

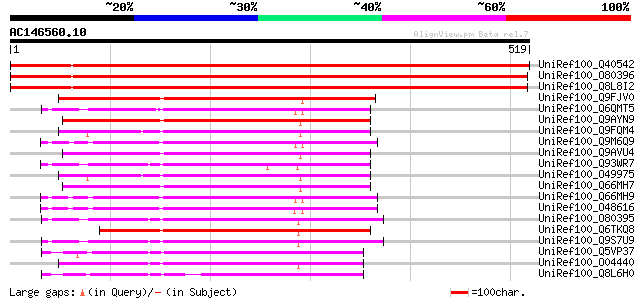
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146560.10 + phase: 0
(519 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q40542 NPK2 [Nicotiana tabacum] 867 0.0
UniRef100_O80396 Putative MAP kinase kinase 3 ATMKK3 [Arabidopsi... 833 0.0
UniRef100_Q8L8I2 Mitogen-activated protein kinase kinase [Suaeda... 831 0.0
UniRef100_Q9FJV0 Protein kinase MEK1 homolog [Arabidopsis thaliana] 248 2e-64
UniRef100_Q6QMT5 Mitogen-activated protein kinase kinase 2 [Petr... 245 3e-63
UniRef100_Q9AYN9 NQK1 MAPKK [Nicotiana tabacum] 244 6e-63
UniRef100_Q9FQM4 MAP kinase kinase 1 [Oryza sativa] 243 1e-62
UniRef100_Q9M6Q9 MAP kinase kinase [Nicotiana tabacum] 241 3e-62
UniRef100_Q9AVU4 MAP kinase [Nicotiana tabacum] 239 1e-61
UniRef100_Q93WR7 MAP kinase kinase [Medicago varia] 239 1e-61
UniRef100_O49975 Protein kinase ZmMEK1 [Zea mays] 238 3e-61
UniRef100_Q66MH7 MAPKK [Lycopersicon esculentum] 238 3e-61
UniRef100_Q66MH9 MAPKK [Lycopersicon esculentum] 236 2e-60
UniRef100_O48616 MAP kinase kinase [Lycopersicon esculentum] 235 2e-60
UniRef100_O80395 MAP kinase kinase 2 [Arabidopsis thaliana] 234 3e-60
UniRef100_Q6TKQ8 Putative mitogen-activated protein kinase kinas... 233 8e-60
UniRef100_Q9S7U9 MAP2k beta protein [Arabidopsis thaliana] 233 1e-59
UniRef100_Q5VP37 Putative MAP kinase kinase [Oryza sativa] 232 2e-59
UniRef100_O04440 Putative mitogen activated protein kinase kinas... 221 3e-56
UniRef100_Q8L6H0 Putative mitogen-activated protein kinase kinas... 212 2e-53
>UniRef100_Q40542 NPK2 [Nicotiana tabacum]
Length = 518
Score = 867 bits (2239), Expect = 0.0
Identities = 424/519 (81%), Positives = 467/519 (89%), Gaps = 1/519 (0%)
Query: 1 MSGLEELRKKLTPLFDAENGFSSTSSFDPNDSYTFSDGGTVNLLSRSYGVYNINELGLQK 60
M+GLEEL+KKL PLFDAE GFS S+ DP DSY+ SDGGTVNLLS+SYGVYNINELGLQK
Sbjct: 1 MAGLEELKKKLVPLFDAEKGFSPASTSDPFDSYSLSDGGTVNLLSQSYGVYNINELGLQK 60
Query: 61 CSTSRSVDESDDNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIALKKINIFEK 120
C TS VD++D EKTY+C SHEMR+FGAIGSGASSVVQRA+HIPTHR+IALKKINIFEK
Sbjct: 61 C-TSWPVDDADHGEKTYKCASHEMRVFGAIGSGASSVVQRAIHIPTHRIIALKKINIFEK 119
Query: 121 EKRQQLLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIP 180
EKRQQLLTEIRTLCEAPC +GLVEF+GAFYTPDSGQISIALEYMDGGSLADI+++ ++IP
Sbjct: 120 EKRQQLLTEIRTLCEAPCCQGLVEFYGAFYTPDSGQISIALEYMDGGSLADIIKVRKSIP 179
Query: 181 EPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMC 240
E ILS M QKLL GLSYLHGVR+LVHRDIKPANLLVNLKGEPKITDFGISAGLE+S+AMC
Sbjct: 180 EAILSPMVQKLLNGLSYLHGVRHLVHRDIKPANLLVNLKGEPKITDFGISAGLESSIAMC 239
Query: 241 ATFVGTVTYMSPERIRNESYSYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDP 300
ATFVGTVTYMSPERIRNE+YSYPADIWSLGLAL E GTGEFPYTANEGPVNLMLQILDDP
Sbjct: 240 ATFVGTVTYMSPERIRNENYSYPADIWSLGLALFECGTGEFPYTANEGPVNLMLQILDDP 299
Query: 301 SPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFDP 360
SPS S +FSPEFCSF+DACL+K+PD+R TAEQLL HPFITKY + VDL FVRS+FDP
Sbjct: 300 SPSLSGHEFSPEFCSFIDACLKKNPDDRLTAEQLLSHPFITKYTDSAVDLGAFVRSIFDP 359
Query: 361 TQRMKDLADMLTIHYYLLFDGPDDSWQHTRNFYNENSIFSFSGKQHIGPNNIFTTLSSIR 420
TQRMKDLADMLTIHYYLLFDG D+ WQHT+ YNE S FSF GK+ IGP+NIF+T+S+IR
Sbjct: 360 TQRMKDLADMLTIHYYLLFDGSDEFWQHTKTLYNECSTFSFGGKESIGPSNIFSTMSNIR 419
Query: 421 TTLIGDWPPEKLVHVVEKLQCRAHGEDGVAIRVSGSFIIGNQFLICGDGVQVEGLPNFKD 480
TL G+WPPEKLVHVVEK+QCR HG+DGVAIRVSGSFI+GNQFLICGDG+QVEGLPN KD
Sbjct: 420 KTLAGEWPPEKLVHVVEKVQCRTHGQDGVAIRVSGSFIVGNQFLICGDGMQVEGLPNLKD 479
Query: 481 LGIDISSKRMGTFHEQFIVEPTSQIGCYTIVKQELYINQ 519
L IDI SKRMGTFHEQFIVE + IG Y I KQEL+I Q
Sbjct: 480 LSIDIPSKRMGTFHEQFIVEQANIIGRYFITKQELFITQ 518
>UniRef100_O80396 Putative MAP kinase kinase 3 ATMKK3 [Arabidopsis thaliana]
Length = 520
Score = 833 bits (2152), Expect = 0.0
Identities = 403/517 (77%), Positives = 453/517 (86%), Gaps = 1/517 (0%)
Query: 1 MSGLEELRKKLTPLFDAENGFSSTSSFDPNDSYTFSDGGTVNLLSRSYGVYNINELGLQK 60
M+ LEEL+KKL+PLFDAE GFSS+SS DPNDSY SDGGTVNLLSRSYGVYN NELGLQK
