

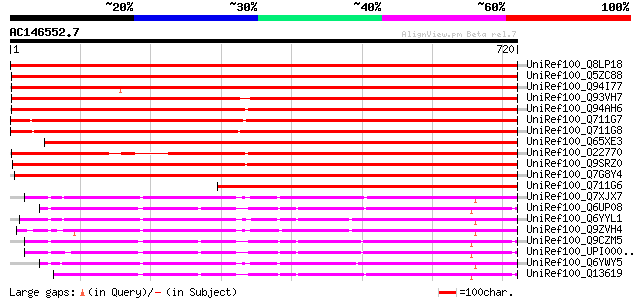
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146552.7 + phase: 0 /pseudo
(720 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LP18 Cullin-like protein1 [Pisum sativum] 1338 0.0
UniRef100_Q5ZC88 CUL1 [Oryza sativa] 1236 0.0
UniRef100_Q94I77 Putative cullin [Oryza sativa] 1229 0.0
UniRef100_Q93VH7 Cullin-like protein [Oryza sativa] 1211 0.0
UniRef100_Q94AH6 Cullin 1 [Arabidopsis thaliana] 1203 0.0
UniRef100_Q711G7 Cullin 1B [Nicotiana tabacum] 1176 0.0
UniRef100_Q711G8 Cullin 1A [Nicotiana tabacum] 1150 0.0
UniRef100_Q65XE3 Putative cullin 1 [Oryza sativa] 1108 0.0
UniRef100_O22770 Putative cullin-like 1 protein [Arabidopsis tha... 1074 0.0
UniRef100_Q9SRZ0 F22D16.2 protein [Arabidopsis thaliana] 976 0.0
UniRef100_Q7G8Y4 Putative cullin [Oryza sativa] 972 0.0
UniRef100_Q711G6 Cullin 1C [Nicotiana tabacum] 785 0.0
UniRef100_Q7XJX7 OSJNBa0063C18.16 protein [Oryza sativa] 403 e-110
UniRef100_Q6UP08 Cullin 4A [Homo sapiens] 392 e-107
UniRef100_Q6YYL1 Putative cullin 3B [Oryza sativa] 390 e-106
UniRef100_Q9ZVH4 T2P11.2 protein [Arabidopsis thaliana] 387 e-106
UniRef100_Q9CZM5 Mus musculus 11 days embryo whole body cDNA, RI... 386 e-105
UniRef100_UPI00001D1662 UPI00001D1662 UniRef100 entry 385 e-105
UniRef100_Q6YWY5 Putative cullin 3 [Oryza sativa] 385 e-105
UniRef100_Q13619 Cullin homolog 4A [Homo sapiens] 385 e-105
>UniRef100_Q8LP18 Cullin-like protein1 [Pisum sativum]
Length = 742
Score = 1338 bits (3463), Expect = 0.0
Identities = 661/720 (91%), Positives = 695/720 (95%)
Query: 1 MSERKTIDLEQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYS 60
MSERKTIDL+QGWDFM +GI KLKNILEGLPEPQF+ +DYMMLYTTIYNMCTQKPP+DYS
Sbjct: 1 MSERKTIDLDQGWDFMLKGIKKLKNILEGLPEPQFTSDDYMMLYTTIYNMCTQKPPHDYS 60
Query: 61 QPLYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYF 120
QPLYDKYKE+FEEYI+STVLPSLREKHDEFMLRELV+RWANHKIMVRWLSRFFHYLDRYF
Sbjct: 61 QPLYDKYKESFEEYIISTVLPSLREKHDEFMLRELVRRWANHKIMVRWLSRFFHYLDRYF 120
Query: 121 IARRSLPPLNEVGLACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVE 180
IARRSLPPLNEVGL CFRDLVYKE++GK+RDA+ISLIDQEREGEQIDRALLKNVLDIFVE
Sbjct: 121 IARRSLPPLNEVGLTCFRDLVYKEINGKVRDAVISLIDQEREGEQIDRALLKNVLDIFVE 180
Query: 181 IGMGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAHYL 240
IGMG+MD Y+NDFEA MLKDTSAYYSRKASNWILEDSCPDYMLKAEECL+REKDRVAHYL
Sbjct: 181 IGMGQMDQYDNDFEAAMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLKREKDRVAHYL 240
Query: 241 HSSSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLDP 300
HSSSE KLLEKVQ+ELLSVYA+QLLEKEHSGCH+LL DDK EDLSRMFRLFSKIPRGL+P
Sbjct: 241 HSSSESKLLEKVQHELLSVYANQLLEKEHSGCHSLLTDDKVEDLSRMFRLFSKIPRGLEP 300
Query: 301 VSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVNSC 360
VS IFKQHVT EG ALVK AEDAASN+KAEKRDIVG QEQ+FVRKVIELHDKYLAYV+ C
Sbjct: 301 VSCIFKQHVTAEGTALVKLAEDAASNRKAEKRDIVGLQEQIFVRKVIELHDKYLAYVSDC 360
Query: 361 FQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLEKV 420
FQNHTLFHKALKEAFE+FCNKGVAG+SSAELLATFCDNILKKGGSEKLSDEAIE+T EKV
Sbjct: 361 FQNHTLFHKALKEAFEIFCNKGVAGSSSAELLATFCDNILKKGGSEKLSDEAIEDTFEKV 420
Query: 421 VKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVT 480
VKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVT
Sbjct: 421 VKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVT 480
Query: 481 DLTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFK 540
DLTLAKENQTSFEEYLSN PN DPGIDLTVTVLTTGF PSYKSFDLNLPAEMV+CVEVFK
Sbjct: 481 DLTLAKENQTSFEEYLSNNPNIDPGIDLTVTVLTTGFGPSYKSFDLNLPAEMVRCVEVFK 540
Query: 541 EFYSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYSEIMT 600
EFY TKTKHRKLTWIYSLGTCN+SGKF+PKT+ELVVTTYQASALLLFNSSDRLSYSEIMT
Sbjct: 541 EFYQTKTKHRKLTWIYSLGTCNVSGKFEPKTMELVVTTYQASALLLFNSSDRLSYSEIMT 600
Query: 601 QLNLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVD 660
QLNL D+DV+RLLHSLSCAKYKIL KEP+TKTI PTD+FEFN+KFTDKMRRIKIPLPPVD
Sbjct: 601 QLNLTDDDVVRLLHSLSCAKYKILTKEPSTKTISPTDHFEFNSKFTDKMRRIKIPLPPVD 660
Query: 661 EKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI*KRIE 720
EKKKVIEDVDKDRRYAIDASIVRIMKSRKVL YQQLVMECVEQLGRMFKPDVKAI KRIE
Sbjct: 661 EKKKVIEDVDKDRRYAIDASIVRIMKSRKVLSYQQLVMECVEQLGRMFKPDVKAIKKRIE 720
>UniRef100_Q5ZC88 CUL1 [Oryza sativa]
Length = 744
Score = 1236 bits (3197), Expect = 0.0
Identities = 603/718 (83%), Positives = 664/718 (91%)
Query: 3 ERKTIDLEQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYSQP 62
ERKTIDLEQGW+FM +GI KLKNILEG PEPQFS EDYMMLYTTIYNMCTQKPP+DYSQ
Sbjct: 5 ERKTIDLEQGWEFMQKGITKLKNILEGKPEPQFSSEDYMMLYTTIYNMCTQKPPHDYSQQ 64
Query: 63 LYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYFIA 122
LY+KY+E+FEEYI S VLPSLREKHDEFMLRELVKRW+NHK+MVRWLSRFFHYLDRYFI+
Sbjct: 65 LYEKYRESFEEYITSMVLPSLREKHDEFMLRELVKRWSNHKVMVRWLSRFFHYLDRYFIS 124
Query: 123 RRSLPPLNEVGLACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVEIG 182
RRSLP L+EVGL+CFRDLVY+E+ GK++ A+ISLIDQEREGEQIDRALLKNVLDIFVEIG
Sbjct: 125 RRSLPQLSEVGLSCFRDLVYQEIKGKVKSAVISLIDQEREGEQIDRALLKNVLDIFVEIG 184
Query: 183 MGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAHYLHS 242
+ MD+YENDFE +LKDT+ YYS KA WILEDSCPDYMLKAEECL+REK+RVAHYLHS
Sbjct: 185 LTSMDYYENDFEDFLLKDTADYYSIKAQTWILEDSCPDYMLKAEECLKREKERVAHYLHS 244
Query: 243 SSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLDPVS 302
SSE KLLEKVQ+ELL+ YASQLLEKEHSGCHALLRDDK +DLSRM+RLFS+I RGL+PVS
Sbjct: 245 SSEQKLLEKVQHELLTQYASQLLEKEHSGCHALLRDDKVDDLSRMYRLFSRITRGLEPVS 304
Query: 303 SIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVNSCFQ 362
IFKQHVT EG ALVK AEDAASNKK EK++IVG QEQVFVRK+IELHDKY+AYV CFQ
Sbjct: 305 QIFKQHVTNEGTALVKQAEDAASNKKPEKKEIVGLQEQVFVRKIIELHDKYVAYVTDCFQ 364
Query: 363 NHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLEKVVK 422
HTLFHKALKEAFEVFCNKGV+G+SSAELLATFCDNILKKGGSEKLSDEAIE+TLEKVV+
Sbjct: 365 GHTLFHKALKEAFEVFCNKGVSGSSSAELLATFCDNILKKGGSEKLSDEAIEDTLEKVVR 424
Query: 423 LLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVTDL 482
LLAYISDKDLFAEFYRKKLARRLLFDKSAND+HERSILTKLKQQCGGQFTSKMEGMVTDL
Sbjct: 425 LLAYISDKDLFAEFYRKKLARRLLFDKSANDEHERSILTKLKQQCGGQFTSKMEGMVTDL 484
Query: 483 TLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEF 542
T+A+++Q FEE++S +PGI L VTVLTTGFWPSYKSFD+NLPAEMVKCVEVFKEF
Sbjct: 485 TVARDHQAKFEEFISTHSELNPGIALAVTVLTTGFWPSYKSFDINLPAEMVKCVEVFKEF 544
Query: 543 YSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYSEIMTQL 602
Y T+TKHRKLTWIYSLGTCNI+ KF+ KT+EL+VTTYQA+ LLLFN DRLSYSEI+TQL
Sbjct: 545 YQTRTKHRKLTWIYSLGTCNINAKFEAKTIELIVTTYQAALLLLFNGVDRLSYSEIVTQL 604
Query: 603 NLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVDEK 662
NL D+DV+RLLHSLSCAKYKIL KEPN ++I P D FEFN+KFTDK+RR+KIPLPPVDEK
Sbjct: 605 NLSDDDVVRLLHSLSCAKYKILSKEPNNRSISPNDVFEFNSKFTDKLRRLKIPLPPVDEK 664
Query: 663 KKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI*KRIE 720
KKV+EDVDKDRRYAIDASIVRIMKSRKVLG+QQLVMECVEQLGRMFKPD KAI KRIE
Sbjct: 665 KKVVEDVDKDRRYAIDASIVRIMKSRKVLGHQQLVMECVEQLGRMFKPDFKAIKKRIE 722
>UniRef100_Q94I77 Putative cullin [Oryza sativa]
Length = 750
