

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146549.11 + phase: 0
(597 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
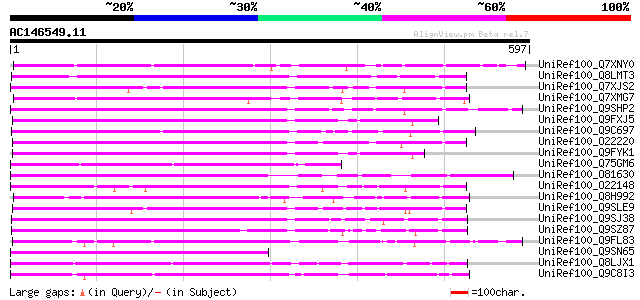
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XNY0 OSJNBb0015N08.11 protein [Oryza sativa] 283 1e-74
UniRef100_Q8LMT3 Putative retroelement [Oryza sativa] 260 9e-68
UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcrip... 243 2e-62
UniRef100_Q7XMG7 OSJNBa0028I23.15 protein [Oryza sativa] 231 3e-59
UniRef100_Q9SHP2 Putative non-LTR retroelement reverse transcrip... 230 8e-59
UniRef100_Q9FXJ5 F5A9.24 [Arabidopsis thaliana] 230 1e-58
UniRef100_Q9C697 Reverse transcriptase, putative; 16838-20266 [A... 226 2e-57
UniRef100_O22220 Putative non-LTR retroelement reverse transcrip... 226 2e-57
UniRef100_Q9FYK1 F21J9.30 [Arabidopsis thaliana] 221 5e-56
UniRef100_Q75GM6 Putative non-LTR retroelement reverse transcrip... 218 4e-55
UniRef100_O81630 F8M12.22 protein [Arabidopsis thaliana] 217 9e-55
UniRef100_O22148 Putative non-LTR retroelement reverse transcrip... 217 9e-55
UniRef100_Q8H992 Oryza sativa (japonica cultivar-group) retrotra... 215 3e-54
UniRef100_Q9SLE9 Putative non-LTR retroelement reverse transcrip... 214 8e-54
UniRef100_Q9SJ38 Putative non-LTR retroelement reverse transcrip... 212 3e-53
UniRef100_Q9SZ87 RNA-directed DNA polymerase-like protein [Arabi... 209 1e-52
UniRef100_Q9FL83 Non-LTR retroelement reverse transcriptase-like... 209 2e-52
UniRef100_Q9SN65 Hypothetical protein F25I24.40 [Arabidopsis tha... 208 3e-52
UniRef100_Q8LJX1 Putative reverse transcriptase [Sorghum bicolor] 204 8e-51
UniRef100_Q9C8I3 Hypothetical protein F19C24.27 [Arabidopsis tha... 203 1e-50
>UniRef100_Q7XNY0 OSJNBb0015N08.11 protein [Oryza sativa]
Length = 1026
Score = 283 bits (724), Expect = 1e-74
Identities = 195/603 (32%), Positives = 301/603 (49%), Gaps = 62/603 (10%)
Query: 5 GFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVLMT 64
GFP W W+ + V V+V VN F +GLRQGDPLSP LF LAAE L +L+
Sbjct: 405 GFPDQWCDWIMKVVMGGKVAVKVNDQIGSFFKTHKGLRQGDPLSPLLFNLAAEALTLLVQ 464
Query: 65 SAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSGLKVN 124
A ++L G +G + ++ LQ+ADDT+ L + + L+ IL +FE +SGLK+N
Sbjct: 465 RAEENSLIEG--LGTNGDNKIAILQYADDTIFLINDKLDHAKNLKYILCLFEQLSGLKIN 522
Query: 125 YNKSLLVGINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCF--WDPVLERLRS 182
+NKS + A+ ++I +CK+G P YLG+PI D +R+ W ++
Sbjct: 523 FNKSEVFCFGEAKEKQDLYSNIFTCKVGSLPLKYLGIPI--DQKRILNKDWKLAENKMEH 580
Query: 183 RLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGSEDHR 242
+L W+ R S GGRLILL S LSS+P+Y +SF+R P G+ I+ +F W + R
Sbjct: 581 KLGCWQGRLQSIGGRLILLNSTLSSVPMYMISFYRLPKGVQERIDYFRKRFLWQEDQGIR 640
Query: 243 KIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSSRYGE--- 299
K V+W +C ++ GGLGV L N A+LGKW WR L + EG W ++ ++Y
Sbjct: 641 KYHLVNWPLVCSPRDQGGLGVLDLEAMNKAMLGKWIWR-LENEEGWWQEIIYAKYCSDKP 699
Query: 300 -QGGRLKEGGMTGSAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDCWLGTE 358
G RLK G S +W+ ++++++ F +K +GNG T FW D WLG +
Sbjct: 700 LSGLRLKAG---SSHFWQGVMEVKDD--------FFSFCTKIVGNGEKTLFWEDSWLGGK 748
Query: 359 SFMVRFRRLYDLSIHKDLSVGEMNILG-------WGENGEAWR-WRRRLLAWEEELVVEI 410
++F LY + I K +++ ++N G +G+ R WR+ + +WE +VE
Sbjct: 749 PLAIQFPSLYGIVITKRITIADLNRKGIDCMKFRRDLHGDKLRDWRKIVNSWEGLNLVE- 807
Query: 411 RILLTNVTLQDTESDVWLWRPNIGDGYTVRGVYQMLMRQERHNYDVVSDAPWHKSAPLKV 470
+ + +W W + +TVR Y+ L Q+ + W PLK+
Sbjct: 808 ----------NCKDKLW-WTLSKDGKFTVRSFYRALKLQQT---SFPNKKIWKFRVPLKI 853
Query: 471 SIWAWRLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQNETIDLLIIHCPIFGALWQQIKT 530
I+ W +N+ TKDNL++RG D + C + ET+ L CP+ +W I
Sbjct: 854 RIFIWFFTKNKILTKDNLLKRGWRKGDNK--CQFCDKVETVQHLFFDCPLARLIWNIIAC 911
Query: 531 WIGVFFVDPHQVLDHYYQFVYSSANTTEQLLEKVKITSLKWLKAKNVCFPFGYHLWRQRP 590
+ V V Q D + ++ S T+ L+ V I ++ W +W+ R
Sbjct: 912 ALNVKPVLSRQ--DLFGSWIQSMDKFTKNLV-IVGIAAVLW------------SIWKCRN 956
Query: 591 LAC 593
AC
Sbjct: 957 KAC 959
>UniRef100_Q8LMT3 Putative retroelement [Oryza sativa]
Length = 764
Score = 260 bits (664), Expect = 9e-68
Identities = 166/525 (31%), Positives = 252/525 (47%), Gaps = 28/525 (5%)
Query: 3 KMGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVL 62
K GF W +W+K ++V +NG D F RG+RQGDPLSP LF L A+ L+ +
Sbjct: 139 KKGFSDRWIQWVKMATIGGKMAVNINGEVKDFFKTYRGVRQGDPLSPLLFNLVADALSEM 198
Query: 63 MTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSGLK 122
+ +A + + P L+HLQ+ADDT+L + N+ ++ +L +E MSGLK
Sbjct: 199 LNNAKQA-------VPHLVPGGLTHLQYADDTILFMTNTEENIVTVKFLLYCYEAMSGLK 251
Query: 123 VNYNKSLLVGINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCFWDPVLERLRS 182
+NY KS ++ I E Q A + +C+ GK PF YLG+PI + D ++
Sbjct: 252 INYQKSEIMVIGGDEMETQRVADLFNCQAGKMPFTYLGIPISMNKLTNADLDIPPNKIEK 311
Query: 183 RLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGSEDHR 242
RL+ WK LSYGG+ IL+ S LSS+P+Y + + P G+ + ++SI +FFW G E R
Sbjct: 312 RLATWKCGYLSYGGKAILINSCLSSIPLYMMGVYLLPEGVHNKMDSIRARFFWEGLEKKR 371
Query: 243 KIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSSRYGEQGG 302
K + W +C KE GGLG + N+ALL KW +R +E +L ++Y + GG
Sbjct: 372 KYHMIKWEALCRPKEFGGLGFIDTRKMNIALLCKWIYRLESGKEDPCCVLLRNKYMKDGG 431
Query: 303 R-LKEGGMTGSAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDCWLGTESFM 361