Sbjct: 1 MAALEELKKKLSPLFDAEKGFSSSSSLDPNDSYLLSDGGTVNLLSRSYGVYNFNELGLQK 60
Query: 61 CSTSRSVDESDDNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIALKKINIFEK 120
C TS VDES+ +E TY+C SHEMR+FGAIGSGASSVVQRA+HIP HR++ALKKINIFE+
Sbjct: 61 C-TSSHVDESESSETTYQCASHEMRVFGAIGSGASSVVQRAIHIPNHRILALKKINIFER 119
Query: 121 EKRQQLLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIP 180
EKRQQLLTEIRTLCEAPC+EGLV+FHGAFY+PDSGQISIALEYM+GGSLADIL++ + IP
Sbjct: 120 EKRQQLLTEIRTLCEAPCHEGLVDFHGAFYSPDSGQISIALEYMNGGSLADILKVTKKIP 179
Query: 181 EPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMC 240
EP+LSS+F KLL+GLSYLHGVR+LVHRDIKPANLL+NLKGEPKITDFGISAGLENS+AMC
Sbjct: 180 EPVLSSLFHKLLQGLSYLHGVRHLVHRDIKPANLLINLKGEPKITDFGISAGLENSMAMC 239
Query: 241 ATFVGTVTYMSPERIRNESYSYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDP 300
ATFVGTVTYMSPERIRN+SYSYPADIWSLGLAL E GTGEFPY ANEGPVNLMLQILDDP
Sbjct: 240 ATFVGTVTYMSPERIRNDSYSYPADIWSLGLALFECGTGEFPYIANEGPVNLMLQILDDP 299
Query: 301 SPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFDP 360
SP+P K++FSPEFCSF+DACLQKDPD RPTA+QLL HPFITK+E +VDL FV+S+FDP
Sbjct: 300 SPTPPKQEFSPEFCSFIDACLQKDPDARPTADQLLSHPFITKHEKERVDLATFVQSIFDP 359
Query: 361 TQRMKDLADMLTIHYYLLFDGPDDSWQHTRNFYNENSIFSFSGKQHIGPNNIFTTLSSIR 420
TQR+KDLADMLTIHYY LFDG DD W H ++ Y E S+FSFSGK + G IF+ LS IR
Sbjct: 360 TQRLKDLADMLTIHYYSLFDGFDDLWHHAKSLYTETSVFSFSGKHNTGSTEIFSALSDIR 419
Query: 421 TTLIGDWPPEKLVHVVEKLQCRAHGEDGVAIRVSGSFIIGNQFLICGDGVQVEGLPNFKD 480
TL GD P EKLVHVVEKL C+ G GV IR GSFI+GNQFLICGDGVQ EGLP+FKD
Sbjct: 420 NTLTGDLPSEKLVHVVEKLHCKPCGSGGVIIRAVGSFIVGNQFLICGDGVQAEGLPSFKD 479
Query: 481 LGIDISSKRMGTFHEQFIVEPTSQIGCYTIVKQELYI 517
LG D++S+R+G F EQF+VE IG Y + KQELYI
Sbjct: 480 LGFDVASRRVGRFQEQFVVESGDLIGKYFLAKQELYI 516
>UniRef100_Q8L8I2 Mitogen-activated protein kinase kinase [Suaeda salsa]
Length = 520
Score = 831 bits (2146), Expect = 0.0
Identities = 401/518 (77%), Positives = 456/518 (87%), Gaps = 2/518 (0%)
Query: 1 MSGLEELRKKLTPLFDAENGFSSTSSFDPNDSYTFSDGGTVNLLSRSYGVYNINELGLQK 60
M+ LEEL+KKLTPLFDAE GFS+ ++ + DS SD GT+NLLSRSYGVYN NE GLQK
Sbjct: 1 MAALEELKKKLTPLFDAEKGFSAGTTLECGDSDMLSDSGTINLLSRSYGVYNFNEQGLQK 60
Query: 61 CSTSRSVDESDDNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIALKKINIFEK 120
C TS VDE+ +EKTYR S EMR+FGAIGSGASS+VQRA+HIPTHR+IALKKINIFEK
Sbjct: 61 C-TSSPVDENSPSEKTYRISSREMRVFGAIGSGASSIVQRAIHIPTHRIIALKKINIFEK 119
Query: 121 EKRQQLLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLADILRMHRTIP 180
EKRQQLLTEIRTLCEAPC EGLVEF+GAFY PDSGQISIALEYMDGGSLAD++++ + IP
Sbjct: 120 EKRQQLLTEIRTLCEAPCSEGLVEFYGAFYIPDSGQISIALEYMDGGSLADVIQVQKCIP 179
Query: 181 EPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISAGLENSVAMC 240
E +LS++ +KLL GL+YLHGVR+LVHRDIKPANLL++LKGEPKITDFGISAGLENS+AMC
Sbjct: 180 ESVLSAIVRKLLHGLNYLHGVRHLVHRDIKPANLLMDLKGEPKITDFGISAGLENSMAMC 239
Query: 241 ATFVGTVTYMSPERIRNESYSYPADIWSLGLALLESGTGEFPYTANEGPVNLMLQILDDP 300
ATF+GTVTYMSPERIRNESYSYPADIWSLGLAL E GTGEFPY AN+GPVNLMLQILDDP
Sbjct: 240 ATFIGTVTYMSPERIRNESYSYPADIWSLGLALFECGTGEFPYIANDGPVNLMLQILDDP 299
Query: 301 SPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFDP 360
SPSPSKEKFS EFCSFVD CLQKDP+ RPTAEQLL HPFITKY+ ++VDL FV+S+FDP
Sbjct: 300 SPSPSKEKFSSEFCSFVDVCLQKDPEARPTAEQLLSHPFITKYKDSQVDLATFVQSIFDP 359
Query: 361 TQRMKDLADMLTIHYYLLFDGPD-DSWQHTRNFYNENSIFSFSGKQHIGPNNIFTTLSSI 419
QR+KDLADMLTIHYYLLFDGPD + WQ ++ YNE S+FSFSGKQ+ G N+IF+ LS +
Sbjct: 360 VQRLKDLADMLTIHYYLLFDGPDEEEWQQAKSLYNEGSVFSFSGKQYAGTNDIFSVLSDV 419
Query: 420 RTTLIGDWPPEKLVHVVEKLQCRAHGEDGVAIRVSGSFIIGNQFLICGDGVQVEGLPNFK 479
R L G WPPEKL+HVVEKLQC+AHG+ G+AIRVSGSFI GNQF+ICGDG+QVEGLPNF+
Sbjct: 420 RRKLAGKWPPEKLIHVVEKLQCKAHGQGGIAIRVSGSFIAGNQFIICGDGMQVEGLPNFR 479
Query: 480 DLGIDISSKRMGTFHEQFIVEPTSQIGCYTIVKQELYI 517
DL +DI+SKRMGTF EQFI+EP IGCY I QELYI
Sbjct: 480 DLNVDIASKRMGTFKEQFIIEPGDAIGCYNISNQELYI 517
>UniRef100_Q9FJV0 Protein kinase MEK1 homolog [Arabidopsis thaliana]
