

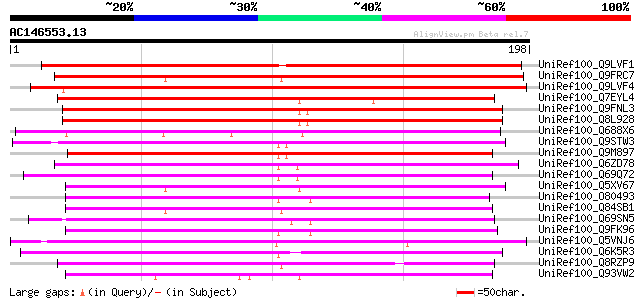
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146553.13 + phase: 0
(198 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LVF1 Emb|CAB45066.1 [Arabidopsis thaliana] 244 9e-64
UniRef100_Q9FRC7 Hypothetical protein OSJNBa0013M12.6 [Oryza sat... 198 6e-50
UniRef100_Q9LVF4 Emb|CAB45066.1 [Arabidopsis thaliana] 194 1e-48
UniRef100_Q7EYL4 Hypothetical protein P0503D09.101 [Oryza sativa] 184 9e-46
UniRef100_Q9FNL3 Arabidopsis thaliana genomic DNA, chromosome 5,... 162 6e-39
UniRef100_Q8L928 Hypothetical protein [Arabidopsis thaliana] 156 3e-37
UniRef100_Q688X6 Hypothetical protein OJ1115_B06.18 [Oryza sativa] 152 6e-36
UniRef100_Q9STW3 Hypothetical protein T22A6.140 [Arabidopsis tha... 151 1e-35
UniRef100_Q9M897 F16B3.6 protein [Arabidopsis thaliana] 147 2e-34
UniRef100_Q6ZD78 Hypothetical protein P0450B04.8 [Oryza sativa] 144 2e-33
UniRef100_Q69Q72 Hypothetical protein OJ1723_B06.129 [Oryza sativa] 139 3e-32
UniRef100_Q5XV67 Hypothetical protein [Arabidopsis thaliana] 138 9e-32
UniRef100_O80493 T12M4.16 [Arabidopsis thaliana] 132 7e-30
UniRef100_Q84SB1 Hypothetical protein OSJNBb0008D07.40 [Oryza sa... 131 1e-29
UniRef100_Q69SN5 Hypothetical protein OSJNBa0022O02.10 [Oryza sa... 128 7e-29
UniRef100_Q9FK96 Gb|AAC24089.1 [Arabidopsis thaliana] 125 6e-28
UniRef100_Q5VNJ6 Hypothetical protein OJ1123_G09.11 [Oryza sativa] 124 2e-27
UniRef100_Q6K5R3 Hypothetical protein P0527E02.39 [Oryza sativa] 110 2e-23
UniRef100_Q8RZP9 B1065E10.15 protein [Oryza sativa] 108 6e-23
UniRef100_Q93VW2 P0560B06.26 protein [Oryza sativa] 107 2e-22
>UniRef100_Q9LVF1 Emb|CAB45066.1 [Arabidopsis thaliana]
Length = 184
Score = 244 bits (623), Expect = 9e-64
Identities = 115/183 (62%), Positives = 142/183 (76%), Gaps = 2/183 (1%)
Query: 13 NKGASTMTGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGILLVICGF 72
+K + +T+SGVG+L+KLLPTGTVFLFQ+L+PV+TNNGHC INKYL+G+L+VIC F
Sbjct: 2 SKTFKAIRDRTYSGVGDLIKLLPTGTVFLFQFLNPVLTNNGHCLLINKYLTGVLIVICAF 61
Query: 73 NCAFTSFTDSYTGSDGQRHYGIVTMNGLWPSPGSDSVDLSAYKLRFGDFVHAFLSVIVFA 132
+C FT FTDSY DG HYG+ T+ GLWP S SVDLS+ +LR GDFVHAF S+IVF+
Sbjct: 62 SCCFTCFTDSYRTRDGYVHYGVATVKGLWPD--SSSVDLSSKRLRVGDFVHAFFSLIVFS 119
Query: 133 VLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGYPTSSDTND 192
V+ LLD N V+CFYP F S+ KI + VLPPVIGV+SGAVF +FPS RHGIG P+ +D
