

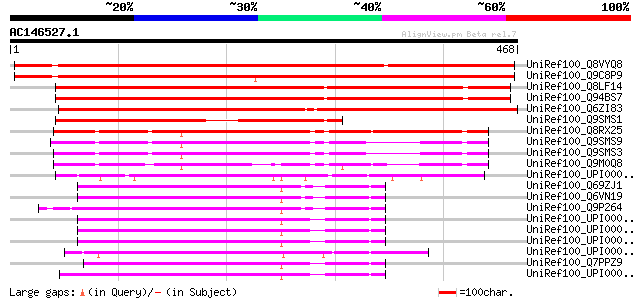
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146527.1 - phase: 0
(468 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8VYQ8 Hypothetical protein At1g35470 [Arabidopsis tha... 610 e-173
UniRef100_Q9C8P9 Hypothetical protein F12A4.1 [Arabidopsis thali... 608 e-172
UniRef100_Q8LF14 Hypothetical protein [Arabidopsis thaliana] 582 e-165
UniRef100_Q94BS7 Hypothetical protein At4g09340 [Arabidopsis tha... 582 e-164
UniRef100_Q6ZI83 RanBPM-like [Oryza sativa] 541 e-152
UniRef100_Q9SMS1 Hypothetical protein AT4g09340 [Arabidopsis tha... 317 4e-85
UniRef100_Q8RX25 Hypothetical protein At4g09200 [Arabidopsis tha... 302 2e-80
UniRef100_Q9SMS9 Hypothetical protein AT4g09250 [Arabidopsis tha... 250 5e-65
UniRef100_Q9SMS3 Hypothetical protein AT4g09310 [Arabidopsis tha... 249 1e-64
UniRef100_Q9M0Q8 Hypothetical protein AT4g09200 [Arabidopsis tha... 225 2e-57
UniRef100_UPI000023D443 UPI000023D443 UniRef100 entry 182 1e-44
UniRef100_Q69ZJ1 MKIAA1464 protein [Mus musculus] 179 1e-43
UniRef100_Q6VN19 Ran-binding protein 10 [Mus musculus] 179 1e-43
UniRef100_Q9P264 KIAA1464 protein [Homo sapiens] 178 3e-43
UniRef100_UPI0000437010 UPI0000437010 UniRef100 entry 177 8e-43
UniRef100_UPI000043700F UPI000043700F UniRef100 entry 177 8e-43
UniRef100_UPI000043700E UPI000043700E UniRef100 entry 177 8e-43
UniRef100_UPI000021B34D UPI000021B34D UniRef100 entry 176 1e-42
UniRef100_Q7PPZ9 ENSANGP00000003407 [Anopheles gambiae str. PEST] 176 1e-42
UniRef100_UPI0000361052 UPI0000361052 UniRef100 entry 174 4e-42
>UniRef100_Q8VYQ8 Hypothetical protein At1g35470 [Arabidopsis thaliana]
Length = 465
Score = 610 bits (1574), Expect = e-173
Identities = 298/464 (64%), Positives = 376/464 (80%), Gaps = 9/464 (1%)
Query: 5 NGKNTIEKPDQDQNQDLGFHFLELSRRKAKLDSEDFEVDDEEKEESPSELNTINSSGGFV 64
N N + + QDL +FL+ R AK D+++ +E EE P+ELNTINS+GGF+
Sbjct: 9 NSANGDTTNNGENGQDLNLNFLDKIRLSAKRDAKE-----DEGEELPTELNTINSAGGFL 63
Query: 65 VVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIVYYFEIFVKDAGVKGQVSIGFTTES 124
VV+ DKLSVKYT+ NLHGHDVGV+QANK AP+K + YYFEIFVKD+G+KGQ++IGFT ES
Sbjct: 64 VVSPDKLSVKYTNTNLHGHDVGVVQANKPAPIKCLTYYFEIFVKDSGIKGQIAIGFTKES 123
Query: 125 FKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAFGPTYTTGDIVGAGINYAAQEFFFTKNG 184
FKMRRQPGWE NSCGYHGDDG+LYRGQGKGE FGP +T D VG GINYA+QEFFFTKNG
Sbjct: 124 FKMRRQPGWEVNSCGYHGDDGYLYRGQGKGEPFGPKFTKDDAVGGGINYASQEFFFTKNG 183
Query: 185 QVVGTVFKEMKGPLFPTVAVHSQNEEVHVNFGQKPFTFDLKEYEAQERMKQQIKIEEVPV 244
+VG + K+++G LFPTVAVHSQNEEV VNFG+K F FD+K YEA ER KQQ+ IE++ +
Sbjct: 184 TIVGKIPKDIRGHLFPTVAVHSQNEEVLVNFGKKKFAFDIKGYEASERNKQQLAIEKISI 243
Query: 245 SPNASYGIVRSYLLHYGYEDTLNSFDEASRSTIPPINIVQENGIDDQETTYALNHRKTLR 304
PN YG+V++YLLHYGYE+TL++F+ A+++T+PPI+I QEN ID+ +++YAL RK LR
Sbjct: 244 PPNIGYGLVKTYLLHYGYEETLDAFNLATKNTVPPIHIDQENAIDEDDSSYALKQRKNLR 303
Query: 305 QLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELVRVGALEEAVKYGRIELS 364
QL+R+GEID A +L++ YPQI +D+ S +CFLLHCQKFIEL VG LEE V YGR+EL+
Sbjct: 304 QLVRNGEIDTALAELQKLYPQIVQDDKSVVCFLLHCQKFIEL--VGKLEEGVNYGRLELA 361
Query: 365 SFFGLSLFEDIVQDCVALLAYERPLESAVGYLLKDSQREVVADAVNAMILSTNPNIK-VT 423
F GL+ F+DIV+DC ALLAYE+P ES+V Y L+DSQRE+VADAVNA ILSTNPN K V
Sbjct: 362 KFVGLTGFQDIVEDCFALLAYEKPEESSVWYFLEDSQRELVADAVNAAILSTNPNKKDVQ 421
Query: 424 KNC-LHSNLERLLRQLTACCLERRSLSGEQGEAFQLQRVLSSGK 466
++C L S+LE+LLRQLT CCLERRSL+G+QGE F+L+ VL++ +
Sbjct: 422 RSCHLQSHLEKLLRQLTVCCLERRSLNGDQGETFRLRHVLNNNR 465
>UniRef100_Q9C8P9 Hypothetical protein F12A4.1 [Arabidopsis thaliana]
Length = 484
Score = 608 bits (1568), Expect = e-172
Identities = 300/481 (62%), Positives = 378/481 (78%), Gaps = 24/481 (4%)
Query: 5 NGKNTIEKPDQDQNQDLGFHFLELSRRKAKLDSEDFEVDDEEKEESPSELNTINSSGGFV 64
N N + + QDL +FL+ R AK D+++ +E EE P+ELNTINS+GGF+
Sbjct: 9 NSANGDTTNNGENGQDLNLNFLDKIRLSAKRDAKE-----DEGEELPTELNTINSAGGFL 63
Query: 65 VVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIVYYFEIFVKDAGVKGQVSIGFTTES 124
VV+ DKLSVKYT+ NLHGHDVGV+QANK AP+K + YYFEIFVKD+G+KGQ++IGFT ES
Sbjct: 64 VVSPDKLSVKYTNTNLHGHDVGVVQANKPAPIKCLTYYFEIFVKDSGIKGQIAIGFTKES 123
Query: 125 FKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAFGPTYTTGDIVGAGINYAAQEFFFTKNG 184
FKMRRQPGWE NSCGYHGDDG+LYRGQGKGE FGP +T D VG GINYA+QEFFFTKNG