Length = 356
Score = 248 bits (634), Expect = 2e-64
Identities = 133/325 (40%), Positives = 199/325 (60%), Gaps = 11/325 (3%)
Query: 49 GVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHR 108
G + +N+ GL+ S + +SD E + + ++ IG G+ VVQ H +
Sbjct: 35 GDFLLNQKGLRLTSDEKQSRQSDSKELDFEITAEDLETVKVIGKGSGGVVQLVRHKWVGK 94
Query: 109 VIALKKINI-FEKEKRQQLLTEIR-TLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDG 166
A+K I + ++E R+Q++ E++ + C +V +H ++ +G S+ LEYMD
Sbjct: 95 FFAMKVIQMNIQEEIRKQIVQELKINQASSQCPHVVVCYHSFYH---NGAFSLVLEYMDR 151
Query: 167 GSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITD 226
GSLAD++R +TI EP L+ + +++L GL YLH R+++HRDIKP+NLLVN KGE KI+D
Sbjct: 152 GSLADVIRQVKTILEPYLAVVCKQVLLGLVYLHNERHVIHRDIKPSNLLVNHKGEVKISD 211
Query: 227 FGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADIWSLGLALLESGTGEFPYTAN 286
FG+SA L +S+ TFVGT YMSPERI +Y Y +DIWSLG+++LE G FPY +
Sbjct: 212 FGVSASLASSMGQRDTFVGTYNYMSPERISGSTYDYSSDIWSLGMSVLECAIGRFPYLES 271
Query: 287 EGPVN------LMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFI 340
E N L+ I+++P P+ ++FSPEFCSFV AC+QKDP R ++ LL HPFI
Sbjct: 272 EDQQNPPSFYELLAAIVENPPPTAPSDQFSPEFCSFVSACIQKDPPARASSLDLLSHPFI 331
Query: 341 TKYETAKVDLPGFVRSVFDPTQRMK 365
K+E +DL V ++ P ++
Sbjct: 332 KKFEDKDIDLGILVGTLEPPVNYLR 356
>UniRef100_Q6QMT5 Mitogen-activated protein kinase kinase 2 [Petroselinum crispum]
Length = 354
Score = 245 bits (625), Expect = 3e-63
Identities = 141/336 (41%), Positives = 200/336 (58%), Gaps = 21/336 (6%)
Query: 32 SYTFSDGGTVNLLSRSYGVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAIG 91
S TF DG +LL GV +++ +Q ++ D + + +G
Sbjct: 29 SGTFMDG---DLLVNRDGVRIVSQTDVQAAPPIQATDN--------QLSLADFDAIKVVG 77
Query: 92 SGASSVVQRAMHIPTHRVIALKKINI-FEKEKRQQLLTEIRTLCEAPCYEGLVEFHGAFY 150
G+ +V+ H T + ALK I + ++ R+Q+ E++ + C +V + +FY
Sbjct: 78 KGSGGIVRLVQHKWTGQFFALKVIQMNIQESARKQIAQELKINQSSQCLNVVVCYQ-SFY 136
Query: 151 TPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIK 210
D+G ISI LEYMDGGSLAD L++ + IPEP L+++F+++L+GL YLH ++++HRD+K
Sbjct: 137 --DNGAISIILEYMDGGSLADFLKIVKNIPEPYLAAIFKQVLKGLWYLHHEKHIIHRDLK 194
Query: 211 PANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADIWSLG 270
P+NLL+N +GE KITDFG+SA L + TFVGT YMSPERI + Y +DIWSLG
Sbjct: 195 PSNLLINHRGEVKITDFGVSAILATTSGQANTFVGTYNYMSPERISGGQHGYSSDIWSLG 254
Query: 271 LALLESGTGEFPYT---ANEGPVN---LMLQILDDPSPSPSKEKFSPEFCSFVDACLQKD 324
L LLE TG FPY+ +EG N LM I++ P ++FSPEFCSFV AC+QKD
Sbjct: 255 LVLLECATGYFPYSPPEQDEGWSNVFELMDTIVNQAPPCAPPDEFSPEFCSFVSACVQKD 314
Query: 325 PDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFDP 360
P RP+A +LL HPFI YE +DL G+ P
Sbjct: 315 PRKRPSANELLRHPFINMYEHLNIDLAGYFTEAGSP 350
>UniRef100_Q9AYN9 NQK1 MAPKK [Nicotiana tabacum]
Length = 354
Score = 244 bits (622), Expect = 6e-63
Identities = 133/315 (42%), Positives = 190/315 (60%), Gaps = 10/315 (3%)
Query: 53 INELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIAL 112
+N+ GL+ S S+ E + ++ IG G+ VVQ H + AL
Sbjct: 38 LNQKGLRLISEENESPASETKEIDLQFSLEDLETIKVIGKGSGGVVQLVRHKWVGTLFAL 97
Query: 113 KKINI-FEKEKRQQLLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLAD 171
K I + +++ R+Q++ E++ + C +V +H ++ +G IS+ LEYMD GSLAD
Sbjct: 98 KVIQMTIQEDIRKQIVQELKINQASQCSHVVVCYHSFYH---NGAISLVLEYMDRGSLAD 154
Query: 172 ILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISA 231
++R +TI EP L+ + +++L+GL YLH R+++HRDIKP+NLLVN KGE KITDFG+SA
Sbjct: 155 VIRQLKTILEPYLAVVCKQVLQGLVYLHNERHVIHRDIKPSNLLVNHKGEVKITDFGVSA 214
Query: 232 GLENSVAMCATFVGTVTYMSPERIRNESYSYPADIWSLGLALLESGTGEFPYTANEGP-- 289
L +S+ TFVGT YM+PERI +Y Y +DIWSLG+ +LE G FPY +E
Sbjct: 215 MLASSMGQRDTFVGTYNYMAPERISGSTYDYKSDIWSLGMVILECAIGRFPYIQSEDQQA 274
Query: 290 ----VNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITKYET 345
L+ I+ P PS ++FSPEFCSFV AC+QKDP +R +A LL HPFI K+E
Sbjct: 275 WPSFYELLEAIVSSPPPSAPADQFSPEFCSFVSACIQKDPRDRSSALDLLSHPFIKKFED 334
Query: 346 AKVDLPGFVRSVFDP 360