+ S +W+ + +++ W + S ++GNG T FWSD W+G
Sbjct: 432 GFFQSKAEESSQFWKGLHEVK--------KWMDLGSSYKVGNGKATNFWSDVWIGETPLK 483
Query: 362 VRFRRLYDLSIHKDLSVGEMNILG-WGENGEAWRWRRRLLAWEEELVVEIRILLTNVTLQ 420
++ +Y + K+ +V +M + G W R L W + I L
Sbjct: 484 TQYPNIYRMCADKEKTVSQMCLEGDWYIELRRSLGERDLNEWNDLHNTPREIHLKE---- 539
Query: 421 DTESDVWLWRPNIGDGYTVRGVYQMLMRQERHNYDVVSDAPWHKSAPLKVSIWAWRLLRN 480
E D +W+ Y + +YQ L D V W S PLKV I W +L+
Sbjct: 540 --ERDCIIWKLTKNGFYKAKTLYQAL--SFGGVKDKVMQDLWRSSIPLKVKILFWLMLKG 595
Query: 481 RWPTKDNLVRRGVISYDTQFCVTGCGQNETIDLLIIHCPIFGALW 525
R L + + + CGQ E +D L+ C I +W
Sbjct: 596 RIQAAGQLKK---MKWSGSPNCKLCGQTEDVDHLMFRCHISQFMW 637
>UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1344
Score = 243 bits (619), Expect = 2e-62
Identities = 172/549 (31%), Positives = 265/549 (47%), Gaps = 57/549 (10%)
Query: 1 MGKMGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLN 60
M +GF W W+ CV++ + SVL+NG P RG+RQGDPLSP LF+L E L
Sbjct: 571 MTALGFNNKWISWIMNCVTSVSYSVLINGQPYGHIIPTRGIRQGDPLSPALFVLCTEALI 630
Query: 61 VLMTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSG 120
++ A + TG +V +HL FADDTLL+ + L L + +SG
Sbjct: 631 HILNKAEQAGKITGIQFQDKKVSV-NHLLFADDTLLMCKATKQECEELMQCLSQYGQLSG 689
Query: 121 LKVNYNKSLLV-GINI---AESWLQEAASI-LSCKLGKTPFMYLGLP--IGGDARRLCFW 173
+N NKS + G N+ + W++ + I L GK YLGLP + G R L +
Sbjct: 690 QMINLNKSAITFGKNVDIQIKDWIKSRSGISLEGGTGK----YLGLPECLSGSKRDLFGF 745
Query: 174 DPVLERLRSRLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKF 233
+ E+L+SRL+ W ++ LS GG+ +LLKS+ +LPVY +S F+ P + + ++++ F
Sbjct: 746 --IKEKLQSRLTGWYAKTLSQGGKEVLLKSIALALPVYVMSCFKLPKNLCQKLTTVMMDF 803
Query: 234 FWGGSEDHRKIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVL 293
+W + RKI W+ W + + K+ GG G + L FN ALL K WR L ++ L+ RV
Sbjct: 804 WWNSMQQKRKIHWLSWQRLTLPKDQGGFGFKDLQCFNQALLAKQAWRVLQEKGSLFSRVF 863
Query: 294 SSRYGEQGGRLK-EGGMTGSAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSD 352
SRY L G S WR I+ G + + +GNG TF W+D
Sbjct: 864 QSRYFSNSDFLSATRGSRPSYAWRSIL--------FGRELLMQGLRTVIGNGQKTFVWTD 915
Query: 353 CWL--GTESFMVRFRRLYDLSIHKDLSVGEM---NILGWGENGEAWRWRRRLLAWEE-EL 406
WL G+ + RR I+ DL V ++ W N R L W++ E+
Sbjct: 916 KWLHDGSNRRPLNRRRF----INVDLKVSQLIDPTSRNWNLN-----MLRDLFPWKDVEI 966
Query: 407 VVEIRILLTNVTLQDTESDVWLWRPNIGDGYTVRGVYQMLMRQERH----------NYDV 456
+++ R L + D + W + Y+V+ Y+ L +Q H + +
Sbjct: 967 ILKQRPLF-------FKEDSFCWLHSHNGLYSVKTGYEFLSKQVHHRLYQEAKVKPSVNS 1019
Query: 457 VSDAPWHKSAPLKVSIWAWRLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQNETIDLLII 516
+ D W+ K+ I+ W+ L P +D L RG+ S D C+ +NETI+ ++
Sbjct: 1020 LFDKIWNLHTAPKIRIFLWKALHGAIPVEDRLRTRGIRSDDG--CLMCDTENETINHILF 1077
Query: 517 HCPIFGALW 525
CP+ +W
Sbjct: 1078 ECPLARQVW 1086
>UniRef100_Q7XMG7 OSJNBa0028I23.15 protein [Oryza sativa]
Length = 593
Score = 231 bits (590), Expect = 3e-59
Identities = 169/536 (31%), Positives = 252/536 (46%), Gaps = 47/536 (8%)
Query: 5 GFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVLMT 64
GF W KW+ + V +V+V VN + F ++GLRQGDPLSP LF L A+ L VL+
Sbjct: 48 GFEPRWCKWIDQVVRGGSVAVKVNDEIGNFFQTKKGLRQGDPLSPLLFNLVADMLAVLIQ 107
Query: 65 SAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSGLKVN 124
G I LS LQ+ADDT+L + + L+ +L FE +SGLK+N
Sbjct: 108 RTSEHGKIHGV-IPHLVDDGLSILQYADDTILFMEHDLEDAKNLKLVLSAFERLSGLKIN 166
Query: 125 YNKSLLVGINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCFWDPVLERLRSRL 184
++KS L+ A +E A + CK G YLGLP+ W V ER + RL
Sbjct: 167 FHKSELLCFGKAIEVEREYALLFGCKTGSYTLKYLGLPMHYRKLSNKDWKEVEERFQKRL 226
Query: 185 SEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGSEDHRKI 244
WK + LS GGRL+L+ SVLSSL +Y LSFF P GII ++ +FFW E +K
Sbjct: 227 GSWKGKLLSVGGRLVLINSVLSSLAMYMLSFFEVPKGIIKKLDYYRSRFFWQSDEHKKKY 286
Query: 245 AWVDWNTICMNKEAGGLGVRRLLEFNVAL---LGKWCWRCLVDREGLWFRVLSSRYGEQG 301
W+ +C KE GGLG++ L N L L K W+ L+ ++ L + + ++G
Sbjct: 287 RLARWSVLCKPKECGGLGIQNLEVQNKCLLRGLAKLVWQNLLRKKYLAKKTIMQVQKQRG 346
Query: 302 GRLKEGGMTGSAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDCWLGTESFM 361
S +W ++ +++ F S +L +G FW DCWLG +S
Sbjct: 347 ---------DSHFWTGLMGVKD--------TFSSFGSFKLQDGLQIRFWEDCWLGNQSLE 389
Query: 362 VRFRRLYDLSIHKDLSVGE------MNILGWGENGEAWRWRRRLLAWEEELVVEIRILLT 415
+ LY+L +K++ V + +N+ +RR ++ + EI +
Sbjct: 390 KIYPSLYNLVRNKNVVVAKVLERVPLNV----------SFRRAIIGDNLKAWHEIVANVV 439
Query: 416 NVTLQDTESDVWLWRPNIGDGYTVRGVYQMLMRQERHNYDVVSDAPWHKSAPLKVSIWAW 475
++ L+ + D + W + +TV +Y+ LM N W PLK+ I+ W
Sbjct: 440 DINLRG-QKDRFTWDASKNGTFTVNSMYKKLM---FGNVIPRQSFIWKLRLPLKIKIFLW 495
Query: 476 RLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQNETIDLLIIHCPIF---GALWQQI 528
L TKDNL +R + C +ETI L CP+ G W ++
Sbjct: 496 YLKEGVILTKDNLAKR---RWKGSLRCCFCRSHETIQYLFFDCPMVIFRGTHWARL 548
>UniRef100_Q9SHP2 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 773
Score = 230 bits (587), Expect = 8e-59
Identities = 167/606 (27%), Positives = 283/606 (46%), Gaps = 57/606 (9%)
Query: 1 MGKMGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLN 60
M +MGF W W+ C++T T S+L+NG P +RG+RQGDP+SP+L+LL EGL+
Sbjct: 48 MKQMGFSEKWCNWIMTCITTTTYSILINGQPVRRIIPKRGIRQGDPISPYLYLLCTEGLS 107
Query: 61 VLMTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSG 120
L+ +++ + G+ + P + SHL FA D+L+ + L ++L ++E SG
Sbjct: 108 ALIQASIKAKQLHGFKASRNGPAI-SHLLFAHDSLVFCKATLEECMTLVNVLKLYEKASG 166
