N YK +PRL TA +LL + +LE+ L +VS+P +V+HGE D +TD S
Sbjct: 181 EPHIRNQVRENKYCYKGRPRLNTAYQLLLVSLDLEKNLHQVSIPFIVLHGEDDKVTDKSI 240
Query: 282 SKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHSSTKN 334
SK LY+ A DK LY +H LL GE +E V DII+WL+D ++ N
Sbjct: 241 SKMLYEVASSSDKTFKLYPKMWHALLYGETNENSEIVFGDIINWLEDRATDSN 293
>UniRef100_Q9M3D1 Lipase-like protein [Arabidopsis thaliana]
Length = 312
Score = 223 bits (569), Expect = 4e-57
Identities = 118/295 (40%), Positives = 175/295 (59%), Gaps = 15/295 (5%)
Query: 55 VKMEEIYEVNSRGLKVFSKSWIPEKSPM-KGIVYYCHGYADTCTFYFEGVARKLASSGFG 113
V +E Y NSRG+++F+ SW E+ K +++ CHGYA + A +LA++GF
Sbjct: 2 VMYKEDYVSNSRGIQLFTCSWKQEEQQEPKALIFLCHGYAMESSITMSSTAVRLANAGFS 61
Query: 114 VFALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKI-------------KGESMGGAIAL 160
V+ +DY G G S GL+GY+ F++LV DV H+S I GESMGGA+ L
Sbjct: 62 VYGMDYEGHGKSGGLNGYVKKFDDLVQDVSSHYSSICELEENKGKMRFLMGESMGGAVVL 121
Query: 161 NIHFKQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIY 220
+ K+P WDGA L+AP+CK AED+ PH +V L + + +P K+VP + + + +
Sbjct: 122 LLERKKPNFWDGAVLVAPMCKLAEDIKPHPMVISFLTKLTRFIPTWKIVPSNDII-DVAF 180
Query: 221 RDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPS 280
++ R+ N YK +PRL TA +LL + +LE+ L++VS+P +V+HGE D +TD +
Sbjct: 181 KETHIRKQVRDNEYCYKGRPRLKTAHQLLMVSLDLEKNLDQVSMPFIVLHGEDDKVTDKN 240
Query: 281 ASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHSSTKNK 335
SK LY+ A DK LY + +H LL GE E + V DIISWL + +S N+
Sbjct: 241 VSKLLYEVASSSDKTFKLYPNMWHGLLYGESPENLEIVFSDIISWLKERASVTNQ 295
>UniRef100_Q8H4S9 Putative lysophospholipase homolog [Oryza sativa]
Length = 334
Score = 212 bits (540), Expect = 1e-53
Identities = 115/299 (38%), Positives = 172/299 (57%), Gaps = 16/299 (5%)
Query: 52 ADGVKMEEIYEVNSRGLKVFSKSWIPE-KSPMKGIVYYCHGYADTCTFYFEGVARKLASS 110
A G E Y G ++F+++W P + +V+ HGY + ++ F+ A LA S
Sbjct: 29 AHGADGESSYFTPPGGRRLFTRAWRPRGDGAPRALVFMVHGYGNDISWTFQSTAVFLARS 88
Query: 111 GFGVFALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKIK-------------GESMGGA 157
GF FA D PG G S GL ++P ++ + D++ F ++ GESMGGA
Sbjct: 89 GFACFAADLPGHGRSHGLRAFVPDLDSAIADLLAFFRSVRRREEHAGLPCFLFGESMGGA 148
Query: 158 IALNIHFKQPTA-WDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVK 216
I L IH + P W GA L+AP+CK ++ + P W + QIL VA+ P +VP + ++
Sbjct: 149 ICLLIHLRTPPEEWAGAVLVAPMCKISDRIRPPWPLPQILTFVARFAPTLAIVPTADLIE 208
Query: 217 ENIYRDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADII 276
+++ A KR +A N + Y +PRLGT +ELL+AT EL RL EV++P LV+HG AD +
Sbjct: 209 KSVKVPA-KRLIAARNPMRYSGRPRLGTVVELLRATDELGARLGEVTVPFLVVHGSADEV 267