Sbjct: 124 FKMRRQPGWEVNSCGYHGDDGYLYRGQGKGEPFGPKFTKDDAVGGGINYASQEFFFTKNG 183
Query: 185 QVVGTVFKEMKGPLFPTVAVHSQNEEVHVNFGQKPFTFDLK-----------------EY 227
+VG + K+++G LFPTVAVHSQNEEV VNFG+K F FD+K Y
Sbjct: 184 TIVGKIPKDIRGHLFPTVAVHSQNEEVLVNFGKKKFAFDIKVCLMSVLGTGVTESIAVGY 243
Query: 228 EAQERMKQQIKIEEVPVSPNASYGIVRSYLLHYGYEDTLNSFDEASRSTIPPINIVQENG 287
EA ER KQQ+ IE++ + PN YG+V++YLLHYGYE+TL++F+ A+++T+PPI+I QEN
Sbjct: 244 EASERNKQQLAIEKISIPPNIGYGLVKTYLLHYGYEETLDAFNLATKNTVPPIHIDQENA 303
Query: 288 IDDQETTYALNHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELV 347
ID+ +++YAL RK LRQL+R+GEID A +L++ YPQI +D+ S +CFLLHCQKFIELV
Sbjct: 304 IDEDDSSYALKQRKNLRQLVRNGEIDTALAELQKLYPQIVQDDKSVVCFLLHCQKFIELV 363
Query: 348 RVGALEEAVKYGRIELSSFFGLSLFEDIVQDCVALLAYERPLESAVGYLLKDSQREVVAD 407
RVG LEE V YGR+EL+ F GL+ F+DIV+DC ALLAYE+P ES+V Y L+DSQRE+VAD
Sbjct: 364 RVGKLEEGVNYGRLELAKFVGLTGFQDIVEDCFALLAYEKPEESSVWYFLEDSQRELVAD 423
Query: 408 AVNAMILSTNPNIK-VTKNC-LHSNLERLLRQLTACCLERRSLSGEQGEAFQLQRVLSSG 465
AVNA ILSTNPN K V ++C L S+LE+LLRQLT CCLERRSL+G+QGE F+L+ VL++
Sbjct: 424 AVNAAILSTNPNKKDVQRSCHLQSHLEKLLRQLTVCCLERRSLNGDQGETFRLRHVLNNN 483
Query: 466 K 466
+
Sbjct: 484 R 484
>UniRef100_Q8LF14 Hypothetical protein [Arabidopsis thaliana]
Length = 429
Score = 582 bits (1501), Expect = e-165
Identities = 287/420 (68%), Positives = 342/420 (81%), Gaps = 5/420 (1%)
Query: 43 DDEEKEESPSELNTINSSGGFVVVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIVYY 102
D+EE+EESP+ELNTI GFVV++ DKLS+KYT V+ HGHDVGV+QANK AP KR YY
Sbjct: 3 DNEEEEESPTELNTITGLSGFVVISPDKLSLKYTGVSQHGHDVGVVQANKPAPCKRFAYY 62
Query: 103 FEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAFGPTYT 162
FEI+VKDAGVKG+VSIGFTT+SF++ R PGWE N+CGYHG+DG +Y G+ +GEAFGPTYT
Sbjct: 63 FEIYVKDAGVKGRVSIGFTTDSFRIARHPGWELNTCGYHGEDGLIYLGKRQGEAFGPTYT 122
Query: 163 TGDIVGAGINYAAQEFFFTKNGQVVGTVFKEMKGPLFPTVAVHSQNEEVHVNFGQKPFTF 222
TGD VG GINY +QEFFFT NG +VGTV K +KGPLFPTVAVHSQNEEV VNFGQ+ F F
Sbjct: 123 TGDTVGGGINYDSQEFFFTVNGTLVGTVSKYIKGPLFPTVAVHSQNEEVTVNFGQEKFAF 182
Query: 223 DLKEYEAQERMKQQIKIEEVPVSPNASYGIVRSYLLHYGYEDTLNSFDEASRSTIPPINI 282
D K YE+ ER KQQI IE++ + N S G+V++YLLHYGYE+T ++ D A+ ST+PPIN
Sbjct: 183 DFKGYESSERNKQQIAIEKISIPSNMSLGLVKNYLLHYGYEETHHALDLATNSTLPPING 242
Query: 283 VQENGIDDQETTYALNHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQK 342
QENGIDD T+YAL+ RK LRQLIR GEIDAA KLR+ YPQ+ +++ S +CFLLHCQK
Sbjct: 243 AQENGIDD--TSYALHERKILRQLIRKGEIDAALAKLRDSYPQLVQNDKSEVCFLLHCQK 300
Query: 343 FIELVRVGALEEAVKYGRIELSSFFGLSLFEDIVQDCVALLAYERPLESAVGYLLKDSQR 402
FIELVR+GALEEAVKYGR+EL+ F GL+ F+DI++DC ALL YERP ES VGY L+++QR
Sbjct: 301 FIELVRIGALEEAVKYGRMELAKFIGLTTFQDILEDCFALLVYERPEESNVGYFLEETQR 360
Query: 403 EVVADAVNAMILSTNPNIKVTKNCLHSNLERLLRQLTACCLERRSLSGEQGEAFQLQRVL 462
EVVAD VNA ILST P KN HS+LE LLRQLTACCLE RSL+ QGEAF L R+L
Sbjct: 361 EVVADTVNAAILSTKPK---GKNQSHSHLETLLRQLTACCLELRSLNDGQGEAFSLNRLL 417
>UniRef100_Q94BS7 Hypothetical protein At4g09340 [Arabidopsis thaliana]
Length = 447
Score = 582 bits (1499), Expect = e-164
Identities = 287/420 (68%), Positives = 341/420 (80%), Gaps = 5/420 (1%)
Query: 43 DDEEKEESPSELNTINSSGGFVVVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIVYY 102
D+EE+EESP+ELNTI GFVV++ DKLS+KYT V+ HGHDVGV+QANK AP KR YY
Sbjct: 21 DNEEEEESPTELNTITGLSGFVVISPDKLSLKYTGVSQHGHDVGVVQANKPAPCKRFAYY 80
Query: 103 FEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAFGPTYT 162
FEI+VKDAGVKG+VSIGFTT SF++ R PGWE N+CGYHG+DG +Y G+ +GEAFGPTYT
Sbjct: 81 FEIYVKDAGVKGRVSIGFTTNSFRIARHPGWELNTCGYHGEDGLIYLGKRQGEAFGPTYT 140
Query: 163 TGDIVGAGINYAAQEFFFTKNGQVVGTVFKEMKGPLFPTVAVHSQNEEVHVNFGQKPFTF 222
TGD VG GINY +QEFFFT NG +VGTV K +KGPLFPTVAVHSQNEEV VNFGQ+ F F
Sbjct: 141 TGDTVGGGINYDSQEFFFTVNGTLVGTVSKYIKGPLFPTVAVHSQNEEVTVNFGQEKFAF 200
Query: 223 DLKEYEAQERMKQQIKIEEVPVSPNASYGIVRSYLLHYGYEDTLNSFDEASRSTIPPINI 282
D K YE+ ER KQQI IE++ + N S G+V++YLLHYGYE+T ++ D A+ ST+PPIN
Sbjct: 201 DFKGYESSERNKQQIAIEKISIPSNMSLGLVKNYLLHYGYEETHHALDLATNSTLPPING 260
Query: 283 VQENGIDDQETTYALNHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQK 342
QENGIDD T+YAL+ RK LRQLIR GEIDAA KLR+ YPQ+ +++ S +CFLLHCQK
Sbjct: 261 AQENGIDD--TSYALHERKILRQLIRKGEIDAALAKLRDSYPQLVQNDKSEVCFLLHCQK 318
Query: 343 FIELVRVGALEEAVKYGRIELSSFFGLSLFEDIVQDCVALLAYERPLESAVGYLLKDSQR 402