Query: 121 LKVNYNKS-LLVGINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCFWDPVLER 179
VN+ KS +L G + ++ + +L + YLGLP + + + +
Sbjct: 167 QAVNFQKSAILFGKGLDFRTSEQLSQLLGIYKTEGFGRYLGLPEFVGRNKTNAFSFIAQT 226
Query: 180 LRSRLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGSE 239
+ ++ W ++ LS G+ +L+KS+++++P Y++S F P +I I S + F+W ++
Sbjct: 227 MDQKMDNWYNKLLSPAGKEVLIKSIVTAIPTYSMSCFLLPMRLIHQITSAMRWFWWSNTK 286
Query: 240 DHRKIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSSRYGE 299
KI WV W+ + K+ GGL +R L +FN+ALL K WR L L RV ++Y
Sbjct: 287 VKHKIPWVAWSKLNDPKKMGGLAIRDLKDFNIALLAKQSWRILQQPFSLMARVFKAKYFP 346
Query: 300 QGGRLKEGGMTGSAW-WREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDCWLGTE 358
+ L + S++ W+ I+ G + GNG N W D WL
Sbjct: 347 KERLLDAKATSQSSYAWKSIL--------HGTKLISRGLKYIAGNGNNIQLWKDNWLPLN 398
Query: 359 SFMVRFRRLYDLSIHKDLSVGEMNILGWGENGEAWRWRRRLLA--WEEELVVEIRILLTN 416
SI+ L V ++ I G RW LL + + IR + +
Sbjct: 399 PPRPPVGTCD--SIYSQLKVSDLLIEG--------RWNEDLLCKLIHQNDIPHIRAIRPS 448
Query: 417 VTLQDTESDVWLWRPNIGDGYTVRGVYQML--MRQERH---------NYDVVSDAPWHKS 465
+T +D W Y+V+ Y +L + Q++H + V W ++
Sbjct: 449 IT---GANDAITWIYTHDGNYSVKSGYHLLRKLSQQQHASLPSPNEVSAQTVFTNIWKQN 505
Query: 466 APLKVSIWAWRLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQ-NETIDLLIIHCPIFGAL 524
AP K+ + WR N PT NL RR +I+ DT CG+ +E ++ L+ C + +
Sbjct: 506 APPKIKHFWWRSAHNALPTAGNLKRRRLITDDT---CQRCGEASEDVNHLLFQCRVSKEI 562
Query: 525 WQQIKTWIGVFFVDPHQVLDHYYQFVYSSANTTEQLLEKVKITSLKWLKAKNVCFPF-GY 583
W+Q H L + +S N + ++K+ ++ K + FPF G+
Sbjct: 563 WEQ-----------AHIKLCPGDSLMSNSFNQNLESIQKLNQSARKDVS----LFPFIGW 607
Query: 584 HLWRQR 589
+W+ R
Sbjct: 608 RIWKMR 613
>UniRef100_Q9FXJ5 F5A9.24 [Arabidopsis thaliana]
Length = 1254
Score = 230 bits (586), Expect = 1e-58
Identities = 155/504 (30%), Positives = 238/504 (46%), Gaps = 35/504 (6%)
Query: 4 MGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVLM 63
+GF W W+ CVS+ T SVL+N P +QRGLRQGDPLSPFLF+L EGL L+
Sbjct: 562 LGFHLKWVNWIMVCVSSVTYSVLINDCPFGLIILQRGLRQGDPLSPFLFVLCTEGLTHLL 621
Query: 64 TSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSGLKV 123
A G + P V HL FADD+L L S L+ IL ++ N +G +
Sbjct: 622 NKAQWEGALEGIQFSENGPMV-HHLLFADDSLFLCKASREQSLVLQKILKVYGNATGQTI 680
Query: 124 NYNKSLLV-GINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCFWDPVLERLRS 182
N NKS + G + E + L YLGLP ++ + +RL+
Sbjct: 681 NLNKSSITFGEKVDEQLKGTIRTCLGIFTEGGAGTYLGLPECFSGSKVDMLHYLKDRLKE 740
Query: 183 RLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGSEDHR 242
+L W +R LS GG+ +LLKSV ++PV+A+S F+ P ++ES + F+W + R
Sbjct: 741 KLDVWFTRCLSQGGKEVLLKSVALAMPVFAMSCFKLPITTCENLESAMASFWWDSCDHSR 800
Query: 243 KIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSSRYGEQGG 302
KI W W +C+ K++GGLG R + FN ALL K WR L + L R+L SRY +
Sbjct: 801 KIHWQSWERLCLPKDSGGLGFRDIQSFNQALLAKQAWRLLHFPDCLLSRLLKSRYFDATD 860
Query: 303 RLKEG-GMTGSAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDCWLGTESFM 361
L S WR I+ G + + KR+G+G + F W D W+ F
Sbjct: 861 FLDAALSQRPSFGWRSIL--------FGRELLSKGLQKRVGDGASLFVWIDPWIDDNGFR 912
Query: 362 VRFRR--LYDLSIHKDLSVGEMNILGWGENGEAWRWRRRLLAWEEELVVEIRILLTNVTL 419
+R+ +YD+++ + N W +L + + E + + +
Sbjct: 913 APWRKNLIYDVTLKVKALL----------NPRTGFWDEEVL--HDLFLPEDILRIKAIKP 960
Query: 420 QDTESDVWLWRPNIGDGYTVRGVYQMLMRQERHNY-DVVSDAP---------WHKSAPLK 469
+++D ++W+ N ++V+ Y + + + N VS P W+ K
Sbjct: 961 VISQADFFVWKLNKSGDFSVKSAYWLAYQTKSQNLRSEVSMQPSTLGLKTQVWNLQTDPK 1020
Query: 470 VSIWAWRLLRNRWPTKDNLVRRGV 493
+ I+ W++L P +NL RG+
Sbjct: 1021 IKIFLWKVLSGILPVAENLNGRGM 1044
>UniRef100_Q9C697 Reverse transcriptase, putative; 16838-20266 [Arabidopsis thaliana]
Length = 1142
Score = 226 bits (575), Expect = 2e-57
Identities = 170/551 (30%), Positives = 264/551 (47%), Gaps = 45/551 (8%)
Query: 3 KMGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVL 62
KMGF W W+ C++T VL+NG P +RGLRQGDPLSP+LF+L E L
Sbjct: 379 KMGFCEKWISWIMWCITTVQYKVLINGQPKGLIIPERGLRQGDPLSPYLFILCTEVLIAN 438
Query: 63 MTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSGLK 122
+ A NL TG + SP V SHL FADD+L + + IL +E++SG +
Sbjct: 439 IRKAERQNLITGIKVATPSPAV-SHLLFADDSLFFCKANKEQCGIILEILKQYESVSGQQ 497
Query: 123 VNYNKSLL-VGINIAESWLQEAASILSCKLGKTPFMYLGLP--IGGDARRLCFWDPVLER 179
+N++KS + G + +S + IL YLGLP +GG ++ + V +R
Sbjct: 498 INFSKSSIQFGHKVEDSIKADIKLILGIHNLGGMGSYLGLPESLGGSKTKVFSF--VRDR 555
Query: 180 LRSRLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGSE 239
L+SR++ W ++ LS GG+ +++KSV ++LP Y +S FR P I S + S + KF+W +
Sbjct: 556 LQSRINGWSAKFLSKGGKEVMIKSVAATLPRYVMSCFRLPKAITSKLTSAVAKFWWSSNG 615
Query: 240 DHRKIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSSRYGE 299
D R + W+ W+ +C +K GGLG R + +FN ALL K WR + + L+ +V RY
Sbjct: 616 DSRGMHWMAWDKLCSSKSDGGLGFRNVDDFNSALLAKQLWRLITAPDSLFAKVFKGRYFR 675
Query: 300 QGGRLKE-GGMTGSAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDCWLGTE 358
+ L + S WR +I + S + + KR+G+G + W+D W+ +
Sbjct: 676 KSNPLDSIKSYSPSYGWRSMISAR--------SLVYKGLIKRVGSGASISVWNDPWIPAQ 727
Query: 359 SFMVRFRR--LYDLSIHKDLSVGEMNILGWGENGEAWRWRRRLL--AWEEELVVEIRIL- 413
F R Y SI D S+ +++ N W LL ++ E V I L
Sbjct: 728 -----FPRPAKYGGSI-VDPSLKVKSLIDSRSN----FWNIDLLKELFDPEDVPLISALP 777
Query: 414 LTNVTLQDTESDVWLWRPNIGDGYTVRGVYQMLMRQERHNYDVVSD-------APWHKSA 466