+D V S+ P
Sbjct: 335 KDIDFGILVSSLEPP 349
>UniRef100_Q9FQM4 MAP kinase kinase 1 [Oryza sativa]
Length = 355
Score = 243 bits (619), Expect = 1e-62
Identities = 137/320 (42%), Positives = 194/320 (59%), Gaps = 11/320 (3%)
Query: 49 GVYNINELGLQKCSTSRSVDESDDNEKT--YRCGSHEMRIFGAIGSGASSVVQRAMHIPT 106
G +N+ GLQ S + + N K + ++ + IG G+ +VQ H
Sbjct: 34 GELRLNQRGLQLISEETADEPQSTNLKVEDVQLSMDDLEMIQVIGKGSGGIVQLVRHKWV 93
Query: 107 HRVIALKKINI-FEKEKRQQLLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMD 165
+ ALK I + ++ R+Q++ E++ + +A +V H +FY +G I + LEYMD
Sbjct: 94 GTLYALKGIQMNIQEAVRKQIVQELK-INQATQNAHIVLCHQSFY--HNGVIYLVLEYMD 150
Query: 166 GGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKIT 225
GSLADI++ +TI EP L+ + +++L GL YLH R+++HRDIKP+NLLVN KGE KIT
Sbjct: 151 RGSLADIIKQVKTILEPYLAVLCKQVLEGLLYLHHERHVIHRDIKPSNLLVNRKGEVKIT 210
Query: 226 DFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADIWSLGLALLESGTGEFPYTA 285
DFG+SA L +S+ TFVGT YM+PERI SY Y +DIWSLGL +LE G FPY
Sbjct: 211 DFGVSAVLASSMGQRDTFVGTYNYMAPERISGSSYDYKSDIWSLGLVILECAIGRFPYIP 270
Query: 286 NEGP-----VNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFI 340
+EG L+ I+D P PS ++FSPEFC+F+ +C+QKDP R +A +LL HPFI
Sbjct: 271 SEGEGWLSFYELLEAIVDQPPPSAPADQFSPEFCAFISSCIQKDPAERMSASELLNHPFI 330
Query: 341 TKYETAKVDLPGFVRSVFDP 360
K+E +DL V S+ P
Sbjct: 331 KKFEDKDLDLRILVESLEPP 350
>UniRef100_Q9M6Q9 MAP kinase kinase [Nicotiana tabacum]
Length = 357
Score = 241 bits (616), Expect = 3e-62
Identities = 142/344 (41%), Positives = 207/344 (59%), Gaps = 21/344 (6%)
Query: 31 DSYTFSDGGTVNLLSRSYGVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAI 90
+S TF DG +LL GV +++ ++ + SV + DN+ + I
Sbjct: 28 ESGTFKDG---DLLVNRDGVRIVSQSEVE----APSVIQPSDNQLCLA----DFEAVKVI 76
Query: 91 GSGASSVVQRAMHIPTHRVIALKKINI-FEKEKRQQLLTEIRTLCEAPCYEGLVEFHGAF 149
G G +V+ H T + ALK I + E+ R+ + E+R + ++ + F
Sbjct: 77 GKGNGGIVRLVQHKWTGQFFALKAIQMNIEESMRKHIAQELRINQSSQVPYVVISYQSFF 136
Query: 150 YTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDI 209
D+G ISI LEYMDGGSLAD L+ +TIPE L+++ +++L+GL YLH ++++HRD+
Sbjct: 137 ---DNGAISIILEYMDGGSLADFLKKVKTIPERYLAAICKQVLKGLWYLHHEKHIIHRDL 193
Query: 210 KPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADIWSL 269
KP+NLL+N G+ KITDFG+SA L ++ + TFVGT YMSPERI +Y Y +DIWSL
Sbjct: 194 KPSNLLINHIGDVKITDFGVSAVLASTSGLANTFVGTYNYMSPERILGGAYGYRSDIWSL 253
Query: 270 GLALLESGTGEFPYT---ANEGPVN---LMLQILDDPSPSPSKEKFSPEFCSFVDACLQK 323
GL LLE TG FPY+ A+EG VN LM I+D P+PS ++FSP+FCSF+ AC+QK
Sbjct: 254 GLVLLECATGVFPYSPPQADEGWVNVYELMETIVDQPAPSAPPDQFSPQFCSFISACVQK 313
Query: 324 DPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFDPTQRMKDL 367
D +R +A +L+ HPFIT Y+ +DL + S P + +L
Sbjct: 314 DQKDRLSANELMRHPFITMYDDLDIDLGSYFTSAGPPLATLTEL 357
>UniRef100_Q9AVU4 MAP kinase [Nicotiana tabacum]
Length = 354
Score = 239 bits (611), Expect = 1e-61
Identities = 132/315 (41%), Positives = 188/315 (58%), Gaps = 10/315 (3%)
Query: 53 INELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIAL 112
+N+ GL+ S S+ E + ++ IG G+ VVQ H + AL
Sbjct: 38 LNQKGLRLISEENESPASETKEIDLQFSLEDLETIKVIGKGSGGVVQLVRHKWVGTLFAL 97
Query: 113 KKINI-FEKEKRQQLLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLAD 171
K I + +++ R+Q++ E++ + C +V +H ++ +G IS+ LEYMD GSLAD
Sbjct: 98 KVIQMTIQEDIRKQIVQELKINQASQCSHVVVCYHSFYH---NGAISLVLEYMDRGSLAD 154
Query: 172 ILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISA 231
++R +TI EP L+ + +++L+GL YLH R+++HRDIKP+NLLVN KGE KITDF +SA
Sbjct: 155 VIRQLKTILEPYLAVVCKQVLQGLVYLHNERHVIHRDIKPSNLLVNHKGEVKITDFXVSA 214
Query: 232 GLENSVAMCATFVGTVTYMSPERIRNESYSYPADIWSLGLALLESGTGEFPYTANEGP-- 289
L +S+ TFVGT YM+PERI +Y Y +DIWSLG+ +LE G FPY +E
Sbjct: 215 MLASSMGQRDTFVGTYNYMAPERISGSTYDYKSDIWSLGMVILECAIGRFPYIQSEDQQA 274
Query: 290 ----VNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITKYET 345
L+ I+ P PS +FSPEFCSFV AC+QKDP +R +A LL HPFI K+E
Sbjct: 275 WPSFYELLEAIVSSPPPSAPAVQFSPEFCSFVSACIQKDPRDRSSALDLLSHPFIKKFED 334