Score = 1229 bits (3180), Expect = 0.0
Identities = 603/724 (83%), Positives = 664/724 (91%), Gaps = 6/724 (0%)
Query: 3 ERKTIDLEQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYSQP 62
ERKTIDLEQGW+FM +GI KLKNILEG PEPQFS EDYMMLYTTIYNMCTQKPP+DYSQ
Sbjct: 5 ERKTIDLEQGWEFMQKGITKLKNILEGKPEPQFSSEDYMMLYTTIYNMCTQKPPHDYSQQ 64
Query: 63 LYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYFIA 122
LY+KY+E+FEEYI S VLPSLREKHDEFMLRELVKRW+NHK+MVRWLSRFFHYLDRYFI+
Sbjct: 65 LYEKYRESFEEYITSMVLPSLREKHDEFMLRELVKRWSNHKVMVRWLSRFFHYLDRYFIS 124
Query: 123 RRSLPPLNEVGLACFRDLVYKELHGKMRDAIISL------IDQEREGEQIDRALLKNVLD 176
RRSLP L+EVGL+CFRDLVY+E+ GK++ A+ISL IDQEREGEQIDRALLKNVLD
Sbjct: 125 RRSLPQLSEVGLSCFRDLVYQEIKGKVKSAVISLTYFLEQIDQEREGEQIDRALLKNVLD 184
Query: 177 IFVEIGMGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRV 236
IFVEIG+ MD+YENDFE +LKDT+ YYS KA WILEDSCPDYMLKAEECL+REK+RV
Sbjct: 185 IFVEIGLTSMDYYENDFEDFLLKDTADYYSIKAQTWILEDSCPDYMLKAEECLKREKERV 244
Query: 237 AHYLHSSSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPR 296
AHYLHSSSE KLLEKVQ+ELL+ YASQLLEKEHSGCHALLRDDK +DLSRM+RLFS+I R
Sbjct: 245 AHYLHSSSEQKLLEKVQHELLTQYASQLLEKEHSGCHALLRDDKVDDLSRMYRLFSRITR 304
Query: 297 GLDPVSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAY 356
GL+PVS IFKQHVT EG ALVK AEDAASNKK EK++IVG QEQVFVRK+IELHDKY+AY
Sbjct: 305 GLEPVSQIFKQHVTNEGTALVKQAEDAASNKKPEKKEIVGLQEQVFVRKIIELHDKYVAY 364
Query: 357 VNSCFQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEET 416
V CFQ HTLFHKALKEAFEVFCNKGV+G+SSAELLATFCDNILKKGGSEKLSDEAIE+T
Sbjct: 365 VTDCFQGHTLFHKALKEAFEVFCNKGVSGSSSAELLATFCDNILKKGGSEKLSDEAIEDT 424
Query: 417 LEKVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKME 476
LEKVV+LLAYISDKDLFAEFYRKKLARRLLFDKSAND+HERSILTKLKQQCGGQFTSKME
Sbjct: 425 LEKVVRLLAYISDKDLFAEFYRKKLARRLLFDKSANDEHERSILTKLKQQCGGQFTSKME 484
Query: 477 GMVTDLTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCV 536
GMVTDLT+A+++Q FEE++S +PGI L VTVLTTGFWPSYKSFD+NLPAEMVKCV
Sbjct: 485 GMVTDLTVARDHQAKFEEFISTHSELNPGIALAVTVLTTGFWPSYKSFDINLPAEMVKCV 544
Query: 537 EVFKEFYSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYS 596
EVFKEFY T+TKHRKLTWIYSLGTCNI+ KF+ KT+EL+VTTYQA+ LLLFN DRLSYS
Sbjct: 545 EVFKEFYQTRTKHRKLTWIYSLGTCNINAKFEAKTIELIVTTYQAALLLLFNGVDRLSYS 604
Query: 597 EIMTQLNLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPL 656
EI+TQLNL D+DV+RLLHSLSCAKYKIL KEPN ++I P D FEFN+KFTDK+RR+KIPL
Sbjct: 605 EIVTQLNLSDDDVVRLLHSLSCAKYKILSKEPNNRSISPNDVFEFNSKFTDKLRRLKIPL 664
Query: 657 PPVDEKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI* 716
PPVDEKKKV+EDVDKDRRYAIDASIVRIMKSRKVLG+QQLVMECVEQLGRMFKPD KAI
Sbjct: 665 PPVDEKKKVVEDVDKDRRYAIDASIVRIMKSRKVLGHQQLVMECVEQLGRMFKPDFKAIK 724
Query: 717 KRIE 720
KRIE
Sbjct: 725 KRIE 728
>UniRef100_Q93VH7 Cullin-like protein [Oryza sativa]
Length = 732
Score = 1211 bits (3133), Expect = 0.0
Identities = 595/718 (82%), Positives = 654/718 (90%), Gaps = 12/718 (1%)
Query: 3 ERKTIDLEQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYSQP 62
ERKTIDLEQGW+FM +GI KLKNILEG PEPQFS EDYMMLYTTIYNMCTQKPP+DYSQ
Sbjct: 5 ERKTIDLEQGWEFMQKGITKLKNILEGKPEPQFSSEDYMMLYTTIYNMCTQKPPHDYSQQ 64
Query: 63 LYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYFIA 122
LY+KY+E+FEEYI S VLPSLREKHDEFMLRELVKRW+NHK+MVRWLSRFFHYLDRYFI+
Sbjct: 65 LYEKYRESFEEYITSMVLPSLREKHDEFMLRELVKRWSNHKVMVRWLSRFFHYLDRYFIS 124
Query: 123 RRSLPPLNEVGLACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVEIG 182
RRSLP L+EVGL+CFRDLVY+E+ GK++ A+ISLIDQEREGEQIDRALLKNVLDIFVEIG
Sbjct: 125 RRSLPQLSEVGLSCFRDLVYQEIKGKVKSAVISLIDQEREGEQIDRALLKNVLDIFVEIG 184
Query: 183 MGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAHYLHS 242
+ MD+YENDFE +LKDT+ YYS KA WILEDSCPDYMLKAEECL+REK+RVAHYLHS
Sbjct: 185 LTSMDYYENDFEDFLLKDTADYYSIKAQTWILEDSCPDYMLKAEECLKREKERVAHYLHS 244
Query: 243 SSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLDPVS 302
SSE KLLEKVQ+ELL+ YASQLLEKEHSGCHALLRDDK +DLSRM+RLFS+I RGL+PVS
Sbjct: 245 SSEQKLLEKVQHELLTQYASQLLEKEHSGCHALLRDDKVDDLSRMYRLFSRITRGLEPVS 304
Query: 303 SIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVNSCFQ 362
IFKQHVT EG ALVK AEDAASNKK VFVRK+IELHDKY+AYV CFQ
Sbjct: 305 QIFKQHVTNEGTALVKQAEDAASNKK------------VFVRKIIELHDKYVAYVTDCFQ 352
Query: 363 NHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLEKVVK 422
HTLFHKALKEAFEVFCNKGV+G+SSAELLATFCDNILKKGGSEKLSDEAIE+TLEKVV+
Sbjct: 353 GHTLFHKALKEAFEVFCNKGVSGSSSAELLATFCDNILKKGGSEKLSDEAIEDTLEKVVR 412
Query: 423 LLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVTDL 482
LLAYISDKDLFAEFYRKKLARRLLFDKSAND+HERSILTKLKQQCGGQFTSKMEGMVTDL
Sbjct: 413 LLAYISDKDLFAEFYRKKLARRLLFDKSANDEHERSILTKLKQQCGGQFTSKMEGMVTDL 472
Query: 483 TLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEF 542
T+A+++Q FEE++S +PGI L VTVLTTGFWPSYKSFD+NLPAEMVKCVEVFKEF
Sbjct: 473 TVARDHQAKFEEFISTHSELNPGIALAVTVLTTGFWPSYKSFDINLPAEMVKCVEVFKEF 532
Query: 543 YSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYSEIMTQL 602
Y T+TKHRKLTWIYSLGTCNI+ KF+ KT+EL+VTTYQA+ LLLFN DRLSYSEI+TQL
Sbjct: 533 YQTRTKHRKLTWIYSLGTCNINAKFEAKTIELIVTTYQAALLLLFNGVDRLSYSEIVTQL 592
Query: 603 NLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVDEK 662
NL D+DV+RLLHSLSCAKYKIL KEPN ++I P D FEFN+KFTDK+RR+KIPLPPVDEK
Sbjct: 593 NLSDDDVVRLLHSLSCAKYKILSKEPNNRSISPNDVFEFNSKFTDKLRRLKIPLPPVDEK 652
Query: 663 KKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI*KRIE 720
KKV+EDVDKDRRYAIDASIVRIMKSRKVLG+QQLVMECVEQLGRMFKPD KAI KRIE
Sbjct: 653 KKVVEDVDKDRRYAIDASIVRIMKSRKVLGHQQLVMECVEQLGRMFKPDFKAIKKRIE 710
>UniRef100_Q94AH6 Cullin 1 [Arabidopsis thaliana]
Length = 738
Score = 1203 bits (3112), Expect = 0.0
Identities = 583/718 (81%), Positives = 657/718 (91%), Gaps = 3/718 (0%)
Query: 3 ERKTIDLEQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYSQP 62
ERKTIDLEQGWD+M GI KLK ILEGL EP F E YMMLYTTIYNMCTQKPP+DYSQ
Sbjct: 2 ERKTIDLEQGWDYMQTGITKLKRILEGLNEPAFDSEQYMMLYTTIYNMCTQKPPHDYSQQ 61
Query: 63 LYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYFIA 122
LYDKY+EAFEEYI STVLP+LREKHDEFMLREL KRW+NHK+MVRWLSRFF+YLDRYFIA
Sbjct: 62 LYDKYREAFEEYINSTVLPALREKHDEFMLRELFKRWSNHKVMVRWLSRFFYYLDRYFIA 121
Query: 123 RRSLPPLNEVGLACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVEIG 182
RRSLPPLNEVGL CFRDLVY ELH K++ A+I+L+D+EREGEQIDRALLKNVLDI+VEIG
Sbjct: 122 RRSLPPLNEVGLTCFRDLVYNELHSKVKQAVIALVDKEREGEQIDRALLKNVLDIYVEIG 181
Query: 183 MGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAHYLHS 242
MG+M+ YE DFE+ ML+DTS+YYSRKAS+WI EDSCPDYMLK+EECL++E++RVAHYLHS
Sbjct: 182 MGQMERYEEDFESFMLQDTSSYYSRKASSWIQEDSCPDYMLKSEECLKKERERVAHYLHS 241
Query: 243 SSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLDPVS 302
SSEPKL+EKVQ+ELL V+ASQLLEKEHSGC ALLRDDK +DLSRM+RL+ KI RGL+PV+
Sbjct: 242 SSEPKLVEKVQHELLVVFASQLLEKEHSGCRALLRDDKVDDLSRMYRLYHKILRGLEPVA 301
Query: 303 SIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVNSCFQ 362
+IFKQHVT EG ALV+ AED A+N+ A + QEQV +RKVIELHDKY+ YV CFQ
Sbjct: 302 NIFKQHVTAEGNALVQQAEDTATNQVANTASV---QEQVLIRKVIELHDKYMVYVTECFQ 358