+ N ++DT W YTV+ Y ++ W
Sbjct: 778 IGNPNMEDTLG----WHFTKAGNYTVKSGYHTARLDLNEGTTLIGPDLTTLKAYIWKVQC 833
Query: 467 PLKVSIWAWRLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQNETIDLLIIHCPIFGALW- 525
P K+ + W++L P +NL +RG++ + CV+ E+I+ + C +W
Sbjct: 834 PPKLRHFLWQILSGCVPVSENLRKRGILC--DKGCVSCGASEESINHTLFQCHPARQIWA 891
Query: 526 -QQIKTWIGVF 535
QI T G+F
Sbjct: 892 LSQIPTAPGIF 902
>UniRef100_O22220 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1094
Score = 226 bits (575), Expect = 2e-57
Identities = 158/535 (29%), Positives = 250/535 (46%), Gaps = 35/535 (6%)
Query: 4 MGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVLM 63
+GF + W WM CVS+ + SVL+NG P RGLRQGDPLSPFLF+L E L ++
Sbjct: 325 LGFNSTWINWMMACVSSVSYSVLINGQPFGHITPHRGLRQGDPLSPFLFVLCTEALIHIL 384
Query: 64 TSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSGLKV 123
A +G P+V +HL FADDTLL+ S + L + ++SG +
Sbjct: 385 NQAEKIGKISGIQFNGTGPSV-NHLLFADDTLLICKASQLECAEIMHCLSQYGHISGQMI 443
Query: 124 NYNKSLLV-GINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCFWDPVLERLRS 182
N KS + G + E Q + + YLGLP + + + E+L+S
Sbjct: 444 NSEKSAITFGAKVNEETKQWIMNRSGIQTEGGTGKYLGLPECFQGSKQVLFGFIKEKLQS 503
Query: 183 RLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGSEDHR 242
RLS W ++ LS GG+ ILLKS+ + PVYA++ FR + + + S+++ F+W +D +
Sbjct: 504 RLSGWYAKTLSQGGKDILLKSIAMAFPVYAMTCFRLSKTLCTKLTSVMMDFWWNSVQDKK 563
Query: 243 KIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSSRYGEQGG 302
KI W+ + + K GG G + L FN ALL K R D + L ++L SRY
Sbjct: 564 KIHWIGAQKLMLPKFLGGFGFKDLQCFNQALLAKQASRLHTDSDSLLSQILKSRYYMNSD 623
Query: 303 RLK-EGGMTGSAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDCWLGTESFM 361
L G S W+ I+ G + K +GNG NT+ W D W+ F
Sbjct: 624 FLSATKGTRPSYAWQSIL--------YGRELLVSGLKKIIGNGENTYVWMDNWI----FD 671
Query: 362 VRFRRLYDLSIHKDLSVGEMNILGWGENGEAWRWRRRLLAWEE-ELVVEIRILLTNVTLQ 420
+ RR L I D+ + ++ R L W+E +++ + R +
Sbjct: 672 DKPRRPESLQIMVDIQLKVSQLIDPFSRNWNLNMLRDLFPWKEIQIICQQRPMA------ 725
Query: 421 DTESDVWLWRPNIGDGYTVRGVYQMLMRQ----------ERHNYDVVSDAPWHKSAPLKV 470
+ D + W YTV+ Y + RQ E+ + + + W+ ++ K+
Sbjct: 726 -SRQDSFCWFGTNHGLYTVKSEYDLCSRQVHKQMFKEAEEQPSLNPLFGKIWNLNSAPKI 784
Query: 471 SIWAWRLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQNETIDLLIIHCPIFGALW 525
++ W++L+ +D L RGV+ D C +NET++ ++ CP+ +W
Sbjct: 785 KVFLWKVLKGAVAVEDRLRTRGVLIEDG--CSMCPEKNETLNHILFQCPLARQVW 837
>UniRef100_Q9FYK1 F21J9.30 [Arabidopsis thaliana]
Length = 1270
Score = 221 bits (563), Expect = 5e-56
Identities = 149/488 (30%), Positives = 230/488 (46%), Gaps = 35/488 (7%)
Query: 4 MGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVLM 63
+GF W W+ CVS+ T SVL+N P +QRGLRQGDPLSPFLF+L EGL L+
Sbjct: 559 LGFHLKWVNWIMVCVSSVTYSVLINDCPFGLIILQRGLRQGDPLSPFLFVLCTEGLTHLL 618
Query: 64 TSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSGLKV 123
A G + P V HL FADD+L L S L+ IL ++ N +G +
Sbjct: 619 NKAQWEGALEGIQFSENGPMV-HHLLFADDSLFLCKASREQSLVLQKILKVYGNATGQTI 677
Query: 124 NYNKSLLV-GINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCFWDPVLERLRS 182
N NKS + G + E + L YLGLP ++ + +RL+
Sbjct: 678 NLNKSSITFGEKVDEQLKGTIRTCLGIFTEGGAGTYLGLPECFSGSKVDMLHYLKDRLKE 737
Query: 183 RLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGSEDHR 242
+L W +R LS GG+ +LLKSV ++PV+A+S F+ P ++ES + F+W + R
Sbjct: 738 KLDVWFTRCLSQGGKEVLLKSVALAMPVFAMSCFKLPITTCENLESAMASFWWDSCDHSR 797
Query: 243 KIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSSRYGEQGG 302
KI W W +C+ K++GGLG R + FN ALL K WR L + L R+L SRY +
Sbjct: 798 KIHWQSWERLCLPKDSGGLGFRDIQSFNQALLAKQAWRLLHFPDCLLSRLLKSRYFDATD 857
Query: 303 RLKEG-GMTGSAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDCWLGTESFM 361
L S WR I+ G + + KR+G+G + F W D W+ F
Sbjct: 858 FLDAALSQRPSFGWRSIL--------FGRELLSKGLQKRVGDGASLFVWIDPWIDDNGFR 909
Query: 362 VRFRR--LYDLSIHKDLSVGEMNILGWGENGEAWRWRRRLLAWEEELVVEIRILLTNVTL 419
+R+ +YD+++ + N W +L + + E + + +
Sbjct: 910 APWRKNLIYDVTLKVKALL----------NPRTGFWDEEVL--HDLFLPEDILRIKAIKP 957
Query: 420 QDTESDVWLWRPNIGDGYTVRGVYQMLMRQERHNY-DVVSDAP---------WHKSAPLK 469
+++D ++W+ N ++V+ Y + + + N VS P W+ K
Sbjct: 958 VISQADFFVWKLNKSGDFSVKSAYWLAYQTKSQNLRSEVSMQPSTLGLKTQVWNLQTDPK 1017
Query: 470 VSIWAWRL 477
+ I+ W++
Sbjct: 1018 IKIFLWKV 1025
>UniRef100_Q75GM6 Putative non-LTR retroelement reverse transcriptase [Oryza sativa]
Length = 1614
Score = 218 bits (555), Expect = 4e-55
Identities = 123/381 (32%), Positives = 198/381 (51%), Gaps = 10/381 (2%)
Query: 1 MGKMGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLN 60
M + GFP+ W W+K CV V + +NG TD F RGLRQGDPLSP LF L ++ L
Sbjct: 1220 MVRKGFPSKWVNWIKTCVMGGRVCININGERTDFFRTFRGLRQGDPLSPLLFNLISDALA 1279
Query: 61 VLMTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSG 120
++ SA + +G + P ++HLQ+ADDT+L + A + IL FE M+
Sbjct: 1280 AMLDSAKREGVLSGLVPDIF-PGGITHLQYADDTVLFVANDDKQIVATKFILYCFEEMAV 1338
Query: 121 LKVNYNKSLLVGINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCFWDPVLERL 180
LKVNY+KS + + ++++ A + +C +G+ P YLGLPIG D +D + ++L
Sbjct: 1339 LKVNYHKSEIFTLGLSDNDTNRVAMMFNCPVGQFPMKYLGLPIGPDKILNLGFDFLGQKL 1398
Query: 181 RSRLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGSED 240
RL+ W NLS+ GR + + + LSS+P YA+ F++ P G+ SI +++W +
Sbjct: 1399 EKRLNSW-GNNLSHAGRAVQINTCLSSIPSYAMCFYQLPEGVHQKFGSIRGRYYWARNRL 1457