Sbjct: 120 VISLLDANTVNCFYPGFGSAGKIFLMVLPPVIGVISGAVFTVFPSRRHGIGNPSDHSEDD 179
Query: 193 TSE 195
SE
Sbjct: 180 ASE 182
>UniRef100_Q9FRC7 Hypothetical protein OSJNBa0013M12.6 [Oryza sativa]
Length = 193
Score = 198 bits (504), Expect = 6e-50
Identities = 98/183 (53%), Positives = 128/183 (69%), Gaps = 4/183 (2%)
Query: 18 TMTGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTI-NKYLSGILLVICGFNCAF 76
++ + GV +L+KLLP+GTVFLFQ+LSP+VTNNGHC+ ++ LS LL +CG CAF
Sbjct: 10 SVADRALRGVADLIKLLPSGTVFLFQFLSPLVTNNGHCAAAYSRVLSAALLALCGAFCAF 69
Query: 77 TSFTDSYTGSDGQRHYGIVTMNGLWP---SPGSDSVDLSAYKLRFGDFVHAFLSVIVFAV 133
+SFTDSY GSDG+ +YG+VT GL P + + DLS Y+LR GDFVHA LS++VFA
Sbjct: 70 SSFTDSYVGSDGRVYYGVVTARGLRTFAADPDAAARDLSGYRLRAGDFVHAALSLLVFAT 129
Query: 134 LGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGYPTSSDTNDT 193
+ LLD + V C YP E SE+ +M VLPPV+G V+ FM+FP+ RHGIGY + T D
Sbjct: 130 IALLDADTVACLYPALEVSERTMMAVLPPVVGGVASYAFMVFPNNRHGIGYQPTRATEDF 189
Query: 194 SEK 196
K
Sbjct: 190 EHK 192
>UniRef100_Q9LVF4 Emb|CAB45066.1 [Arabidopsis thaliana]
Length = 207
Score = 194 bits (492), Expect = 1e-48
Identities = 93/191 (48%), Positives = 125/191 (64%), Gaps = 2/191 (1%)
Query: 9 KTSTNKGASTM--TGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGIL 66
KT++ + M T K+ +G+ +L+KLLPTGT+F++ L+PV+TN+G CST NK +S IL
Sbjct: 10 KTNSPASSENMANTNKSLTGLESLIKLLPTGTLFIYLLLNPVLTNDGECSTGNKVMSSIL 69
Query: 67 LVICGFNCAFTSFTDSYTGSDGQRHYGIVTMNGLWPSPGSDSVDLSAYKLRFGDFVHAFL 126
+ +C F+C F+ FTDS+ G DG R +GIVT GLW SVDLS YKLR DFVHA
Sbjct: 70 VALCSFSCVFSCFTDSFKGVDGSRKFGIVTKKGLWTYAEPGSVDLSKYKLRIADFVHAGF 129
Query: 127 SVIVFAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGYPT 186
+ VF L LLD N CFYP+F ++K L+ LPP +GV S +F +FPS R GIGY
Sbjct: 130 VLAVFGTLVLLDANTASCFYPRFRETQKTLVMALPPAVGVASATIFALFPSKRSGIGYAP 189
Query: 187 SSDTNDTSEKT 197
++ E+T
Sbjct: 190 IAEEVGAEEET 200
>UniRef100_Q7EYL4 Hypothetical protein P0503D09.101 [Oryza sativa]
Length = 188
Score = 184 bits (468), Expect = 9e-46
Identities = 87/176 (49%), Positives = 116/176 (65%), Gaps = 9/176 (5%)
Query: 19 MTGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGILLVICGFNCAFTS 78
M TF +G+++KLLPT TV +++ L+P+VTN G C NK ++ ++LV+C F CAF+
Sbjct: 1 MADTTFKSIGDVLKLLPTATVIVYEVLTPIVTNTGDCHVANKVVTPVILVLCAFFCAFSQ 60
Query: 79 FTDSYTGSDGQRHYGIVTMNGLWPSPGSDSV--------DLSAYKLRFGDFVHAFLSVIV 130
FTDSY G+DG+ YG+VT GL P G D S Y+LRFGDFVHAF SV V