Query: 363 NHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLEKVVK 422
NHTLFHKALKEAFE+FCNK VAG+SSAELLATFCDNILKKGGSEKLSDEAIE+TLEKVVK
Sbjct: 359 NHTLFHKALKEAFEIFCNKTVAGSSSAELLATFCDNILKKGGSEKLSDEAIEDTLEKVVK 418
Query: 423 LLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVTDL 482
LLAYISDKDLFAEFYRKKLARRLLFD+SANDDHERSILTKLKQQCGGQFTSKMEGMVTDL
Sbjct: 419 LLAYISDKDLFAEFYRKKLARRLLFDRSANDDHERSILTKLKQQCGGQFTSKMEGMVTDL 478
Query: 483 TLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEF 542
TLA+ENQ SFE+YL + P A+PGIDLTVTVLTTGFWPSYKSFD+NLP+EM+KCVEVFK F
Sbjct: 479 TLARENQNSFEDYLGSNPAANPGIDLTVTVLTTGFWPSYKSFDINLPSEMIKCVEVFKGF 538
Query: 543 YSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYSEIMTQL 602
Y TKTKHRKLTWIYSLGTC+I+GKFD K +EL+V+TYQA+ LLLFN++D+LSY+EI+ QL
Sbjct: 539 YETKTKHRKLTWIYSLGTCHINGKFDQKAIELIVSTYQAAVLLLFNTTDKLSYTEILAQL 598
Query: 603 NLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVDEK 662
NL ED++RLLHSLSCAKYKIL+KEPNTKT+ D FEFN+KFTD+MRRIKIPLPPVDE+
Sbjct: 599 NLSHEDLVRLLHSLSCAKYKILLKEPNTKTVSQNDAFEFNSKFTDRMRRIKIPLPPVDER 658
Query: 663 KKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI*KRIE 720
KKV+EDVDKDRRYAIDA+IVRIMKSRKVLG+QQLV ECVEQL RMFKPD+KAI KR+E
Sbjct: 659 KKVVEDVDKDRRYAIDAAIVRIMKSRKVLGHQQLVSECVEQLSRMFKPDIKAIKKRME 716
>UniRef100_Q711G7 Cullin 1B [Nicotiana tabacum]
Length = 739
Score = 1176 bits (3042), Expect = 0.0
Identities = 577/720 (80%), Positives = 651/720 (90%), Gaps = 5/720 (0%)
Query: 1 MSERKTIDLEQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYS 60
MS+ K I+LE+GW+FM +G+ KLK ILEG + F+ E+YMMLYTTIYNMCTQKPP+DYS
Sbjct: 3 MSQMKIIELEEGWEFMQKGVTKLKKILEG-QQDSFNSEEYMMLYTTIYNMCTQKPPHDYS 61
Query: 61 QPLYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYF 120
Q LY+KYKEAFEEYI STVLP+LRE+HDEFMLRE VKRWANHK+MVRWLSRFF+YLDRYF
Sbjct: 62 QQLYEKYKEAFEEYINSTVLPALRERHDEFMLREFVKRWANHKLMVRWLSRFFYYLDRYF 121
Query: 121 IARRSLPPLNEVGLACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVE 180
IARRSLP LNEVGL CFRDLVY+EL+ K RDA+I LIDQEREGEQIDRALLKNVLDIFV
Sbjct: 122 IARRSLPALNEVGLTCFRDLVYQELNSKARDAVIVLIDQEREGEQIDRALLKNVLDIFVG 181
Query: 181 IGMGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAHYL 240
IGMG+M++YENDFE MLKDT+AYYSRKAS+WI+EDSCPDYMLKAEECL++EKDRV+HYL
Sbjct: 182 IGMGQMEYYENDFEDAMLKDTAAYYSRKASSWIVEDSCPDYMLKAEECLKKEKDRVSHYL 241
Query: 241 HSSSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLDP 300
H SSE KLLEKVQNELL VY +QLLEKEHSGC ALLRDDK EDLSRM+RLF +IP+GL+P
Sbjct: 242 HVSSETKLLEKVQNELLVVYTNQLLEKEHSGCRALLRDDKVEDLSRMYRLFHRIPKGLEP 301
Query: 301 VSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVNSC 360
V+++FKQHVT+EGM LV+ AED ASNK G+ EQVFVRK+IELHDKY+AYV C
Sbjct: 302 VANMFKQHVTSEGMVLVQQAEDTASNKAESS----GSGEQVFVRKLIELHDKYMAYVTEC 357
Query: 361 FQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLEKV 420
F N++LFHKALKEAFEVFCNK V+G SSAELLA++CDNILKKGGSEKLSD+AIEETL+KV
Sbjct: 358 FTNNSLFHKALKEAFEVFCNKIVSGCSSAELLASYCDNILKKGGSEKLSDDAIEETLDKV 417
Query: 421 VKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVT 480
VKLLAYISDKDL+AEFYRKKL+RRLLFDKSANDDHER ILTKLKQQCGGQFTS MEGMVT
Sbjct: 418 VKLLAYISDKDLYAEFYRKKLSRRLLFDKSANDDHERLILTKLKQQCGGQFTSXMEGMVT 477
Query: 481 DLTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFK 540
DLTLA+ENQ F+EYLSN P A PGIDLTVTVLTTGFWPSYKS DL+LP EMVK VEVFK
Sbjct: 478 DLTLARENQNHFQEYLSNNPAASPGIDLTVTVLTTGFWPSYKSSDLSLPVEMVKSVEVFK 537
Query: 541 EFYSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYSEIMT 600
EFY TKTKHRKLTWIYSLGTCNI+GKF PKT+EL+V TYQA+ALLLFN+SDRLSYSEI +
Sbjct: 538 EFYQTKTKHRKLTWIYSLGTCNINGKFAPKTIELIVGTYQAAALLLFNASDRLSYSEIKS 597
Query: 601 QLNLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVD 660
QLNL D+D++RLLHSLSCAKYKIL KEP+ +T+ P+D+FEFN+KFTD+MRRI++PLPP D
Sbjct: 598 QLNLADDDLVRLLHSLSCAKYKILTKEPSNRTVSPSDHFEFNSKFTDRMRRIRVPLPPAD 657
Query: 661 EKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI*KRIE 720
E+KKV+EDVDKDRRYAIDA IVRIMKSRKVL +QQLV+ECVEQL RMFKPD KAI KRIE
Sbjct: 658 ERKKVVEDVDKDRRYAIDACIVRIMKSRKVLPHQQLVLECVEQLSRMFKPDFKAIKKRIE 717
>UniRef100_Q711G8 Cullin 1A [Nicotiana tabacum]
Length = 741
Score = 1150 bits (2976), Expect = 0.0
Identities = 571/720 (79%), Positives = 642/720 (88%), Gaps = 3/720 (0%)
Query: 1 MSERKTIDLEQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYS 60
M++ KTI+LE+GW+FM +GI KLK ILEG P+ FS E+YMMLYTTIYNMCTQKPP+DYS
Sbjct: 3 MNQMKTIELEEGWEFMQKGITKLKIILEGSPD-SFSSEEYMMLYTTIYNMCTQKPPHDYS 61
Query: 61 QPLYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYF 120
Q LY+KYKEAFEEYI STVL SLREKHDEFMLRELVKRWANHK+MVRWLSRFFHYLDRYF
Sbjct: 62 QQLYEKYKEAFEEYINSTVLSSLREKHDEFMLRELVKRWANHKLMVRWLSRFFHYLDRYF 121
Query: 121 IARRSLPPLNEVGLACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVE 180
IARRSLP LNEVGL CFRDLVY+EL K RDA+I+LIDQEREGEQIDRALLKNVL IFVE
Sbjct: 122 IARRSLPALNEVGLTCFRDLVYQELKSKARDAVIALIDQEREGEQIDRALLKNVLGIFVE 181
Query: 181 IGMGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAHYL 240
IGMG+M++YENDFE MLKDT+AYYSRKASNWI+EDSCPDYMLKAEECL++EKDRV+HYL
Sbjct: 182 IGMGEMEYYENDFEDAMLKDTAAYYSRKASNWIVEDSCPDYMLKAEECLKKEKDRVSHYL 241
Query: 241 HSSSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLDP 300
HSSSE KLLEKVQNELL VY +QLLEKEHSGC ALL DDK EDLSRM+RLF +IP+GL+P
Sbjct: 242 HSSSEAKLLEKVQNELLVVYTNQLLEKEHSGCRALLIDDKVEDLSRMYRLFHRIPKGLEP 301
Query: 301 VSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVNSC 360
V+++FKQHVT EGM LV+ A + K + V + + + +++ DKY+AYV +C
Sbjct: 302 VANMFKQHVTAEGMVLVQQARRLS--KLTRLKVPVVHRSRYLLGRLLSCLDKYMAYVTNC 359
Query: 361 FQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLEKV 420
F N++LFHKALKEAFEVFCNK VAG SSAELLA++CDNILKKGGSEKLSD+AIEETL+KV
Sbjct: 360 FANNSLFHKALKEAFEVFCNKVVAGCSSAELLASYCDNILKKGGSEKLSDDAIEETLDKV 419
Query: 421 VKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVT 480
VKLLAYISDKDLFAEFYRKKL+RRLLFDKSANDDHER ILTKLKQQCGGQFTSKMEGMVT
Sbjct: 420 VKLLAYISDKDLFAEFYRKKLSRRLLFDKSANDDHERLILTKLKQQCGGQFTSKMEGMVT 479
Query: 481 DLTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFK 540
DLTLAKENQ F+EYLSN A+PGIDLTVTVLTTGFWPSYKS DL+LP EMVKCVEVFK
Sbjct: 480 DLTLAKENQNHFQEYLSNNSAANPGIDLTVTVLTTGFWPSYKSSDLSLPVEMVKCVEVFK 539
Query: 541 EFYSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYSEIMT 600
EFY TKTKHRKLTWIYSLGTCNI+GKF+PKT+EL+V TYQA+ALLLFN+SDRLSYS I +
Sbjct: 540 EFYQTKTKHRKLTWIYSLGTCNINGKFEPKTIELIVGTYQAAALLLFNASDRLSYSHIKS 599
Query: 601 QLNLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVD 660
QLNL D+D++RLL SLSCAKYKIL KEP ++T+ TD+FEFN+KFTD+MRRI+IPLPPVD
Sbjct: 600 QLNLADDDLVRLLQSLSCAKYKILTKEPTSRTVSSTDHFEFNSKFTDRMRRIRIPLPPVD 659
Query: 661 EKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI*KRIE 720
E+KKV+EDVDKDRRYAIDA IVRIMKSRKVL + QLV ECVEQL RMFKPD KAI KRIE
Sbjct: 660 ERKKVVEDVDKDRRYAIDACIVRIMKSRKVLPHSQLVSECVEQLSRMFKPDFKAIKKRIE 719
>UniRef100_Q65XE3 Putative cullin 1 [Oryza sativa]
Length = 693
Score = 1108 bits (2867), Expect = 0.0
Identities = 541/671 (80%), Positives = 611/671 (90%)
Query: 50 MCTQKPPNDYSQPLYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWL 109
MCTQKPP+DYSQ LYDKY+E+FEEYI S VLPSLR+KHDEFMLRELVKRW+NHKIMVRWL
Sbjct: 1 MCTQKPPHDYSQQLYDKYRESFEEYITSMVLPSLRDKHDEFMLRELVKRWSNHKIMVRWL 60
Query: 110 SRFFHYLDRYFIARRSLPPLNEVGLACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRA 169
SRFF YLDRYFI+RRSL PL +VGL CFRDL+Y+E+ G+++ A+I+LID+EREGEQIDRA
Sbjct: 61 SRFFFYLDRYFISRRSLIPLEQVGLTCFRDLIYQEIKGQVKGAVIALIDKEREGEQIDRA 120
Query: 170 LLKNVLDIFVEIGMGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECL 229
LLKNVL IFVEIG+G M+ YENDFE +LKDT+ YYS KA +WILEDSCPDYM+KAEECL
Sbjct: 121 LLKNVLGIFVEIGLGSMECYENDFEDFLLKDTTDYYSLKAQSWILEDSCPDYMIKAEECL 180
Query: 230 RREKDRVAHYLHSSSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFR 289
++EK+RV HYLH SSE KLLEKVQNELL+ YA+ LLEKEHSGC ALLRDDK EDLSRM+R
Sbjct: 181 KKEKERVGHYLHISSEQKLLEKVQNELLAQYATPLLEKEHSGCFALLRDDKEEDLSRMYR 240
Query: 290 LFSKIPRGLDPVSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIEL 349
LFSKI RGL+P++++FK HVT EG ALVK AED+ASNKK EK+D+VG QEQVFV K+IEL
Sbjct: 241 LFSKINRGLEPIANMFKTHVTNEGTALVKQAEDSASNKKPEKKDMVGMQEQVFVWKIIEL 300
Query: 350 HDKYLAYVNSCFQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLS 409
HDKY+AYV CFQ HTLFHKALKEAFEVFCNKGV+G+SSAELLATFCDNILKKG SEKLS
Sbjct: 301 HDKYVAYVTECFQGHTLFHKALKEAFEVFCNKGVSGSSSAELLATFCDNILKKGCSEKLS 360
Query: 410 DEAIEETLEKVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGG 469
DEAIE+ LEKVV+LLAYISDKDLFAEFYRKKLARRLLFDKSAND+HERSILTKLKQQCGG
Sbjct: 361 DEAIEDALEKVVRLLAYISDKDLFAEFYRKKLARRLLFDKSANDEHERSILTKLKQQCGG 420
Query: 470 QFTSKMEGMVTDLTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLP 529
QFTSKMEGMVTDLT+A+++QT FEE+++ +PGIDL VTVLTTGFWPSYK+FD+NLP
Sbjct: 421 QFTSKMEGMVTDLTVARDHQTKFEEFVAAHQELNPGIDLAVTVLTTGFWPSYKTFDINLP 480
Query: 530 AEMVKCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLFNS 589
AEMVKCVEVFKEFY T+TKHRKLTWIYSLGTCNI+ KF+ KT+EL+VTTYQA+ LLLFN
Sbjct: 481 AEMVKCVEVFKEFYQTRTKHRKLTWIYSLGTCNINAKFEAKTIELIVTTYQAALLLLFNG 540
Query: 590 SDRLSYSEIMTQLNLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTDKM 649
SDRL+YSEI+TQLNL D+DV+RLLHSLSCAKYKIL KEP ++I P D FEFN+KFTD+M
Sbjct: 541 SDRLTYSEIVTQLNLSDDDVVRLLHSLSCAKYKILNKEPANRSISPNDVFEFNSKFTDRM 600
Query: 650 RRIKIPLPPVDEKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFK 709
RRIKIPLPPVDEKKKV+EDVDKDRRYAIDASIVRIMKSRKV+G+QQLV ECVEQL RMFK
Sbjct: 601 RRIKIPLPPVDEKKKVVEDVDKDRRYAIDASIVRIMKSRKVMGHQQLVAECVEQLSRMFK 660
Query: 710 PDVKAI*KRIE 720
PD KAI KRIE
Sbjct: 661 PDFKAIKKRIE 671
>UniRef100_O22770 Putative cullin-like 1 protein [Arabidopsis thaliana]
Length = 676
Score = 1074 bits (2778), Expect = 0.0
Identities = 539/718 (75%), Positives = 602/718 (83%), Gaps = 65/718 (9%)
Query: 3 ERKTIDLEQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYSQP 62
ERKTIDLEQGWD+M GI KLK ILEGL EP F E YMMLYTTIYNMCTQKPP+DYSQ
Sbjct: 2 ERKTIDLEQGWDYMQTGITKLKRILEGLNEPAFDSEQYMMLYTTIYNMCTQKPPHDYSQQ 61
Query: 63 LYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYFIA 122
LYDKY+EAFEEYI STVLP+LREKHDEFMLREL KRW+NHK+MVRWLSRFF+YLDRYFIA
Sbjct: 62 LYDKYREAFEEYINSTVLPALREKHDEFMLRELFKRWSNHKVMVRWLSRFFYYLDRYFIA 121
Query: 123 RRSLPPLNEVGLACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVEIG 182
RRSLPPLNEVGL CFRDLV D+EREGEQIDRALLKNVLDI+
Sbjct: 122 RRSLPPLNEVGLTCFRDLV----------------DKEREGEQIDRALLKNVLDIY---- 161
Query: 183 MGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAHYLHS 242
+EECL++E++RVAHYLHS
Sbjct: 162 ------------------------------------------SEECLKKERERVAHYLHS 179
Query: 243 SSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLDPVS 302
SSEPKL+EKVQ+ELL V+ASQLLEKEHSGC ALLRDDK +DLSRM+RL+ KI RGL+PV+
Sbjct: 180 SSEPKLVEKVQHELLVVFASQLLEKEHSGCRALLRDDKVDDLSRMYRLYHKILRGLEPVA 239
Query: 303 SIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVNSCFQ 362
+IFKQHVT EG ALV+ AED A+N+ A + QEQV +RKVIELHDKY+ YV CFQ
Sbjct: 240 NIFKQHVTAEGNALVQQAEDTATNQVANTASV---QEQVLIRKVIELHDKYMVYVTECFQ 296
Query: 363 NHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLEKVVK 422
NHTLFHKALKEAFE+FCNK VAG+SSAELLATFCDNILKKGGSEKLSDEAIE+TLEKVVK
Sbjct: 297 NHTLFHKALKEAFEIFCNKTVAGSSSAELLATFCDNILKKGGSEKLSDEAIEDTLEKVVK 356
Query: 423 LLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVTDL 482
LLAYISDKDLFAEFYRKKLARRLLFD+SANDDHERSILTKLKQQCGGQFTSKMEGMVTDL
Sbjct: 357 LLAYISDKDLFAEFYRKKLARRLLFDRSANDDHERSILTKLKQQCGGQFTSKMEGMVTDL 416
Query: 483 TLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEF 542
TLA+ENQ SFE+YL + P A+PGIDLTVTVLTTGFWPSYKSFD+NLP+EM+KCVEVFK F
Sbjct: 417 TLARENQNSFEDYLGSNPAANPGIDLTVTVLTTGFWPSYKSFDINLPSEMIKCVEVFKGF 476
Query: 543 YSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYSEIMTQL 602
Y TKTKHRKLTWIYSLGTC+I+GKFD K +EL+V+TYQA+ LLLFN++D+LSY+EI+ QL
Sbjct: 477 YETKTKHRKLTWIYSLGTCHINGKFDQKAIELIVSTYQAAVLLLFNTTDKLSYTEILAQL 536
Query: 603 NLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVDEK 662
NL ED++RLLHSLSCAKYKIL+KEPNTKT+ D FEFN+KFTD+MRRIKIPLPPVDE+
Sbjct: 537 NLSHEDLVRLLHSLSCAKYKILLKEPNTKTVSQNDAFEFNSKFTDRMRRIKIPLPPVDER 596
Query: 663 KKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI*KRIE 720
KKV+EDVDKDRRYAIDA+IVRIMKSRKVLG+QQLV ECVEQL RMFKPD+KAI KR+E
Sbjct: 597 KKVVEDVDKDRRYAIDAAIVRIMKSRKVLGHQQLVSECVEQLSRMFKPDIKAIKKRME 654
>UniRef100_Q9SRZ0 F22D16.2 protein [Arabidopsis thaliana]
Length = 742
Score = 976 bits (2522), Expect = 0.0
Identities = 476/719 (66%), Positives = 585/719 (81%), Gaps = 3/719 (0%)
Query: 4 RKTIDLEQGWDFMHRGIMKLKNILEGLP-EPQFSPEDYMMLYTTIYNMCTQKPPNDYSQP 62
+K LE GW M G+ KL+ ILE +P EP F P M LYTT++N+CTQKPPNDYSQ
Sbjct: 3 KKDSVLEAGWSVMEAGVAKLQKILEEVPDEPPFDPVQRMQLYTTVHNLCTQKPPNDYSQQ 62
Query: 63 LYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYFIA 122
+YD+Y + +Y TVLP++REKH E+MLRELVKRWAN KI+VRWLS FF YLDR++
Sbjct: 63 IYDRYGGVYVDYNKQTVLPAIREKHGEYMLRELVKRWANQKILVRWLSHFFEYLDRFYTR 122
Query: 123 RRSLPPLNEVGLACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVEIG 182
R S P L+ VG FRDLVY+EL K +DA+++LI +EREGEQIDRALLKNV+D++ G
Sbjct: 123 RGSHPTLSAVGFISFRDLVYQELQSKAKDAVLALIHKEREGEQIDRALLKNVIDVYCGNG 182
Query: 183 MGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAHYLHS 242
MG++ YE DFE+ +L+D+++YYSR AS W E+SCPDYM+KAEE LR EK+RV +YLHS
Sbjct: 183 MGELVKYEEDFESFLLEDSASYYSRNASRWNQENSCPDYMIKAEESLRLEKERVTNYLHS 242
Query: 243 SSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLDPVS 302
++EPKL+ KVQNELL V A QL+E EHSGC ALLRDDK +DL+RM+RL+ IP+GLDPV+
Sbjct: 243 TTEPKLVAKVQNELLVVVAKQLIENEHSGCRALLRDDKMDDLARMYRLYHPIPQGLDPVA 302
Query: 303 SIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVNSCFQ 362
+FKQH+T EG AL+K A +AA++K A + Q+QV +R++I+LHDK++ YV+ CFQ
Sbjct: 303 DLFKQHITVEGSALIKQATEAATDKAASTSGLK-VQDQVLIRQLIDLHDKFMVYVDECFQ 361
Query: 363 NHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGS-EKLSDEAIEETLEKVV 421
H+LFHKALKEAFEVFCNK VAG SSAE+LAT+CDNILK GG EKL +E +E TLEKVV
Sbjct: 362 KHSLFHKALKEAFEVFCNKTVAGVSSAEILATYCDNILKTGGGIEKLENEDLELTLEKVV 421
Query: 422 KLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVTD 481