Query: 241 HRKIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSSRYGEQ 300
K V W + K+ GGLG N ALL KW + + + L +L +Y +
Sbjct: 1458 KGKYHMVKWEDLAFPKDYGGLGFTETRRMNTALLAKWIMKIESEDDSLCIELLRRKYLQD 1517
Query: 301 GGRLKEGGMTGSAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDCWLGTESF 360
GG + S +W+ ++ I+ + + G W ++G+G + FW D W G
Sbjct: 1518 GGFFQCKERYASQFWKGLLNIRRWLSL-GSVW-------QVGDGSHISFWRDVWWGQCPL 1569
Query: 361 MVRFRRLYDLSIHKDLSVGEM 381
F ++ + +D+ V ++
Sbjct: 1570 RTLFPAIFAVCNQQDILVSQV 1590
>UniRef100_O81630 F8M12.22 protein [Arabidopsis thaliana]
Length = 1662
Score = 217 bits (552), Expect = 9e-55
Identities = 162/586 (27%), Positives = 258/586 (43%), Gaps = 86/586 (14%)
Query: 1 MGKMGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLN 60
M GF W KW+ V + SVLVNG P QRG+RQGDPLSP+LF+L A+ LN
Sbjct: 975 MRLFGFSETWIKWIMGAVKSVNYSVLVNGIPHGTIQPQRGIRQGDPLSPYLFILCADILN 1034
Query: 61 VLMTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSG 120
L+ + V G IG P V +HLQFADD+L + N +AL+ + ++E SG
Sbjct: 1035 HLIKNRVAEGDIRGIRIGNGVPGV-THLQFADDSLFFCQSNVRNCQALKDVFDVYEYYSG 1093
Query: 121 LKVNYNKSLLV-GINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCFWDPVLER 179
K+N +KS++ G + + +IL + YLGLP ++ ++ ++ER
Sbjct: 1094 QKINMSKSMITFGSRVHGTTQNRLKNILGIQSHGGGGKYLGLPEQFGRKKRDMFNYIIER 1153
Query: 180 LRSRLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGSE 239
++ R S W ++ LS G+ I+LKSV S+PVYA+S F+ P I+S IE++L+ F+W +
Sbjct: 1154 VKKRTSSWSAKYLSPAGKEIMLKSVAMSMPVYAMSCFKLPLNIVSEIEALLMNFWWEKNA 1213
Query: 240 DHRKIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSSRYGE 299
R+I W+ W + +K+ GGLG R L +FN ALL K WR + + L+ R++ +RY
Sbjct: 1214 KKREIPWIAWKRLQYSKKEGGLGFRDLAKFNDALLAKQVWRMINNPNSLFARIMKARY-- 1271
Query: 300 QGGRLKEGGMTGSAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDCWLGTES 359
+ E+SI + ++ W+ G +
Sbjct: 1272 --------------------------------FREDSILDAKRQRYQSYGWTSMLAGLDV 1299
Query: 360 FMVRFRRLYDLSIHKDLSVGEMNILGWGENGEAWRWRRRLLAWEEELVV--EIRILLTNV 417
+ G I+G G+ G W L++ +LV + R ++ +
Sbjct: 1300 ----------------IKKGSRFIVGDGKTGSYRYWNAHLIS---QLVSPDDHRFVMNHH 1340
Query: 418 TLQDTESDVWLWRPNIGDGYTVRGVYQMLMRQERHNYDVVSDAPWHKSAPLKVSIWAWRL 477
+ D +W + YT+ W K+ WR
Sbjct: 1341 LSRIVHQDKLVWNYSSSGDYTL----------------------WKLPIIPKIKYMLWRT 1378
Query: 478 LRNRWPTKDNLVRRGV-ISYDTQFCVTGCGQNETIDLLIIHCPIFGALWQQIK-TWI--G 533
+ PT+ L+ RG+ I C T + ETI+ ++ CP ++W W+
Sbjct: 1379 ISKALPTRSRLLTRGMDIDPHCPRCPT---EEETINHVLFTCPYAASIWGLSNFPWLPGH 1435
Query: 534 VFFVDPHQVLDHYYQFVYSSANTTEQLLEKVKITSLKWLKAKNVCF 579
F D + + ++ TEQ L + W N+ F
Sbjct: 1436 TFSQDTEENISFLINSFSNNTLNTEQRLAPFWLIWRLWKARNNLVF 1481
>UniRef100_O22148 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1374
Score = 217 bits (552), Expect = 9e-55
Identities = 164/554 (29%), Positives = 252/554 (44%), Gaps = 59/554 (10%)
Query: 1 MGKMGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLN 60
M +GF W + + ECV + VL+NG+P E RGLRQGDPLSP+LF++ E L
Sbjct: 593 MRGLGFADHWIRLIMECVKSVRYQVLINGTPHGEIIPSRGLRQGDPLSPYLFVICTEMLV 652
Query: 61 VLMTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMS- 119
++ SA N TG + +P + SHL FADD++ N AL I+ I E S
Sbjct: 653 KMLQSAEQKNQITGLKVARGAPPI-SHLLFADDSMFY---CKVNDEALGQIIRIIEEYSL 708
Query: 120 --GLKVNYNKS-LLVGINIAESWLQEAASILSCKLGKT----PFMYLGLPIGGDARRLCF 172
G +VNY KS + G +I+E E ++ KLG +YLGLP ++
Sbjct: 709 ASGQRVNYLKSSIYFGKHISE----ERRCLVKRKLGIEREGGEGVYLGLPESFQGSKVAT 764
Query: 173 WDPVLERLRSRLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIK 232
+ +RL ++ W+S LS GG+ ILLK+V +LP Y +S F+ P I IES++ +
Sbjct: 765 LSYLKDRLGKKVLGWQSNFLSPGGKEILLKAVAMALPTYTMSCFKIPKTICQQIESVMAE 824
Query: 233 FFWGGSEDHRKIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRV 292
F+W ++ R + W W + K GGLG + + FN+ALLGK WR + +++ L +V
Sbjct: 825 FWWKNKKEGRGLHWKAWCHLSRPKAVGGLGFKEIEAFNIALLGKQLWRMITEKDSLMAKV 884
Query: 293 LSSRYGEQGGRLKEG-GMTGSAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWS 351
SRY + L G S W+ I + Q ++ I +GNG W+
Sbjct: 885 FKSRYFSKSDPLNAPLGSRPSFAWKSIYEAQ--------VLIKQGIRAVIGNGETINVWT 936
Query: 352 DCWLGTE----------SFMVRFRRLYDLSIHKDLSVGEMNILGWGENGEAWRWRRRLLA 401
D W+G + S +V + + KDL + +G W W L
Sbjct: 937 DPWIGAKPAKAAQAVKRSHLVSQYAANSIHVVKDLLL---------PDGRDWNWNLVSLL 987
Query: 402 WEEELVVEIRILLTNVTLQDTESDVWLWRPNIGDGYTVRGVYQMLMR--QERHN------ 453
+ + + IL ++T D + W + Y+V+ Y ++ +R+N
Sbjct: 988 FPDN--TQENILALRPGGKETR-DRFTWEYSRSGHYSVKSGYWVMTEIINQRNNPQEVLQ 1044
Query: 454 --YDVVSDAPWHKSAPLKVSIWAWRLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQNETI 511
D + W P K+ + WR + N NL R + + CV ET+
Sbjct: 1045 PSLDPIFQQIWKLDVPPKIHHFLWRCVNNCLSVASNLAYRHLAR--EKSCVRCPSHGETV 1102
Query: 512 DLLIIHCPIFGALW 525
+ L+ CP W
Sbjct: 1103 NHLLFKCPFARLTW 1116
>UniRef100_Q8H992 Oryza sativa (japonica cultivar-group) retrotransposon Karma DNA,
complete sequence [Oryza sativa]
Length = 1197
Score = 215 bits (547), Expect = 3e-54
Identities = 165/539 (30%), Positives = 256/539 (46%), Gaps = 59/539 (10%)
Query: 5 GFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVLMT 64
GFP W W+K + T+ +VL+NG P ++GLRQGDPLSP+LF+L A+ L L+
Sbjct: 607 GFPNNWISWIKSLLQTSKSAVLLNGIPGKWINCKKGLRQGDPLSPYLFILVADVLQRLL- 665
Query: 65 SAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSGLKVN 124
