

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146548.3 + phase: 0
(173 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
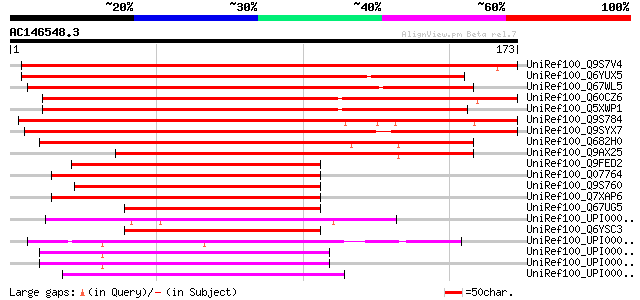
UniRef100_Q9S7V4 HVA22-like protein a [Arabidopsis thaliana] 268 6e-71
UniRef100_Q6YUX5 Putative ABA-responsive protein [Oryza sativa] 232 3e-60
UniRef100_Q67WL5 Putative abscisic acid-induced protein [Oryza s... 231 8e-60
UniRef100_Q60CZ6 Putative TB2/DP1, HVA22 family protein [Solanum... 218 4e-56
UniRef100_Q5XWP1 P0431G06.4-like [Solanum tuberosum] 213 1e-54
UniRef100_Q9S784 HVA22-like protein c [Arabidopsis thaliana] 211 5e-54
UniRef100_Q9SYX7 HVA22-like protein b [Arabidopsis thaliana] 199 2e-50
UniRef100_Q682H0 HVA22-like protein f [Arabidopsis thaliana] 140 1e-32
UniRef100_Q9AX25 P0456A01.10 protein [Oryza sativa] 129 3e-29
UniRef100_Q9FED2 HVA22-like protein e [Arabidopsis thaliana] 120 1e-26
UniRef100_Q07764 HVA22 protein [Hordeum vulgare] 112 4e-24
UniRef100_Q9S760 HVA22-like protein d [Arabidopsis thaliana] 112 4e-24
UniRef100_Q7XAP6 Hypothetical protein Wrab15 [Triticum aestivum] 112 5e-24
UniRef100_Q67UG5 Putative abscisic acid-induced protein HVA22 [O... 94 2e-18
UniRef100_UPI00003C21AA UPI00003C21AA UniRef100 entry 87 2e-16
UniRef100_Q6YSC3 Putative abscisic acid-induced protein [Oryza s... 85 9e-16
UniRef100_UPI000042DA88 UPI000042DA88 UniRef100 entry 82 6e-15
UniRef100_UPI0000453626 UPI0000453626 UniRef100 entry 81 1e-14
UniRef100_UPI000042C0A4 UPI000042C0A4 UniRef100 entry 81 1e-14
UniRef100_UPI000023F3A2 UPI000023F3A2 UniRef100 entry 79 5e-14
>UniRef100_Q9S7V4 HVA22-like protein a [Arabidopsis thaliana]
Length = 177
Score = 268 bits (684), Expect = 6e-71
Identities = 124/174 (71%), Positives = 147/174 (84%), Gaps = 5/174 (2%)
Query: 5 GSGAGSFLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLF 64
GSGAG+FL VLLRNFDVLAGPV+SLVYPLYASV+AIE++S DD+QWLTYWVLYSL+TL
Sbjct: 2 GSGAGNFLKVLLRNFDVLAGPVVSLVYPLYASVQAIETQSHADDKQWLTYWVLYSLLTLI 61
Query: 65 ELTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQTINIWYVPRKK 124
ELTFAK++EW+PIW Y KLILTCWLV+PYF+GAAYVYEH+VRP NP++INIWYVP+K
Sbjct: 62 ELTFAKLIEWLPIWSYMKLILTCWLVIPYFSGAAYVYEHFVRPVFVNPRSINIWYVPKKM 121
Query: 125 DVFTKQDDIITAAEKYIKENGTEAFENLIHRADKSKWGSSH-----HTMYDETY 173
D+F K DD++TAAEKYI ENG +AFE ++ RADKSK + H TMY E Y
Sbjct: 122 DIFRKPDDVLTAAEKYIAENGPDAFEKILSRADKSKRYNKHEYESYETMYGEGY 175
>UniRef100_Q6YUX5 Putative ABA-responsive protein [Oryza sativa]
Length = 192
Score = 232 bits (592), Expect = 3e-60