KLL YISDKDLFAEF+RKK ARRLLFD++ ND HERS+LTK K+ G QFTSKMEGM+TD
Sbjct: 422 KLLVYISDKDLFAEFFRKKQARRLLFDRNGNDYHERSLLTKFKELLGAQFTSKMEGMLTD 481
Query: 482 LTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKE 541
+TLAKE+QT+F E+LS G+D TVTVLTTGFWPSYK+ DLNLP EMV CVE FK
Sbjct: 482 MTLAKEHQTNFVEFLSVNKTKKLGMDFTVTVLTTGFWPSYKTTDLNLPIEMVNCVEAFKA 541
Query: 542 FYSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYSEIMTQ 601
+Y TKT R+L+WIYSLGTC ++GKFD KT+E+VVTTYQA+ LLLFN+++RLSY+EI+ Q
Sbjct: 542 YYGTKTNSRRLSWIYSLGTCQLAGKFDKKTIEIVVTTYQAAVLLLFNNTERLSYTEILEQ 601
Query: 602 LNLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVDE 661
LNL ED+ RLLHSLSC KYKILIKEP ++ I TD FEFN+KFTDKMRRI++PLPP+DE
Sbjct: 602 LNLGHEDLARLLHSLSCLKYKILIKEPMSRNISNTDTFEFNSKFTDKMRRIRVPLPPMDE 661
Query: 662 KKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI*KRIE 720
+KK++EDVDKDRRYAIDA++VRIMKSRKVLG+QQLV ECVE L +MFKPD+K I KRIE
Sbjct: 662 RKKIVEDVDKDRRYAIDAALVRIMKSRKVLGHQQLVSECVEHLSKMFKPDIKMIKKRIE 720
>UniRef100_Q7G8Y4 Putative cullin [Oryza sativa]
Length = 746
Score = 972 bits (2513), Expect = 0.0
Identities = 471/715 (65%), Positives = 583/715 (80%), Gaps = 1/715 (0%)
Query: 7 IDLEQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYSQPLYDK 66
+DLE GW + G+ KLK IL+G F P++YM LYTT+YNMCTQKPPNDYSQ LYD+
Sbjct: 10 VDLEDGWRDVLAGVAKLKCILDGSNVVHFVPDEYMHLYTTVYNMCTQKPPNDYSQVLYDR 69
Query: 67 YKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYFIARRSL 126
YK+A +++I S VLPSL EKH F+LRE+V+RW HK+MVRWL RFF YLDRY++ RRSL
Sbjct: 70 YKQALDDHIESVVLPSLNEKHGVFLLREIVQRWEKHKLMVRWLRRFFDYLDRYYVTRRSL 129
Query: 127 PPLNEVGLACFRDLVYKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVEIGMGKM 186
L ++G + FRDLV+ +L + +I +ID EREG IDR LLKN LDI+VEIG ++
Sbjct: 130 DSLKDLGWSSFRDLVFDKLKSTVATIMIGMIDDEREGNLIDRPLLKNALDIYVEIGDSQL 189
Query: 187 DHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAHYLHSSSEP 246
++Y +DFE L T+ YYS+KA WILE+SCP+YMLKAEECL++EKDRVA+YLHS++EP
Sbjct: 190 NYYSDDFEQSFLNGTTDYYSKKAQTWILENSCPEYMLKAEECLQKEKDRVANYLHSTTEP 249
Query: 247 KLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLDPVSSIFK 306
KL EL+ A ++L KE+SGC LL D+K EDL+RMFRLFS+I GL PVS IFK
Sbjct: 250 KLFAAALFELIDRRAEEILNKENSGCKVLLCDEKTEDLARMFRLFSRITDGLLPVSKIFK 309
Query: 307 QHVTTEGMALVKHAEDAASNKKAEKRDIV-GTQEQVFVRKVIELHDKYLAYVNSCFQNHT 365
+HV EGM+L+KHA DAA+++K EK+ +V G EQ FVR VIELHDKY+AYV +CFQ+++
Sbjct: 310 EHVIAEGMSLLKHATDAANSRKDEKKGVVVGLPEQDFVRSVIELHDKYMAYVTNCFQSNS 369
Query: 366 LFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLEKVVKLLA 425
+FHKALKEAFEV CNK V G SSAEL A +CD+ILK+GGSEKLSDEAI+E+LEKVVKLL
Sbjct: 370 VFHKALKEAFEVICNKDVVGCSSAELFAAYCDSILKRGGSEKLSDEAIDESLEKVVKLLT 429
Query: 426 YISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVTDLTLA 485
Y+SDKDLF EF+RKKL RRLLFDK+ ND+HER +L+KLKQ GGQFTSKMEGM+ D+TLA
Sbjct: 430 YLSDKDLFVEFHRKKLGRRLLFDKNTNDEHERILLSKLKQFFGGQFTSKMEGMLKDITLA 489
Query: 486 KENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEFYST 545
KE+Q+SFEEY+SN P ++P IDL VTVLTTG+WP+YK+ D+NLP EMVKCVEVFKE+Y +
Sbjct: 490 KEHQSSFEEYVSNNPESNPLIDLNVTVLTTGYWPTYKNSDINLPLEMVKCVEVFKEYYRS 549
Query: 546 KTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYSEIMTQLNLL 605
+HRKLTWI+SLG C + G FD K VE V+ TYQA+ LLLFN +D+LSYS+I++QL L
Sbjct: 550 DKQHRKLTWIFSLGNCVVIGNFDAKPVEFVLNTYQAALLLLFNEADKLSYSDIVSQLKLS 609
Query: 606 DEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVDEKKKV 665
D+D +RLLHSLSCAKYKIL KEP+ + I P D FEFN+KFTD+MRRIK+PLP +DEKKKV
Sbjct: 610 DDDAVRLLHSLSCAKYKILNKEPSNRVISPEDEFEFNSKFTDRMRRIKVPLPQIDEKKKV 669
Query: 666 IEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI*KRIE 720
++DV+KDRR+AIDAS+VRIMKSRKVLG+QQLV ECVEQL RMFKPD+K I +RIE
Sbjct: 670 VDDVNKDRRFAIDASLVRIMKSRKVLGHQQLVAECVEQLSRMFKPDIKIIKRRIE 724
>UniRef100_Q711G6 Cullin 1C [Nicotiana tabacum]
Length = 447
Score = 785 bits (2026), Expect = 0.0
Identities = 392/425 (92%), Positives = 409/425 (96%)
Query: 296 RGLDPVSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLA 355
RGLDPV+SIFKQHVT EG ALVK AEDAASNKKAEKRD+VG QEQVFVRKVIELHDKYLA
Sbjct: 1 RGLDPVASIFKQHVTAEGTALVKQAEDAASNKKAEKRDVVGLQEQVFVRKVIELHDKYLA 60
Query: 356 YVNSCFQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEE 415
YVN+CFQNHTLFHKALKEAFEVFCNKGVAG+SSAELLATFCDNILKKGGSEKLSDEAIEE
Sbjct: 61 YVNNCFQNHTLFHKALKEAFEVFCNKGVAGSSSAELLATFCDNILKKGGSEKLSDEAIEE 120
Query: 416 TLEKVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKM 475
TLEKVVKLLAYISDKDLFAEFYRKKLARRLLFDKSAND+HERSILTKLKQQCGGQFTSKM
Sbjct: 121 TLEKVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDEHERSILTKLKQQCGGQFTSKM 180
Query: 476 EGMVTDLTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKC 535
EGMVTDLTLA+ENQ SFEEYLSN P A+PGIDLTVTVLTTGFWPSYKSFDLNLPAEMV+C
Sbjct: 181 EGMVTDLTLARENQASFEEYLSNNPAANPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVRC 240
Query: 536 VEVFKEFYSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSY 595
VEVFKEFY TKTKHRKLTWIYSLGTCNI+GKF+PKT+ELVVTTYQASALLLFN+SDRLSY
Sbjct: 241 VEVFKEFYQTKTKHRKLTWIYSLGTCNINGKFEPKTIELVVTTYQASALLLFNASDRLSY 300
Query: 596 SEIMTQLNLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIP 655
EIMTQLNL D+DV+RLLHSLSCAKYKIL KEP+TKTI PTD FEFN++F DKMRRIKIP
Sbjct: 301 QEIMTQLNLSDDDVVRLLHSLSCAKYKILFKEPSTKTISPTDVFEFNSRFADKMRRIKIP 360
Query: 656 LPPVDEKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI 715
LPP DEKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI
Sbjct: 361 LPPEDEKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI 420
Query: 716 *KRIE 720
KRIE
Sbjct: 421 KKRIE 425
>UniRef100_Q7XJX7 OSJNBa0063C18.16 protein [Oryza sativa]
Length = 731
Score = 403 bits (1035), Expect = e-110
Identities = 247/707 (34%), Positives = 391/707 (54%), Gaps = 36/707 (5%)
Query: 22 KLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYSQPLYDKYKEAFEEYIVSTVLP 81
KL++ + + S + LY T YN+ K + LYDK E + ++ +
Sbjct: 31 KLEDAIREIYNHNASGLSFEELYRTAYNLVLHK----HGLKLYDKLTENLKGHL-KEMCR 85
Query: 82 SLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYFIARRSLPPLNEVGLACFRDLV 141
S+ + L EL +RWA+H ++ + Y+DR FIA P+ ++GL +RD+V
Sbjct: 86 SIEDAQGSLFLEELQRRWADHNKALQMIRDILMYMDRTFIATNKKTPVFDLGLELWRDIV 145
Query: 142 YK--ELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVEIGMGKMDHYENDFEADMLK 199
+ ++HG++ D ++ LI +ER GE I+R L+++ + +++G Y +DFE L+
Sbjct: 146 VRTPKIHGRLLDTLLELIHRERMGEMINRGLMRSTTKMLMDLGSSV---YHDDFEKPFLE 202
Query: 200 DTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAHYLHSSSEPKLLEKVQNELLSV 259
++++YS ++ +I C +Y+ KAE L E +RV+ Y+ + + K+ V E+L+
Sbjct: 203 VSASFYSGESQQFIECCDCGEYLKKAERRLAEELERVSQYMDAKTADKITSVVDTEMLAN 262
Query: 260 YASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLDPVSSIFKQHVTTEGMALVKH 319
+ +L+ E+SG +L DDK EDLSRM+ LF ++P G + S+ HV G ALV
Sbjct: 263 HMQRLILMENSGLVNMLVDDKHEDLSRMYNLFKRVPDGHSTIRSVMASHVKESGKALVSD 322
Query: 320 AEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVNSCFQNHTLFHKALKEAFEVFC 379
E + +D V FV++++ DKY ++ F N F AL +FE F