N + I P + +Q+ADDTL++ +V ALRS L+ F +GL++N
Sbjct: 666 ---EKNFQIRHPIYQDRP--CATIQYADDTLVICRAEEDDVLALRSTLLQFSKATGLQIN 720
Query: 125 YNKSLLVGINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCFWDPVLERLRSRL 184
+ KS ++ ++I S + +L CKL P YLGLP+ P++ ++ S L
Sbjct: 721 FAKSTMISLHIDRSKESSLSELLQCKLESLPMSYLGLPLSLHKLTNNDLQPIVVKVDSFL 780
Query: 185 SEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGSED-HRK 243
+ W++ LS RLIL+ +VLSS+PVYA+S F+ P +I +I+ FFW G +
Sbjct: 781 TGWEASLLSQAERLILVNAVLSSVPVYAMSAFKLPPKVIEAIDKRRRAFFWTGDDTCSGA 840
Query: 244 IAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSSRYGEQGGR 303
V W+ +C KE GGLG++ L N ALL K + D W + + G
Sbjct: 841 KCLVAWDEVCTAKEKGGLGIKSLKTQNEALLLKRLFNLFSDNSS-WTNWIWKEF--DGRS 897
Query: 304 LKEGGMTGSAW------WREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDCWLGT 357
L + G W E+ KI + +G+G T FW D W G
Sbjct: 898 LLKSLPLGQHWSVFQKLLPELFKI---------------TTVHIGDGSRTSFWHDRWTGN 942
Query: 358 ESFMVRFRRLYDLS------IHKDLSVGEMNILGWGENGEAWRWRRRLLAWEEELVVEIR 411
+ +F L+ S + + LS G +IL RL + + + E++
Sbjct: 943 MTLAAQFESLFSHSTDDLATVEQILSFGIYDIL-----------TPRLSSAAQNDLQELQ 991
Query: 412 ILLTNVTLQDTESDVWLWRPNIGDGYTVRGVYQMLMRQ--ERHNYDVVSDAPWHKSAPLK 469
+L N +L + ESD L N T + +Y + N+ + W PLK
Sbjct: 992 TILQNFSLTN-ESDTRL--NNRLGNLTTKTIYDLRSPPGFPSPNWKFI----WDCRMPLK 1044
Query: 470 VSIWAWRLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQNETIDLLIIHCPIFGALWQQI 528
+ ++AW L+R+R TK NL+++ ++ T C +ET D L +CP + WQ +
Sbjct: 1045 IKLFAWLLVRDRLSTKLNLLKKKIV--QTATCDICSTTDETADHLSFNCPFAISFWQAL 1101
>UniRef100_Q9SLE9 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1319
Score = 214 bits (544), Expect = 8e-54
Identities = 156/538 (28%), Positives = 251/538 (45%), Gaps = 41/538 (7%)
Query: 4 MGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVLM 63
+GF W W+ CV++ T SVL+NG +RG+RQGDP+SPFLF+L E L ++
Sbjct: 548 LGFNDKWISWIMGCVTSVTYSVLINGQHFGHITPERGIRQGDPISPFLFVLCTEALIHIL 607
Query: 64 TSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSGLKV 123
A NS +G P+V +HL F DDT L+ + ++ + L + ++SG +
Sbjct: 608 QQAENSKKVSGIQFNGSGPSV-NHLLFVDDTQLVCRATKSDCEQMMLCLSQYGHISGQLI 666
Query: 124 NYNKSLLV-GINIAES---WLQEAASI-LSCKLGKTPFMYLGLPIGGDARRLCFWDPVLE 178
N KS + G+ + E W++ + I L GK YLGLP + + + E
Sbjct: 667 NVEKSSITFGVKVDEDTKRWIKNRSGIHLEGGTGK----YLGLPENLSGSKQDLFGYIKE 722
Query: 179 RLRSRLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGS 238
+L+S LS W + LS GG+ ILLKS+ +LPVY ++ FR P G+ + + S+++ F+W
Sbjct: 723 KLQSHLSGWYDKTLSQGGKEILLKSIALALPVYIMTCFRLPKGLCTKLTSVMMDFWWNSM 782
Query: 239 EDHRKIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSSRYG 298
E KI W+ + + K GG G + L FN ALL K WR D + + ++ SRY
Sbjct: 783 EFSNKIHWIGGKKLTLPKSLGGFGFKDLQCFNQALLAKQAWRLFSDSKSIVSQIFKSRYF 842
Query: 299 EQGGRLK-EGGMTGSAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDCWLGT 357
L G S WR I+ G + + +GNG T W D WL
Sbjct: 843 MNTDFLNARQGTRPSYTWRSIL--------YGRELLNGGLKRLIGNGEQTNVWIDKWL-- 892
Query: 358 ESFMVRFRRLYDLSIHKDLSVGEMNILGWGENGEAWRWRRRLLAWEEELVVEIRILLTNV 417
F RR +L ++ + +++ W ++ + E+ ++++++
Sbjct: 893 --FDGHSRRPMNLHSLMNIHMKVSHLI--DPLTRNWNLKKLTELFHEK---DVQLIMHQR 945
Query: 418 TLQDTESDVWLWRPNIGDGYTVRGVYQMLMRQERHN-------YDVVS---DAPWHKSAP 467
L +E D + W YTV+ Y+ R+ N Y V+ D W
Sbjct: 946 PLISSE-DSYCWAGTNNGLYTVKSGYERSSRETFKNLFKEADVYPSVNPLFDKVWSLETV 1004
Query: 468 LKVSIWAWRLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQNETIDLLIIHCPIFGALW 525
K+ ++ W+ L+ +D L RG+ + D C+ + ETI+ L+ CP +W
Sbjct: 1005 PKIKVFMWKALKGALAVEDRLRSRGIRTADG--CLFCKEEIETINHLLFQCPFARQVW 1060
>UniRef100_Q9SJ38 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1229
Score = 212 bits (539), Expect = 3e-53
Identities = 153/532 (28%), Positives = 239/532 (44%), Gaps = 33/532 (6%)
Query: 3 KMGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVL 62
+ GF ++W W+ ECV++ + S L+NG+P + RGLRQGDPLSP LF+L E L+ L
Sbjct: 490 RFGFHSIWIDWVLECVTSVSYSFLINGTPQGKVVPTRGLRQGDPLSPCLFILCTEVLSGL 549
Query: 63 MTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSGLK 122
T A G + ++ P V +HL FADDT+ + L IL + SG
Sbjct: 550 CTRAQRLRQLPGVRVSINGPRV-NHLLFADDTMFFSKSDPESCNKLSEILSRYGKASGQS 608
Query: 123 VNYNKSLLV-GINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCFWDPVLERLR 181
+N++KS + S + IL + YLGLP R+ + +++++R
Sbjct: 609 INFHKSSVTFSSKTPRSVKGQVKRILKIRKEGGTGKYLGLPEHFGRRKRDIFGAIIDKIR 668
Query: 182 SRLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGSEDH 241
+ W SR LS G+ ++LK+VL+S+P+Y++S F+ P+ + I+S+L +F+W D
Sbjct: 669 QKSHSWASRFLSQAGKQVMLKAVLASMPLYSMSCFKLPSALCRKIQSLLTRFWWDTKPDV 728
Query: 242 RKIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSSRYGEQG 301
RK +WV W+ + K AGGLG R + N +LL K WR L E L R+L +Y
Sbjct: 729 RKTSWVAWSKLTNPKNAGGLGFRDIERCNDSLLAKLGWRLLNSPESLLSRILLGKYCHSS 788
Query: 302 GRLK-EGGMTGSAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDCWLGTESF 360
++ + S WR II G +E + + NG W+D WL
Sbjct: 789 SFMECKLPSQPSHGWRSII--------AGREILKEGLGWLITNGEKVSIWNDPWLSISKP 840
Query: 361 MVRFRRLYDLSIHKDLSVGEMNILGWGENGEAWRWRR--RLLAWEEELVVEIRILLTNVT 418
+V L H+DL V + +N W W + +L E L+ + L +
Sbjct: 841 LVPIGPA--LREHQDLRVSAL----INQNTLQWDWNKIAVILPNYENLIKQ----LPAPS 890
Query: 419 LQDTESDVWL----WRPNIGDGYTVRGVYQMLMRQERHNYDVVSDAPWHKSAPLKVSIWA 474
+ + WL + GY + V + + Q + N+ W K+