Sbjct: 61 FTDSYVGADGKVRYGLVTARGLLPFSGGGGADGGDAAGRDFSKYRLRFGDFVHAFFSVAV 120
Query: 131 FAVLGLL-DTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGYP 185
FA + LL D N V CFYP + +K ++ LP V+G ++ VF++FPS RHGIGYP
Sbjct: 121 FAAVALLADANTVSCFYPSLKDQQKKVVMALPVVVGALASVVFVVFPSTRHGIGYP 176
>UniRef100_Q9FNL3 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MCL19
[Arabidopsis thaliana]
Length = 214
Score = 162 bits (409), Expect = 6e-39
Identities = 82/171 (47%), Positives = 109/171 (62%), Gaps = 3/171 (1%)
Query: 21 GKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGILLVICGFNCAFTSFT 80
GKTF NL LLPTGTV FQ LSP+ TN G C +++++ +L+ ICGF+C SFT
Sbjct: 43 GKTFQTTANLANLLPTGTVLAFQILSPICTNVGRCDLTSRFMTALLVSICGFSCFILSFT 102
Query: 81 DSYTGSDGQRHYGIVTMNGLWPSPGSDSV--DLS-AYKLRFGDFVHAFLSVIVFAVLGLL 137
DSY +G YG T++G W GS ++ +LS +YKLRF DFVHA +S +VF + L
Sbjct: 103 DSYKDLNGSVCYGFATIHGFWIIDGSATLPQELSKSYKLRFIDFVHAIMSFLVFGAVVLF 162
Query: 138 DTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGYPTSS 188
D NVV+CFYP+ + L+ LP +GV VF FP+ RHGIG+P S+
Sbjct: 163 DQNVVNCFYPEPSAEVVELLTTLPVAVGVFCSMVFAKFPTTRHGIGFPLSA 213
>UniRef100_Q8L928 Hypothetical protein [Arabidopsis thaliana]
Length = 213
Score = 156 bits (395), Expect = 3e-37
Identities = 78/171 (45%), Positives = 108/171 (62%), Gaps = 3/171 (1%)
Query: 21 GKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGILLVICGFNCAFTSFT 80
G+TF +L LLPTGTV FQ LSP+ +N G C ++K ++ L+ ICGF+C SFT
Sbjct: 42 GQTFQTTAHLANLLPTGTVLAFQLLSPIFSNGGQCDLVSKIMTSTLVAICGFSCFILSFT 101
Query: 81 DSYTGSDGQRHYGIVTMNGLWPSPGSDSV--DLS-AYKLRFGDFVHAFLSVIVFAVLGLL 137
DSY +G YG+ T++G W GS ++ +LS YKLRF DFVHAF+S+ VF + L
Sbjct: 102 DSYKDKNGTICYGLATIHGFWIIDGSTTLPQELSKRYKLRFIDFVHAFMSLFVFGAVVLF 161
Query: 138 DTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGYPTSS 188
D N V+CF+P + ++ LP +GV S +F FP+ R+GIG+P SS
Sbjct: 162 DRNAVNCFFPSPSAEALEVLTALPVGVGVFSSMLFATFPTTRNGIGFPLSS 212
>UniRef100_Q688X6 Hypothetical protein OJ1115_B06.18 [Oryza sativa]
Length = 238
Score = 152 bits (383), Expect = 6e-36
Identities = 83/197 (42%), Positives = 111/197 (56%), Gaps = 12/197 (6%)
Query: 3 EKASSSKTSTNKGASTMT------GKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCS 56
EK S+ G + M+ +T+ +L KLLPTGTV FQ LSP+VTN GHC
Sbjct: 40 EKRSAGDGGGGSGGNGMSPVQRAISQTYQSTAHLAKLLPTGTVLAFQLLSPIVTNQGHCD 99
Query: 57 T-INKYLSGILLVICGFNCAFTSFTDSY--TGSDGQRHYGIVTMNGLWPSPGSDSVD--- 110