Identities = 102/151 (67%), Positives = 130/151 (85%), Gaps = 1/151 (0%)
Query: 5 GSGAGSFLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLF 64
GSG+GSFL V+++N DVLAGP++SL YPLYASVRAIE+KS VDDQQWLTYWVLYS ITLF
Sbjct: 2 GSGSGSFLKVVVKNLDVLAGPIVSLAYPLYASVRAIETKSAVDDQQWLTYWVLYSFITLF 61
Query: 65 ELTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQTINIWYVPRKK 124
ELTF+ +LEW+P+W YAKL CWLVLPYF GAA+VYEH+VRP + N Q +NIWY+PR K
Sbjct: 62 ELTFSPVLEWLPLWSYAKLFFNCWLVLPYFNGAAHVYEHFVRPMVVNQQIVNIWYIPR-K 120
Query: 125 DVFTKQDDIITAAEKYIKENGTEAFENLIHR 155
D + DD+I+AA++YI++NG+ AFE+L+++
Sbjct: 121 DESDRPDDVISAAQRYIEQNGSRAFESLVNK 151
>UniRef100_Q67WL5 Putative abscisic acid-induced protein [Oryza sativa]
Length = 193
Score = 231 bits (588), Expect = 8e-60
Identities = 103/152 (67%), Positives = 128/152 (83%), Gaps = 1/152 (0%)
Query: 7 GAGSFLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFEL 66
G+GSFL +L NFDVLAGP++SL YPLYASVRAIE+KSPVDDQQWLTYWVLYS ITLFEL
Sbjct: 2 GSGSFLKLLANNFDVLAGPLVSLAYPLYASVRAIETKSPVDDQQWLTYWVLYSFITLFEL 61
Query: 67 TFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQTINIWYVPRKKDV 126
TFA ++EW+P W YAKL CWLVLP F GAAYVY+H+VRP N Q +N+WYVPRK+++
Sbjct: 62 TFAPVIEWLPFWSYAKLFFNCWLVLPCFHGAAYVYDHFVRPMFVNRQIVNVWYVPRKENL 121
Query: 127 FTKQDDIITAAEKYIKENGTEAFENLIHRADK 158
+K DD+++AAE+YI++NG EAFE LI ++ +
Sbjct: 122 -SKPDDVLSAAERYIEQNGPEAFEKLISKSTR 152
>UniRef100_Q60CZ6 Putative TB2/DP1, HVA22 family protein [Solanum demissum]
Length = 171
Score = 218 bits (556), Expect = 4e-56
Identities = 98/163 (60%), Positives = 133/163 (81%), Gaps = 2/163 (1%)
Query: 12 LMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKI 71
+ V+ +N DVLA P++SLVYPLYAS++AIE+KS DD+QWLTYWVLYSLITLFEL+F+K+
Sbjct: 8 IAVIAKNIDVLALPLVSLVYPLYASIKAIETKSRADDRQWLTYWVLYSLITLFELSFSKL 67
Query: 72 LEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQTINIWYVPRKKDVFTKQD 131
+EW PIW YAKL CWLVLPYF GA YVYE+++RPF NP + IWYVP KKD+F+K D
Sbjct: 68 IEWFPIWSYAKLGAICWLVLPYFNGACYVYENFIRPFYRNP-LVKIWYVPLKKDIFSKPD 126
Query: 132 DIITAAEKYIKENGTEAFENLIHRADK-SKWGSSHHTMYDETY 173
D++TAAEKYI+++G +AFE L+ +AD+ ++ +++ +D+ Y
Sbjct: 127 DVLTAAEKYIEKHGPQAFERLLAKADRDARTRRNNYMTFDDDY 169
>UniRef100_Q5XWP1 P0431G06.4-like [Solanum tuberosum]
Length = 1979
Score = 213 bits (543), Expect = 1e-54
Identities = 95/145 (65%), Positives = 122/145 (83%), Gaps = 1/145 (0%)
Query: 12 LMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKI 71
+ V+ +N DVLA P++SLVYPLYAS++AIE+KS DD+QWLTYWVLYSLITLFEL+F+K+
