

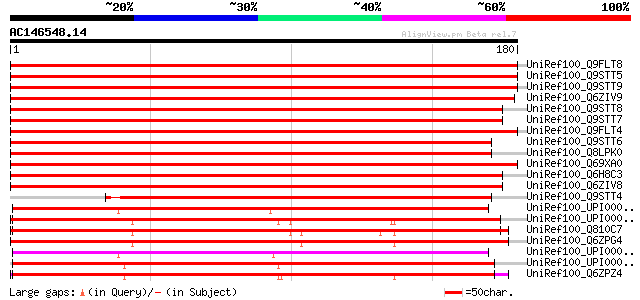
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146548.14 - phase: 0
(180 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FLT8 Similarity to ABC transporter [Arabidopsis thal... 289 3e-77
UniRef100_Q9STT5 ABC transporter-like protein [Arabidopsis thali... 287 8e-77
UniRef100_Q9STT9 ABC transporter-like protein [Arabidopsis thali... 283 2e-75
UniRef100_Q6ZIV9 Putative ABC transporter [Oryza sativa] 281 4e-75
UniRef100_Q9STT8 ABC transporter-like protein [Arabidopsis thali... 276 1e-73
UniRef100_Q9STT7 ABC transporter-like protein [Arabidopsis thali... 276 1e-73
UniRef100_Q9FLT4 Similarity to ABC transporter [Arabidopsis thal... 276 2e-73
UniRef100_Q9STT6 ABC transporter-like protein [Arabidopsis thali... 271 6e-72
UniRef100_Q8LPK0 ABC-type transport protein-like protein [Arabid... 259 2e-68
UniRef100_Q69XA0 Putative ABC family transporter [Oryza sativa] 259 3e-68
UniRef100_Q6H8C3 Putative ABC transporter [Oryza sativa] 246 1e-64
UniRef100_Q6ZIV8 Putative ABC transporter [Oryza sativa] 221 9e-57
UniRef100_Q9STT4 ABC-type transport-like protein [Arabidopsis th... 182 3e-45
UniRef100_UPI00001CEA6C UPI00001CEA6C UniRef100 entry 134 8e-31
UniRef100_UPI00003AB565 UPI00003AB565 UniRef100 entry 133 2e-30
UniRef100_Q810C7 ABC transporter subfamily A mABCA5 [Mus musculus] 133 2e-30
UniRef100_Q6ZPG4 MKIAA1888 protein [Mus musculus] 133 2e-30
UniRef100_UPI00001CE9F6 UPI00001CE9F6 UniRef100 entry 132 5e-30
UniRef100_UPI00003690F8 UPI00003690F8 UniRef100 entry 132 5e-30
UniRef100_Q6ZPZ4 MKIAA1062 protein [Mus musculus] 132 5e-30
>UniRef100_Q9FLT8 Similarity to ABC transporter [Arabidopsis thaliana]
Length = 917
Score = 289 bits (739), Expect = 3e-77
Identities = 141/180 (78%), Positives = 162/180 (89%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVAISLIG+PKVVY+DEPSTGLDPASRK LWNVIK AKQ+ AIILTTHSMEEA+
Sbjct: 738 MKRRLSVAISLIGNPKVVYLDEPSTGLDPASRKNLWNVIKRAKQNTAIILTTHSMEEAEF 797
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
LCDRLGIFVDG LQC+G KELK RYGG+YVF MTTSS HE++VE +V+ ++PNA KIYH
Sbjct: 798 LCDRLGIFVDGGLQCIGNSKELKSRYGGSYVFTMTTSSKHEEEVERLVESVSPNAKKIYH 857
Query: 121 LSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREAQAFDTLT 180
L+GTQKFELPK+E+++A VFRA+E AK NFTVFAWGL+DTTLEDVFIKVAR AQAF +L+
Sbjct: 858 LAGTQKFELPKQEVRIAEVFRAVEKAKANFTVFAWGLADTTLEDVFIKVARTAQAFISLS 917
>UniRef100_Q9STT5 ABC transporter-like protein [Arabidopsis thaliana]
Length = 900
Score = 287 bits (735), Expect = 8e-77
Identities = 137/180 (76%), Positives = 161/180 (89%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVAISLIG+PKVVYMDEPSTGLDPASRK LW VIK AKQ+ AIILTTHSMEEA+