N+ ++G L+ +C +C SFTDS+ + G YG T GLW G +D
Sbjct: 100 VEANRAMAGALIALCALSCFVLSFTDSFRDAATGGAVRYGFATPAGLWVIDGGAPLDPQA 159
Query: 111 LSAYKLRFGDFVHAFLSVIVFAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGA 170
+AY+LR D VHA +SV+VFA + L D NVV CFYP + ++ LP IGVV
Sbjct: 160 AAAYRLRLLDLVHAVVSVMVFAAVALFDQNVVSCFYPVPSEGTRQVLTALPIAIGVVGSM 219
Query: 171 VFMIFPSYRHGIGYPTS 187
+F+ FP+ RHGIG+P S
Sbjct: 220 LFVSFPTTRHGIGFPLS 236
>UniRef100_Q9STW3 Hypothetical protein T22A6.140 [Arabidopsis thaliana]
Length = 213
Score = 151 bits (381), Expect = 1e-35
Identities = 77/193 (39%), Positives = 111/193 (56%), Gaps = 7/193 (3%)
Query: 2 SEKASSSKTSTNKGASTMTGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKY 61
SE + T++ A + +T + NL LLPTGT+ F L PV T+NG C +
Sbjct: 19 SESVPQLRRQTSQHA--VMSQTLTSAANLANLLPTGTLLAFTLLIPVFTSNGSCDYPTQV 76
Query: 62 LSGILLVICGFNCAFTSFTDSYTGSDGQRHYGIVTMNGLW----PSP-GSDSVDLSAYKL 116
L+ +LL + +C +SFTDS DG +YG T G+W P P G +LS Y++
Sbjct: 77 LTIVLLTLLSISCFLSSFTDSVKAEDGNVYYGFATRKGMWVFDYPDPDGLGLPNLSKYRI 136
Query: 117 RFGDFVHAFLSVIVFAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFP 176
R D++HA LSV+VF + L D N V CFYP E K ++ ++P +GV+ G +F++FP
Sbjct: 137 RIIDWIHAVLSVLVFGAVALRDKNAVSCFYPAPEQETKKVLDIVPMGVGVICGMLFLVFP 196
Query: 177 SYRHGIGYPTSSD 189
+ RHGIGYP + D
Sbjct: 197 ARRHGIGYPVTGD 209
>UniRef100_Q9M897 F16B3.6 protein [Arabidopsis thaliana]
Length = 219
Score = 147 bits (371), Expect = 2e-34
Identities = 71/167 (42%), Positives = 103/167 (61%), Gaps = 5/167 (2%)
Query: 23 TFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGILLVICGFNCAFTSFTDS 82
T + NL LLPTGT+ FQ L+PV T+NG C ++L+ +LL + +C +SFTDS
Sbjct: 44 TLTSAANLSNLLPTGTLLAFQLLTPVFTSNGVCDHATRFLTAVLLFLLAASCFVSSFTDS 103
Query: 83 YTGSDGQRHYGIVTMNGLW----PSP-GSDSVDLSAYKLRFGDFVHAFLSVIVFAVLGLL 137
DG ++G VT G+W P P G DL+ Y++RF D++HA LSV+VF + L
Sbjct: 104 VKADDGTIYFGFVTFKGMWVVDYPDPSGLGLPDLAKYRMRFVDWIHATLSVLVFGAVALR 163
Query: 138 DTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGY 184
D + CFYP E+ K ++ ++P +GV+ +FM+FP+ RHGIGY
Sbjct: 164 DKYITDCFYPSPEAETKHVLDIVPVGVGVMCSLLFMVFPARRHGIGY 210
>UniRef100_Q6ZD78 Hypothetical protein P0450B04.8 [Oryza sativa]
Length = 224
Score = 144 bits (362), Expect = 2e-33
Identities = 73/184 (39%), Positives = 106/184 (56%), Gaps = 7/184 (3%)
Query: 18 TMTGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGILLVICGFNCAFT 77