Query: 346 AKVDLPGFVRSVFDP 360
+D V S+ P
Sbjct: 335 KDIDFGILVSSLEPP 349
>UniRef100_Q93WR7 MAP kinase kinase [Medicago varia]
Length = 356
Score = 239 bits (611), Expect = 1e-61
Identities = 142/339 (41%), Positives = 202/339 (58%), Gaps = 23/339 (6%)
Query: 31 DSYTFSDGGTVNLLSRSYGVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAI 90
+S TF DG +LL GV ++E ++ ++ D + ++ I +
Sbjct: 28 ESGTFKDG---DLLVNRDGVRIVSETEVEAPPPIKATDN--------QLSLADIDIVKVV 76
Query: 91 GSGASSVVQRAMHIPTHRVIALKKINI-FEKEKRQQLLTEIRTLCEAPCYEGLVEFHGAF 149
G G VVQ H T++ ALK I + E+ R+++ E++ A C +V + +F
Sbjct: 77 GKGNGGVVQLVQHKWTNQFFALKIIQMNIEESVRKRIAKELKINQAAQC-PYVVVCYQSF 135
Query: 150 YTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDI 209
Y D+G ISI LEYMDGGS+AD+L+ +TIPEP LS++ +++L+GL YLH R+++HRD+
Sbjct: 136 Y--DNGVISIILEYMDGGSMADLLKKVKTIPEPYLSAICKQVLKGLIYLHHERHIIHRDL 193
Query: 210 KPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIR--NESYSYPADIW 267
KP+NLL+N GE KITDFG+SA +E++ TF+GT YMSPERI Y+Y +DIW
Sbjct: 194 KPSNLLINHTGEVKITDFGVSAIMESTSGQANTFIGTYNYMSPERINGSQRGYNYKSDIW 253
Query: 268 SLGLALLESGTGEFPYTAN------EGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACL 321
SLGL LLE G FPYT E L+ I+D P PS E+FS EFCSF+ ACL
Sbjct: 254 SLGLILLECAMGRFPYTPPDQSERWESIFELIETIVDKPPPSAPSEQFSSEFCSFISACL 313
Query: 322 QKDPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFDP 360
QKDP +R +A++L+ PFI+ Y+ VDL + P
Sbjct: 314 QKDPGSRLSAQELMELPFISMYDDLHVDLSAYFSDAGSP 352
>UniRef100_O49975 Protein kinase ZmMEK1 [Zea mays]
Length = 355
Score = 238 bits (607), Expect = 3e-61
Identities = 133/320 (41%), Positives = 193/320 (59%), Gaps = 11/320 (3%)
Query: 49 GVYNINELGLQKCSTSRSVDESDDNEKT--YRCGSHEMRIFGAIGSGASSVVQRAMHIPT 106
G +N+ GL+ S ++ K + ++ + IG G+ VVQ H
Sbjct: 34 GELRLNQSGLRLISEENGDEDESTKLKVEDVQLSMDDLEMIQVIGKGSGGVVQLVRHKWV 93
Query: 107 HRVIALKKINI-FEKEKRQQLLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMD 165
+ ALK I + ++ R+Q++ E++ + +A +V H +FY +G I + LEYMD
Sbjct: 94 GTLFALKGIQMNIQESVRKQIVQELK-INQATQSPHIVMCHQSFY--HNGVIYLVLEYMD 150
Query: 166 GGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKIT 225
GSLADI++ +TI EP L+ + +++L GL YLH R+++HRDIKP+NLLVN KGE KIT
Sbjct: 151 RGSLADIVKQVKTILEPYLAVLCKQVLEGLLYLHHQRHVIHRDIKPSNLLVNRKGEVKIT 210
Query: 226 DFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADIWSLGLALLESGTGEFPYTA 285
DFG+SA L +S+ TFVGT YM+PERI +Y Y +DIWSLGL +LE G FPY
Sbjct: 211 DFGVSAVLASSIGQRDTFVGTYNYMAPERISGSTYDYKSDIWSLGLVILECAIGRFPYIP 270
Query: 286 NEGP-----VNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFI 340
+EG L+ I+D P PS ++FSPEFCSF+ +C+QKDP R +A +LL HPF+
Sbjct: 271 SEGEGWLSFYELLEAIVDQPPPSAPADQFSPEFCSFISSCIQKDPAQRMSASELLNHPFL 330
Query: 341 TKYETAKVDLPGFVRSVFDP 360
K+E ++L V ++ P
Sbjct: 331 KKFEDKDLNLGILVENLEPP 350
>UniRef100_Q66MH7 MAPKK [Lycopersicon esculentum]
Length = 354
Score = 238 bits (607), Expect = 3e-61
Identities = 131/315 (41%), Positives = 189/315 (59%), Gaps = 10/315 (3%)
Query: 53 INELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHRVIAL 112
+N+ GL+ S S+ E + ++ IG G+ VVQ H + AL
Sbjct: 38 LNQKGLRLISEENESLPSETKEIDLQFSLEDLETIKVIGKGSGGVVQLVRHKWVGTLFAL 97
Query: 113 KKINI-FEKEKRQQLLTEIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGGSLAD 171
K I + +++ R+Q++ E++ + C +V +H ++ +G IS+ LEYMD GSL D
Sbjct: 98 KVIQMNIQEDIRKQIVQELKINQASQCPHVVVCYHSFYH---NGAISLVLEYMDRGSLVD 154
Query: 172 ILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDFGISA 231
++ +TI EP L+ + +++L+GL YLH R+++HRDIKP+NLLVN KGE KITDFG+SA
Sbjct: 155 VIGQLKTILEPYLAVVCKQVLQGLVYLHHERHVIHRDIKPSNLLVNHKGEVKITDFGVSA 214
Query: 232 GLENSVAMCATFVGTVTYMSPERIRNESYSYPADIWSLGLALLESGTGEFPYTANEGP-- 289
L +S+ TFVGT YM+PERI +Y Y +DIWSLG+ +LE G FPY +E
Sbjct: 215 MLASSMGQRDTFVGTYNYMAPERISGSTYDYKSDIWSLGMVILECAIGRFPYIQSEDQQA 274
Query: 290 ----VNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFITKYET 345
