

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146554.5 + phase: 0
(236 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
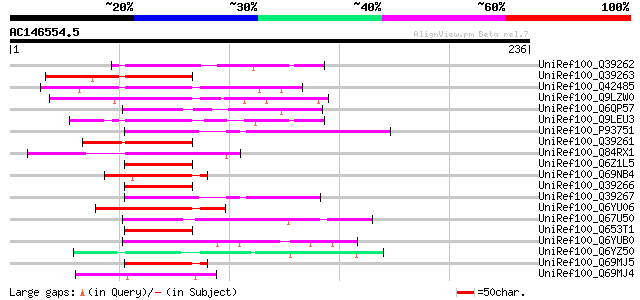
UniRef100_Q39262 Zinc finger protein 3 [Arabidopsis thaliana] 66 6e-10
UniRef100_Q39263 Zinc finger protein 4 [Arabidopsis thaliana] 63 7e-09
UniRef100_Q42485 Zinc finger protein 1 [Arabidopsis thaliana] 62 9e-09
UniRef100_Q9LZW0 Putative zinc finger protein [Arabidopsis thali... 62 2e-08
UniRef100_Q6QP57 Putative zinc finger protein [Zea mays] 62 2e-08
UniRef100_Q9LEU3 Zinc finger-like protein [Arabidopsis thaliana] 61 3e-08
UniRef100_P93751 Putative C2H2-type zinc finger protein [Arabido... 59 1e-07
UniRef100_Q39261 Zinc finger protein 2 [Arabidopsis thaliana] 59 1e-07
UniRef100_Q84RX1 Hypothetical protein OJ1710_H11.113 [Oryza sativa] 58 2e-07
UniRef100_Q6Z1L5 Hypothetical protein OSJNBa0091M20.24 [Oryza sa... 58 2e-07
UniRef100_Q69NB4 Hypothetical protein OSJNBa0016E03.15 [Oryza sa... 58 2e-07
UniRef100_Q39266 Zinc finger protein 7 [Arabidopsis thaliana] 57 3e-07
UniRef100_Q39267 Zinc finger protein [Arabidopsis thaliana] 57 4e-07
UniRef100_Q6YU06 Hypothetical protein P0435E12.40 [Oryza sativa] 57 5e-07
UniRef100_Q67U50 Zinc finger protein-like [Oryza sativa] 57 5e-07
UniRef100_Q653T1 Hypothetical protein OJ1065_E04.11 [Oryza sativa] 56 9e-07
UniRef100_Q6YUB0 Putative zinc finger protein [Oryza sativa] 55 1e-06
UniRef100_Q6YZ50 Putative zinc finger protein [Oryza sativa] 55 2e-06
UniRef100_Q69MJ5 Hypothetical protein OSJNBa0039E17.11 [Oryza sa... 55 2e-06
UniRef100_Q69MJ4 Hypothetical protein OSJNBa0039E17.14 [Oryza sa... 55 2e-06
>UniRef100_Q39262 Zinc finger protein 3 [Arabidopsis thaliana]
Length = 235
Score = 66.2 bits (160), Expect = 6e-10
Identities = 43/100 (43%), Positives = 53/100 (53%), Gaps = 14/100 (14%)
Query: 47 EQKVVEFSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRREEEMDLTLRFSSFIHYQ 106
EQK+ FSCNYC + F +SQALGGHQNAHKRER L K R + S+F H
Sbjct: 57 EQKL--FSCNYCQRTFYSSQALGGHQNAHKRERTLAKRGQR-------MAASASAFGHPY 107
Query: 107 GHS---YFRSANLHNPIGVHVTNAFPSWIGSPYGGYGGVY 143
G S + N H +G+ + S S Y G+GG Y
Sbjct: 108 GFSPLPFHGQYNNHRSLGIQAHSI--SHKLSSYNGFGGHY 145
>UniRef100_Q39263 Zinc finger protein 4 [Arabidopsis thaliana]
Length = 260
Score = 62.8 bits (151), Expect = 7e-09
Identities = 34/68 (50%), Positives = 43/68 (63%), Gaps = 3/68 (4%)
Query: 17 LGTHTKSTSKPLENGGHSSS-FQNTLDEPLPEQKVVEFSCNYCDKKFSTSQALGGHQNAH 75
L + S + PL + SS+ Q +P ++V FSCNYC +KF +SQALGGHQNAH