++ + + +L LLPTGTV FQ L+P TNNG C L+ LL + +C
Sbjct: 41 SLLSQALASTASLANLLPTGTVMAFQLLAPTFTNNGACDATTSLLTAALLALLALSCVLA 100
Query: 78 SFTDSYTGSDGQRHYGIVTMNGLW----PSPGSDS---VDLSAYKLRFGDFVHAFLSVIV 130
SFTDS G DG+ +YG+ T GLW P G+ + D S Y++R D VHA LSV V
Sbjct: 101 SFTDSVRGPDGRVYYGLATPRGLWLLDYPPAGAGAPPQPDTSRYRMRAIDGVHALLSVGV 160
Query: 131 FAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGYPTSSDT 190
F V+ D NVV CF+P + ++ ++P +GV+ +F++FP+ RHGIGYP +++T
Sbjct: 161 FGVVAARDKNVVGCFWPSPAKGTEEVLGIVPLGVGVMCSLLFVVFPTTRHGIGYPVTNNT 220
Query: 191 NDTS 194
+S
Sbjct: 221 TSSS 224
>UniRef100_Q69Q72 Hypothetical protein OJ1723_B06.129 [Oryza sativa]
Length = 262
Score = 139 bits (351), Expect = 3e-32
Identities = 78/196 (39%), Positives = 105/196 (52%), Gaps = 17/196 (8%)
Query: 6 SSSKTSTNKGASTMTGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGI 65
+ S +S +T KT S V NL KLLPTGTV FQ LSP TN G C T N+YL+
Sbjct: 61 AGSSSSPPPPPTTAMDKTLSSVANLAKLLPTGTVLAFQSLSPSFTNRGACLTSNRYLTAA 120
Query: 66 LLVICGFNCAFTSFTDSYTGSDGQRHYGIVTMNGLW--------PSPGSDS--------- 108
LL +C +C F SFTDS+ G DG+ +YG+ T G S G D
Sbjct: 121 LLYLCVLSCIFFSFTDSFVGGDGKLYYGVATAKGFLVFNYDAGSSSDGDDDDQRRRREVF 180
Query: 109 VDLSAYKLRFGDFVHAFLSVIVFAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVS 168
DL ++R+ D+VHA + +VF + T V C++P+ + K L+ LP G +S
Sbjct: 181 KDLRRLRIRWVDYVHAVFTALVFMTVAFSSTAVQSCYFPEAGDNVKQLLTNLPLGAGFLS 240
Query: 169 GAVFMIFPSYRHGIGY 184
VF++FP+ R GIGY
Sbjct: 241 TTVFLVFPTTRKGIGY 256
>UniRef100_Q5XV67 Hypothetical protein [Arabidopsis thaliana]
Length = 200
Score = 138 bits (347), Expect = 9e-32
Identities = 70/172 (40%), Positives = 104/172 (59%), Gaps = 4/172 (2%)
Query: 22 KTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTI-NKYLSGILLVICGFNCAFTSFT 80
KTF G +L LLPTG+V FQ + PV+T+ G C TI +++L+ L+ +C +C SFT
Sbjct: 29 KTFKGTAHLSNLLPTGSVMSFQIMCPVLTHQGQCPTITSRWLTCFLVSLCAISCFLFSFT 88
Query: 81 DSYTGSDGQRHYGIVTMNGLWPSPGSDSV---DLSAYKLRFGDFVHAFLSVIVFAVLGLL 137
DS +G+ YG+ T +GL GS ++ + YKL+ DF+HA +S++VF + +
Sbjct: 89 DSIRDPNGKVRYGLATWSGLLVMDGSITLTEEEKEKYKLKILDFIHAIMSMLVFFAVSMF 148
Query: 138 DTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGYPTSSD 189
D NV C +P K ++ LP VIGV+ GA F+ FP+ RHGIG P + +
Sbjct: 149 DQNVTRCLFPVPSEETKEILTSLPFVIGVICGAFFLAFPTRRHGIGSPLTKE 200
>UniRef100_O80493 T12M4.16 [Arabidopsis thaliana]
Length = 243
Score = 132 bits (331), Expect = 7e-30