Query: 277 TDPSASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHSSTKNK 335
TDP S+ALY A KDK + +Y H++L GEPDE I V DI++WL++ + + +
Sbjct: 268 TDPDISRALYDAAASKDKTIKIYDGMMHSMLFGEPDENIERVRADILAWLNERCTPREE 326
>UniRef100_Q9M3D0 Lipase-like protein [Arabidopsis thaliana]
Length = 319
Score = 211 bits (538), Expect = 2e-53
Identities = 112/288 (38%), Positives = 162/288 (55%), Gaps = 14/288 (4%)
Query: 58 EEIYEVNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFAL 117
EE E NSRG+++ + W P + ++++CHGYA C+ F+ +A K A GF V +
Sbjct: 12 EEFIE-NSRGMQLLTCKWFPVNQEPRALIFFCHGYAIDCSTTFKDIAPKFAKEGFAVHGI 70
Query: 118 DYPGFGLSDGLHGYIPSFENLVNDVIEHFSKIK-------------GESMGGAIALNIHF 164
+Y G G S GL YI +F+ L++DV HFSKI GESMGGA+ L +H
Sbjct: 71 EYEGHGRSSGLSVYIDNFDLLIDDVSSHFSKISEMGDNTKKKRFLMGESMGGAVVLLLHR 130
Query: 165 KQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDAR 224
K+P WDG LIAP+CK AE+M P +V ++ V ++P K + ++ + +
Sbjct: 131 KKPEFWDGGILIAPMCKIAEEMKPSRMVISMINMVTNLIPSWKSIIHGPDILNSAIKLPE 190
Query: 225 KRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKA 284
KR N Y PR+ T EL + + +LE RL EV++P +V+HGE D +TD SK
Sbjct: 191 KRHEIRTNPNCYNGWPRMKTMSELFRISLDLENRLNEVTMPFIVLHGEDDKVTDKGGSKL 250
Query: 285 LYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHSST 332
LY+ A DK L LY + +H+LL GEP E V +DI+ W+ +T
Sbjct: 251 LYEVALSNDKTLKLYPEMWHSLLFGEPPENSEIVFNDIVQWMQTRITT 298
>UniRef100_Q8LBE6 Putative phospholipase [Arabidopsis thaliana]
Length = 306
Score = 210 bits (534), Expect = 5e-53
Identities = 107/235 (45%), Positives = 148/235 (62%), Gaps = 14/235 (5%)
Query: 106 KLASSGFGVFALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKI-------------KGE 152
+LAS+G+ VF +DY G G S G YI F N+VND ++++ I GE
Sbjct: 13 RLASAGYAVFGMDYEGHGRSMGSRCYIKKFANVVNDCYDYYTSICAQEEYMDKGRFLYGE 72
Query: 153 SMGGAIALNIHFKQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQK 212
SMGGA+ L +H K P W+GA L+AP+CK +E + PH +V +L V +++PK K+VP K
Sbjct: 73 SMGGAVTLLLHKKDPLFWNGAILVAPMCKISEKVKPHPIVINLLTRVEEIIPKWKIVPTK 132
Query: 213 EEVKENIYRDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGE 272
+ + + ++D KRE N L Y+DKPRL TALE+L+ + LE L E+++P V+HGE
Sbjct: 133 DVI-DAAFKDLVKREEVRNNKLIYQDKPRLKTALEMLRTSMNLEDTLHEITMPFFVLHGE 191
Query: 273 ADIITDPSASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLD 327
AD +TDP SKALY+KA +DK L LY +H L GEPD + V DII+WLD
Sbjct: 192 ADTVTDPEVSKALYEKASTRDKTLKLYPGMWHALTSGEPDCNVDLVFADIINWLD 246
>UniRef100_Q94D81 Phospholipase-like protein [Oryza sativa]
Length = 329
Score = 176 bits (445), Expect = 1e-42