FIELVR+GALEEAVKYGR+EL+ F GL+ F+DI++DC ALL YERP ES VGY L+++QR
Sbjct: 319 FIELVRIGALEEAVKYGRMELAKFIGLTTFQDILEDCFALLVYERPEESNVGYFLEETQR 378
Query: 403 EVVADAVNAMILSTNPNIKVTKNCLHSNLERLLRQLTACCLERRSLSGEQGEAFQLQRVL 462
EVVAD VNA ILST P KN HS+LE LLRQLTACCLE RSL+ QGEAF L R+L
Sbjct: 379 EVVADTVNAAILSTKPK---GKNQSHSHLETLLRQLTACCLELRSLNDGQGEAFSLNRLL 435
>UniRef100_Q6ZI83 RanBPM-like [Oryza sativa]
Length = 469
Score = 541 bits (1394), Expect = e-152
Identities = 263/423 (62%), Positives = 331/423 (78%), Gaps = 3/423 (0%)
Query: 46 EKEESPSELNTINSSGGFVVVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIVYYFEI 105
E E +PSELNT NSSG F VV+TD++SV+Y VN HGHDVGV+QAN+ AP +R VYYFE+
Sbjct: 47 EAEPAPSELNTTNSSGLFAVVSTDRMSVRYLGVNQHGHDVGVVQANRPAPTRRAVYYFEM 106
Query: 106 FVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAFGPTYTTGD 165
VK+AG KGQ SIGFTTE+FKMRRQPGWE+NSCGYHGDDG+LYRG GK E+FGP +T+GD
Sbjct: 107 GVKNAGQKGQTSIGFTTENFKMRRQPGWESNSCGYHGDDGYLYRGPGKSESFGPKFTSGD 166
Query: 166 IVGAGINYAAQEFFFTKNGQVVGTVFKEMKGPLFPTVAVHSQNEEVHVNFGQKPFTFDLK 225
+GAGINY +QEFFFTKNG +VG+ KE+KGPL+PT+AVHSQ+EEV VNFG++PF FD++
Sbjct: 167 TIGAGINYFSQEFFFTKNGSLVGSFQKEIKGPLYPTIAVHSQDEEVTVNFGKEPFCFDIE 226
Query: 226 EYEAQERMKQQIKIEEVPVSPNASYGIVRSYLLHYGYEDTLNSFDEASRSTIPPINIVQE 285
Y +E+MKQQ +++ + P+ S+ IVRSYLLHYGY+DTLNSFD AS T PP N +
Sbjct: 227 GYIFEEKMKQQSVSDKLDLQPDISHWIVRSYLLHYGYQDTLNSFDMAS-ETDPPSN--HQ 283
Query: 286 NGIDDQETTYALNHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIE 345
NG + Y L+HRK LRQLI G+ID+AF KL EWYPQ+ +D TS +CFLLH Q+FIE
Sbjct: 284 NGYGEPPEMYGLSHRKLLRQLIMSGDIDSAFKKLGEWYPQVIKDETSIICFLLHSQRFIE 343
Query: 346 LVRVGALEEAVKYGRIELSSFFGLSLFEDIVQDCVALLAYERPLESAVGYLLKDSQREVV 405
+ G LE+AVKY R L++F F+ ++++ VALLAYE+P ES +GYLL QRE V
Sbjct: 344 FIGAGQLEDAVKYARSNLANFLTHKAFDGLLKESVALLAYEKPAESCIGYLLDSPQREFV 403
Query: 406 ADAVNAMILSTNPNIKVTKNCLHSNLERLLRQLTACCLERRSLSGEQGEAFQLQRVLSSG 465
ADAVNA +LSTNPN+K ++CL+S LE+LLRQLT C ERR+ SG+QG+AF L + + S
Sbjct: 404 ADAVNAAVLSTNPNMKDPESCLYSCLEKLLRQLTVCSFERRAFSGDQGDAFLLHKEVQSC 463
Query: 466 KRS 468
RS
Sbjct: 464 DRS 466
>UniRef100_Q9SMS1 Hypothetical protein AT4g09340 [Arabidopsis thaliana]
Length = 237
Score = 317 bits (813), Expect = 4e-85
Identities = 157/265 (59%), Positives = 192/265 (72%), Gaps = 31/265 (11%)
Query: 43 DDEEKEESPSELNTINSSGGFVVVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIVYY 102
D+EE+EESP+ELNTI GFVV++ DKLS+KYT V+ HGHDVGV+QANK AP KR YY
Sbjct: 3 DNEEEEESPTELNTITGLSGFVVISPDKLSLKYTGVSQHGHDVGVVQANKPAPCKRFAYY 62
Query: 103 FEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAFGPTYT 162
FEI+VKDAGVKG+VSIGFTT SF++ R PGWE N+CGYHG+DG +Y G+ +GEAFGPTYT
Sbjct: 63 FEIYVKDAGVKGRVSIGFTTNSFRIARHPGWELNTCGYHGEDGLIYLGKRQGEAFGPTYT 122
Query: 163 TGDIVGAGINYAAQEFFFTKNGQVVGTVFKEMKGPLFPTVAVHSQNEEVHVNFGQKPFTF 222
TGD VG GINY +QEFFFT V VNFGQ+ F F
Sbjct: 123 TGDTVGGGINYDSQEFFFT-----------------------------VTVNFGQEKFAF 153
Query: 223 DLKEYEAQERMKQQIKIEEVPVSPNASYGIVRSYLLHYGYEDTLNSFDEASRSTIPPINI 282
D K YE+ ER KQQI IE++ + N S G+V++YLLHYGYE+T ++ D A+ ST+PPIN
Sbjct: 154 DFKGYESSERNKQQIAIEKISIPSNMSLGLVKNYLLHYGYEETHHALDLATNSTLPPING 213
Query: 283 VQENGIDDQETTYALNHRKTLRQLI 307
QENGIDD T+YAL+ RK LRQ++
Sbjct: 214 AQENGIDD--TSYALHERKILRQVL 236
>UniRef100_Q8RX25 Hypothetical protein At4g09200 [Arabidopsis thaliana]
Length = 397
Score = 302 bits (773), Expect = 2e-80
Identities = 173/408 (42%), Positives = 249/408 (60%), Gaps = 24/408 (5%)
Query: 41 EVDDEEKEESPSELNTINSSGGFVVVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIV 100
E ++EE+E P+ L T+ S F V+ D LS +YT + HD V+Q AP +
Sbjct: 2 EENEEEEELRPTALYTVGGSDEFTFVSPDGLSGQYTGPD---HDGVVLQTENPAPTNCLA 58
Query: 101 YYFEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAF--- 157
YYFE+ + +AG KG+++IGF+ E G+ SC Y G+ G + G G+
Sbjct: 59 YYFEVHIMNAGEKGEIAIGFSKEHIYSE---GYTVRSCAYIGNSGLICSQNGDGDRTVAK 115
Query: 158 -GPTYTTGDIVGAGINYAAQEFFFTKNGQVVGTVFKEMKGPLFPTVAVHSQNEEVHVNF- 215
TYTTGD+VG GI+ +QEFFFTKNG +VGT+ ++ + P++PT+ +HSQNE V VNF
Sbjct: 116 ASDTYTTGDLVGCGIDSVSQEFFFTKNGTIVGTIPRQFRRPVYPTIVLHSQNEAVTVNFG 175
Query: 216 GQKPFTFDLKEYEAQERMKQQIKIEEVPVSPNASYGIVRSYLLHYGYEDTLNSFD-EASR 274
GQ F+FD + +E R+K++ +IE++ +S + S+G+V++YLL YGYED+ +F+ ASR