Identities = 68/177 (38%), Positives = 100/177 (56%), Gaps = 15/177 (8%)
Query: 22 KTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGILLVICGFNCAFTSFTD 81
KT S LV LPTGT+ +F+ + P + +G C+ IN + +LL++C +C F FTD
Sbjct: 63 KTVSKTSMLVNFLPTGTLLMFEMVLPTIYRDGDCNGINTLMIHLLLLLCAMSCFFFHFTD 122
Query: 82 SYTGSDGQRHYGIVTMNGLW-----PSPGSDSVDLSA----------YKLRFGDFVHAFL 126
S+ SDG+ +YG VT GL PSPG D+ A YKLR DFVH+ +
Sbjct: 123 SFKASDGKIYYGFVTPRGLAVFMKPPSPGFGGGDVIAEKEIPVTDERYKLRVNDFVHSVM 182
Query: 127 SVIVFAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIG 183
SV+VF + D V C +P E +M+ P ++G+V A+F++FP+ R+G+G
Sbjct: 183 SVLVFMAIAFSDRRVTGCLFPGKEKEMDQVMESFPLMVGIVCSALFLVFPTSRYGVG 239
>UniRef100_Q84SB1 Hypothetical protein OSJNBb0008D07.40 [Oryza sativa]
Length = 225
Score = 131 bits (329), Expect = 1e-29
Identities = 72/172 (41%), Positives = 98/172 (56%), Gaps = 9/172 (5%)
Query: 22 KTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTI-NKYLSGILLVICGFNCAFTSFT 80
KT SG +L+KLLPTGTV FQ L+P +N+G C + N+YL L+ C +C SFT
Sbjct: 46 KTLSGASDLLKLLPTGTVLAFQALAPSFSNHGVCHAVANRYLVLALIGACAASCMLLSFT 105
Query: 81 DSYTGSDGQRHYGIVTMNGLWP--------SPGSDSVDLSAYKLRFGDFVHAFLSVIVFA 132
DS G DG+ +YG+ T+ G P G+ DLS +++ DFVHAF S +VF
Sbjct: 106 DSLIGHDGKLYYGVATLRGFRPFNFAGTREEHGTVFKDLSRFRITALDFVHAFFSAVVFL 165
Query: 133 VLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGY 184
+ D V C +P+ E+ + L+ LP G +S VFMIFP+ R IGY
Sbjct: 166 AVAFADAAVQTCLFPEAEADMRELLVNLPLGAGFLSSMVFMIFPTTRKSIGY 217
>UniRef100_Q69SN5 Hypothetical protein OSJNBa0022O02.10 [Oryza sativa]
Length = 253
Score = 128 bits (322), Expect = 7e-29
Identities = 72/181 (39%), Positives = 99/181 (53%), Gaps = 4/181 (2%)
Query: 8 SKTSTNKGASTMTGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGILL 67
S T + T+T + F L K LPTG V +F+ LSPV TN G C +N+ ++ L+
Sbjct: 70 SDDETTRLERTIT-RAFRSTAELAKHLPTGAVLVFEVLSPVFTNGGKCQDVNRVMTAWLV 128
Query: 68 VICGFNCAFTSFTDSYTGSDGQRHYGIVTMNGLWPSPGS--DSVDLSA-YKLRFGDFVHA 124
+C C F FTDS+ G Y + T GLW G+ D++A Y+LRF DF HA
Sbjct: 129 GLCAAACFFLCFTDSFHDGKGTVRYVVATRAGLWVIDGTAPPPPDVAATYRLRFIDFFHA 188
Query: 125 FLSVIVFAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGY 184
LS+IVF + + D NV CFYP + ++ +P G+V +F FPS RHGIG+
Sbjct: 189 VLSLIVFLSVAMFDHNVGACFYPVMSYDTRQVLTDVPLAGGLVGTMLFATFPSTRHGIGF 248
Query: 185 P 185
P
Sbjct: 249 P 249
>UniRef100_Q9FK96 Gb|AAC24089.1 [Arabidopsis thaliana]
Length = 244