Identities = 98/284 (34%), Positives = 154/284 (53%), Gaps = 16/284 (5%)
Query: 63 VNSRGLKVFSKSWIPE--KSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYP 120
VN RGL++F++ W+P +P+ G + HG+ ++ + A A +GF V A+D+
Sbjct: 36 VNPRGLRIFTQRWVPAGGDAPLLGAIAVVHGFTGESSWTVQLTAVHFAKAGFAVAAVDHQ 95
Query: 121 GFGLSDGLHGYIPSFENLVNDVIEHFSKIK------------GESMGGAIALNIHFKQPT 168
G G S+GL G+IP ++ D F+ + GES+GGAIAL +H +
Sbjct: 96 GHGFSEGLQGHIPDIVPVLEDCEAAFAPFRADYPPPLPCFLYGESLGGAIALLLHLRDKE 155
Query: 169 AW-DGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRE 227
W DGA L +C + +P W ++ +L A V P +L + + + ++ KR
Sbjct: 156 RWRDGAVLNGAMCGVSPRFMPPWPLEHLLWAAAAVAPTWRLAFTRGNIPDRSFKVPWKRA 215
Query: 228 LAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQ 287
LA + PR TALELL+ +EL+ R EEV LPLLV+HG D + DP ++ L++
Sbjct: 216 LAVASPRRTTAPPRAATALELLRVCRELQSRFEEVELPLLVVHGGEDTVCDPGCAEELHR 275
Query: 288 KAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHSS 331
+A KDK L +Y +H L+ GEP+E + V D++ WL H++
Sbjct: 276 RAGSKDKTLRVYPGMWHQLV-GEPEENVDKVFGDVLDWLKSHAA 318
>UniRef100_Q9LZI0 Hypothetical protein F26K9_290 [Arabidopsis thaliana]
Length = 226
Score = 174 bits (442), Expect = 2e-42
Identities = 84/174 (48%), Positives = 119/174 (68%), Gaps = 1/174 (0%)
Query: 154 MGGAIALNIHFKQPTAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKE 213
MGGA+AL +H K P+ W+GA L+AP+CK +E + PH +V +L V ++PK K+VP K+
Sbjct: 1 MGGAVALLLHKKDPSFWNGALLVAPMCKISEKVKPHPVVINLLTRVEDIIPKWKIVPTKD 60
Query: 214 EVKENIYRDARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEA 273
+ + ++D KRE N L Y+DKPRL TALE+L+ + +LE L E++LP V+HGEA
Sbjct: 61 VI-DAAFKDPVKREEIRNNKLIYQDKPRLKTALEMLRTSMDLEDTLHEITLPFFVLHGEA 119
Query: 274 DIITDPSASKALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLD 327
DI+TDP SKAL++KA +DK + LY +H L GEPD + V DI++WLD
Sbjct: 120 DIVTDPEISKALFEKASTRDKTIKLYPGMWHGLTSGEPDANVDLVFADIVNWLD 173
>UniRef100_Q9LFQ7 Lysophospholipase-like protein [Arabidopsis thaliana]
Length = 327
Score = 170 bits (431), Expect = 4e-41
Identities = 88/283 (31%), Positives = 151/283 (53%), Gaps = 14/283 (4%)
Query: 61 YEVNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGVFALDYP 120
Y N GLK+F++ W P P G++ HG+ +++ + + A SG+ A+D+
Sbjct: 35 YVTNPTGLKLFTQWWTPLNRPPLGLIAVVHGFTGESSWFLQLTSVLFAKSGYLTCAIDHQ 94
Query: 121 GFGLSDGLHGYIPSFENLVNDVIEHFSKIK-------------GESMGGAIALNIHFKQP 167
G G SDGL +IP+ +V+D I F + ES+GGAIAL I +Q
Sbjct: 95 GHGFSDGLTAHIPNINLIVDDCISFFDDFRKRHASSFLPAFLYSESLGGAIALYITLRQK 154
Query: 168 TAWDGAALIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRDARKRE 227