Sbjct: 971 IAVIAKNIDVLALPLVSLVYPLYASIKAIETKSRADDRQWLTYWVLYSLITLFELSFSKL 1030
Query: 72 LEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQTINIWYVPRKKDVFTKQD 131
+EW PIW YAKL CWLVLPYF GA YVYE+++RPF NP + IWYVP KKD+F+K D
Sbjct: 1031 IEWFPIWSYAKLGAICWLVLPYFNGACYVYENFIRPFYRNP-LVKIWYVPLKKDIFSKPD 1089
Query: 132 DIITAAEKYIKENGTEAFENLIHRA 156
D++TAAEKYI+++G +AFE L+ +A
Sbjct: 1090 DVLTAAEKYIEKHGPQAFERLLAKA 1114
>UniRef100_Q9S784 HVA22-like protein c [Arabidopsis thaliana]
Length = 184
Score = 211 bits (538), Expect = 5e-54
Identities = 103/180 (57%), Positives = 138/180 (76%), Gaps = 10/180 (5%)
Query: 4 SGSGAGSFLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITL 63
S SG + L VL++NFDVLA P+++LVYPLYASV+AIE++S +D+QWLTYWVLY+LI+L
Sbjct: 3 SNSGDDNVLQVLIKNFDVLALPLVTLVYPLYASVKAIETRSLPEDEQWLTYWVLYALISL 62
Query: 64 FELTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQ--TINIWYVP 121
FELTF+K LEW PIWPY KL CWLVLP F GA ++Y+H++RPF +PQ T IWYVP
Sbjct: 63 FELTFSKPLEWFPIWPYMKLFGICWLVLPQFNGAEHIYKHFIRPFYRDPQRATTKIWYVP 122
Query: 122 RKK-DVFTKQ--DDIITAAEKYIKENGTEAFENLIHRAD-----KSKWGSSHHTMYDETY 173
KK + F K+ DDI+TAAEKY++++GTEAFE +I + D +S G ++H ++D+ Y
Sbjct: 123 HKKFNFFPKRDDDDILTAAEKYMEQHGTEAFERMIVKKDSYERGRSSRGINNHMIFDDDY 182
>UniRef100_Q9SYX7 HVA22-like protein b [Arabidopsis thaliana]
Length = 167
Score = 199 bits (506), Expect = 2e-50
Identities = 98/168 (58%), Positives = 121/168 (71%), Gaps = 5/168 (2%)
Query: 6 SGAGSFLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFE 65
SG GS + V+ +NFDV+AGPVISLVYPLYASVRAIES+S DD+QWLTYW LYSLI LFE
Sbjct: 3 SGIGSLVKVIFKNFDVIAGPVISLVYPLYASVRAIESRSHGDDKQWLTYWALYSLIKLFE 62
Query: 66 LTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQTINIWYVPRKKD 125
LTF ++LEWIP++PYAKL LT WLVLP GAAY+YEHYVR FL +P T+N+WYVP KKD
Sbjct: 63 LTFFRLLEWIPLYPYAKLALTSWLVLPGMNGAAYLYEHYVRSFLLSPHTVNVWYVPAKKD 122
Query: 126 VFTKQDDIITAAEKYIKENGTEAFENLIHRADKSKWGSSHHTMYDETY 173
DD+ A K+ N + A + I + + H+ +D+ Y
Sbjct: 123 -----DDLGATAGKFTPVNDSGAPQEKIVSSVDTSAKYVGHSAFDDAY 165
>UniRef100_Q682H0 HVA22-like protein f [Arabidopsis thaliana]
Length = 158
Score = 140 bits (353), Expect = 1e-32
Identities = 65/151 (43%), Positives = 96/151 (63%), Gaps = 3/151 (1%)
Query: 11 FLMVLLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAK 70
F++ + + FD L GP + L+YPLYAS RAIES + +DDQQWLTYW++YSLIT+FEL+ +
Sbjct: 3 FIIAIAKRFDALVGPGVMLLYPLYASFRAIESPTMLDDQQWLTYWIIYSLITIFELSVWR 62
Query: 71 ILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQTI--NIWYVPRKKDVFT 128