L+ I+ P PS ++FSPEFCSFV AC+QKDP +R +A LL HPF+ K+E
Sbjct: 275 RPSFYELLDAIVSSPPPSAPVDQFSPEFCSFVSACIQKDPRDRSSALDLLSHPFVKKFED 334
Query: 346 AKVDLPGFVRSVFDP 360
+DL V S+ P
Sbjct: 335 KDIDLSILVSSLEPP 349
>UniRef100_Q66MH9 MAPKK [Lycopersicon esculentum]
Length = 357
Score = 236 bits (601), Expect = 2e-60
Identities = 138/344 (40%), Positives = 201/344 (58%), Gaps = 21/344 (6%)
Query: 31 DSYTFSDGGTVNLLSRSYGVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAI 90
+S TF DG +LL GV +++ + + SV + DN+ + I
Sbjct: 28 ESGTFKDG---DLLVNRDGVRIVSQSEV----AAPSVIQPSDNQLCLA----DFEAVKVI 76
Query: 91 GSGASSVVQRAMHIPTHRVIALKKINI-FEKEKRQQLLTEIRTLCEAPCYEGLVEFHGAF 149
G G +V+ H T + ALK I + ++ R+ + E+R + C ++ + F
Sbjct: 77 GKGNGGIVRLVQHKWTGQFFALKVIQMNIDESMRKHIAQELRINQSSQCPYVVICYQSFF 136
Query: 150 YTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDI 209
D+G IS+ LEYMDGGSLAD L+ +TIPE L+ + +++L+GL YLH ++++HRD+
Sbjct: 137 ---DNGAISLILEYMDGGSLADFLKKVKTIPERFLAVICKQVLKGLWYLHHEKHIIHRDL 193
Query: 210 KPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADIWSL 269
KP+NLL+N +G+ KITDFG+SA L ++ + TFVGT YMSPERI +Y Y +DIWSL
Sbjct: 194 KPSNLLINHRGDVKITDFGVSAVLASTSGLANTFVGTYNYMSPERISGGAYDYKSDIWSL 253
Query: 270 GLALLESGTGEFPYT---ANEGPVN---LMLQILDDPSPSPSKEKFSPEFCSFVDACLQK 323
GL LLE TG FPY +EG VN LM I+D P P ++FSP+FCSF+ AC+QK
Sbjct: 254 GLVLLECATGHFPYNPPEGDEGWVNVYELMETIVDQPEPCAPPDQFSPQFCSFISACVQK 313
Query: 324 DPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFDPTQRMKDL 367
+R +A L+ HPFIT Y+ +DL + S P + +L
Sbjct: 314 HQKDRLSANDLMSHPFITMYDDQDIDLGSYFTSAGPPLATLTEL 357
>UniRef100_O48616 MAP kinase kinase [Lycopersicon esculentum]
Length = 357
Score = 235 bits (600), Expect = 2e-60
Identities = 138/344 (40%), Positives = 201/344 (58%), Gaps = 21/344 (6%)
Query: 31 DSYTFSDGGTVNLLSRSYGVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAI 90
+S TF DG +LL GV +++ + + SV + DN+ + I
Sbjct: 28 ESGTFKDG---DLLVNRDGVRIVSQSEV----AAPSVIQPSDNQLCLA----DFEAVKVI 76
Query: 91 GSGASSVVQRAMHIPTHRVIALKKINI-FEKEKRQQLLTEIRTLCEAPCYEGLVEFHGAF 149
G G +V+ H T + ALK I + ++ R+ + E+R + C ++ + F
Sbjct: 77 GKGNGGIVRLVQHKWTGQFFALKVIQMNIDESMRKHIAQELRINQSSQCPYVVICYQSFF 136
Query: 150 YTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDI 209
D+G IS+ LEYMDGGSLAD L+ +TIPE L+ + +++L+GL YLH ++++HRD+
Sbjct: 137 ---DNGAISLILEYMDGGSLADFLKKVKTIPERFLAVICKQVLKGLWYLHHEKHIIHRDL 193
Query: 210 KPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADIWSL 269
KP+NLL+N +G+ KITDFG+SA L ++ + TFVGT YMSPERI +Y Y +DIWSL
Sbjct: 194 KPSNLLINHRGDVKITDFGVSAVLASTSGLANTFVGTYNYMSPERISGGAYDYKSDIWSL 253
Query: 270 GLALLESGTGEFPY---TANEGPVN---LMLQILDDPSPSPSKEKFSPEFCSFVDACLQK 323
GL LLE TG FPY +EG VN LM I+D P P ++FSP+FCSF+ AC+QK
Sbjct: 254 GLVLLECATGHFPYKPPEGDEGWVNVYELMETIVDQPEPCAPPDQFSPQFCSFISACVQK 313
Query: 324 DPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFDPTQRMKDL 367
+R +A L+ HPFIT Y+ +DL + S P + +L
Sbjct: 314 HQKDRLSANDLMSHPFITMYDDQDIDLGSYFTSAGPPLATLTEL 357
>UniRef100_O80395 MAP kinase kinase 2 [Arabidopsis thaliana]
Length = 363
Score = 234 bits (598), Expect = 3e-60
Identities = 135/349 (38%), Positives = 202/349 (57%), Gaps = 21/349 (6%)
Query: 32 SYTFSDGGTVNLLSRSYGVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAIG 91
S TF DG +L GV I++L + S + D+ + ++ + IG
Sbjct: 29 SGTFKDG---DLRVNKDGVRIISQLEPEVLSPIKPADD--------QLSLSDLDMVKVIG 77
Query: 92 SGASSVVQRAMHIPTHRVIALKKINI-FEKEKRQQLLTEIRTLCEAPCYEGLVEFHGAFY 150
G+S VVQ H T + ALK I + ++ R+ + E++ + C LV + +FY
Sbjct: 78 KGSSGVVQLVQHKWTGQFFALKVIQLNIDEAIRKAIAQELKINQSSQC-PNLVTSYQSFY 136
Query: 151 TPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIK 210
D+G IS+ LEYMDGGSLAD L+ + IP+ LS++F+++L+GL YLH R+++HRD+K
Sbjct: 137 --DNGAISLILEYMDGGSLADFLKSVKAIPDSYLSAIFRQVLQGLIYLHHDRHIIHRDLK 194
Query: 211 PANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADIWSLG 270
P+NLL+N +GE KITDFG+S + N+ + TFVGT YMSPERI Y +DIWSLG