Sbjct: 721 MKRRLSVAISLIGNPKVVYMDEPSTGLDPASRKNLWTVIKRAKQNTAIILTTHSMEEAEF 780
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
LCDRLGIFVDG LQC+G PKELKGRYGG+YVF MTTSS+HE++VE +++ ++PNA KIYH
Sbjct: 781 LCDRLGIFVDGGLQCIGNPKELKGRYGGSYVFTMTTSSEHEQNVEKLIKDVSPNAKKIYH 840
Query: 121 LSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREAQAFDTLT 180
++GTQKFELPKEE++++ VF+A+E AK NFTVFAWGL+DTTLEDVFIKV R QAF+ +
Sbjct: 841 IAGTQKFELPKEEVRISEVFQAVEKAKSNFTVFAWGLADTTLEDVFIKVVRNGQAFNVFS 900
>UniRef100_Q9STT9 ABC transporter-like protein [Arabidopsis thaliana]
Length = 925
Score = 283 bits (723), Expect = 2e-75
Identities = 136/180 (75%), Positives = 158/180 (87%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVAISLIG+PKVVYMDEPSTGLDPASRK LW VIK AK+ AIILTTHSMEEA+
Sbjct: 746 MKRRLSVAISLIGNPKVVYMDEPSTGLDPASRKNLWTVIKNAKRHTAIILTTHSMEEAEF 805
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
LCDRLGIFVDG LQC+G PKELKGRYGG+YV MTTSS+HEKDVE +VQ ++PN KIYH
Sbjct: 806 LCDRLGIFVDGRLQCIGNPKELKGRYGGSYVLTMTTSSEHEKDVEMLVQEVSPNVKKIYH 865
Query: 121 LSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREAQAFDTLT 180
++GTQKFE+PK+E++++ VF+ +E AK NF VFAWGL+DTTLEDVFIKVAR AQAF+ +
Sbjct: 866 IAGTQKFEIPKDEVRISEVFQVVEKAKSNFKVFAWGLADTTLEDVFIKVARTAQAFNVFS 925
>UniRef100_Q6ZIV9 Putative ABC transporter [Oryza sativa]
Length = 929
Score = 281 bits (720), Expect = 4e-75
Identities = 134/179 (74%), Positives = 159/179 (87%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVAISLIGDPKVV+MDEPSTGLDPASR LWNV+K AK++RAIILTTHSMEEA+
Sbjct: 750 MKRRLSVAISLIGDPKVVFMDEPSTGLDPASRNNLWNVVKEAKKNRAIILTTHSMEEAEV 809
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
LCDRLGIFVDG QC+G PKELK RYGGTYVF MTTSS+HE++V+ +VQ L+P+AN+IYH
Sbjct: 810 LCDRLGIFVDGGFQCLGNPKELKARYGGTYVFTMTTSSEHEQEVKQLVQHLSPSANRIYH 869
Query: 121 LSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREAQAFDTL 179
+SGTQKFELPK+E+K+A+VF A+E AKR F++ AWGL DTTLEDVFIKVA+ AQ + +
Sbjct: 870 ISGTQKFELPKQEVKIADVFHAVEKAKRQFSIHAWGLVDTTLEDVFIKVAKGAQGVNVI 928
>UniRef100_Q9STT8 ABC transporter-like protein [Arabidopsis thaliana]
Length = 895
Score = 276 bits (707), Expect = 1e-73
Identities = 134/175 (76%), Positives = 154/175 (87%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVAISLIG PKVVYMDEPSTGLDPASR+ LW IK AK+ AIILTTHSMEEA+
Sbjct: 716 MKRRLSVAISLIGSPKVVYMDEPSTGLDPASRRSLWTAIKGAKKHTAIILTTHSMEEAEF 775
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
LCDRLGIFVDG LQCVG PKELK RYGG+YV MTTSS+HEKDVE ++Q ++PNA KIYH
Sbjct: 776 LCDRLGIFVDGRLQCVGNPKELKARYGGSYVLTMTTSSEHEKDVEMLIQDVSPNAKKIYH 835