Sbjct: 176 GQIAFSFDFEHFEESLRVKKEREIEKISMSRSISHGLVKTYLLRYGYEDSFRAFNLAASR 235
Query: 275 STIPPINIVQENGIDDQETTYALNHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAM 334
T+ I QEN D+ Y L+ RK LR+LI EID A L++ YPQ+ E + A
Sbjct: 236 HTV----IAQENSFDE----YELHQRKKLRELIMTAEIDDAIAALKDRYPQLIEGGSEAY 287
Query: 335 CFLLHCQKFIELVRVGALEEAVKYGRIELSSFFGLSLFEDIVQDCVALLAYERPLESAVG 394
FLL CQK +ELVR GA+ EA +G + F SL +++ DC AL E ES
Sbjct: 288 -FLLICQKIVELVRKGAIAEAKSFGNQDFKDFRDSSLLKNLFDDCSALFECETIEESGAA 346
Query: 395 YLLKDSQREVVADAVNAMILSTNPNIKVTKNCLHSNLERLLRQLTACC 442
YLL ++Q+ +VA AV+ ILSTNP T++ ++LER+LRQ A C
Sbjct: 347 YLLGETQKNIVAAAVSEAILSTNP---ATRDQQSASLERVLRQFEATC 391
>UniRef100_Q9SMS9 Hypothetical protein AT4g09250 [Arabidopsis thaliana]
Length = 427
Score = 250 bits (639), Expect = 5e-65
Identities = 155/411 (37%), Positives = 223/411 (53%), Gaps = 72/411 (17%)
Query: 38 EDFEVDDEEKEESPSELNTINSSGGFVVVATDKLSVKYTSVNLHGHDVGVIQANKVAPMK 97
E E ++EE+E P+ L T+ S F V+ D LS +YT + HD V+Q AP
Sbjct: 77 EAMEENEEEEELRPTALYTVGGSDEFTFVSPDGLSGQYTGPD---HDGVVLQTENPAPTN 133
Query: 98 RIVYYFEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAF 157
+ YYFE+ + +AG KG+++IGF+ E G+ SC Y G+ G + G G+
Sbjct: 134 CLAYYFEVHIMNAGEKGEIAIGFSKEHIYSE---GYTVRSCAYIGNSGLICSQNGDGDRT 190
Query: 158 ----GPTYTTGDIVGAGINYAAQEFFFTKNGQVVGTVFKEMKGPLFPTVAVHSQNEEVHV 213
TYTTGD+VG GI+ +QEFFFTKNG +VGT+ ++ + P++PT+ +HSQNE V V
Sbjct: 191 VAKASDTYTTGDLVGCGIDSVSQEFFFTKNGTIVGTIPRQFRRPVYPTIVLHSQNEAVTV 250
Query: 214 NF-GQKPFTFDLKEYEAQERMKQQIKIEEVPVSPNASYGIVRSYLLHYGYEDTLNSFD-E 271
NF GQ F+FD + +E R+K++ +IE++ +S + S+G+V++YLL YGYED+ +F+
Sbjct: 251 NFGGQIAFSFDFEHFEESLRVKKEREIEKISMSRSISHGLVKTYLLRYGYEDSFRAFNLA 310
Query: 272 ASRSTIPPINIVQENGIDDQETTYALNHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNT 331
ASR T+ I QEN D+ Y L+ RK LR+LI EID A L++ YPQ+ ED
Sbjct: 311 ASRHTV----IAQENSFDE----YELHQRKKLRELIMTAEIDDAIAALKDRYPQLIED-- 360
Query: 332 SAMCFLLHCQKFIELVRVGALEEAVKYGRIELSSFFGLSLFEDIVQDCVALLAYERPLES 391
C AL E ES
Sbjct: 361 -----------------------------------------------CSALFECETIEES 373
Query: 392 AVGYLLKDSQREVVADAVNAMILSTNPNIKVTKNCLHSNLERLLRQLTACC 442
YLL ++Q+ +VA AV+ ILSTNP T++ ++LER+LRQ A C
Sbjct: 374 GAAYLLGETQKNIVAAAVSEAILSTNP---ATRDQQSASLERVLRQFEATC 421
>UniRef100_Q9SMS3 Hypothetical protein AT4g09310 [Arabidopsis thaliana]
Length = 349
Score = 249 bits (636), Expect = 1e-64
Identities = 154/408 (37%), Positives = 222/408 (53%), Gaps = 72/408 (17%)
Query: 41 EVDDEEKEESPSELNTINSSGGFVVVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIV 100
E ++EE+E P+ L T+ S F V+ D LS +YT + HD V+Q AP +
Sbjct: 2 EENEEEEELRPTALYTVGGSDEFTFVSPDGLSGQYTGPD---HDGVVLQTENPAPTNCLA 58
Query: 101 YYFEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAF--- 157
YYFE+ + +AG KG+++IGF+ E G+ SC Y G+ G + G G+
Sbjct: 59 YYFEVHIMNAGEKGEIAIGFSKEHIYSE---GYTVRSCAYIGNSGLICSQNGDGDRTVAK 115
Query: 158 -GPTYTTGDIVGAGINYAAQEFFFTKNGQVVGTVFKEMKGPLFPTVAVHSQNEEVHVNF- 215
TYTTGD+VG GI+ +QEFFFTKNG +VGT+ ++ + P++PT+ +HSQNE V VNF
Sbjct: 116 ASDTYTTGDLVGCGIDSVSQEFFFTKNGTIVGTIPRQFRRPVYPTIVLHSQNEAVTVNFG 175
Query: 216 GQKPFTFDLKEYEAQERMKQQIKIEEVPVSPNASYGIVRSYLLHYGYEDTLNSFD-EASR 274
GQ F+FD + +E R+K++ +IE++ +S + S+G+V++YLL YGYED+ +F+ ASR
Sbjct: 176 GQIAFSFDFEHFEESLRVKKEREIEKISMSRSISHGLVKTYLLRYGYEDSFRAFNLAASR 235
Query: 275 STIPPINIVQENGIDDQETTYALNHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAM 334
T+ I QEN D+ Y L+ RK LR+LI EID A L++ YPQ+ ED
Sbjct: 236 HTV----IAQENSFDE----YELHQRKKLRELIMTAEIDDAIAALKDRYPQLIED----- 282
Query: 335 CFLLHCQKFIELVRVGALEEAVKYGRIELSSFFGLSLFEDIVQDCVALLAYERPLESAVG 394
C AL E ES
Sbjct: 283 --------------------------------------------CSALFECETIEESGAA 298
Query: 395 YLLKDSQREVVADAVNAMILSTNPNIKVTKNCLHSNLERLLRQLTACC 442
YLL ++Q+ +VA AV+ ILSTNP T++ ++LER+LRQ A C
Sbjct: 299 YLLGETQKNIVAAAVSEAILSTNP---ATRDQQSASLERVLRQFEATC 343
>UniRef100_Q9M0Q8 Hypothetical protein AT4g09200 [Arabidopsis thaliana]
Length = 377
Score = 225 bits (573), Expect = 2e-57
Identities = 159/442 (35%), Positives = 219/442 (48%), Gaps = 112/442 (25%)
Query: 41 EVDDEEKEESPSELNTINSSGGFVVVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIV 100
E ++EE+E P+ L T+ S F V+ D LS +YT + HD V+Q AP +
Sbjct: 2 EENEEEEELRPTALYTVGGSDEFTFVSPDGLSGQYTGPD---HDGVVLQTENPAPTNCLA 58