+L W+P WPY KL+ WLVLP F+GAAY+Y ++VR ++ + Y ++ V
Sbjct: 63 VLAWLPFWPYLKLLFCMWLVLPMFSGAAYIYSNFVRQYVKIGMNVGGGTNYTDEQRRVLQ 122
Query: 129 KQD-DIITAAEKYIKENGTEAFENLIHRADK 158
D + + Y+ G ++ E I A+K
Sbjct: 123 MMSLDARKSVQDYVDRFGWDSVEKAIKAAEK 153
>UniRef100_Q9AX25 P0456A01.10 protein [Oryza sativa]
Length = 128
Score = 129 bits (324), Expect = 3e-29
Identities = 60/123 (48%), Positives = 84/123 (67%), Gaps = 1/123 (0%)
Query: 37 VRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILEWIPIWPYAKLILTCWLVLPYFTG 96
+RAIES S +DDQQWLTYWVLYSLITLFEL+ K+L+W P+WPY KL+ CWLVLP F G
Sbjct: 1 MRAIESPSTLDDQQWLTYWVLYSLITLFELSCWKVLQWFPLWPYMKLLFCCWLVLPIFNG 60
Query: 97 AAYVYEHYVRPFLANPQTINIWYVPRKKDVFTKQD-DIITAAEKYIKENGTEAFENLIHR 155
AAY+YE +VR + Q ++ Y R++ D + E++I+ +G +A + +I
Sbjct: 61 AAYIYETHVRRYFKIGQYVSPNYNERQRKALQMMSLDARKSVERFIESHGPDALDKIIRA 120
Query: 156 ADK 158
A++
Sbjct: 121 AEE 123
>UniRef100_Q9FED2 HVA22-like protein e [Arabidopsis thaliana]
Length = 116
Score = 120 bits (302), Expect = 1e-26
Identities = 55/85 (64%), Positives = 64/85 (74%)
Query: 22 LAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILEWIPIWPYA 81
LAGPV+ L+YPLYASV AIES S VDD+QWL YW+LYS +TL EL +LEWIPIW A
Sbjct: 14 LAGPVVMLLYPLYASVIAIESPSKVDDEQWLAYWILYSFLTLSELILQSLLEWIPIWYTA 73
Query: 82 KLILTCWLVLPYFTGAAYVYEHYVR 106
KL+ WLVLP F GAA++Y VR
Sbjct: 74 KLVFVAWLVLPQFRGAAFIYNKVVR 98
>UniRef100_Q07764 HVA22 protein [Hordeum vulgare]
Length = 130
Score = 112 bits (280), Expect = 4e-24
Identities = 50/92 (54%), Positives = 65/92 (70%)
Query: 15 LLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILEW 74
LL + +AGP I+L+YPLYASV A+ES S VDD+QWL YW+LYS ITL E+ +L W
Sbjct: 7 LLTHLHSVAGPSITLLYPLYASVCAMESPSKVDDEQWLAYWILYSFITLLEMVAEPVLYW 66
Query: 75 IPIWPYAKLILTCWLVLPYFTGAAYVYEHYVR 106
IP+W KL+ WL LP F GA+++Y+ VR
Sbjct: 67 IPVWYPVKLLFVAWLALPQFKGASFIYDKVVR 98
>UniRef100_Q9S760 HVA22-like protein d [Arabidopsis thaliana]
Length = 135
Score = 112 bits (280), Expect = 4e-24
Identities = 47/84 (55%), Positives = 62/84 (72%)
Query: 23 AGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILEWIPIWPYAK 82
AGP++ L+YPLYASV A+ES + VDD+QWL YW++YS ++L EL ++EWIPIW K
Sbjct: 15 AGPIVMLLYPLYASVIAMESTTKVDDEQWLAYWIIYSFLSLTELILQSLIEWIPIWYTVK 74
Query: 83 LILTCWLVLPYFTGAAYVYEHYVR 106
L+ WLVLP F GAA++Y VR
Sbjct: 75 LVFVAWLVLPQFQGAAFIYNRVVR 98
>UniRef100_Q7XAP6 Hypothetical protein Wrab15 [Triticum aestivum]
Length = 130
Score = 112 bits (279), Expect = 5e-24
Identities = 50/92 (54%), Positives = 65/92 (70%)