Query: 121 LSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREAQA 175
++GTQKFE+PK+E+++A +F+A+E AK NF VFAWGL+DTTLEDVFIKVAR AQA
Sbjct: 836 IAGTQKFEIPKDEVRIAELFQAVEKAKGNFRVFAWGLADTTLEDVFIKVARTAQA 890
>UniRef100_Q9STT7 ABC transporter-like protein [Arabidopsis thaliana]
Length = 664
Score = 276 bits (707), Expect = 1e-73
Identities = 135/175 (77%), Positives = 152/175 (86%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVAISLIG PKVVYMDEPSTGLDPASR+ LW IK AK AIILTTHSMEEA+
Sbjct: 485 MKRRLSVAISLIGSPKVVYMDEPSTGLDPASRRSLWTAIKRAKNHTAIILTTHSMEEAEF 544
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
LCDRLGIFVDG LQCVG PKELK RYGG+YV MTT S+HEKDVE +VQ ++PNA KIYH
Sbjct: 545 LCDRLGIFVDGRLQCVGNPKELKARYGGSYVLTMTTPSEHEKDVEMLVQDVSPNAKKIYH 604
Query: 121 LSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREAQA 175
++GTQKFE+PKEE++++ VF+A+E AK NF VFAWGL+DTTLEDVFIKVAR AQA
Sbjct: 605 IAGTQKFEIPKEEVRISEVFQAVEKAKDNFRVFAWGLADTTLEDVFIKVARTAQA 659
>UniRef100_Q9FLT4 Similarity to ABC transporter [Arabidopsis thaliana]
Length = 907
Score = 276 bits (705), Expect = 2e-73
Identities = 133/180 (73%), Positives = 157/180 (86%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVAISLIG+PKVVYMDEPSTGLDPASRK LW VI+ AKQ+ AIILTTHSMEEA+
Sbjct: 728 MKRRLSVAISLIGNPKVVYMDEPSTGLDPASRKDLWTVIQRAKQNTAIILTTHSMEEAEF 787
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
LCDRLGIFVDG LQCVG PKELKGRYGG+YVF MTTS +HE+ VE MV+ ++PN+ ++YH
Sbjct: 788 LCDRLGIFVDGGLQCVGNPKELKGRYGGSYVFTMTTSVEHEEKVERMVKHISPNSKRVYH 847
Query: 121 LSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREAQAFDTLT 180
L+GTQKFE+PK+E+ +A+VF +E K FTVFAWGL+DTTLEDVF KVA AQAF++L+
Sbjct: 848 LAGTQKFEIPKQEVMIADVFFMVEKVKSKFTVFAWGLADTTLEDVFFKVATTAQAFNSLS 907
>UniRef100_Q9STT6 ABC transporter-like protein [Arabidopsis thaliana]
Length = 722
Score = 271 bits (693), Expect = 6e-72
Identities = 131/171 (76%), Positives = 150/171 (87%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVAISLIG PKVVYMDEPSTGLDPASR LW VIK AK+ AIILTTHSMEEA+
Sbjct: 549 MKRRLSVAISLIGSPKVVYMDEPSTGLDPASRINLWTVIKRAKKHAAIILTTHSMEEAEF 608
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
LCDRLGIFVDG LQC+G PKELKGRYGG+YV +TTS +HEKDVE +VQ ++ NA KIYH
Sbjct: 609 LCDRLGIFVDGRLQCIGNPKELKGRYGGSYVLTITTSPEHEKDVETLVQEVSSNARKIYH 668
Query: 121 LSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAR 171
++GTQKFE PKEE++++ VF+A+E AKRNFTVFAWG +DTTLEDVFIKVA+
Sbjct: 669 IAGTQKFEFPKEEVRISEVFQAVENAKRNFTVFAWGFADTTLEDVFIKVAK 719
>UniRef100_Q8LPK0 ABC-type transport protein-like protein [Arabidopsis thaliana]
Length = 901
Score = 259 bits (662), Expect = 2e-68