Sbjct: 891 SRGVDKLAWLPVKSGQYTSRSGYGIASVASIPIPQTQFNW---QSNLWKLQTLPKIKHLM 947
Query: 475 WRLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQNETIDLLIIHCPIFGALWQ 526
W+ P LVRR + CG E+ L HC +W+
Sbjct: 948 WKAAMEALPVGIQLVRRHI---SPSAACHRCGAPESTTHLFFHCEFAAQVWE 996
>UniRef100_Q9SZ87 RNA-directed DNA polymerase-like protein [Arabidopsis thaliana]
Length = 1274
Score = 209 bits (533), Expect = 1e-52
Identities = 158/536 (29%), Positives = 246/536 (45%), Gaps = 47/536 (8%)
Query: 3 KMGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVL 62
++GF W +W+ +CV T + S L+NGSP RGLRQGDPLSP+LF+L E L+ L
Sbjct: 536 RLGFHDKWIRWVMQCVCTVSYSFLINGSPQGSVVPSRGLRQGDPLSPYLFILCTEVLSGL 595
Query: 63 MTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSGLK 122
A + G + SP V +HL FADDT+ + AL +IL +E SG
Sbjct: 596 CRKAQEKGVMVGIRVARGSPQV-NHLLFADDTMFFCKTNPTCCGALSNILKKYELASGQS 654
Query: 123 VNYNKSLLVGINIAESWLQEAASILSCKLGKTPFM--YLGLPIGGDARRLCFWDPVLERL 180
+N KS + + ++ LS ++ + YLGLP R+ + +++R+
Sbjct: 655 INLAKSAITFSSKTPQDIKRRVK-LSLRIDNEGGIGKYLGLPEHFGRRKRDIFSSIVDRI 713
Query: 181 RSRLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGSED 240
R R W R LS G+ ILLK+VLSS+P YA+ F+ PA + I+S+L +F+W D
Sbjct: 714 RQRSHSWSIRFLSSAGKQILLKAVLSSMPSYAMMCFKLPASLCKQIQSVLTRFWWDSKPD 773
Query: 241 HRKIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSSRYGEQ 300
RK+AWV W+ + + GGLG R + K WR L + L RVL +Y
Sbjct: 774 KRKMAWVSWDKLTLPINEGGLGFRE-------IEAKLSWRILKEPHSLLSRVLLGKYCNT 826
Query: 301 GGRL--KEGGMTGSAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDCWLGTE 358
+ S WR I+ G + + +G G + W++ WL
Sbjct: 827 SSFMDCSASPSFASHGWRGIL--------AGRDLLRKGLGWSIGQGDSINVWTEAWLSPS 878
Query: 359 SFMVRFRRLYDLSIHKDLSVGEM---NILGWGENGEAWRWRRRLLAWEEELVVEIRILLT 415
S +KDLSV ++ ++ W N EA R+ L +E+++ +I +
Sbjct: 879 SPQTPIGP--PTETNKDLSVHDLICHDVKSW--NVEA--IRKHLPQYEDQI---RKITIN 929
Query: 416 NVTLQDTESDVWLWRPNIGDGYTVRGVYQMLMRQERHNYDVVSDAPWHKS-----APLKV 470
+ LQD+ +W P YT + Y + + D W K+ KV
Sbjct: 930 ALPLQDS----LVWLPVKSGEYTTKTGYALAKLNSFPASQL--DFNWQKNIWKIHTSPKV 983
Query: 471 SIWAWRLLRNRWPTKDNLVRRGVISYDTQFCVTGCGQNETIDLLIIHCPIFGALWQ 526
+ W+ ++ P + L RR + + + CGQ E+ L++ CP +W+
Sbjct: 984 KHFLWKAMKGALPVGEALSRRNI---EAEVTCKRCGQTESSLHLMLLCPYAKKVWE 1036
>UniRef100_Q9FL83 Non-LTR retroelement reverse transcriptase-like protein [Arabidopsis
thaliana]
Length = 1223
Score = 209 bits (532), Expect = 2e-52
Identities = 177/606 (29%), Positives = 269/606 (44%), Gaps = 71/606 (11%)
Query: 4 MGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLNVLM 63
+GFP + W+ C++TA+ SV VNG F RGLRQG LSP+LF++ + L+ ++
Sbjct: 619 LGFPREFIHWINICITTASFSVQVNGELAGYFQSSRGLRQGCALSPYLFVICMDVLSKML 678
Query: 64 TSAVNSNLFTGYSIGMHSPTV---LSHLQFADDTLLLGVKSWANVRALRSILVIFENM-- 118
A + F G H L+HL FADD ++L S +R++ I+ +F+
Sbjct: 679 DKAAAARHF-----GYHPKCKTMGLTHLSFADDLMVL---SDGKIRSIERIIKVFDEFAK 730
Query: 119 -SGLKVNYNKSLLVGINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCFWD--P 175
SGL+++ KS + ++ + E A G+ P YLGLP+ +RL D P
Sbjct: 731 WSGLRISLEKSTVYLAGLSATARNEVADRFPFSSGQLPVRYLGLPL--ITKRLSTTDCLP 788
Query: 176 VLERLRSRLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFW 235
+LE++R R+ W SR LSY GRL L+ SVL S+ + L+ FR P I +E + F W
Sbjct: 789 LLEQVRKRIGSWTSRFLSYAGRLNLISSVLWSICNFWLAAFRLPRKCIRELEKMCSAFLW 848
Query: 236 GGSEDHRKIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSS 295
G+E + A + W+ +C K+ GGLG+R L E N K W+ + LW + +
Sbjct: 849 SGTEMNSNKAKISWHMVCKPKDEGGLGLRSLKEANDVCCLKLVWKIVSHSNSLWVKWVDQ 908
Query: 296 RYGEQGG--RLKEGGMTGSAWWREIIKIQNGVGVEGGSWFEESISK-RLGNGFNTFFWSD 352
+K+ GS W++++K + +++SK +GNG T FW D
Sbjct: 909 HLLRNASFWEVKQTVSQGSWIWKKLLKYRE---------VAKTLSKVEVGNGKQTSFWYD 959
Query: 353 CWLGTESFMVRF--RRLYDLSIHKDLSVGEMNILGWGENGEAWRWRRRLLAWEEEL-VVE 409
W + R R L DL I + ++V EAW RR+ + V+E
Sbjct: 960 NWSDLGQLLERTGDRGLIDLGISRRMTV-----------EEAWTNRRQRRHRNDVYNVIE 1008
Query: 410 IRILLTNVTLQDTESDVWLWRPNIGDGYTVRGVYQMLMRQERHNYDVVSDAPWHK----- 464
+ + T +TE V LWR G R + H + PWHK
Sbjct: 1009 DALKKSWDTRTETEDKV-LWR---GKSDVFRTTFS-TRDTWHHTRSTSARVPWHKVIWFS 1063
Query: 465 SAPLKVSIWAWRLLRNRWPTKDNLVR-RGVISYDTQFCVTGCGQNETIDLLIIHCPIFGA 523
A K S +W R PT D ++ I+ D FC G ET D L C
Sbjct: 1064 HATPKYSFCSWLAAHGRLPTGDRMINWANGIATDCIFCQ---GTLETRDHLFFTCSFTSV 1120
Query: 524 LWQQIKTWIGVFFVDPHQVLDHYYQFVYSSANTTEQLLEKVKITSLKWLKAKNVCFPFGY 583
+W + G+F Q H+ + + N+ +E W + V Y
Sbjct: 1121 IWVDLAR--GIF---KTQYTSHWQSIIEAITNSQHHRVE--------WFLRRYVFQATIY 1167
Query: 584 HLWRQR 589
+WR+R
Sbjct: 1168 IVWRER 1173
>UniRef100_Q9SN65 Hypothetical protein F25I24.40 [Arabidopsis thaliana]
Length = 1294
Score = 208 bits (530), Expect = 3e-52
Identities = 115/298 (38%), Positives = 172/298 (57%), Gaps = 2/298 (0%)
Query: 1 MGKMGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLN 60
M GF W KW+ V + SVLVNG P QRG+RQGDPLSP+LF+L A+ LN
Sbjct: 955 MRLFGFSETWIKWIMGAVKSVNYSVLVNGIPHGTIQPQRGIRQGDPLSPYLFILCADILN 1014
Query: 61 VLMTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSG 120
L+ + V G IG P V +HLQFADD+L + N +AL+ + ++E SG
Sbjct: 1015 HLIKNRVAEGDIRGIRIGNGVPGV-THLQFADDSLFFCQSNVRNCQALKDVFDVYEYYSG 1073
Query: 121 LKVNYNKSLLV-GINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCFWDPVLER 179
K+N +KS++ G + + +IL + YLGLP ++ ++ ++ER
Sbjct: 1074 QKINMSKSMITFGSRVHGTTQNRLKNILGIQSHGGGGKYLGLPEQFGRKKRDMFNYIIER 1133