Query: 15 LLRNFDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILEW 74
LL + +AGP I+L+YPLYASV A+ES S VDD+QWL YW+LYS ITL E+ +L W
Sbjct: 7 LLTHLHSVAGPSITLLYPLYASVCAMESPSKVDDEQWLAYWILYSFITLMEMLAEPVLYW 66
Query: 75 IPIWPYAKLILTCWLVLPYFTGAAYVYEHYVR 106
IP+W KL+ WL LP F GA+++Y+ VR
Sbjct: 67 IPVWYPVKLLFVAWLALPQFKGASFIYDKVVR 98
>UniRef100_Q67UG5 Putative abscisic acid-induced protein HVA22 [Oryza sativa]
Length = 99
Score = 93.6 bits (231), Expect = 2e-18
Identities = 38/67 (56%), Positives = 51/67 (75%)
Query: 40 IESKSPVDDQQWLTYWVLYSLITLFELTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAY 99
+ES S VDD+QWL YW+LYSLITL E+ K+L WIP+W AK++ WLVLP F GA++
Sbjct: 1 MESTSKVDDEQWLVYWILYSLITLMEMALHKVLYWIPLWYEAKVLFVAWLVLPQFRGASF 60
Query: 100 VYEHYVR 106
+Y+ +VR
Sbjct: 61 IYDKFVR 67
>UniRef100_UPI00003C21AA UPI00003C21AA UniRef100 entry
Length = 1161
Score = 87.0 bits (214), Expect = 2e-16
Identities = 45/123 (36%), Positives = 70/123 (56%), Gaps = 3/123 (2%)
Query: 13 MVLLRNFDVLAGPVISLVYPLYASVRAI-ESKSPVDDQQ-WLTYWVLYSLITLFELTFAK 70
+VLL + +LVYPLY+S +A+ SK+ + D + WL YW +++ TL E F
Sbjct: 936 LVLLHLPTLALNAAATLVYPLYSSYKAVTSSKTSLPDMEVWLVYWSVFACWTLLESVFGF 995
Query: 71 ILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFL-ANPQTINIWYVPRKKDVFTK 129
+ W+P + +L+ WLV P GA Y+Y +++ PFL +N Q I+ W K++V K
Sbjct: 996 LWSWLPFYYEFRLLFNIWLVAPQTRGATYIYTNHLHPFLQSNQQQIDAWIEDAKRNVKAK 1055
Query: 130 QDD 132
DD
Sbjct: 1056 MDD 1058
>UniRef100_Q6YSC3 Putative abscisic acid-induced protein [Oryza sativa]
Length = 102
Score = 84.7 bits (208), Expect = 9e-16
Identities = 34/67 (50%), Positives = 47/67 (69%)
Query: 40 IESKSPVDDQQWLTYWVLYSLITLFELTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAY 99
+ES + VDD+QWL YW+LYS ITL E+ +L WIP+W K++ WLVLP F GA++
Sbjct: 1 MESPTKVDDEQWLAYWILYSFITLLEMVAEPVLYWIPVWYPVKVLFVAWLVLPQFKGASF 60
Query: 100 VYEHYVR 106
+Y+ VR
Sbjct: 61 IYKKLVR 67
>UniRef100_UPI000042DA88 UPI000042DA88 UniRef100 entry
Length = 206
Score = 82.0 bits (201), Expect = 6e-15
Identities = 51/152 (33%), Positives = 77/152 (50%), Gaps = 14/152 (9%)
Query: 7 GAGSFLMVLLRNFDVLAGPVISLV---YPLYASVRAIESKSPVDDQQWLTYWVLYSLITL 63
G S L++ F LA P+ +L+ P Y S+ AIES DD+QWLTYWV++ + L
Sbjct: 61 GFSSVLLIFFNMFG-LAQPISNLIGWALPAYLSILAIESPQTNDDKQWLTYWVVFGSLNL 119
Query: 64 FE-LTFAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQTINIWYVPR 122
E + +L W+P++ K + T WL+LP GA +Y H++RP + N ++ R
Sbjct: 120 VESMGLRAVLYWVPMYFVFKTLFTIWLMLPATRGAEILYFHFLRPMVGNVKS-------R 172