Identities = 125/171 (73%), Positives = 146/171 (85%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVAISLIG PKVVYMDEPSTGLDPASRK LW+V+K AK+ AIILTTHSMEEA+
Sbjct: 729 MKRRLSVAISLIGSPKVVYMDEPSTGLDPASRKSLWDVVKRAKRKGAIILTTHSMEEAEI 788
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
LCDR+GIFVDGSLQC+G PKELK RYGG+YV +TTS +HEK+VE +V ++ NA KIY
Sbjct: 789 LCDRIGIFVDGSLQCIGNPKELKSRYGGSYVLTVTTSEEHEKEVEQLVHNISTNAKKIYR 848
Query: 121 LSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAR 171
+GTQKFELPK+E+K+ VF+A+E AK F V AWGL+DTTLEDVFIKVA+
Sbjct: 849 TAGTQKFELPKQEVKIGEVFKALEKAKTMFPVVAWGLADTTLEDVFIKVAQ 899
>UniRef100_Q69XA0 Putative ABC family transporter [Oryza sativa]
Length = 949
Score = 259 bits (661), Expect = 3e-68
Identities = 124/180 (68%), Positives = 153/180 (84%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVAISLIGD KVVYMDEPSTGLDPASRK LW+ +K AK+DRAI+LTTHSMEEA+
Sbjct: 770 MKRRLSVAISLIGDAKVVYMDEPSTGLDPASRKSLWDAVKQAKRDRAIVLTTHSMEEAEV 829
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
LCDRL I VDGSLQC+G PKEL RYGG YV MTTS + E++VEN+ ++L+PNA K+YH
Sbjct: 830 LCDRLCIMVDGSLQCIGTPKELIARYGGYYVLTMTTSPEFEQEVENLARKLSPNARKVYH 889
Query: 121 LSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREAQAFDTLT 180
LSGTQK+ELPK+++++A+VF A+E KR V AWGL+DTT+EDVF+KVA+ AQ+ + L+
Sbjct: 890 LSGTQKYELPKQQVRIADVFMAVENFKRRTEVQAWGLADTTMEDVFVKVAKGAQSSEELS 949
>UniRef100_Q6H8C3 Putative ABC transporter [Oryza sativa]
Length = 919
Score = 246 bits (629), Expect = 1e-64
Identities = 116/175 (66%), Positives = 146/175 (83%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVAISLIGDPKVVYMDEPS+GLDPASRK LWN +K AKQDRAIILTTHSMEEA+
Sbjct: 739 MKRRLSVAISLIGDPKVVYMDEPSSGLDPASRKDLWNAVKSAKQDRAIILTTHSMEEAEF 798
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
LCDR+GI +GSLQC+G KELK +YGG+YV +TT++ +++ +VQ ++P N +YH
Sbjct: 799 LCDRIGIIANGSLQCIGNSKELKAKYGGSYVLTVTTATGEAEEMRRLVQSISPTMNIVYH 858
Query: 121 LSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREAQA 175
+SGTQKFE+ K+E++++ VFRA+E AK V AWGL+DTTLEDVFI+VARE+ +
Sbjct: 859 ISGTQKFEMAKQEVRISQVFRAMEHAKLRMNVLAWGLADTTLEDVFIRVARESDS 913
>UniRef100_Q6ZIV8 Putative ABC transporter [Oryza sativa]
Length = 956
Score = 221 bits (562), Expect = 9e-57
Identities = 108/175 (61%), Positives = 135/175 (76%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKRRLSVAI+LIG+PKVVYMDEPSTGLD SR LWNVIK AK++ IILTTHSMEEA+
Sbjct: 776 MKRRLSVAIALIGNPKVVYMDEPSTGLDTTSRSNLWNVIKRAKKNCTIILTTHSMEEAEE 835
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEKDVENMVQRLTPNANKIYH 120
LCDR+GIFVDG+ QC+G PKELK RYGG +TT++ HE+ VE V R P A K+Y