Score = 125 bits (314), Expect = 6e-28
Identities = 67/180 (37%), Positives = 98/180 (54%), Gaps = 15/180 (8%)
Query: 22 KTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGILLVICGFNCAFTSFTD 81
KT S LV LPTGT+ +F+ + P + +G C+ IN + +LL++C +C F FTD
Sbjct: 64 KTVSKTSMLVNFLPTGTLLMFEMVLPSIYRDGDCNGINTLMIHLLLLLCAMSCFFFHFTD 123
Query: 82 SYTGSDGQRHYGIVTMNGLW-----PSPGSDSVDLSA----------YKLRFGDFVHAFL 126
S+ SDG+ +YG VT GL P P D+ A YKL DFVHA +
Sbjct: 124 SFKASDGKIYYGFVTPRGLAVFMKPPPPEFGGGDVIAEAEIPVTDDRYKLTVNDFVHAVM 183
Query: 127 SVIVFAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGYPT 186
SV+VF + D V C +P E +M+ P ++G+V A+F++FP+ R+G+G T
Sbjct: 184 SVLVFMAIAFSDRRVTGCLFPGKEKEMDQVMESFPIMVGIVCSALFLVFPTTRYGVGCMT 243
>UniRef100_Q5VNJ6 Hypothetical protein OJ1123_G09.11 [Oryza sativa]
Length = 240
Score = 124 bits (310), Expect = 2e-27
Identities = 72/204 (35%), Positives = 109/204 (53%), Gaps = 9/204 (4%)
Query: 1 MSEKASSSKTSTNKGASTMTGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINK 60
+ E +SSKT T+T K + NL +LLPTGTV +Q LSP TN+G C+ NK
Sbjct: 37 VDESTTSSKTDV--ATKTVTDKVMASTANLAQLLPTGTVLAYQALSPSFTNHGECNAANK 94
Query: 61 YLSGILLVICGFNCAFTSFTDSYTGSDGQRHYGIVTMNGL------WPSPGSDSVDLSAY 114
+L+ +L+ + F SFTDS G DG+ +YG+ T GL + + S
Sbjct: 95 WLTAVLVGVLAGLSLFFSFTDSVVGQDGKLYYGVATRRGLNVFNMSREEEEAKKLSHSEL 154
Query: 115 KLRFGDFVHAFLSVIVFAVLGLLDTNVVHCFYPKFE-SSEKILMQVLPPVIGVVSGAVFM 173
+LR DFVH+F + +VF + D + +CF+ + + K L++ LP + +S VF+
Sbjct: 155 RLRPLDFVHSFFTAMVFLTVAFSDVGLQNCFFGQNPGGNTKELLKNLPLGMAFLSSFVFL 214
Query: 174 IFPSYRHGIGYPTSSDTNDTSEKT 197
IFP+ R GIGY ++ + T
Sbjct: 215 IFPTKRKGIGYNDNTPNRKAEDVT 238
>UniRef100_Q6K5R3 Hypothetical protein P0527E02.39 [Oryza sativa]
Length = 216
Score = 110 bits (276), Expect = 2e-23
Identities = 64/197 (32%), Positives = 93/197 (46%), Gaps = 17/197 (8%)
Query: 5 ASSSKTSTNKGASTMTGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSG 64
+S +T +++ K + +L K LPTG V F+ LSP T +G C+ N+ L+
Sbjct: 22 SSDDGQTTQATQASLVCKALNSTADLAKHLPTGAVLAFEVLSPSFTADGSCTAANRALTA 81
Query: 65 ILLVICGFNCAFTSFTDSYTGSDGQRHYGIVTMNGLW-------------PSPGSDSVDL 111
L+ C C FTDSY + G YG VT +G P P D
Sbjct: 82 CLVGACALCCFLLCFTDSYRDATGSVRYGFVTPSGSLRLIDSGSGSGSPPPPPPRD---- 137
Query: 112 SAYKLRFGDFVHAFLSVIVFAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAV 171
Y+L D +H LS VF + ++D NVV CFYP + + L+ +P G +
Sbjct: 138 DRYRLGARDVLHGALSFAVFLAVAMVDRNVVACFYPVESPATRQLLAAVPMAAGAAGSFL 197