Query: 123 KKDVFTKQDDIITAAEKYIKENGTEAFENLIH 154
+ F D + A E GT A + H
Sbjct: 173 SQSSFGTSDPL--AKETGFNPAGTTAPSSFAH 202
>UniRef100_UPI0000453626 UPI0000453626 UniRef100 entry
Length = 205
Score = 81.3 bits (199), Expect = 1e-14
Identities = 39/102 (38%), Positives = 59/102 (57%), Gaps = 3/102 (2%)
Query: 11 FLMVLLRNFDVLAGPVISLV---YPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELT 67
F++VL+ + + +LV YP Y S +A+E+ +DD+QWLTYWV+Y++ + E+
Sbjct: 64 FILVLIVSMGYAGALICNLVGFIYPAYMSFKALETPGKLDDKQWLTYWVVYAIFNILEVF 123
Query: 68 FAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFL 109
IL W+P + KL WL LP TGA +Y + RP L
Sbjct: 124 IDIILFWMPFYYLFKLCFLFWLFLPQTTGAVTLYSNIFRPLL 165
>UniRef100_UPI000042C0A4 UPI000042C0A4 UniRef100 entry
Length = 205
Score = 81.3 bits (199), Expect = 1e-14
Identities = 39/102 (38%), Positives = 59/102 (57%), Gaps = 3/102 (2%)
Query: 11 FLMVLLRNFDVLAGPVISLV---YPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELT 67
F++VL+ + + +LV YP Y S +A+E+ +DD+QWLTYWV+Y++ + E+
Sbjct: 64 FILVLIVSMGYAGALICNLVGFIYPAYMSFKALETPGKLDDKQWLTYWVVYAIFNILEVF 123
Query: 68 FAKILEWIPIWPYAKLILTCWLVLPYFTGAAYVYEHYVRPFL 109
IL W+P + KL WL LP TGA +Y + RP L
Sbjct: 124 IDIILFWMPFYYLFKLCFLFWLFLPQTTGAVTLYNNIFRPLL 165
>UniRef100_UPI000023F3A2 UPI000023F3A2 UniRef100 entry
Length = 360
Score = 79.0 bits (193), Expect = 5e-14
Identities = 35/96 (36%), Positives = 56/96 (57%)
Query: 19 FDVLAGPVISLVYPLYASVRAIESKSPVDDQQWLTYWVLYSLITLFELTFAKILEWIPIW 78
F +L V S ++P++AS +A+++ P WL YWV+ S L E I+ WIP +
Sbjct: 5 FPMLLSSVASFLFPIFASYKALKTSDPTQLTPWLMYWVVLSCCLLVESWVWFIISWIPFY 64
Query: 79 PYAKLILTCWLVLPYFTGAAYVYEHYVRPFLANPQT 114
Y +L+ +L+LP GAA++Y+ YV P+L +T
Sbjct: 65 GYFRLMFLLYLILPQTQGAAFLYDEYVHPYLEKNET 100
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.139 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 314,276,288
Number of Sequences: 2790947
Number of extensions: 12937774
Number of successful extensions: 30328
Number of sequences better than 10.0: 173
Number of HSP's better than 10.0 without gapping: 153
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 30145
Number of HSP's gapped (non-prelim): 173
length of query: 173
length of database: 848,049,833
effective HSP length: 118
effective length of query: 55
effective length of database: 518,718,087
effective search space: 28529494785
effective search space used: 28529494785
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 70 (31.6 bits)
Medicago: description of AC146548.3