Query: 101 YYFEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAF--- 157
YYFE+ + +AG KG+++IGF+ E + SC Y G+ G + G G+
Sbjct: 59 YYFEVHIMNAGEKGEIAIGFSKEHI-------YSEGSCAYIGNSGLICSQNGDGDRTVAK 111
Query: 158 -GPTYTTGDIVGAGINYAAQEFFFTKNGQVVGTVFKEMKGPLFPTVAVHSQNEEVHVNF- 215
TYTTGD+VG GI+ +QEFFFTKNG +VGT+ ++ + P++PT+ +HSQNE V VNF
Sbjct: 112 ASDTYTTGDLVGCGIDSVSQEFFFTKNGTIVGTIPRQFRRPVYPTIVLHSQNEAVTVNFG 171
Query: 216 GQKPFTFDLKEYEAQERMKQQIKIEEVPVSPNASYGIVRSYLLHYGYEDTLNSFD-EASR 274
GQ F+FD E+P S +V++YLL YGYED+ +F+ ASR
Sbjct: 172 GQIAFSFDF----------------ELPFS-----RLVKTYLLRYGYEDSFRAFNLAASR 210
Query: 275 STIPPINIVQENGIDDQETTYALNHRKTLRQ----------------------------- 305
T+ I QEN D+ Y L+ RK LR+
Sbjct: 211 HTV----IAQENSFDE----YELHQRKKLREVLFTAMVQNQPKASYIATSFLFHIELIHA 262
Query: 306 -----LIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELVRVGALEEAVKYGR 360
LI EID A L++ YPQ+ E + A FLL CQK +ELVR
Sbjct: 263 FCLLKLIMTAEIDDAIAALKDRYPQLIEGGSEAY-FLLICQKIVELVR------------ 309
Query: 361 IELSSFFGLSLFEDIVQDCVALLAYERPLESAVGYLLKDSQREVVADAVNAMILSTNPNI 420
DC AL E ES YLL ++Q+ +VA AV+ ILSTNP
Sbjct: 310 -----------------DCSALFECETIEESGAAYLLGETQKNIVAAAVSEAILSTNP-- 350
Query: 421 KVTKNCLHSNLERLLRQLTACC 442
T++ ++LER+LRQ A C
Sbjct: 351 -ATRDQQSASLERVLRQFEATC 371
>UniRef100_UPI000023D443 UPI000023D443 UniRef100 entry
Length = 674
Score = 182 bits (463), Expect = 1e-44
Identities = 142/465 (30%), Positives = 217/465 (46%), Gaps = 76/465 (16%)
Query: 43 DDEEKEESPSELNTINSSGGFVVVATDKLSVKYTSV-NLHG--HDVGVIQANKVAPMKRI 99
++++ P+ N + G + V D LSVK+ N H H+ I+A+ P +
Sbjct: 192 ENDDLAPLPTRWNKEDQMTG-IEVQQDGLSVKHVGPKNQHDRDHEASAIRADHYMPPQCG 250
Query: 100 VYYFEIFVKDAGVK---GQVSIGFTTESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEA 156
+YYFE+ + VK VSIGF+T++ + R GWE S GYHGDDG + GQ G+
Sbjct: 251 IYYFEVQI--LAVKRDDATVSIGFSTKTAALSRPVGWEPESWGYHGDDGRCFTGQNIGKH 308
Query: 157 FGPTYTTGDIVGAGINYAAQEFFFTKNGQVVGTVFKEM-KGPLFPTVAVHSQNEEVHVNF 215
FGP Y D++G G+N+ FFTKNG +GT F ++ +G L+PT+ + E V VNF
Sbjct: 309 FGPVYNVNDVIGCGVNFKENSAFFTKNGVKIGTAFHDVGRGKLYPTIGLKKPGEFVRVNF 368
Query: 216 GQKPFTFDLKEYEAQERMKQQIKIEEV---PVSPNAS-----YGIVRSYLLHYGYEDTLN 267
GQ PF +++ + +ER K + I+ + P+ S +V +L H GY +T
Sbjct: 369 GQTPFVYNIDDMMREEREKVRKAIQATDGSAIQPDWSETDLIQHLVLQFLQHDGYVETAR 428
Query: 268 SFDE-----ASRSTIPPINIVQENGIDDQETTYALNHRKTLRQLIRDGEIDAAFGKLREW 322
+F E + P +V+ D E N+R+ +R+ I G+ID A +
Sbjct: 429 AFAEDMAIQKEALNLDPNTVVEHQTDRDDEDA---NNRQRIRRAILGGDIDRALKYTNAY 485
Query: 323 YPQIAEDNTSAMCFLLHCQKFIELVRVGAL------------------------------ 352
YP + EDN + F L C+KFIELVR A
Sbjct: 486 YPHVLEDN-DHVYFKLRCRKFIELVRKAAQLSMLNEAKGNKHVNNQTMDIDLNGSENSSW 544
Query: 353 -----EEAVKYGRIELSSF-FGLSLFEDIVQD-----------CVALLAYERPL-ESAVG 394
E A + +E S +G L E+ D AL+AY+ PL E V
Sbjct: 545 ETDGGENATELAELERSMLEYGQKLQEEYANDPRVEVSKALEEIWALVAYQNPLKEPQVS 604
Query: 395 YLLKDSQREVVADAVN-AMILSTNPNIKVTKNCLHSNLERLLRQL 438
+LL + R VA+ +N A++LS + + +++ LL +L
Sbjct: 605 HLLDRNGRVTVAEELNSAILLSLGKSSRAALELIYAQNTVLLEEL 649
>UniRef100_Q69ZJ1 MKIAA1464 protein [Mus musculus]
Length = 613
Score = 179 bits (455), Expect = 1e-43
Identities = 94/291 (32%), Positives = 155/291 (52%), Gaps = 20/291 (6%)
Query: 63 FVVVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIVYYFEIFVKDAGVKGQVSIGFTT 122
++ ++ L V Y + D ++A P +YYFE+ + G G + IG +
Sbjct: 58 YIGLSQGNLRVHYKGHGKNHKDAASVRATHPIPAACGIYYFEVKIVSKGRDGYMGIGLSA 117
Query: 123 ESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAFGPTYTTGDIVGAGINYAAQEFFFTK 182
+ M R PGW+ +S GYHGDDG + G G+ +GPT+TTGD++G +N F+TK
Sbjct: 118 QGVNMNRLPGWDKHSYGYHGDDGHSFCSSGTGQPYGPTFTTGDVIGCCVNLINGTCFYTK 177
Query: 183 NGQVVGTVFKEMKGPLFPTVAVHSQNEEVHVNFGQKPFTFDLKEYEAQERMKQQIKIEEV 242
NG +G F ++ L+PTV + + E V NFGQ+PF FD+++Y + R K Q +
Sbjct: 178 NGHSLGIAFTDLPANLYPTVGLQTPGEIVDANFGQQPFLFDIEDYMREWRAKVQGTVHGF 237
Query: 243 PVSPNAS------YGIVRSYLLHYGYEDTLNSFDEASRSTIPPINIVQENGIDDQETTYA 296
P+S +V SYL+H+GY T +F +R T PI QE +
Sbjct: 238 PISARLGEWQAVLQNMVSSYLVHHGYCSTATAF---ARMTETPI----------QEEQAS 284
Query: 297 LNHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELV 347
+ +R+ +++L+ +G + A + +YP + E N + + F+L C++F+E+V
Sbjct: 285 IKNRQKIQKLVLEGRVGEAIETTQRFYPGLLEHNPN-LLFMLKCRQFVEMV 334
Score = 36.2 bits (82), Expect = 2.1