Query: 172 FMIFPSYRHGIGYPTSS 188
F +FPS R GIG+P ++
Sbjct: 198 FAMFPSTRRGIGFPVAA 214
>UniRef100_Q8RZP9 B1065E10.15 protein [Oryza sativa]
Length = 592
Score = 108 bits (271), Expect = 6e-23
Identities = 56/178 (31%), Positives = 93/178 (51%), Gaps = 14/178 (7%)
Query: 19 MTGKTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGHCSTINKYLSGILLVICGFNCAFTS 78
+T SG L LLPT T+ F +P++T++G C+ +N+ L+G L+++C +C F +
Sbjct: 406 LTNLILSGTARLNVLLPTATILAFAIFAPLLTDDGKCTRLNRALTGALMLLCAASCVFFT 465
Query: 79 FTDSYTGSDGQRHYGIVTMNGLWP-----------SPGSDSVDLSAYKLRFGDFVHAFLS 127
TDS+ G+ YGI T +G+ + + Y+LR+ D H L+
Sbjct: 466 LTDSFRSPTGRLRYGIATTSGIRTFCVGGRRRRRGGGKAGPREPERYRLRWSDLFHTALA 525
Query: 128 VIVFAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGYP 185
++ F ++V C+YP + ++ +P VIG V +F++FPS R GIGYP
Sbjct: 526 LVAFVTFAASHHDIVLCYYP---GVPRKVVNTVPLVIGFVVSLLFVLFPSKRRGIGYP 580
>UniRef100_Q93VW2 P0560B06.26 protein [Oryza sativa]
Length = 223
Score = 107 bits (266), Expect = 2e-22
Identities = 64/174 (36%), Positives = 94/174 (53%), Gaps = 11/174 (6%)
Query: 22 KTFSGVGNLVKLLPTGTVFLFQYLSPVVTNNGH-CSTINKYLSGILLVICGFNCAFTSFT 80
KT +++KLLPTGTV F L+P TN+G C ++Y + L+ C +C SFT
Sbjct: 17 KTMCAACDILKLLPTGTVLAFHELAPSFTNHGGACGAASRYTTAALIAACTASCVLLSFT 76
Query: 81 DSYTGS-DGQR-HYGIVTMNGLWPSPGSDSV--------DLSAYKLRFGDFVHAFLSVIV 130
DS DG+R +YG+ T+ G P + DL K+R DFVHA +S +V
Sbjct: 77 DSLVSHVDGRRLYYGVATLRGFRPFNFEGTREEMEERFGDLPGMKVRALDFVHAHVSAVV 136
Query: 131 FAVLGLLDTNVVHCFYPKFESSEKILMQVLPPVIGVVSGAVFMIFPSYRHGIGY 184
F V+ L + +V C +P + + + LP +G+++ VFMIFP+ R IGY
Sbjct: 137 FVVVALGNADVQGCLFPDAGTGFTEMFRNLPMGLGLLASMVFMIFPTTRKSIGY 190
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.136 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 340,315,271
Number of Sequences: 2790947
Number of extensions: 14338195
Number of successful extensions: 29402
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 37
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 29329
Number of HSP's gapped (non-prelim): 47
length of query: 198
length of database: 848,049,833
effective HSP length: 121
effective length of query: 77
effective length of database: 510,345,246
effective search space: 39296583942
effective search space used: 39296583942
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 71 (32.0 bits)
Medicago: description of AC146553.13