Sbjct: 323 PE--------KIKDPV-----EFVQRLLNEKDKYDEIISISFSNDKAFQNALNSSFENFI 369
Query: 380 NKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLEKVVKLLAYISDKDLFAEFYRK 439
N N S E ++ F D+ L+K G + ++E +E L+KV+ L Y+ +KD+F ++Y++
Sbjct: 370 N---LNNRSPEFISLFVDDKLRK-GVKGANEEDVETVLDKVMMLFRYLQEKDVFEKYYKQ 425
Query: 440 KLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVTDLTLAKENQTSFEEYLSNT 499
LA+RLL K+ +D+ ERS+L KLK +CG QFTSK+EGM DL + + SF LS
Sbjct: 426 HLAKRLLSGKTTSDEAERSMLVKLKTECGYQFTSKLEGMFNDLKTSHDTMQSFYANLSGD 485
Query: 500 PNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEFYSTKTKHRKLTWIYSLG 559
++ ++V +LTTG WP+ LP E+V E F+ FY R+LTW ++G
Sbjct: 486 TDSP---TISVQILTTGSWPTQPCTPCKLPPEIVDISEKFRAFYLGTHNGRRLTWQTNMG 542
Query: 560 TCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYSEIMTQLNLLDEDVIRLLHSLSCA 619
+I F + EL V+TYQ L+LFNS+D L+Y +I + D+ R L SL+C
Sbjct: 543 NADIKATFGGRRHELNVSTYQMCVLMLFNSADGLTYGDIEQATGIPHADLKRCLQSLACV 602
Query: 620 KYK-ILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVD-----EKKKVIEDVDKDR 673
K K +L KEP +K I D F +N KFT K+ ++KI EK + + V++DR
Sbjct: 603 KGKNVLRKEPMSKDISEDDTFYYNDKFTSKLVKVKIGTVVAQKETEPEKLETRQRVEEDR 662
Query: 674 RYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI*KRIE 720
+ I+A+IVRIMKSR+VL + ++ E +QL F P+ I KRIE
Sbjct: 663 KPQIEAAIVRIMKSRRVLDHNSIITEVTKQLQSRFLPNPVVIKKRIE 709
>UniRef100_Q6UP08 Cullin 4A [Homo sapiens]
Length = 759
Score = 392 bits (1007), Expect = e-107
Identities = 242/687 (35%), Positives = 386/687 (55%), Gaps = 44/687 (6%)
Query: 43 LYTTIYNMCTQKPPNDYSQPLYDKYKEAFEEYIVSTVLPSLREKHDEFM-LRELVKRWAN 101
LY + N+C+ K S LY + ++A E+++ + +LP + D + L+++ W +
Sbjct: 86 LYQAVENLCSHK----VSPMLYKQLRQACEDHVQAQILPFREDSLDSVLFLKKINTCWQD 141
Query: 102 HKIMVRWLSRFFHYLDRYFIARRS-LPPLNEVGLACFRDLVY--KELHGKMRDAIISLID 158
H + + F +LDR ++ + S LP + ++GL FR + K + K D I+ LI+
Sbjct: 142 HCRQMIMIRSIFLFLDRTYVLQNSTLPSIWDMGLELFRTHIISDKMVQSKTIDGILLLIE 201
Query: 159 QEREGEQIDRALLKNVLDIFVEIGMGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSC 218
+ER GE +DR+LL+++L + ++ + Y++ FE L++T+ Y+ + + E
Sbjct: 202 RERSGEAVDRSLLRSLLGMLSDLQV-----YKDSFELKFLEETNCLYAAEGQRLMQEREV 256
Query: 219 PDYMLKAEECLRREKDRVAHYLHSSSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRD 278
P+Y+ + L E DRV YL S++ L+ V+ +LL + + +L+K G LL +
Sbjct: 257 PEYLNHVSKRLEEEGDRVITYLDHSTQKPLIACVEKQLLGEHLTAILQK---GLDHLLDE 313
Query: 279 DKCEDLSRMFRLFSKIPRGLDPVSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQ 338
++ DL++M++LFS++ G + + +++ T G A+V + E +
Sbjct: 314 NRVPDLAQMYQLFSRVRGGQQALLQHWSEYIKTFGTAIVINPE----------------K 357
Query: 339 EQVFVRKVIELHDKYLAYVNSCFQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDN 398
++ V+ +++ DK + CFQ + F +KE+FE F NK N AEL+A D+
Sbjct: 358 DKDMVQDLLDFKDKVDHVIEVCFQKNERFVNLMKESFETFINK--RPNKPAELIAKHVDS 415
Query: 399 ILKKGGSEKLSDEAIEETLEKVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERS 458
L+ G E +DE +E TL+K++ L +I KD+F FY+K LA+RLL KSA+ D E+S
Sbjct: 416 KLRAGNKEA-TDEELERTLDKIMILFRFIHGKDVFEAFYKKDLAKRLLVGKSASVDAEKS 474
Query: 459 ILTKLKQQCGGQFTSKMEGMVTDLTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFW 518
+L+KLK +CG FTSK+EGM D+ L+K+ F++++ N ++ P IDLTV +LT G+W
Sbjct: 475 MLSKLKHECGAAFTSKLEGMFKDMELSKDIMVHFKQHMQNQSDSGP-IDLTVNILTMGYW 533
Query: 519 PSYKSFDLNLPAEMVKCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTT 578
P+Y +++L EM+K EVFK FY K RKL W +LG + +F E V+
Sbjct: 534 PTYTPMEVHLTPEMIKLQEVFKAFYLGKHSGRKLQWQTTLGHAVLKAEFKEGKKEFQVSL 593
Query: 579 YQASALLLFNSSDRLSYSEIMTQLNLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDY 638
+Q LL+FN D S+ EI + D ++ R L SL+C K ++LIK P K + D
Sbjct: 594 FQTLVLLMFNEGDGFSFEEIKMATGIEDSELRRTLQSLACGKARVLIKSPKGKEVEDGDK 653
Query: 639 FEFNAKFTDKMRRIKI----PLPPVDEKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQ 694
F FN +F K+ RIKI V+E+ E V +DR+Y IDA+IVRIMK RK LG+
Sbjct: 654 FIFNGEFKHKLFRIKINQIQMKETVEEQVSTTERVFQDRQYQIDAAIVRIMKMRKTLGHN 713
Query: 695 QLVMECVEQLGRMFKP-DVKAI*KRIE 720
LV E QL KP D+K KRIE
Sbjct: 714 LLVSELYNQLKFPVKPGDLK---KRIE 737
>UniRef100_Q6YYL1 Putative cullin 3B [Oryza sativa]
Length = 731
Score = 390 bits (1001), Expect = e-106
Identities = 242/715 (33%), Positives = 394/715 (54%), Gaps = 37/715 (5%)
Query: 15 FMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYSQPLYDKYKEAFEEY 74
F + KL + + + S + LY T YN+ K + LYDK E E++
Sbjct: 23 FFEKAWRKLDDAIREIYNHNASGLSFEELYRTAYNLVLHK----HGPKLYDKLTENMEDH 78
Query: 75 IVSTVLPSLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYFIARRSLPPLNEVGL 134
+ + S+ L EL ++W +H ++ + Y+DR FI P+ ++GL
Sbjct: 79 L-QEMRVSIEAAQGGLFLVELQRKWDDHNKALQMIRDILMYMDRVFIPTNKKTPVFDLGL 137
Query: 135 ACFRDLVYK--ELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVEIGMGKMDHYEND 192
+RD + + ++HG++ D ++ LI +ER GE I+R+L+++ + +++G Y++D
Sbjct: 138 DLWRDTIVRSPKIHGRLLDTLLDLIHRERTGEVINRSLMRSTTKMLMDLGSSV---YQDD 194
Query: 193 FEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAHYLHSSSEPKLLEKV 252
FE L+ ++++YS ++ +I SC +Y+ KA++ L E +RV+ Y+ + ++ K+ V
Sbjct: 195 FERPFLEVSASFYSGESQKFIECCSCGEYLKKAQQRLDEEAERVSQYMDAKTDEKITAVV 254
Query: 253 QNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLDPVSSIFKQHVTTE 312
E+L+ + +L+ E+SG +L +DK EDL+ M+ LF ++P G + S+ HV
Sbjct: 255 VKEMLANHMQRLILMENSGLVNMLVEDKYEDLTMMYSLFQRVPDGHSTIKSVMNSHVKET 314
Query: 313 GMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVNSCFQNHTLFHKALK 372
G +V E +D V FV++++ DKY + V + F N F AL
Sbjct: 315 GKDMVMDPE--------RLKDPVD-----FVQRLLNEKDKYDSIVTTSFSNDKSFQNALN 361
Query: 373 EAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLEKVVKLLAYISDKDL 432
+FE F N N E ++ + D+ L+KG E ++E +E L+KV+ L Y+ +KDL
Sbjct: 362 SSFEHFIN---LNNRCPEFISLYVDDKLRKGMKEA-NEEDVETVLDKVMMLFRYLQEKDL 417
Query: 433 FAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVTDLTLAKENQTSF 492
F ++Y++ LA+RLL K+A+DD ERS+L KLK +CG QFTSK+EGM DL K + +
Sbjct: 418 FEKYYKQHLAKRLLSGKAASDDSERSMLVKLKTECGYQFTSKLEGMFNDL---KTSHDTT 474
Query: 493 EEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEFYSTKTKHRKL 552
+ + + TP+ ++V +LTTG WP+ NLP E++ E+F+ FY R+L
Sbjct: 475 QRFYAGTPDLGDAPTISVQILTTGSWPTQPCNTCNLPPEILGVSEMFRGFYLGTHNGRRL 534
Query: 553 TWIYSLGTCNISGKF-DPKTVELVVTTYQASALLLFNSSDRLSYSEIMTQLNLLDEDVIR 611
TW ++GT +I F + EL V+TYQ L+LFNS+D LSY +I + D+ R
Sbjct: 535 TWQTNMGTADIKAVFGNGSKHELNVSTYQMCVLMLFNSADCLSYRDIEQTTAIPSADLKR 594
Query: 612 LLHSLSCAKYK-ILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPPVD-----EKKKV 665
L SL+ K K +L KEP ++ I D F N KFT K+ ++KI EK +
Sbjct: 595 CLQSLALVKGKNVLRKEPMSRDISDDDNFYVNDKFTSKLFKVKIGTVATQKESEPEKMET 654
Query: 666 IEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVKAI*KRIE 720
+ V++DR+ I+A+IVRIMKSR+VL + +V E +QL F P+ I KR+E
Sbjct: 655 RQRVEEDRKPQIEAAIVRIMKSRRVLDHNSIVTEVTKQLQPRFMPNPVVIKKRVE 709
>UniRef100_Q9ZVH4 T2P11.2 protein [Arabidopsis thaliana]
Length = 732
Score = 387 bits (995), Expect = e-106
Identities = 247/727 (33%), Positives = 390/727 (52%), Gaps = 58/727 (7%)
Query: 10 EQGWDFMHRGIMKLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYSQPLYDKYKE 69
++ W + R I ++ N S + LY YNM K + + LY +
Sbjct: 26 DKTWQILERAIHQIYN-------QDASGLSFEELYRNAYNMVLHK----FGEKLYTGF-- 72
Query: 70 AFEEYIVSTVLPSLREKHDEF-------MLRELVKRWANHKIMVRWLSRFFHYLDRYFIA 122
++T+ L+EK L EL K+W H + + Y+DR +I
Sbjct: 73 ------IATMTSHLKEKSKLIEAAQGGSFLEELNKKWNEHNKALEMIRDILMYMDRTYIE 126
Query: 123 RRSLPPLNEVGLACFRDLV--YKELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVE 180
++ +GL +RD V + ++H ++ + ++ L+ +ER GE IDR L++NV+ +F++
Sbjct: 127 STKKTHVHPMGLNLWRDNVVHFTKIHTRLLNTLLDLVQKERIGEVIDRGLMRNVIKMFMD 186
Query: 181 IGMGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAHYL 240
+G Y+ DFE L +S +Y ++ +I C DY+ K+E+ L E +RVAHYL
Sbjct: 187 LGESV---YQEDFEKPFLDASSEFYKVESQEFIESCDCGDYLKKSEKRLTEEIERVAHYL 243
Query: 241 HSSSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLDP 300
+ SE K+ V+ E+++ + +L+ E+SG +L +DK EDL RM+ LF ++ GL
Sbjct: 244 DAKSEEKITSVVEKEMIANHMQRLVHMENSGLVNMLLNDKYEDLGRMYNLFRRVTNGLVT 303
Query: 301 VSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVNSC 360
V + H+ G LV E + +D V FV+++++ DKY +N+
Sbjct: 304 VRDVMTSHLREMGKQLVTDPE--------KSKDPV-----EFVQRLLDERDKYDKIINTA 350
Query: 361 FQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLEKV 420
F N F AL +FE F N S E ++ F D+ L+K G + ++D +E L+KV
Sbjct: 351 FGNDKTFQNALNSSFEYFINLNA---RSPEFISLFVDDKLRK-GLKGITDVDVEVILDKV 406
Query: 421 VKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVT 480
+ L Y+ +KD+F ++Y++ LA+RLL K+ +DD ERS++ KLK +CG QFTSK+EGM T
Sbjct: 407 MMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFT 466
Query: 481 DLTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFK 540
D+ K ++ + + + P G L V VLTTG WP+ + NLPAE+ E F+
Sbjct: 467 DM---KTSEDTMRGFYGSHPELSEGPTLIVQVLTTGSWPTQPAVPCNLPAEVSVLCEKFR 523
Query: 541 EFYSTKTKHRKLTWIYSLGTCNISGKFDP-KTVELVVTTYQASALLLFNSSDRLSYSEIM 599
+Y R+L+W ++GT +I F + EL V+T+Q L+LFN+SDRLSY EI
Sbjct: 524 SYYLGTHTGRRLSWQTNMGTADIKAIFGKGQKHELNVSTFQMCVLMLFNNSDRLSYKEIE 583
Query: 600 TQLNLLDEDVIRLLHSLSCAKYK-ILIKEPNTKTILPTDYFEFNAKFTDKMRRIKIPLPP 658
+ D+ R L SL+C K K ++ KEP +K I D F N KFT K ++KI
Sbjct: 584 QATEIPAADLKRCLQSLACVKGKNVIKKEPMSKDIGEEDLFVVNDKFTSKFYKVKIGTVV 643
Query: 659 VD-----EKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKPDVK 713
EK++ + V++DR+ I+A+IVRIMKSRK+L + ++ E +QL F +
Sbjct: 644 AQKETEPEKQETRQRVEEDRKPQIEAAIVRIMKSRKILDHNNIIAEVTKQLQPRFLANPT 703
Query: 714 AI*KRIE 720
I KRIE
Sbjct: 704 EIKKRIE 710
>UniRef100_Q9CZM5 Mus musculus 11 days embryo whole body cDNA, RIKEN full-length
enriched library, clone:2700050M05 product:cullin 4B,
full insert sequence [Mus musculus]
Length = 915
Score = 386 bits (991), Expect = e-105
Identities = 240/708 (33%), Positives = 392/708 (54%), Gaps = 44/708 (6%)
Query: 22 KLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYSQPLYDKYKEAFEEYIVSTVLP 81
KLK +E + + LY + N+C+ K S LY + ++ E++I + +
Sbjct: 221 KLKEAVEAIQNSTSIKYNLEELYQAVENLCSHK----ISANLYKQLRQICEDHIKAQIHQ 276
Query: 82 SLREKHDEFM-LRELVKRWANHKIMVRWLSRFFHYLDRYFIARRS-LPPLNEVGLACFRD 139
+ D + L+++ + W NH + + F +LDR ++ + S LP + ++GL FR
Sbjct: 277 FREDSLDSVLFLKKIDRCWQNHCRQMIMIRSIFLFLDRTYVLQNSMLPSIWDMGLELFRA 336
Query: 140 LVYKE--LHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVEIGMGKMDHYENDFEADM 197
+ + + K D I+ LI++ER GE IDR+LL+++L + ++ + Y++ FE
Sbjct: 337 HIISDQKVQTKTIDGILLLIERERNGEAIDRSLLRSLLSMLSDLQI-----YQDSFEQQF 391
Query: 198 LKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAHYLHSSSEPKLLEKVQNELL 257
L++T+ Y+ + + E P+Y+ + L E DR+ YL +++ L+ V+ +LL
Sbjct: 392 LQETNRLYAAEGQKLMQEREVPEYLHHVNKRLEEEADRLITYLDQTTQKSLIASVEKQLL 451
Query: 258 SVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLDPVSSIFKQHVTTEGMALV 317
+ + +L+K G ++LL +++ +DLS +++LFS++ G+ + + +++ G +V
Sbjct: 452 GEHLTAILQK---GLNSLLDENRIQDLSLLYQLFSRVRGGVQVLLQQWIEYIKAFGSTIV 508
Query: 318 KHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVNSCFQNHTLFHKALKEAFEV 377
+ E +++ V+++++ DK +++CF + F A+KEAFE
Sbjct: 509 INPE----------------KDKTMVQELLDFKDKVDHIIDTCFLENEKFINAMKEAFET 552
Query: 378 FCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLEKVVKLLAYISDKDLFAEFY 437
F NK N AEL+A + D+ L+ G E +DE +E+ L+K++ + +I KD+F FY
Sbjct: 553 FINK--RPNKPAELIAKYVDSKLRAGNKEA-TDEELEKMLDKIMIIFRFIYGKDVFEAFY 609
Query: 438 RKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVTDLTLAKENQTSFEEYLS 497
+K LA+RLL KSA+ D E+S+L+KLK +CG FTSK+EGM D+ L+K+ F++Y+
Sbjct: 610 KKDLAKRLLVGKSASVDAEKSMLSKLKHECGAAFTSKLEGMFKDMELSKDIMIQFKQYMQ 669
Query: 498 NTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEFYSTKTKHRKLTWIYS 557
N N I+LTV +LT G+WP+Y +++LP EMVK E+FK FY K RKL W +
Sbjct: 670 NQ-NVPGNIELTVNILTMGYWPTYVPMEVHLPPEMVKLQEIFKTFYLGKHSGRKLQWQST 728
Query: 558 LGTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYSEIMTQLNLLDEDVIRLLHSLS 617
LG C + +F EL V+ +Q LL+FN + S EI + D ++ R L SL+
Sbjct: 729 LGHCVLKAEFKEGKKELQVSLFQTMVLLMFNEGEEFSLEEIKHATGIEDGELRRTLQSLA 788
Query: 618 CAKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKI----PLPPVDEKKKVIEDVDKDR 673
C K ++L K P K I D F N F K+ RIKI V+E+ E V +DR
Sbjct: 789 CGKARVLAKNPKGKDIEDGDKFICNDDFKHKLFRIKINQIQMKETVEEQASTTERVFQDR 848
Query: 674 RYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKP-DVKAI*KRIE 720
+Y IDA+IVRIMK RK L + LV E QL KP D+K KRIE
Sbjct: 849 QYQIDAAIVRIMKMRKTLSHNLLVSEVYNQLKFPVKPADLK---KRIE 893
>UniRef100_UPI00001D1662 UPI00001D1662 UniRef100 entry
Length = 1012
Score = 385 bits (990), Expect = e-105
Identities = 240/707 (33%), Positives = 391/707 (54%), Gaps = 50/707 (7%)
Query: 22 KLKNILEGLPEPQFSPEDYMMLYTTIYNMCTQKPPNDYSQPLYDKYKEAFEEYIVSTVLP 81
KLK +E + + LY + N+C+ K S LY + ++ E++I + +
Sbjct: 326 KLKEAVEAIQNSTSIKYNLEELYQAVENLCSYK----ISANLYKQLRQICEDHIKAQI-- 379
Query: 82 SLREKHDEFMLRELVKRWANHKIMVRWLSRFFHYLDRYFIARRS-LPPLNEVGLACFRDL 140
H L+++ + W NH + + F +LDR ++ + S LP + ++GL FR
Sbjct: 380 -----HHVLFLKKIDRCWQNHCRQMIMIRSIFLFLDRTYVLQNSMLPSIWDMGLELFRAH 434
Query: 141 VYKE--LHGKMRDAIISLIDQEREGEQIDRALLKNVLDIFVEIGMGKMDHYENDFEADML 198
+ + + K D I+ LI++ER GE IDR+LL+++L++ ++ + Y++ FE L
Sbjct: 435 IISDQKVQTKTIDGILLLIERERNGEAIDRSLLRSLLNMLSDLQI-----YQDSFEQRFL 489
Query: 199 KDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAHYLHSSSEPKLLEKVQNELLS 258
++T+ Y+ + + E P+Y+ + L E DR+ YL +++ L+ V+ +LL
Sbjct: 490 QETNRLYAAEGQKLMQEREVPEYLHHVNKRLEEEADRLITYLDQTTQKSLIASVEKQLLG 549
Query: 259 VYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGLDPVSSIFKQHVTTEGMALVK 318
+ + +L+K G ++LL +++ +DLS +++LFS++ G+ + + +++ G +V
Sbjct: 550 EHLTAILQK---GLNSLLDENRIQDLSLLYQLFSRVRGGVQVLLQHWIEYIKAFGSTIVI 606
Query: 319 HAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVNSCFQNHTLFHKALKEAFEVF 378
+ E +++ V+++++ DK +++CF + F A+KEAFE F
Sbjct: 607 NPE----------------KDKTMVQELLDFKDKVDHIIDTCFLKNEKFINAMKEAFETF 650
Query: 379 CNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLEKVVKLLAYISDKDLFAEFYR 438
NK N AEL+A + D+ L+ G E +DE +E+ L+K++ + +I KD+F FY+
Sbjct: 651 INK--RPNKPAELIAKYVDSKLRAGNKEA-TDEELEKMLDKIMIIFRFIYGKDVFEAFYK 707