Identities = 25/73 (34%), Positives = 38/73 (51%), Gaps = 7/73 (9%)
Query: 351 ALEEAVKYGRI------ELSSFFGLSLFE-DIVQDCVALLAYERPLESAVGYLLKDSQRE 403
A E + +GR +L +G +L +++QD +LLAY P VG+ L QRE
Sbjct: 500 ATERIILFGRELQALSEQLGREYGKNLAHTEMLQDAFSLLAYSDPWSCPVGHQLDPIQRE 559
Query: 404 VVADAVNAMILST 416
V A+N+ IL +
Sbjct: 560 PVCAALNSAILES 572
>UniRef100_Q6VN19 Ran-binding protein 10 [Mus musculus]
Length = 648
Score = 179 bits (455), Expect = 1e-43
Identities = 94/291 (32%), Positives = 155/291 (52%), Gaps = 20/291 (6%)
Query: 63 FVVVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIVYYFEIFVKDAGVKGQVSIGFTT 122
++ ++ L V Y + D ++A P +YYFE+ + G G + IG +
Sbjct: 93 YIGLSQGNLRVHYKGHGKNHKDAASVRATHPIPAACGIYYFEVKIVSKGRDGYMGIGLSA 152
Query: 123 ESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAFGPTYTTGDIVGAGINYAAQEFFFTK 182
+ M R PGW+ +S GYHGDDG + G G+ +GPT+TTGD++G +N F+TK
Sbjct: 153 QGVNMNRLPGWDKHSYGYHGDDGHSFCSSGTGQPYGPTFTTGDVIGCCVNLINGTCFYTK 212
Query: 183 NGQVVGTVFKEMKGPLFPTVAVHSQNEEVHVNFGQKPFTFDLKEYEAQERMKQQIKIEEV 242
NG +G F ++ L+PTV + + E V NFGQ+PF FD+++Y + R K Q +
Sbjct: 213 NGHSLGIAFTDLPANLYPTVGLQTPGEIVDANFGQQPFLFDIEDYMREWRAKVQGTVHGF 272
Query: 243 PVSPNAS------YGIVRSYLLHYGYEDTLNSFDEASRSTIPPINIVQENGIDDQETTYA 296
P+S +V SYL+H+GY T +F +R T PI QE +
Sbjct: 273 PISARLGEWQAVLQNMVSSYLVHHGYCSTATAF---ARMTETPI----------QEEQAS 319
Query: 297 LNHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELV 347
+ +R+ +++L+ +G + A + +YP + E N + + F+L C++F+E+V
Sbjct: 320 IKNRQKIQKLVLEGRVGEAIETTQRFYPGLLEHNPN-LLFMLKCRQFVEMV 369
Score = 36.2 bits (82), Expect = 2.1
Identities = 25/73 (34%), Positives = 38/73 (51%), Gaps = 7/73 (9%)
Query: 351 ALEEAVKYGRI------ELSSFFGLSLFE-DIVQDCVALLAYERPLESAVGYLLKDSQRE 403
A E + +GR +L +G +L +++QD +LLAY P VG+ L QRE
Sbjct: 535 ATERIILFGRELQALSEQLGREYGKNLAHTEMLQDAFSLLAYSDPWSCPVGHQLDPIQRE 594
Query: 404 VVADAVNAMILST 416
V A+N+ IL +
Sbjct: 595 PVCAALNSAILES 607
>UniRef100_Q9P264 KIAA1464 protein [Homo sapiens]
Length = 621
Score = 178 bits (452), Expect = 3e-43
Identities = 101/327 (30%), Positives = 169/327 (50%), Gaps = 25/327 (7%)
Query: 27 ELSRRKAKLDSEDFEVDDEEKEESPSELNTINSSGGFVVVATDKLSVKYTSVNLHGHDVG 86
ELSRR +L + ++++ P + ++ ++ L V Y + D
Sbjct: 35 ELSRRLQRL----YPAVNQQETPLPRSWSP-KDKYNYIGLSQGNLRVHYKGHGKNHKDAA 89
Query: 87 VIQANKVAPMKRIVYYFEIFVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGF 146
++A P +YYFE+ + G G + IG + + M R PGW+ +S GYHGDDG
Sbjct: 90 SVRATHPIPAACGIYYFEVKIVSKGRDGYMGIGLSAQGVNMNRLPGWDKHSYGYHGDDGH 149
Query: 147 LYRGQGKGEAFGPTYTTGDIVGAGINYAAQEFFFTKNGQVVGTVFKEMKGPLFPTVAVHS 206
+ G G+ +GPT+TTGD++G +N F+TKNG +G F ++ L+PTV + +
Sbjct: 150 SFCSSGTGQPYGPTFTTGDVIGCCVNLINGTCFYTKNGHSLGIAFTDLPANLYPTVGLQT 209
Query: 207 QNEEVHVNFGQKPFTFDLKEYEAQERMKQQIKIEEVPVSPNAS------YGIVRSYLLHY 260
E V NFGQ+PF FD+++Y + R K Q + P+S +V SYL+H+
Sbjct: 210 PGEIVDANFGQQPFLFDIEDYMREWRAKVQGTVHCFPISARLGEWQAVLQNMVSSYLVHH 269
Query: 261 GYEDTLNSFDEASRSTIPPINIVQENGIDDQETTYALNHRKTLRQLIRDGEIDAAFGKLR 320
GY T +F +R T PI QE ++ +R+ +++L+ +G + A +
Sbjct: 270 GYCATATAF---ARMTETPI----------QEEQASIKNRQKIQKLVLEGRVGEAIETTQ 316
Query: 321 EWYPQIAEDNTSAMCFLLHCQKFIELV 347
+YP + E N + + F+L C++F+E+V
Sbjct: 317 RFYPGLLEHNPN-LLFMLKCRQFVEMV 342
Score = 35.0 bits (79), Expect = 4.6
Identities = 25/73 (34%), Positives = 37/73 (50%), Gaps = 7/73 (9%)
Query: 351 ALEEAVKYGRI------ELSSFFGLSLFE-DIVQDCVALLAYERPLESAVGYLLKDSQRE 403
A E + +GR +L +G +L +++QD +LLAY P VG L QRE
Sbjct: 508 ATERIILFGRELQALSEQLGREYGKNLAHTEMLQDAFSLLAYSDPWSCPVGQQLDPIQRE 567
Query: 404 VVADAVNAMILST 416
V A+N+ IL +
Sbjct: 568 PVCAALNSAILES 580
>UniRef100_UPI0000437010 UPI0000437010 UniRef100 entry
Length = 557
Score = 177 bits (448), Expect = 8e-43
Identities = 91/291 (31%), Positives = 153/291 (52%), Gaps = 20/291 (6%)
Query: 63 FVVVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIVYYFEIFVKDAGVKGQVSIGFTT 122
++ ++ + L V Y + D ++A P +YYFE+ + G G + IG +
Sbjct: 37 YIGLSQNNLRVHYKGHGKNHKDAASVRATHPIPAACGIYYFEVKIVSKGRDGYMGIGLSA 96
Query: 123 ESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAFGPTYTTGDIVGAGINYAAQEFFFTK 182
+ M R PGW+ +S GYHGDDG + G G+ +GPT+TTGD++G +N F+TK
Sbjct: 97 QGVNMNRLPGWDKHSYGYHGDDGHSFCSSGTGQPYGPTFTTGDVIGCCVNLINNTCFYTK 156
Query: 183 NGQVVGTVFKEMKGPLFPTVAVHSQNEEVHVNFGQKPFTFDLKEYEAQERMKQQIKIEEV 242