Sbjct: 195 PSNLLINHRGEVKITDFGVSTVMTNTAGLANTFVGTYNYMSPERIVGNKYGNKSDIWSLG 254
Query: 271 LALLESGTGEFPYTANE------GPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKD 324
L +LE TG+FPY LM I+D P P+ FSPE SF+ CLQKD
Sbjct: 255 LVVLECATGKFPYAPPNQEETWTSVFELMEAIVDQPPPALPSGNFSPELSSFISTCLQKD 314
Query: 325 PDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFDPTQRMKDLADMLTI 373
P++R +A++L+ HPF+ KY+ + ++L + P + +L+ ++
Sbjct: 315 PNSRSSAKELMEHPFLNKYDYSGINLASYFTDAGSPLATLGNLSGTFSV 363
>UniRef100_Q6TKQ8 Putative mitogen-activated protein kinase kinase [Vitis aestivalis]
Length = 354
Score = 233 bits (595), Expect = 8e-60
Identities = 125/277 (45%), Positives = 172/277 (61%), Gaps = 8/277 (2%)
Query: 90 IGSGASSVVQRAMHIPTHRVIALKKINIFEKEKRQQLLTEIRTLCEAPCYEGLVEFHGAF 149
IG GA VQ H T + ALK I + +E + + + + ++ +V + +F
Sbjct: 76 IGKGAGGTVQLVQHKWTGQFFALKVIQMNIQEAALKHIAQELKINQSSQCPYVVVCYKSF 135
Query: 150 YTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDI 209
Y D+G SI LEYMDGGSL D L+ ++IPEP L+++ ++L+GLSYLH R+++HRD+
Sbjct: 136 Y--DNGAFSIILEYMDGGSLLDFLKKVKSIPEPYLAAICNQVLKGLSYLHHERHIIHRDL 193
Query: 210 KPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADIWSL 269
KP+NLL+N +GE KITDFG+SA L ++ TFVGT YMSPERI Y +DIWSL
Sbjct: 194 KPSNLLINHRGEVKITDFGVSAILTSTSGQANTFVGTYNYMSPERISGGKYGSKSDIWSL 253
Query: 270 GLALLESGTGEFPYTANE------GPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQK 323
GL LLE TG+FPY+ E LM I+D P P S +FS EFCSF+ AC+QK
Sbjct: 254 GLVLLECATGQFPYSPPEQGEGWTSFYELMEAIVDQPPPCASTNQFSAEFCSFISACIQK 313
Query: 324 DPDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFDP 360
DP++R +A +L+ HPFI+ Y VDL + + P
Sbjct: 314 DPNDRKSAHELMAHPFISMYRDLNVDLATYFTNAGSP 350
>UniRef100_Q9S7U9 MAP2k beta protein [Arabidopsis thaliana]
Length = 363
Score = 233 bits (594), Expect = 1e-59
Identities = 134/349 (38%), Positives = 202/349 (57%), Gaps = 21/349 (6%)
Query: 32 SYTFSDGGTVNLLSRSYGVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAIG 91
S TF DG +L GV I++L + S + D+ + ++ + IG
Sbjct: 29 SGTFKDG---DLRVNKDGVRIISQLEPEVLSPIKPADD--------QLSLSDLDMVKVIG 77
Query: 92 SGASSVVQRAMHIPTHRVIALKKINI-FEKEKRQQLLTEIRTLCEAPCYEGLVEFHGAFY 150
G+S VVQ H T + ALK I + ++ R+ + E++ + C LV + +FY
Sbjct: 78 KGSSGVVQLVQHKWTGQFFALKVIQLNIDEAIRKAIAQELKINQSSQC-PNLVTSYQSFY 136
Query: 151 TPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIK 210
D+G IS+ LEYMDGGSLAD L+ + IP+ LS++F+++L+GL YLH R+++HRD+K
Sbjct: 137 --DNGAISLILEYMDGGSLADFLKSVKAIPDSYLSAIFRQVLQGLIYLHHDRHIIHRDLK 194
Query: 211 PANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADIWSLG 270
P+NLL+N +GE KITDFG+S + N+ + TFVGT YMSPERI Y +DIWSLG
Sbjct: 195 PSNLLINHRGEVKITDFGVSTVMTNTAGLANTFVGTYNYMSPERIVGNKYGNKSDIWSLG 254
Query: 271 LALLESGTGEFPYTANE------GPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKD 324
L +LE TG+FPY LM I+D P P+ FSPE SF+ CLQK+
Sbjct: 255 LVVLECATGKFPYAPPNQEETWTSVFELMEAIVDQPPPALPSGNFSPELSSFISTCLQKE 314
Query: 325 PDNRPTAEQLLLHPFITKYETAKVDLPGFVRSVFDPTQRMKDLADMLTI 373
P++R +A++L+ HPF+ KY+ + ++L + P + +L+ ++
Sbjct: 315 PNSRSSAKELMEHPFLNKYDYSGINLASYFTDAGSPLATLGNLSGTFSV 363
>UniRef100_Q5VP37 Putative MAP kinase kinase [Oryza sativa]
Length = 352
Score = 232 bits (591), Expect = 2e-59
Identities = 131/328 (39%), Positives = 195/328 (58%), Gaps = 24/328 (7%)
Query: 32 SYTFSDGGTVNLLSRSYGVYNINELGLQKCSTSRS-----VDESDDNEKTYRCGSHEMRI 86
S TF DG + +N+ GL+ S S ++ D N+ + ++
Sbjct: 23 SGTFKDGDLL-----------VNKDGLRIVSQSEEGEAPPIEPLDHNQLSL----DDLDA 67
Query: 87 FGAIGSGASSVVQRAMHIPTHRVIALKKINIFEKEK-RQQLLTEIRTLCEAPCYEGLVEF 145
IG G+S +VQ H T + ALK I + +E R+Q+ E++ C + +V
Sbjct: 68 IKVIGKGSSGIVQLVRHKWTGQFFALKVIQLNIQENIRRQIAQELKISLSTQC-QYVVAC 126
Query: 146 HGAFYTPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLV 205
FY +G ISI LEYMD GSL+D L+ +TIPEP L+++ +++L+GL YLH ++++
Sbjct: 127 CQCFYV--NGVISIVLEYMDSGSLSDFLKTVKTIPEPYLAAICKQVLKGLMYLHHEKHII 184
Query: 206 HRDIKPANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPAD 265