Query: 180 LRSRLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFWGGSE 239
++ R S W ++ LS G+ I+LKSV S+PVYA+S F+ P I+S IE++L+ F+W +
Sbjct: 1134 VKKRTSSWSAKYLSPAGKEIMLKSVAMSMPVYAMSCFKLPLNIVSEIEALLMNFWWEKNA 1193
Query: 240 DHRKIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSSRY 297
R+I W+ W + +K+ GGLG R L +FN ALL K WR + + L+ R++ +RY
Sbjct: 1194 KKREIPWIAWKRLQYSKKEGGLGFRDLAKFNDALLAKQVWRMINNPNSLFARIMKARY 1251
>UniRef100_Q8LJX1 Putative reverse transcriptase [Sorghum bicolor]
Length = 1998
Score = 204 bits (518), Expect = 8e-51
Identities = 155/530 (29%), Positives = 244/530 (45%), Gaps = 28/530 (5%)
Query: 1 MGKMGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLN 60
M GF W WMK ++ T +VL+NG+P + RG+RQGDPLSP LF+LAA+ L
Sbjct: 1404 MKAKGFGQKWLNWMKSIFTSGTSNVLLNGTPGKTIHCLRGVRQGDPLSPLLFVLAADFLQ 1463
Query: 61 VLMTSAVNSNLFTGYSIGMHSPTVLSHLQFADDTLLLGVKSWANVRALRSILVIFENMSG 120
L+ A +L I M + +Q+ADDTL++ + L+SI+ F +G
Sbjct: 1464 SLINKAKCMDLLK-LPIPMQTNQDFPIIQYADDTLIIAEGDTRQLLILKSIINTFSEATG 1522
Query: 121 LKVNYNKSLLVGINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCFWDPVLERL 180
LKVN KS+++ IN+ E L A C G PF YLGLP+G + + P++ +
Sbjct: 1523 LKVNLQKSMMLPINMDEERLDTLARTFGCSKGSFPFTYLGLPLGITKPTIQDYQPLINKC 1582
Query: 181 RSRLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFW-GGSE 239
+RL + LS GRL L +V ++LP + + P +I I+ W G
Sbjct: 1583 EARLGS-VATFLSEAGRLELTNAVFTALPTFYMCTLAIPKSVIHKIDKFRKHCLWRGNGI 1641
Query: 240 DHRKIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFRVLSSRYGE 299
+ RK W +C K GGLGV + + N ALL K + ++ W +++ ++
Sbjct: 1642 NARKPPKAAWKLVCKPKNEGGLGVIDIEKQNEALLMKNLDKFFNKKDTPWVQMIWDKHYA 1701
Query: 300 QGGRLKEGGMTGSAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDCWLGTES 359
+ G+L GS WWR+I+K+ + ++E ++ G + FW D W G +
Sbjct: 1702 K-GKLPGQIRKGSFWWRDILKLLDP--------YKEMSLIQMKGGESIVFWQDKW-GMQK 1751
Query: 360 FMVRFRRLYDLSIHKDLSVGEMNILGWGENGEAWRWRRRLLAWEEELVVEIRILLTNVTL 419
+ L+ HK LS E+ G+ R L E +++ L T +
Sbjct: 1752 LRLEMPELFSFVQHKFLSAAEV-------LGQEDFTRLLQLPISETAFHQLQQLTTRIDN 1804
Query: 420 QDTESDVWLWRPNIGDGYTVRGVYQMLM-RQERHNYDVVSDAPWHKSAPLKVSIWAWRLL 478
+ + W+ G+ ++ Y LM Q+ H V W K ++ W L+
Sbjct: 1805 LERTQEQDQWKYKWGNNFSATKAYSELMGHQQIHE---VHKWIWKSFCQPKHKVFFWLLI 1861
Query: 479 RNRWPTKDNLVRRGVISYDTQFCVTGCGQN--ETIDLLIIHCPIFGALWQ 526
++R T N+++R + + CV C QN ET L +HC WQ
Sbjct: 1862 KDRLST-CNILKRKNMQLQSYNCVL-CLQNSEETTSHLFLHCAYARQCWQ 1909
>UniRef100_Q9C8I3 Hypothetical protein F19C24.27 [Arabidopsis thaliana]
Length = 629
Score = 203 bits (517), Expect = 1e-50
Identities = 168/542 (30%), Positives = 239/542 (43%), Gaps = 45/542 (8%)
Query: 1 MGKMGFPTLWRKWMKECVSTATVSVLVNGSPTDEFYMQRGLRQGDPLSPFLFLLAAEGLN 60
+ M FP + W+ C++T + SV VNG F RG+RQG LSP+LF+++ E L+
Sbjct: 21 LSAMNFPGEFIHWISRCITTTSFSVQVNGELAGYFRSARGIRQGCALSPYLFVISMEVLS 80
Query: 61 VLMTSAVNSNLFTGYSIGMHSPTV---LSHLQFADDTLLLGVKSWANVRALRSILVIFEN 117
++ A F G H L+HL FADD ++L +V + ++ +F
Sbjct: 81 KMLDQAAGGKRF-----GFHPKCKNLGLTHLCFADDLMILTDGKVRSVDGIVEVMNLFAK 135
Query: 118 MSGLKVNYNKSLLVGINIAESWLQEAASILSCKLGKTPFMYLGLPIGGDARRLCFWD--P 175
SGL++N K+ L +++ S LG+ P YLGLP+ +RL D P
Sbjct: 136 RSGLQINMEKTTLYTAGVSDHNRYMMISRYPFGLGQLPVRYLGLPLV--TKRLTKEDLSP 193
Query: 176 VLERLRSRLSEWKSRNLSYGGRLILLKSVLSSLPVYALSFFRAPAGIISSIESILIKFFW 235
+ E++R+R+ W SR LS+ GRL L+ SVL S + +S FR P+ + I SI F W
Sbjct: 194 LFEQIRNRIGTWTSRYLSFAGRLNLISSVLWSTMNFWMSAFRLPSACLKEINSICSAFLW 253
Query: 236 GGSEDHRKIAWVDWNTICMNKEAGGLGVRRLLEFNVALLGKWCWRCLVDREGLWFR--VL 293
G E HR+ A V W+ IC K+ GGLG+R L E NV + K WR + + LW + +
Sbjct: 254 SGPELHRRKAKVSWDDICKPKQEGGLGLRSLTEANVVSVLKLIWRVTSNDDSLWVKWSKM 313
Query: 294 SSRYGEQGGRLKEGGMTGSAWWREIIKIQNGVGVEGGSWFEESISKRLGNGFNTFFWSDC 353
+ E L GS W++++K + E F + NG T FW D
Sbjct: 314 NLLKQESFWSLTPNSSLGSWMWKKMLKYR-----ETAKPFSR---VEVNNGARTSFWFDN 365
Query: 354 WLGTESFM--VRFRRLYDLSIHKDLSVGEMNILGWGENGEAWRWRRRLLAWEEEL-VVEI 410
W G M R DL I ++ +V EAW RRR E+L +E
Sbjct: 366 WSGMGHLMDVTGQRGQIDLGISRNKTV-----------AEAWSNRRRRKHRTEQLNDIEA 414
Query: 411 RILLTNVTLQDTESDVWLWRPNIGDGYTVRGVYQMLMRQERHNYDVVS--DAPWHKSAPL 468
+ T D LWR GD + + Q R + V+ W +
Sbjct: 415 ALNQKYQTRNLLREDATLWRGK-GDVFKTSFSTKDTWNQVRKKSNEVAWYKGVWFSHSTP 473
Query: 469 KVSIWAWRLLRNRWPT--KDNLVRRGVISYDTQFCVTGCGQNETIDLLIIHCPIFGALWQ 526
K W LRNR T + L G FC T ET D L C A+W
Sbjct: 474 KYQFCTWLALRNRLSTGYRMQLWNNG-SDVKCTFCSTSI---ETRDHLFFSCSYASAIWT 529
Query: 527 QI 528
I
Sbjct: 530 AI 531
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.324 0.140 0.463
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,080,598,698
Number of Sequences: 2790947
Number of extensions: 46984645
Number of successful extensions: 119449
Number of sequences better than 10.0: 671
Number of HSP's better than 10.0 without gapping: 450
Number of HSP's successfully gapped in prelim test: 221
Number of HSP's that attempted gapping in prelim test: 117760
Number of HSP's gapped (non-prelim): 1025
length of query: 597
length of database: 848,049,833
effective HSP length: 133
effective length of query: 464
effective length of database: 476,853,882
effective search space: 221260201248
effective search space used: 221260201248
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 78 (34.7 bits)
Medicago: description of AC146549.11