Sbjct: 50 LDLESSSLTLPLSSTSESSNPEQQQQQQPSVSKRV--FSCNYCQRKFYSSQALGGHQNAH 107
Query: 76 KRERVLKK 83
KRER L K
Sbjct: 108 KRERTLAK 115
>UniRef100_Q42485 Zinc finger protein 1 [Arabidopsis thaliana]
Length = 228
Score = 62.4 bits (150), Expect = 9e-09
Identities = 50/135 (37%), Positives = 67/135 (49%), Gaps = 21/135 (15%)
Query: 15 LRLGTHTKSTSKPLEN---GGHSSSFQNTLDEPLPEQKVVEFSCNYCDKKFSTSQALGGH 71
L GT T S +++ S+S E L + +V FSCNYC +KF +SQALGGH
Sbjct: 29 LSRGTATSSELNLIDSFKTSSSSTSHHQHQQEQLADPRV--FSCNYCQRKFYSSQALGGH 86
Query: 72 QNAHKRERVLKKMEDRRREEEMDLTLRFSSFIHYQGHSYFR-----------SANLHNPI 120
QNAHKRER L K R + +M L+ SS + S R S N + +
Sbjct: 87 QNAHKRERTLAK---RGQYYKMTLSSLPSSAFAFGHGSVSRFASMASLPLHGSVNNRSTL 143
Query: 121 GV--HVTNAFPSWIG 133
G+ H T PS++G
Sbjct: 144 GIQAHSTIHKPSFLG 158
>UniRef100_Q9LZW0 Putative zinc finger protein [Arabidopsis thaliana]
Length = 215
Score = 61.6 bits (148), Expect = 2e-08
Identities = 49/138 (35%), Positives = 70/138 (50%), Gaps = 15/138 (10%)
Query: 19 THTKSTSKPLENGGHSSSFQNTLDEPLP--EQKVVEFSCNYCDKKFSTSQALGGHQNAHK 76
T + T++P + F+ +E E K F+C +C K+FSTSQALGGHQNAHK
Sbjct: 31 TKEEVTAEPSKENQQRLEFRFLFNESSTRNEAKARVFACTFCKKEFSTSQALGGHQNAHK 90
Query: 77 RERVLKKMEDRRREEEMDLTLRFSSFIHY--QGHSYFRSAN------LHNPIGVHVTNAF 128
+ER L K RR+E E++ S + Y G S+ S++ +NP T +
Sbjct: 91 QERSLAK---RRKEIEINYP-ELSIYSQYPPSGLSFSSSSSQYDLGVRYNPNIAKTTKPY 146
Query: 129 PSWIGSPYGGY-GGVYMP 145
P I + GY GG+ P
Sbjct: 147 PFNIFACRFGYRGGLNFP 164
>UniRef100_Q6QP57 Putative zinc finger protein [Zea mays]
Length = 303
Score = 61.6 bits (148), Expect = 2e-08
Identities = 39/98 (39%), Positives = 49/98 (49%), Gaps = 18/98 (18%)
Query: 52 EFSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRREEEMDLTLRFSSFIHYQGHSYF 111
+F C+YC + F TSQALGGHQNAHKRER RR E +SF + G +Y
Sbjct: 71 KFECHYCCRNFPTSQALGGHQNAHKRER----QHARRAHLE-------ASFAAHCGAAYL 119
Query: 112 RSANLHNPIGV-------HVTNAFPSWIGSPYGGYGGV 142
A+L+ G H + W G+ G YGGV
Sbjct: 120 PGAHLYGLFGYGGGGGGGHTAHYPAVWAGAAPGIYGGV 157
>UniRef100_Q9LEU3 Zinc finger-like protein [Arabidopsis thaliana]
Length = 272
Score = 60.8 bits (146), Expect = 3e-08
Identities = 47/122 (38%), Positives = 57/122 (46%), Gaps = 25/122 (20%)
Query: 28 LENGGHSSSFQNTLDEPLPEQKVVEFSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDR 87
LE G + SF + EQK+ FSCNYC + F +SQALGGHQNAHKRER L K
Sbjct: 84 LETGVVTPSFNGSTST---EQKL--FSCNYCQRTFYSSQALGGHQNAHKRERTLAK---- 134
Query: 88 RREEEMDLTLRFSSFIHYQGHSY------FRSANLHNPIGVHVTNAFPSWIGSPYGGYGG 141