Sbjct: 836 LCDRVGIFVDGNFQCLGTPKELKARYGGVRALTITTAAGHEEAVERAVARRCPGAAKVYG 895
Query: 121 LSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREAQA 175
+ GTQ+FE+P+ ++ V A+EAA+R V AWG++D TLEDVF++VA +A+A
Sbjct: 896 VGGTQRFEVPRRGARLDGVLGAVEAARRAAPVVAWGVADATLEDVFVRVAMDARA 950
>UniRef100_Q9STT4 ABC-type transport-like protein [Arabidopsis thaliana]
Length = 727
Score = 182 bits (462), Expect = 3e-45
Identities = 87/137 (63%), Positives = 107/137 (77%), Gaps = 3/137 (2%)
Query: 35 LWNVIKLAKQDRAIILTTHSMEEADALCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAM 94
LW K + R + TTHSMEEA+ LCDR+GIFVDGSLQC+G PKELK RYGG+YV +
Sbjct: 592 LW---KAQESQRFCLNTTHSMEEAEILCDRIGIFVDGSLQCIGNPKELKSRYGGSYVLTV 648
Query: 95 TTSSDHEKDVENMVQRLTPNANKIYHLSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFA 154
TTS +HEK+VE +V ++ NA KIY +GTQKFELPK+E+K+ VF+A+E AK F V A
Sbjct: 649 TTSEEHEKEVEQLVHNISTNAKKIYRTAGTQKFELPKQEVKIGEVFKALEKAKTMFPVVA 708
Query: 155 WGLSDTTLEDVFIKVAR 171
WGL+DTTLEDVFIKVA+
Sbjct: 709 WGLADTTLEDVFIKVAQ 725
>UniRef100_UPI00001CEA6C UPI00001CEA6C UniRef100 entry
Length = 950
Score = 134 bits (338), Expect = 8e-31
Identities = 68/174 (39%), Positives = 105/174 (60%), Gaps = 5/174 (2%)
Query: 2 KRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWN-VIKLAKQDRAIILTTHSMEEADA 60
KRRLS AI+++G+ VV++DEPSTG+DP +R+ LWN VI+ + + II+T+HSMEE +A
Sbjct: 763 KRRLSTAIAIMGNSSVVFLDEPSTGMDPLARRMLWNAVIRTRESGKVIIITSHSMEECEA 822
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYV----FAMTTSSDHEKDVENMVQRLTPNAN 116
LC RL I V G L C+G P+ LK ++G Y F T D +D++N + + P ++
Sbjct: 823 LCTRLAIMVQGKLVCLGSPQHLKNKFGNIYTMNIKFKTGTDDDVVQDLKNYIAEVFPGSD 882
Query: 117 KIYHLSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVA 170
G + +P ++ VF +E AK ++ + + +S TLE VF+ A
Sbjct: 883 LKQENQGILNYYIPSKDNSWGKVFGILEKAKEDYNLEDYSISQITLEQVFLTFA 936
Score = 35.0 bits (79), Expect = 0.89
Identities = 24/93 (25%), Positives = 41/93 (43%), Gaps = 11/93 (11%)
Query: 91 VFAMTTSSDHEKDVENMVQRLTPNANKIYHLSGTQKFELPKEEIK----------MANVF 140
V T SD K + +++ P A +++ F LPKE A +F
Sbjct: 13 VIVKTPDSDDGK-ISQLIKNYIPTAEMETNVAAELSFILPKEHTHRQGSRHATQWFAELF 71
Query: 141 RAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREA 173
+E + + +G+S TT+++VF KV+ A
Sbjct: 72 TDLEERQEELGISGFGVSMTTMDEVFFKVSNLA 104
>UniRef100_UPI00003AB565 UPI00003AB565 UniRef100 entry
Length = 1637
Score = 133 bits (335), Expect = 2e-30
Identities = 66/181 (36%), Positives = 112/181 (61%), Gaps = 7/181 (3%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLA--KQDRAIILTTHSMEEA 58
+KR+L A+S++G P+V +DEPSTG+DP +++ +W I+ A ++RA ILTTH MEEA