W+G L +C + P W ++ +L A ++P ++VP + + +++ KR+
Sbjct: 155 HQWNGLILSGAMCSISHKFKPPWPLQHLLTLAATLIPTWRVVPTRGSIAGVSFKEPWKRK 214
Query: 228 LAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSASKALYQ 287
LA N KPR TA EL++ ++L+ R EEV +PL+++HG D++ DP++ + LY+
Sbjct: 215 LAYANPNRTVGKPRAATAYELVRVCEDLQNRFEEVEVPLMIVHGRDDVVCDPASVEELYR 274
Query: 288 KAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHS 330
+ +DK + +Y +H L+ GE +E + V D++ W+ S
Sbjct: 275 RCSSRDKTIKIYPGMWHQLI-GESEENVDLVFGDVLDWIKTRS 316
>UniRef100_Q75JJ5 Similar to Arabidopsis thaliana (Mouse-ear cress). Putative
phospholipase, alternative splicing isoform
[Dictyostelium discoideum]
Length = 409
Score = 169 bits (429), Expect = 8e-41
Identities = 106/291 (36%), Positives = 160/291 (54%), Gaps = 18/291 (6%)
Query: 55 VKMEEIYEVNSRGLKVFSKSWIPEKSPMKGIVYYCHGYADTCTFYFEGVARKLASSGFGV 114
V+ ++ Y +NSRG+K+ + WIP +GIV HGY D + +A +GF
Sbjct: 116 VQYKKGYFINSRGMKLVCQEWIPHNP--RGIVIVLHGYGDHGQTTLAEDCKIMARNGFAS 173
Query: 115 FALDYPGFGLSDGLHGYIPSFENLVNDVIEHFSKIKGE-----------SMGGAIALNIH 163
F D G GLS+G+ YI F++LV D + S IK SMGGA+ +
Sbjct: 174 FIYDQQGHGLSEGVPAYIRDFDDLVEDSLLFISDIKFRFPRLKRFVCCTSMGGAVGTLVS 233
Query: 164 FKQPTAWDGAA-LIAPLCKFAEDMIPHWLVKQILIGVAKVLPKTKLVPQKEEVKENIYRD 222
++P +DG L+APL K E+MIP+ ++ +L V+K P +VP E V + +D
Sbjct: 234 LRKPEVFDGGLILLAPLIKLDENMIPNPILVSLLTWVSKSFPTLAIVPG-ENVLDRSIKD 292
Query: 223 ARKRELAPYNVLFYKDKPRLGTALELLKATQELEQRLEEVSLPLLVMHGEADIITDPSAS 282
+KR + L YK + R+GT L +LKAT L+ LE++S+PLL++HG D ++ P+ S
Sbjct: 293 PQKRVEHANHPLTYKGRARIGTGLAILKATSFLQSHLEDISVPLLILHGSLDRVSSPTVS 352
Query: 283 KALYQKAKVKDKKLCLYKDAFHTLLEGEPDETIFHVLDDIISWLDDHSSTK 333
+ LY+KA DK L LY +H + E D I V +DII+W+ + + K
Sbjct: 353 EELYKKAISADKTLKLYPTFWHG-ITSEKDADI--VYNDIINWMIERLNPK 400
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.138 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 576,780,777
Number of Sequences: 2790947
Number of extensions: 24751672
Number of successful extensions: 61602
Number of sequences better than 10.0: 711
Number of HSP's better than 10.0 without gapping: 193
Number of HSP's successfully gapped in prelim test: 518
Number of HSP's that attempted gapping in prelim test: 61014
Number of HSP's gapped (non-prelim): 800
length of query: 336
length of database: 848,049,833
effective HSP length: 128
effective length of query: 208
effective length of database: 490,808,617
effective search space: 102088192336
effective search space used: 102088192336
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC146563.12