HRD+KP+N+L+N GE KI+DFG+SA + +S A TF GT YM+PERI + + Y +D
Sbjct: 185 HRDLKPSNILINHMGEVKISDFGVSAIIASSSAQRDTFTGTYNYMAPERISGQKHGYMSD 244
Query: 266 IWSLGLALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDP 325
IWSLGL +LE TGEFPY E L+ ++D P PS ++FS EFCSFV AC+QK+
Sbjct: 245 IWSLGLVMLELATGEFPYPPRESFYELLEAVVDHPPPSAPSDQFSEEFCSFVSACIQKNA 304
Query: 326 DNRPTAEQLLLHPFITKYETAKVDLPGF 353
+R +A+ LL HPF++ Y+ +DL +
Sbjct: 305 SDRSSAQILLNHPFLSMYDDLNIDLASY 332
>UniRef100_O04440 Putative mitogen activated protein kinase kinase nMAPKK
[Arabidopsis thaliana]
Length = 354
Score = 221 bits (564), Expect = 3e-56
Identities = 125/312 (40%), Positives = 185/312 (59%), Gaps = 10/312 (3%)
Query: 49 GVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAIGSGASSVVQRAMHIPTHR 108
G +N+ G+Q S S + ++ + IG G+S VQ H T +
Sbjct: 33 GDLRVNKDGIQTVSLSEPGAPPPIEPLDNQLSLADLEVIKVIGKGSSGNVQLVKHKLTQQ 92
Query: 109 VIALKKINIFEKEKRQQLLT-EIRTLCEAPCYEGLVEFHGAFYTPDSGQISIALEYMDGG 167
ALK I + +E + ++ E+R + C LV + +FY +G +SI LE+MDGG
Sbjct: 93 FFALKVIQLNTEESTCRAISQELRINLSSQC-PYLVSCYQSFY--HNGLVSIILEFMDGG 149
Query: 168 SLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIKPANLLVNLKGEPKITDF 227
SLAD+L+ +PE +LS++ +++LRGL Y+H R ++HRD+KP+NLL+N +GE KITDF
Sbjct: 150 SLADLLKKVGKVPENMLSAICKRVLRGLCYIHHERRIIHRDLKPSNLLINHRGEVKITDF 209
Query: 228 GISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADIWSLGLALLESGTGEFPYTANE 287
G+S L ++ ++ +FVGT YMSPERI YS +DIWSLGL LLE TG+FPYT E
Sbjct: 210 GVSKILTSTSSLANSFVGTYPYMSPERISGSLYSNKSDIWSLGLVLLECATGKFPYTPPE 269
Query: 288 ------GPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPTAEQLLLHPFIT 341
L+ I+++P P FSPEFCSF+ C+QKDP +R +A++LL H F+
Sbjct: 270 HKKGWSSVYELVDAIVENPPPCAPSNLFSPEFCSFISQCVQKDPRDRKSAKELLEHKFVK 329
Query: 342 KYETAKVDLPGF 353
+E + +L +
Sbjct: 330 MFEDSDTNLSAY 341
>UniRef100_Q8L6H0 Putative mitogen-activated protein kinase kinase [Oryza sativa]
Length = 333
Score = 212 bits (540), Expect = 2e-53
Identities = 125/323 (38%), Positives = 182/323 (55%), Gaps = 33/323 (10%)
Query: 32 SYTFSDGGTVNLLSRSYGVYNINELGLQKCSTSRSVDESDDNEKTYRCGSHEMRIFGAIG 91
S TF DG + +N+ GL+ S S +E D+ + ++ IG
Sbjct: 23 SGTFKDGDLL-----------VNKDGLRIVSQSEEGEEPLDHNQL---SLDDLDAIKVIG 68
Query: 92 SGASSVVQRAMHIPTHRVIALKKINIFEKEK-RQQLLTEIRTLCEAPCYEGLVEFHGAFY 150
G+S +VQ H T + ALK I + +E R+Q+ E++ C + +V FY
Sbjct: 69 KGSSGIVQLVRHKWTGQFFALKVIQLNIQENIRRQIAQELKISLSTQC-QYVVACCQCFY 127
Query: 151 TPDSGQISIALEYMDGGSLADILRMHRTIPEPILSSMFQKLLRGLSYLHGVRYLVHRDIK 210
+G ISI LEYMD GSL+D L+ +L+GL YLH ++++HRD+K
Sbjct: 128 V--NGVISIVLEYMDSGSLSDFLKT---------------VLKGLMYLHHEKHIIHRDLK 170
Query: 211 PANLLVNLKGEPKITDFGISAGLENSVAMCATFVGTVTYMSPERIRNESYSYPADIWSLG 270
P+N+L+N GE KI+DFG+SA + +S A TF GT YM+PERI + + Y +DIWSLG
Sbjct: 171 PSNILINHMGEVKISDFGVSAIIASSSAQRDTFTGTYNYMAPERISGQKHGYMSDIWSLG 230
Query: 271 LALLESGTGEFPYTANEGPVNLMLQILDDPSPSPSKEKFSPEFCSFVDACLQKDPDNRPT 330
L +LE TGEFPY E L+ ++D P PS ++FS EFCSFV AC+QK+ +R +
Sbjct: 231 LVMLELATGEFPYPPRESFYELLEAVVDHPPPSAPSDQFSEEFCSFVSACIQKNASDRSS 290
Query: 331 AEQLLLHPFITKYETAKVDLPGF 353
A+ LL HPF++ Y+ +DL +
Sbjct: 291 AQILLNHPFLSMYDDLNIDLASY 313
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.138 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 915,673,585
Number of Sequences: 2790947
Number of extensions: 40736711
Number of successful extensions: 132259
Number of sequences better than 10.0: 18574
Number of HSP's better than 10.0 without gapping: 8247
Number of HSP's successfully gapped in prelim test: 10327
Number of HSP's that attempted gapping in prelim test: 93417
Number of HSP's gapped (non-prelim): 21777
length of query: 519
length of database: 848,049,833
effective HSP length: 132
effective length of query: 387
effective length of database: 479,644,829
effective search space: 185622548823
effective search space used: 185622548823
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC146560.10