Query: 439 KKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGMVTDLTLAKENQTSFEEYLSN 498
K LA+RLL KSA+ D E+S+L+KLK +CG FTSK+EGM D+ L+K+ F++Y+ N
Sbjct: 708 KDLAKRLLVGKSASVDAEKSMLSKLKHECGAAFTSKLEGMFKDMELSKDIMIQFKQYMQN 767
Query: 499 TPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEVFKEFYSTKTKHRKLTWIYSL 558
N I+LTV +LT G+WP+Y +++LP EMVK E+FK FY K RKL W +L
Sbjct: 768 Q-NVPGNIELTVNILTMGYWPTYVPMEVHLPPEMVKLQEIFKTFYLGKHSGRKLQWQSTL 826
Query: 559 GTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYSEIMTQLNLLDEDVIRLLHSLSC 618
G C + +F EL V+ +Q LL+FN + S EI + D ++ R L SL+C
Sbjct: 827 GHCVLKAEFKEGKKELQVSLFQTLVLLMFNEGEEFSLEEIKQATGIEDGELRRTLQSLAC 886
Query: 619 AKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKI----PLPPVDEKKKVIEDVDKDRR 674
K ++L K P K I D F N F K+ RIKI V+E+ E V +DR+
Sbjct: 887 GKARVLAKNPKGKDIEDGDKFICNDDFKHKLFRIKINQIQMKETVEEQASTTERVFQDRQ 946
Query: 675 YAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKP-DVKAI*KRIE 720
Y IDA+IVRIMK RK L + LV E QL KP D+K KRIE
Sbjct: 947 YQIDAAIVRIMKMRKTLSHNLLVSEVYNQLKFPVKPADLK---KRIE 990
>UniRef100_Q6YWY5 Putative cullin 3 [Oryza sativa]
Length = 736
Score = 385 bits (989), Expect = e-105
Identities = 234/687 (34%), Positives = 379/687 (55%), Gaps = 36/687 (5%)
Query: 43 LYTTIYNMCTQKPPNDYSQPLYDKYKEAFEEYIVSTVLPSLREKHDEFMLRELVKRWANH 102
LY + YNM K Y + LYD + + + + S+ L EL +W +H
Sbjct: 55 LYRSAYNMVLHK----YGEKLYDGLERTMT-WRLKEISKSIEAAQGGLFLEELNAKWMDH 109
Query: 103 KIMVRWLSRFFHYLDRYFIARRSLPPLNEVGLACFRDLVYKE--LHGKMRDAIISLIDQE 160
++ + Y+DR ++ + P++E+GL +RD + +H ++ D ++ LI +E
Sbjct: 110 NKALQMIRDILMYMDRTYVPQSRRTPVHELGLNLWRDHIIHSPMIHSRLLDTLLDLIHRE 169
Query: 161 REGEQIDRALLKNVLDIFVEIGMGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPD 220
R GE I+R L++++ + +++G Y++DFE L T+++YS ++ +I C +
Sbjct: 170 RMGEMINRGLMRSITKMLMDLGAAV---YQDDFEKPFLDVTASFYSGESQEFIECCDCGN 226
Query: 221 YMLKAEECLRREKDRVAHYLHSSSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDK 280
Y+ K+E L E +RV+HYL S +E K+ V+ E+++ + +L+ E+SG +L DDK
Sbjct: 227 YLKKSERRLNEEMERVSHYLDSGTEAKITSVVEKEMIANHMHRLVHMENSGLVNMLVDDK 286
Query: 281 CEDLSRMFRLFSKIPRGLDPVSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQ 340
+DL+RM+ LF ++ GL + + ++ G LV E +D V
Sbjct: 287 YDDLARMYNLFRRVFDGLSTIRDVMTSYLRETGKQLVTDPE--------RLKDPV----- 333
Query: 341 VFVRKVIELHDKYLAYVNSCFQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNIL 400
FV++++ DK+ +N F N F AL +FE F N N S E ++ + D+ L
Sbjct: 334 EFVQRLLNEKDKHDKIINVAFGNDKTFQNALNSSFEYFIN---LNNRSPEFISLYVDDKL 390
Query: 401 KKGGSEKLSDEAIEETLEKVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSIL 460
+K G + ++E +E L+KV+ L Y+ +KD+F ++Y++ LA+RLL K+ +DD ERS++
Sbjct: 391 RK-GLKGATEEDVEVILDKVMMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSMI 449
Query: 461 TKLKQQCGGQFTSKMEGMVTDLTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPS 520
KLK +CG QFTSK+EGM TD+ +++ F Y + G L V +LTTG WP+
Sbjct: 450 VKLKTECGYQFTSKLEGMFTDMKTSQDTMIDF--YAKKSEELGDGPTLDVHILTTGSWPT 507
Query: 521 YKSFDLNLPAEMVKCVEVFKEFYSTKTKHRKLTWIYSLGTCNISGKFDP-KTVELVVTTY 579
NLP E++ + F+ +Y R+LTW ++GT +I F + EL V+TY
Sbjct: 508 QPCPPCNLPTEILAICDKFRTYYLGTHSGRRLTWQTNMGTADIKATFGKGQKHELNVSTY 567
Query: 580 QASALLLFNSSDRLSYSEIMTQLNLLDEDVIRLLHSLSCAKYK-ILIKEPNTKTILPTDY 638
Q L+LFNS+D L+Y +I + D+ R L SL+C K K +L KEP +K I D
Sbjct: 568 QMCVLMLFNSTDGLTYKDIEQDTAIPASDLKRCLQSLACVKGKNVLRKEPMSKDISEDDT 627
Query: 639 FEFNAKFTDKMRRIKIPLPPVD-----EKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGY 693
F FN KFT K+ ++KI EK++ + V++DR+ I+A+IVRIMKSR+VL +
Sbjct: 628 FYFNDKFTSKLVKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRVLDH 687
Query: 694 QQLVMECVEQLGRMFKPDVKAI*KRIE 720
+V E +QL F P+ I KRIE
Sbjct: 688 NSIVAEVTKQLQARFMPNPVVIKKRIE 714
>UniRef100_Q13619 Cullin homolog 4A [Homo sapiens]
Length = 659
Score = 385 bits (988), Expect = e-105
Identities = 236/667 (35%), Positives = 377/667 (56%), Gaps = 40/667 (5%)
Query: 63 LYDKYKEAFEEYIVSTVLPSLREKHDEFM-LRELVKRWANHKIMVRWLSRFFHYLDRYFI 121
LY + ++A E+++ + +LP + D + L+++ W +H + + F +LDR ++
Sbjct: 2 LYKQLRQACEDHVQAQILPFREDSLDSVLFLKKINTCWQDHCRQMIMIRSIFLFLDRTYV 61
Query: 122 ARRS-LPPLNEVGLACFRDLVY--KELHGKMRDAIISLIDQEREGEQIDRALLKNVLDIF 178
+ S LP + ++GL FR + K + K D I+ LI++ER GE +DR+LL+++L +
Sbjct: 62 LQNSTLPSIWDMGLELFRTHIISDKMVQSKTIDGILLLIERERSGEAVDRSLLRSLLGML 121
Query: 179 VEIGMGKMDHYENDFEADMLKDTSAYYSRKASNWILEDSCPDYMLKAEECLRREKDRVAH 238
++ + Y++ FE L++T+ Y+ + + E P+Y+ + L E DRV
Sbjct: 122 SDLQV-----YKDSFELKFLEETNCLYAAEGQRLMQEREVPEYLNHVSKRLEEEGDRVIT 176
Query: 239 YLHSSSEPKLLEKVQNELLSVYASQLLEKEHSGCHALLRDDKCEDLSRMFRLFSKIPRGL 298
YL S++ L+ V+ +LL + + +L+K G LL +++ DL++M++LFS++ G
Sbjct: 177 YLDHSTQKPLIACVEKQLLGEHLTAILQK---GLDHLLDENRVPDLAQMYQLFSRVRGGQ 233
Query: 299 DPVSSIFKQHVTTEGMALVKHAEDAASNKKAEKRDIVGTQEQVFVRKVIELHDKYLAYVN 358
+ + +++ T G A+V + E +++ V+ +++ DK +
Sbjct: 234 QALLQHWSEYIKTFGTAIVINPE----------------KDKDMVQDLLDFKDKVDHVIE 277
Query: 359 SCFQNHTLFHKALKEAFEVFCNKGVAGNSSAELLATFCDNILKKGGSEKLSDEAIEETLE 418
CFQ + F +KE+FE F NK N AEL+A D+ L+ G E +DE +E TL+
Sbjct: 278 VCFQKNERFVNLMKESFETFINK--RPNKPAELIAKHVDSKLRAGNKEA-TDEELERTLD 334
Query: 419 KVVKLLAYISDKDLFAEFYRKKLARRLLFDKSANDDHERSILTKLKQQCGGQFTSKMEGM 478
K++ L +I KD+F FY+K LA+RLL KSA+ D E+S+L+KLK +CG FTSK+EGM
Sbjct: 335 KIMILFRFIHGKDVFEAFYKKDLAKRLLVGKSASVDAEKSMLSKLKHECGAAFTSKLEGM 394
Query: 479 VTDLTLAKENQTSFEEYLSNTPNADPGIDLTVTVLTTGFWPSYKSFDLNLPAEMVKCVEV 538
D+ L+K+ F++++ N ++ P IDLTV +LT G+WP+Y +++L EM+K EV
Sbjct: 395 FKDMELSKDIMVHFKQHMQNQSDSGP-IDLTVNILTMGYWPTYTPMEVHLTPEMIKLQEV 453
Query: 539 FKEFYSTKTKHRKLTWIYSLGTCNISGKFDPKTVELVVTTYQASALLLFNSSDRLSYSEI 598
FK FY K RKL W +LG + +F E V+ +Q LL+FN D S+ EI
Sbjct: 454 FKAFYLGKHSGRKLQWQTTLGHAVLKAEFKEGKKEFQVSLFQTLVLLMFNEGDGFSFEEI 513
Query: 599 MTQLNLLDEDVIRLLHSLSCAKYKILIKEPNTKTILPTDYFEFNAKFTDKMRRIKI---- 654
+ D ++ R L SL+C K ++LIK P K + D F FN +F K+ RIKI
Sbjct: 514 KMATGIEDSELRRTLQSLACGKARVLIKSPKGKEVEDGDKFIFNGEFKHKLFRIKINQIQ 573
Query: 655 PLPPVDEKKKVIEDVDKDRRYAIDASIVRIMKSRKVLGYQQLVMECVEQLGRMFKP-DVK 713
V+E+ E V +DR+Y IDA+IVRIMK RK LG+ LV E QL KP D+K
Sbjct: 574 MKETVEEQVSTTERVFQDRQYQIDAAIVRIMKMRKTLGHNLLVSELYNQLKFPVKPGDLK 633
Query: 714 AI*KRIE 720
KRIE
Sbjct: 634 ---KRIE 637
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,172,311,697
Number of Sequences: 2790947
Number of extensions: 50062732
Number of successful extensions: 138955
Number of sequences better than 10.0: 355
Number of HSP's better than 10.0 without gapping: 295
Number of HSP's successfully gapped in prelim test: 60
Number of HSP's that attempted gapping in prelim test: 137030
Number of HSP's gapped (non-prelim): 479
length of query: 720
length of database: 848,049,833
effective HSP length: 135
effective length of query: 585
effective length of database: 471,271,988
effective search space: 275694112980
effective search space used: 275694112980
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 79 (35.0 bits)
Medicago: description of AC146552.7