R + M F GH Y F + +G+ + S S Y YGG
Sbjct: 135 -RGQRMASAAAF-------GHPYGFAPVPFHGQYSNRTLGIQAHSM--SHKPSSYNVYGG 184
Query: 142 VY 143
Y
Sbjct: 185 EY 186
>UniRef100_P93751 Putative C2H2-type zinc finger protein [Arabidopsis thaliana]
Length = 257
Score = 58.5 bits (140), Expect = 1e-07
Identities = 37/121 (30%), Positives = 51/121 (41%), Gaps = 14/121 (11%)
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRREEEMDLTLRFSSFIHYQGHSYFR 112
F C+YC + F TSQALGGHQNAHKRER K +S++H+ H
Sbjct: 95 FECHYCFRNFPTSQALGGHQNAHKRERQHAKRGS------------MTSYLHH--HQPHD 140
Query: 113 SANLHNPIGVHVTNAFPSWIGSPYGGYGGVYMPNTPLATRYVMQMPNSPQATPQFGMTNF 172
+++ + H +PSW YGG +R + P+S T
Sbjct: 141 PHHIYGFLNNHHHRHYPSWTTEARSYYGGGGHQTPSYYSRNTLAPPSSNPPTINGSPLGL 200
Query: 173 W 173
W
Sbjct: 201 W 201
>UniRef100_Q39261 Zinc finger protein 2 [Arabidopsis thaliana]
Length = 150
Score = 58.5 bits (140), Expect = 1e-07
Identities = 31/50 (62%), Positives = 34/50 (68%), Gaps = 1/50 (2%)
Query: 34 SSSFQNTLDEPLPEQKVVEFSCNYCDKKFSTSQALGGHQNAHKRERVLKK 83
SSS +T EQ V FSCNYC +KF +SQALGGHQNAHK ER L K
Sbjct: 34 SSSSSSTNSSSCLEQPRV-FSCNYCQRKFYSSQALGGHQNAHKLERTLAK 82
>UniRef100_Q84RX1 Hypothetical protein OJ1710_H11.113 [Oryza sativa]
Length = 188
Score = 58.2 bits (139), Expect = 2e-07
Identities = 37/100 (37%), Positives = 49/100 (49%), Gaps = 20/100 (20%)
Query: 9 VEDFLTLRLGTHTKSTSKPLENGGHSSSFQNTLDEPLPEQKVVEFSCNYCDKKFSTSQAL 68
V D LT G +S KP H + FSCNYC +KF +SQAL
Sbjct: 32 VVDALTAEAGKLPESNPKPAVAAPHRT-----------------FSCNYCMRKFFSSQAL 74
Query: 69 GGHQNAHKRERVLKKMEDRRREEEMDLTL---RFSSFIHY 105
GGHQNAHKRER + +++ + + L SSF+H+
Sbjct: 75 GGHQNAHKRERCAPRKSHGFQQQHLMVGLSPTAPSSFLHH 114
>UniRef100_Q6Z1L5 Hypothetical protein OSJNBa0091M20.24 [Oryza sativa]
Length = 306
Score = 58.2 bits (139), Expect = 2e-07
Identities = 25/31 (80%), Positives = 26/31 (83%)
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRERVLKK 83
F CNYC +KF TSQALGGHQNAHKRER L K
Sbjct: 107 FKCNYCQRKFYTSQALGGHQNAHKRERSLAK 137
>UniRef100_Q69NB4 Hypothetical protein OSJNBa0016E03.15 [Oryza sativa]
Length = 152
Score = 57.8 bits (138), Expect = 2e-07
Identities = 31/48 (64%), Positives = 34/48 (70%), Gaps = 4/48 (8%)
Query: 44 PLPEQKVVEFS-CNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRRE 90
P PE +V F C YCD+KF +SQALGGHQNAHK ER L K RRRE
Sbjct: 24 PSPEPPLVGFFLCMYCDRKFDSSQALGGHQNAHKYERSLAK---RRRE 68
>UniRef100_Q39266 Zinc finger protein 7 [Arabidopsis thaliana]
Length = 209
Score = 57.4 bits (137), Expect = 3e-07