NG +G F ++ L+PTV + + E V NFGQ+PF FD+++Y ++ R K I
Sbjct: 157 NGHSLGVAFTDLPPNLYPTVGLQTPGEIVDANFGQQPFVFDIEDYMSEWRAKIHSMIARF 216
Query: 243 PVSPNAS------YGIVRSYLLHYGYEDTLNSFDEASRSTIPPINIVQENGIDDQETTYA 296
P+ +V SYL+H+GY T +F A+ + I QE +
Sbjct: 217 PIGERLGDWQAVLQNMVSSYLVHHGYCATAMAFARATETMI-------------QEDQTS 263
Query: 297 LNHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELV 347
+ +R+ +++L+ G + A ++ YP + E N + + F+L C++F+E+V
Sbjct: 264 IKNRQRIQKLVLAGRVGEAIDATQQLYPGLLEHNPN-LLFMLKCRQFVEMV 313
Score = 35.0 bits (79), Expect = 4.6
Identities = 17/42 (40%), Positives = 24/42 (56%)
Query: 375 IVQDCVALLAYERPLESAVGYLLKDSQREVVADAVNAMILST 416
++QD +LLAY P VG L QRE + A+N+ IL +
Sbjct: 478 MLQDAFSLLAYSDPWNCPVGQQLDPMQREAICSALNSAILES 519
>UniRef100_UPI000043700F UPI000043700F UniRef100 entry
Length = 526
Score = 177 bits (448), Expect = 8e-43
Identities = 91/291 (31%), Positives = 153/291 (52%), Gaps = 20/291 (6%)
Query: 63 FVVVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIVYYFEIFVKDAGVKGQVSIGFTT 122
++ ++ + L V Y + D ++A P +YYFE+ + G G + IG +
Sbjct: 14 YIGLSQNNLRVHYKGHGKNHKDAASVRATHPIPAACGIYYFEVKIVSKGRDGYMGIGLSA 73
Query: 123 ESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAFGPTYTTGDIVGAGINYAAQEFFFTK 182
+ M R PGW+ +S GYHGDDG + G G+ +GPT+TTGD++G +N F+TK
Sbjct: 74 QGVNMNRLPGWDKHSYGYHGDDGHSFCSSGTGQPYGPTFTTGDVIGCCVNLINNTCFYTK 133
Query: 183 NGQVVGTVFKEMKGPLFPTVAVHSQNEEVHVNFGQKPFTFDLKEYEAQERMKQQIKIEEV 242
NG +G F ++ L+PTV + + E V NFGQ+PF FD+++Y ++ R K I
Sbjct: 134 NGHSLGVAFTDLPPNLYPTVGLQTPGEIVDANFGQQPFVFDIEDYMSEWRAKIHSMIARF 193
Query: 243 PVSPNAS------YGIVRSYLLHYGYEDTLNSFDEASRSTIPPINIVQENGIDDQETTYA 296
P+ +V SYL+H+GY T +F A+ + I QE +
Sbjct: 194 PIGERLGDWQAVLQNMVSSYLVHHGYCATAMAFARATETMI-------------QEDQTS 240
Query: 297 LNHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELV 347
+ +R+ +++L+ G + A ++ YP + E N + + F+L C++F+E+V
Sbjct: 241 IKNRQRIQKLVLAGRVGEAIDATQQLYPGLLEHNPN-LLFMLKCRQFVEMV 290
Score = 35.0 bits (79), Expect = 4.6
Identities = 17/42 (40%), Positives = 24/42 (56%)
Query: 375 IVQDCVALLAYERPLESAVGYLLKDSQREVVADAVNAMILST 416
++QD +LLAY P VG L QRE + A+N+ IL +
Sbjct: 444 MLQDAFSLLAYSDPWNCPVGQQLDPMQREAICSALNSAILES 485
>UniRef100_UPI000043700E UPI000043700E UniRef100 entry
Length = 598
Score = 177 bits (448), Expect = 8e-43
Identities = 91/291 (31%), Positives = 153/291 (52%), Gaps = 20/291 (6%)
Query: 63 FVVVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIVYYFEIFVKDAGVKGQVSIGFTT 122
++ ++ + L V Y + D ++A P +YYFE+ + G G + IG +
Sbjct: 42 YIGLSQNNLRVHYKGHGKNHKDAASVRATHPIPAACGIYYFEVKIVSKGRDGYMGIGLSA 101
Query: 123 ESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAFGPTYTTGDIVGAGINYAAQEFFFTK 182
+ M R PGW+ +S GYHGDDG + G G+ +GPT+TTGD++G +N F+TK
Sbjct: 102 QGVNMNRLPGWDKHSYGYHGDDGHSFCSSGTGQPYGPTFTTGDVIGCCVNLINNTCFYTK 161
Query: 183 NGQVVGTVFKEMKGPLFPTVAVHSQNEEVHVNFGQKPFTFDLKEYEAQERMKQQIKIEEV 242
NG +G F ++ L+PTV + + E V NFGQ+PF FD+++Y ++ R K I
Sbjct: 162 NGHSLGVAFTDLPPNLYPTVGLQTPGEIVDANFGQQPFVFDIEDYMSEWRAKIHSMIARF 221
Query: 243 PVSPNAS------YGIVRSYLLHYGYEDTLNSFDEASRSTIPPINIVQENGIDDQETTYA 296
P+ +V SYL+H+GY T +F A+ + I QE +
Sbjct: 222 PIGERLGDWQAVLQNMVSSYLVHHGYCATAMAFARATETMI-------------QEDQTS 268
Query: 297 LNHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELV 347
+ +R+ +++L+ G + A ++ YP + E N + + F+L C++F+E+V
Sbjct: 269 IKNRQRIQKLVLAGRVGEAIDATQQLYPGLLEHNPN-LLFMLKCRQFVEMV 318
Score = 35.0 bits (79), Expect = 4.6
Identities = 17/42 (40%), Positives = 24/42 (56%)
Query: 375 IVQDCVALLAYERPLESAVGYLLKDSQREVVADAVNAMILST 416
++QD +LLAY P VG L QRE + A+N+ IL +
Sbjct: 516 MLQDAFSLLAYSDPWNCPVGQQLDPMQREAICSALNSAILES 557
>UniRef100_UPI000021B34D UPI000021B34D UniRef100 entry
Length = 759
Score = 176 bits (447), Expect = 1e-42
Identities = 110/351 (31%), Positives = 182/351 (51%), Gaps = 18/351 (5%)
Query: 51 PSELNTINSSGGFVVVATDKLSVKYTSVNL----HGHDVGVIQANKVAPMKRIVYYFEIF 106
P+E N + G VVA D L VK + H+ I+A+ P + +YYFE+
Sbjct: 283 PTEWNCNDKYGNIEVVA-DGLEVKLSGPRPPDRERDHEAYAIRADHFMPPECGIYYFEVT 341
Query: 107 V-KDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAFGPTYTTGD 165
+ + +++GF+ + ++ R PGWEA S GYHGDDG + G+ +GP Y TGD
Sbjct: 342 ILASKREECTIAVGFSGKQVQLSRAPGWEAESWGYHGDDGRSFATSNVGKDYGPKYGTGD 401