Sbjct: 1437 LKRKLCFALSMLGSPRVTLLDEPSTGMDPKAKQRMWRAIRAAFKNKERAAILTTHYMEEA 1496
Query: 59 DALCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAM----TTSSDHEKDVENMVQRLTPN 114
DA+CDR+ I V G L+C+G + LK ++G Y M T + ++ V + PN
Sbjct: 1497 DAVCDRVAILVAGQLRCIGTVQHLKSKFGRGYFLEMKLKDTADVQQVEYLQRQVLHIFPN 1556
Query: 115 ANKIYHLSGTQKFELPKEEIK-MANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREA 173
AN+ + +++P+E+++ +++ F +E K F + + S TLE VF++++RE
Sbjct: 1557 ANRQESFASILAYKIPREDVQSLSHCFSKLEEVKYAFNIEEYSFSQATLEQVFLELSREQ 1616
Query: 174 Q 174
+
Sbjct: 1617 E 1617
Score = 110 bits (274), Expect = 2e-23
Identities = 61/175 (34%), Positives = 106/175 (59%), Gaps = 3/175 (1%)
Query: 2 KRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADAL 61
KRRLSV I+++G+PKV+ +DEP+ G+DP SR +WN++K K + + +TH M+EAD L
Sbjct: 619 KRRLSVGIAVLGNPKVLLLDEPTAGMDPCSRHIVWNLLKNRKANCVTVFSTHFMDEADIL 678
Query: 62 CDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSS-DHEKDVENMVQRLTPNANKIYH 120
DR + G L+C+G LK ++G Y +M + + + +++++ P A+ I
Sbjct: 679 ADRKAVISQGMLKCLGSSLFLKSKWGIGYRLSMHIDAYCNTEATTSLIRQHIPAASLIQE 738
Query: 121 LSGTQKFELPKEEI-KMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREAQ 174
+ + LP ++ K A +F ++ + V ++G+S TTLEDV++K+ EA+
Sbjct: 739 NTQQLVYTLPLRDMDKFAGLFSDLD-THSHLGVISYGVSMTTLEDVYLKLEVEAE 792
>UniRef100_Q810C7 ABC transporter subfamily A mABCA5 [Mus musculus]
Length = 1642
Score = 133 bits (334), Expect = 2e-30
Identities = 67/184 (36%), Positives = 113/184 (61%), Gaps = 7/184 (3%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLA--KQDRAIILTTHSMEEA 58
+KR+L A+S++G+P+V +DEPSTG+DP +++ +W I+ A + RA +LTTH MEEA
Sbjct: 1437 IKRKLCFALSMLGNPQVTLLDEPSTGMDPRAKQHMWRAIRTAFKNKKRAALLTTHYMEEA 1496
Query: 59 DALCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEK----DVENMVQRLTPN 114
+A+CDR+ I V G L+C+G + LK ++G Y + E ++ +Q + PN
Sbjct: 1497 EAVCDRVAIMVSGQLRCIGTVQHLKSKFGKGYFLEIKLKDWIENLEIDRLQREIQYIFPN 1556
Query: 115 ANKIYHLSGTQKFELPKEEIK-MANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREA 173
A++ S F++PKE+++ ++ F +E AKR F + + S TLE VF+++ +E
Sbjct: 1557 ASRQESFSSILAFKIPKEDVQSLSQSFAKLEEAKRTFAIEEYSFSQATLEQVFVELTKEQ 1616
Query: 174 QAFD 177
+ D
Sbjct: 1617 EEED 1620
Score = 115 bits (288), Expect = 5e-25
Identities = 64/175 (36%), Positives = 105/175 (59%), Gaps = 3/175 (1%)
Query: 2 KRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADAL 61
KR+LSV I+++G+PK++ +DEP+ G+DP SR +WN++K K +R + +TH M+EAD L
Sbjct: 620 KRKLSVGIAVLGNPKILLLDEPTAGMDPCSRHIVWNLLKYRKANRVTVFSTHFMDEADIL 679
Query: 62 CDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSS-DHEKDVENMVQRLTPNANKIYH 120