Identities = 24/31 (77%), Positives = 27/31 (86%)
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRERVLKK 83
FSCNYC +KF +SQALGGHQNAHKRER + K
Sbjct: 59 FSCNYCRRKFYSSQALGGHQNAHKRERTMAK 89
>UniRef100_Q39267 Zinc finger protein [Arabidopsis thaliana]
Length = 239
Score = 57.0 bits (136), Expect = 4e-07
Identities = 32/89 (35%), Positives = 43/89 (47%), Gaps = 14/89 (15%)
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRREEEMDLTLRFSSFIHYQGHSYFR 112
F C+YC + F TSQALGGHQNAHKRER K +S++H+ H
Sbjct: 93 FECHYCFRNFPTSQALGGHQNAHKRERQHAKRGS------------MTSYLHH--HQPHD 138
Query: 113 SANLHNPIGVHVTNAFPSWIGSPYGGYGG 141
+++ + H +PSW YGG
Sbjct: 139 PHHIYGFLNNHHHRHYPSWTTEARSYYGG 167
>UniRef100_Q6YU06 Hypothetical protein P0435E12.40 [Oryza sativa]
Length = 174
Score = 56.6 bits (135), Expect = 5e-07
Identities = 29/59 (49%), Positives = 37/59 (62%), Gaps = 3/59 (5%)
Query: 40 TLDEPLPEQKVVEFSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRREEEMDLTLR 98
+L P + EFSC YC K+F +SQALGGHQNAHK +R L K R RE + ++ R
Sbjct: 46 SLQPSYPSRFQTEFSCCYCPKRFQSSQALGGHQNAHKLQRNLAK---RNREAFLSISQR 101
>UniRef100_Q67U50 Zinc finger protein-like [Oryza sativa]
Length = 296
Score = 56.6 bits (135), Expect = 5e-07
Identities = 36/118 (30%), Positives = 54/118 (45%), Gaps = 12/118 (10%)
Query: 52 EFSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRREEEMDLTLRFSSFIHYQGHSYF 111
+F C+YC + F TSQALGGHQNAHKRER + +R + + + Y ++ +
Sbjct: 71 KFECHYCCRNFPTSQALGGHQNAHKRER-----QHAKRAQFQSAMAMHAHYPAYPAYASY 125
Query: 112 RSANLHNPIGVHVT----NAFPSWIGSPYGGYGGVYMPNTPLATRYVMQMPNSPQATP 165
++ P H+ +PSW + GG P +A RY P + P
Sbjct: 126 YGSHRFGPSPPHMAPPPPPPYPSWSNHHHLPPGG---PAPMVAARYYGPAPPGSVSHP 180
>UniRef100_Q653T1 Hypothetical protein OJ1065_E04.11 [Oryza sativa]
Length = 165
Score = 55.8 bits (133), Expect = 9e-07
Identities = 24/31 (77%), Positives = 26/31 (83%)
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRERVLKK 83
FSCNYC +KF +SQALGGHQNAHK ER L K
Sbjct: 49 FSCNYCHRKFFSSQALGGHQNAHKLERTLAK 79
>UniRef100_Q6YUB0 Putative zinc finger protein [Oryza sativa]
Length = 300
Score = 55.1 bits (131), Expect = 1e-06
Identities = 39/124 (31%), Positives = 56/124 (44%), Gaps = 21/124 (16%)
Query: 52 EFSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRREEEM------------DLTLRF 99
+F C+YC + F TSQALGGHQNAHKRER K + M F
Sbjct: 98 KFECHYCCRNFPTSQALGGHQNAHKRERQRAKHAQFQTAMAMHHGHGQYYPLPDPYAAAF 157
Query: 100 SSFI-HYQGHSYFRSANLHNPIGVHVTNAFPSWIGSP--YGGYGGVYMP--NTPLATRYV 154