Query: 166 IVGAGINYAAQEFFFTKNGQVVGTVFKEMKGPLFPTVAVHSQNEEVHVNFGQKPFTFDLK 225
++G G+N+ F+TKNGQ +GT F+++KG ++P V + + V NFGQ PF F +
Sbjct: 402 VIGCGVNFRTGTAFYTKNGQNLGTAFRDIKGKMYPVVGLRKTGDHVRTNFGQAPFVFAID 461
Query: 226 EYEAQERMKQQIKIEEVPVSPNASYG-------IVRSYLLHYGYEDTLNSFDEASRSTIP 278
ER + + +IE + + G +V +L H GY ++ +F++ +R+
Sbjct: 462 VLMESERSRIREEIELAKPAEISGMGETEFIQQLVVQFLQHDGYIESARAFNQETRAEKN 521
Query: 279 PINIVQENGI---DDQETTYALNHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAMC 335
+ + E+ + D + A+N R+ +R+ I +G+ID A + +YP++ +N + +
Sbjct: 522 ALALKSEDAVEITDIADDEDAIN-RQRIRKAILEGDIDRALKYTKAYYPRVLSENEN-VY 579
Query: 336 FLLHCQKFIELVRVGALEEAVKYGRIELSSFFGLSLFEDIVQDCVALLAYE 386
F L C+KFIE+V A + R S G+S +D D A L E
Sbjct: 580 FRLRCRKFIEMVLKEATQRLEAEKRYSNGSSNGISDHQDTDMDDGAALGTE 630
>UniRef100_Q7PPZ9 ENSANGP00000003407 [Anopheles gambiae str. PEST]
Length = 573
Score = 176 bits (446), Expect = 1e-42
Identities = 93/285 (32%), Positives = 149/285 (51%), Gaps = 20/285 (7%)
Query: 69 DKLSVKYTSVNLHGHDVGVIQANKVAPMKRIVYYFEIFVKDAGVKGQVSIGFTTESFKMR 128
+ L V Y + D ++ P +YYFE+ + G G + IG T +FKM
Sbjct: 36 NNLRVHYKGIGKSHTDAASVRTAYPIPAACGLYYFEVKIISKGRDGYMGIGLTHTNFKMN 95
Query: 129 RQPGWEANSCGYHGDDGFLYRGQGKGEAFGPTYTTGDIVGAGINYAAQEFFFTKNGQVVG 188
R PGW+ S GYHGDDG + G G+ +GPT+TTGDI+G G+N F+TKNG +G
Sbjct: 96 RLPGWDKQSYGYHGDDGHSFCSSGNGQPYGPTFTTGDIIGCGVNLVDNTCFYTKNGHHLG 155
Query: 189 TVFKEMKGPLFPTVAVHSQNEEVHVNFGQKPFTFDLKEYEAQERMKQQIKIEEVPVSPNA 248
F+++ L+PTV + + E V NFGQ+PF FD+++ + R + I P+ +
Sbjct: 156 IAFRDLPPRLYPTVGLQTPGEVVDANFGQEPFKFDIEDMLKELRASTKATIYNFPLPDDQ 215
Query: 249 S------YGIVRSYLLHYGYEDTLNSFDEASRSTIPPINIVQENGIDDQETTYALNHRKT 302
+ +V SYL+H+GY T +F ++ T+ QE ++ +R+
Sbjct: 216 GDWTVILHKMVSSYLVHHGYSSTAETFARSAGQTL-------------QEDMTSIKNRQK 262
Query: 303 LRQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLHCQKFIELV 347
+ +L+ G + A + YP + E N + + F+L C++FIE+V
Sbjct: 263 IIKLVLSGRMGQAIEQTIRLYPGLLESNQN-LLFMLKCRQFIEMV 306
Score = 35.8 bits (81), Expect = 2.7
Identities = 17/44 (38%), Positives = 28/44 (63%)
Query: 373 EDIVQDCVALLAYERPLESAVGYLLKDSQREVVADAVNAMILST 416
+ +++D +LLAY P S +G+ L S+RE V A+N+ IL +
Sbjct: 487 QKMLEDAFSLLAYSNPWASPLGWQLCPSRREAVCAALNSAILES 530
>UniRef100_UPI0000361052 UPI0000361052 UniRef100 entry
Length = 538
Score = 174 bits (442), Expect = 4e-42
Identities = 95/308 (30%), Positives = 160/308 (51%), Gaps = 21/308 (6%)
Query: 47 KEESPSELN-TINSSGGFVVVATDKLSVKYTSVNLHGHDVGVIQANKVAPMKRIVYYFEI 105
+EESP + + ++ ++ + L V Y + D ++A P +YYFE+
Sbjct: 25 EEESPLPRSWSPKDKYSYIGLSQNNLRVHYKGHGKNHKDAASVRATYPIPAACGIYYFEV 84
Query: 106 FVKDAGVKGQVSIGFTTESFKMRRQPGWEANSCGYHGDDGFLYRGQGKGEAFGPTYTTGD 165
+ G G + IG + + M R PGW+ +S GYHGDDG + G G+ +GPT+TTGD
Sbjct: 85 KIVSKGRDGYMGIGLSAQGVNMNRLPGWDKHSYGYHGDDGHSFCSSGTGQPYGPTFTTGD 144
Query: 166 IVGAGINYAAQEFFFTKNGQVVGTVFKEMKGPLFPTVAVHSQNEEVHVNFGQKPFTFDLK 225
++G +N F+TKNG +G F ++ L+PTV + + E V NFGQ+PF FD++
Sbjct: 145 VIGCCVNLINNTCFYTKNGISLGVAFTDLPPNLYPTVGLQTPGEIVDANFGQQPFVFDIE 204
Query: 226 EYEAQERMKQQIKIEEVPVSPNAS------YGIVRSYLLHYGYEDTLNSFDEASRSTIPP 279
+Y ++ R K I P+ +V SYL+H+GY T +F A+ + I
Sbjct: 205 DYMSEWRAKIHGMIARFPIGERLGEWQTVLQNMVSSYLVHHGYCATATAFARATETKI-- 262
Query: 280 INIVQENGIDDQETTYALNHRKTLRQLIRDGEIDAAFGKLREWYPQIAEDNTSAMCFLLH 339
QE ++ +R+ +++L+ G + A ++ YP + E N + + F+L
Sbjct: 263 -----------QEDQTSIKNRQRIQKLVLAGRVGEAIQVTQQLYPGLLEHNPN-LLFVLK 310
Query: 340 CQKFIELV 347
C++F+E+V
Sbjct: 311 CRQFVEMV 318
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.135 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 783,636,665
Number of Sequences: 2790947
Number of extensions: 33952345
Number of successful extensions: 101748
Number of sequences better than 10.0: 339
Number of HSP's better than 10.0 without gapping: 129
Number of HSP's successfully gapped in prelim test: 210
Number of HSP's that attempted gapping in prelim test: 101105
Number of HSP's gapped (non-prelim): 574
length of query: 468
length of database: 848,049,833
effective HSP length: 131
effective length of query: 337
effective length of database: 482,435,776
effective search space: 162580856512
effective search space used: 162580856512
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 77 (34.3 bits)
Medicago: description of AC146527.1