DR + G L+CVG LK ++G Y +M + + ++V++ P A +
Sbjct: 680 ADRKAVISQGMLKCVGSSIFLKSKWGIGYRLSMYIDRYCATESLSSLVRQHIPAAALLQQ 739
Query: 121 LSGTQKFELP-KEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREAQ 174
+ LP K+ K + +F A++ N V ++G+S TTLEDVF+K+ EA+
Sbjct: 740 NDQQLVYSLPFKDMDKFSGLFSALD-IHSNLGVISYGVSMTTLEDVFLKLEVEAE 793
>UniRef100_Q6ZPG4 MKIAA1888 protein [Mus musculus]
Length = 609
Score = 133 bits (334), Expect = 2e-30
Identities = 67/184 (36%), Positives = 113/184 (61%), Gaps = 7/184 (3%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLA--KQDRAIILTTHSMEEA 58
+KR+L A+S++G+P+V +DEPSTG+DP +++ +W I+ A + RA +LTTH MEEA
Sbjct: 404 IKRKLCFALSMLGNPQVTLLDEPSTGMDPRAKQHMWRAIRTAFKNKKRAALLTTHYMEEA 463
Query: 59 DALCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMTTSSDHEK----DVENMVQRLTPN 114
+A+CDR+ I V G L+C+G + LK ++G Y + E ++ +Q + PN
Sbjct: 464 EAVCDRVAIMVSGQLRCIGTVQHLKSKFGKGYFLEIKLKDWIENLEIDRLQREIQYIFPN 523
Query: 115 ANKIYHLSGTQKFELPKEEIK-MANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVAREA 173
A++ S F++PKE+++ ++ F +E AKR F + + S TLE VF+++ +E
Sbjct: 524 ASRQESFSSILAFKIPKEDVQSLSQSFAKLEEAKRTFAIEEYSFSQATLEQVFVELTKEQ 583
Query: 174 QAFD 177
+ D
Sbjct: 584 EEED 587
>UniRef100_UPI00001CE9F6 UPI00001CE9F6 UniRef100 entry
Length = 394
Score = 132 bits (331), Expect = 5e-30
Identities = 66/171 (38%), Positives = 103/171 (59%), Gaps = 2/171 (1%)
Query: 2 KRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWN-VIKLAKQDRAIILTTHSMEEADA 60
KRRLS AI+++G P V+++DEPSTG+DP +R+ LW+ VIK+ + +AII+T+HSMEE +A
Sbjct: 219 KRRLSTAIAMMGKPSVIFLDEPSTGMDPRARRLLWDAVIKIRESGKAIIITSHSMEECEA 278
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVF-AMTTSSDHEKDVENMVQRLTPNANKIY 119
LC RL I V G L C+G P+ LK ++G Y+ S + K+ +N + P +
Sbjct: 279 LCTRLSIMVHGKLTCLGSPQYLKNKFGDIYILKTKVKSGETLKEFKNFITLTFPGSELKQ 338
Query: 120 HLSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVA 170
G + +P ++ VF +E AK + + + +S TL+ VF+ A
Sbjct: 339 ENQGILNYYIPSKDNSWGKVFGILEKAKEQYDLEDYSISQITLDQVFLAFA 389
>UniRef100_UPI00003690F8 UPI00003690F8 UniRef100 entry
Length = 423
Score = 132 bits (331), Expect = 5e-30
Identities = 71/173 (41%), Positives = 106/173 (61%), Gaps = 2/173 (1%)
Query: 2 KRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVI-KLAKQDRAIILTTHSMEEADA 60
KR+LS AI+LIG P +++DEP+TG+DP +R+ LWN+I L K R+++LT+HSMEE +A
Sbjct: 179 KRKLSTAIALIGYPAFIFLDEPTTGMDPKARRFLWNLILDLIKTGRSVVLTSHSMEECEA 238
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAM-TTSSDHEKDVENMVQRLTPNANKIY 119
LC RL I V+G L+C+G + LK R+G Y+ + T SS KDV R P A
Sbjct: 239 LCTRLAIMVNGRLRCLGSIQHLKNRFGDGYMITVRTKSSQSVKDVVRFFNRNFPEAMLKE 298