+++ H+ H + +A P H +PSW Y G G + P +P+A +
Sbjct: 158 AAYPGHHHHHRFAATAAAAMPPPPH----YPSWAAGSRYYSGPGSISQPINGSPVAPAGM 213
Query: 155 MQMP 158
++P
Sbjct: 214 WRLP 217
>UniRef100_Q6YZ50 Putative zinc finger protein [Oryza sativa]
Length = 279
Score = 54.7 bits (130), Expect = 2e-06
Identities = 45/154 (29%), Positives = 61/154 (39%), Gaps = 25/154 (16%)
Query: 30 NGGHSSSFQNTLDEPLPEQKVVEFSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRR 89
NGG + E+K F C+YC + F TSQALGGHQNAHKRE R+
Sbjct: 70 NGGDGGTKNGGAATAAAERK---FECHYCCRNFPTSQALGGHQNAHKRE--------RQH 118
Query: 90 EEEMDLTLRFSSFIHYQGHSYFRSANLHNPIGVHVTN------------AFPSWIGSPYG 137
+ L + + GH Y N H+ IG + +P W + G
Sbjct: 119 AKRAHLQASLAMHRYMPGHMY-GLFNYHHHIGGRFDHHPPPPPPPPPPAHYPMWTSAAPG 177
Query: 138 GYGGVYMPNTPLATRYVMQ-MPNSPQATPQFGMT 170
+ G P+ V + + P T FG T
Sbjct: 178 AFAGPGSMAQPINGSPVQAGLWSVPPPTENFGST 211
>UniRef100_Q69MJ5 Hypothetical protein OSJNBa0039E17.11 [Oryza sativa]
Length = 135
Score = 54.7 bits (130), Expect = 2e-06
Identities = 27/38 (71%), Positives = 29/38 (76%), Gaps = 3/38 (7%)
Query: 53 FSCNYCDKKFSTSQALGGHQNAHKRERVLKKMEDRRRE 90
F C YCD+KF +SQALGGHQNAHK ER L K RRRE
Sbjct: 31 FLCMYCDRKFYSSQALGGHQNAHKYERSLAK---RRRE 65
>UniRef100_Q69MJ4 Hypothetical protein OSJNBa0039E17.14 [Oryza sativa]
Length = 236
Score = 54.7 bits (130), Expect = 2e-06
Identities = 32/72 (44%), Positives = 40/72 (55%), Gaps = 8/72 (11%)
Query: 31 GGHSSSFQNTLDEPLPEQKVVE----FSCNYCDKKFSTSQALGGHQNAHKRERVLKK--- 83
GG Q L+ L + E F+C YCDKKF +SQALGGHQNAHK ER + K
Sbjct: 117 GGEEGDEQLDLNLSLQPSQANEPPGYFTCTYCDKKFYSSQALGGHQNAHKFERSVAKRTR 176
Query: 84 -MEDRRREEEMD 94
+ RR++ D
Sbjct: 177 ELAAARRQQAAD 188
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.315 0.132 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 421,447,614
Number of Sequences: 2790947
Number of extensions: 18660377
Number of successful extensions: 99051
Number of sequences better than 10.0: 677
Number of HSP's better than 10.0 without gapping: 447
Number of HSP's successfully gapped in prelim test: 231
Number of HSP's that attempted gapping in prelim test: 94251
Number of HSP's gapped (non-prelim): 4853
length of query: 236
length of database: 848,049,833
effective HSP length: 124
effective length of query: 112
effective length of database: 501,972,405
effective search space: 56220909360
effective search space used: 56220909360
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.5 bits)
S2: 73 (32.7 bits)
Medicago: description of AC146554.5