Query: 120 HLSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVARE 172
+++L E I +A VF +E + + +S TTL++VF+ A++
Sbjct: 299 RHHTKVQYQLKSEHISLAQVFSKMEQVSGVLGIEDYSVSQTTLDNVFVNFAKK 351
>UniRef100_Q6ZPZ4 MKIAA1062 protein [Mus musculus]
Length = 1416
Score = 132 bits (331), Expect = 5e-30
Identities = 71/173 (41%), Positives = 107/173 (61%), Gaps = 2/173 (1%)
Query: 2 KRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVI-KLAKQDRAIILTTHSMEEADA 60
KR+LS AI+LIG P +++DEP+TG+DP +R+ LWN+I L K R+++LT+HSMEE +A
Sbjct: 1174 KRKLSTAIALIGYPAFIFLDEPTTGMDPKARRFLWNLILDLIKTGRSVVLTSHSMEECEA 1233
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAM-TTSSDHEKDVENMVQRLTPNANKIY 119
LC RL I V+G L+C+G + LK R+G Y+ + T SS + KDV R P A
Sbjct: 1234 LCTRLAIMVNGRLRCLGSIQHLKNRFGDGYMITVRTKSSQNVKDVVRFFNRNFPEAMLKE 1293
Query: 120 HLSGTQKFELPKEEIKMANVFRAIEAAKRNFTVFAWGLSDTTLEDVFIKVARE 172
+++L E I +A VF +E + + +S TTL++VF+ A++
Sbjct: 1294 RHHTKVQYQLKSEHISLAQVFSKMEQVVGVLGIEDYSVSQTTLDNVFVNFAKK 1346
Score = 115 bits (288), Expect = 5e-25
Identities = 67/203 (33%), Positives = 106/203 (52%), Gaps = 26/203 (12%)
Query: 1 MKRRLSVAISLIGDPKVVYMDEPSTGLDPASRKCLWNVIKLAKQDRAIILTTHSMEEADA 60
MKR+LSVAI+ +G + + +DEP+ G+DP +R+ +W++I K R I+L+TH M+EAD
Sbjct: 109 MKRKLSVAIAFVGGSRAIILDEPTAGVDPYARRAIWDLILKYKPGRTILLSTHHMDEADL 168
Query: 61 LCDRLGIFVDGSLQCVGYPKELKGRYGGTYVFAMT-----------------------TS 97
L DR+ I G L+C G P LKG YG Y + S
Sbjct: 169 LGDRIAIISHGKLKCCGSPLFLKGAYGDGYRLTLVKQPAEPGTSQEPGLASSPSGCPRLS 228
Query: 98 SDHEKDVENMVQRLTPNANKIYHLSGTQKFELPKEEIK---MANVFRAIEAAKRNFTVFA 154
S E V +++ ++ + S + LP E +K +F+ +E + + +
Sbjct: 229 SCSEPQVSQFIRKHVASSLLVSDTSTELSYILPSEAVKKGAFERLFQQLEHSLDALHLSS 288
Query: 155 WGLSDTTLEDVFIKVAREAQAFD 177
+GL DTTLE+VF+KV+ E Q+ +
Sbjct: 289 FGLMDTTLEEVFLKVSEEDQSLE 311
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.134 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 284,620,837
Number of Sequences: 2790947
Number of extensions: 10949782
Number of successful extensions: 58364
Number of sequences better than 10.0: 16482
Number of HSP's better than 10.0 without gapping: 8302
Number of HSP's successfully gapped in prelim test: 8181
Number of HSP's that attempted gapping in prelim test: 41219
Number of HSP's gapped (non-prelim): 18489
length of query: 180
length of database: 848,049,833
effective HSP length: 119
effective length of query: 61
effective length of database: 515,927,140
effective search space: 31471555540
effective search space used: 31471555540
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 70 (31.6 bits)
Medicago: description of AC146548.14


