

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146343.8 + phase: 0 /pseudo
(379 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
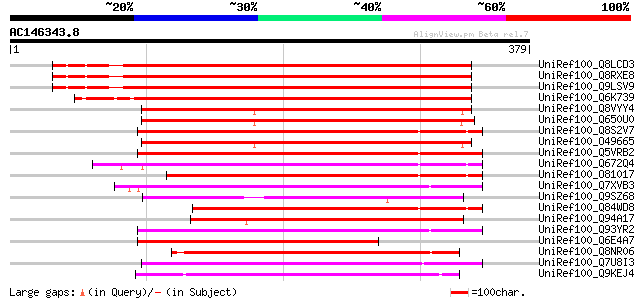
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LCD3 Hypothetical protein [Arabidopsis thaliana] 410 e-113
UniRef100_Q8RXE8 Hypothetical protein At3g25410 [Arabidopsis tha... 409 e-113
UniRef100_Q9LSV9 Similarity to transporter protein [Arabidopsis ... 409 e-113
UniRef100_Q6K739 Bile acid:sodium symporter-like [Oryza sativa] 386 e-106
UniRef100_Q8VYY4 Hypothetical protein At4g22840 [Arabidopsis tha... 201 3e-50
UniRef100_Q650U0 Hypothetical protein P0669G04.12 [Oryza sativa] 196 9e-49
UniRef100_Q8S2V7 Putative sodium-dependent bile acid symporter [... 194 3e-48
UniRef100_O49665 Predicted protein [Arabidopsis thaliana] 193 8e-48
UniRef100_Q5VRB2 Na(+) dependent transporter-like [Oryza sativa] 192 1e-47
UniRef100_Q672Q4 Putative anion:sodium symporter precursor [Lyco... 184 4e-45
UniRef100_O81017 Putative Na+ dependent ileal bile acid transpor... 183 8e-45
UniRef100_Q7XVB3 OSJNBa0072D21.3 protein [Oryza sativa] 172 1e-41
UniRef100_Q9SZ68 Putative transport protein [Arabidopsis thaliana] 170 5e-41
UniRef100_Q84WD8 Putative Na+ dependent ileal bile acid transpor... 170 5e-41
UniRef100_Q94A17 AT4g12030/F16J13_100 [Arabidopsis thaliana] 164 5e-39
UniRef100_Q93YR2 Hypothetical protein T30F21.11 [Arabidopsis tha... 154 4e-36
UniRef100_Q6E4A7 Hypothetical protein [Cynodon dactylon] 150 4e-35
UniRef100_Q8NR06 Predicted Na+-dependent transporter [Corynebact... 150 7e-35
UniRef100_Q7U8I3 Sodium/bile acid cotransporter family [Synechoc... 148 3e-34
UniRef100_Q9KEJ4 Sodium-dependent transporter [Bacillus halodurans] 145 1e-33
>UniRef100_Q8LCD3 Hypothetical protein [Arabidopsis thaliana]
Length = 431
Score = 410 bits (1055), Expect = e-113
Identities = 218/307 (71%), Positives = 252/307 (82%), Gaps = 12/307 (3%)
Query: 32 FSLQRVSKGCSISSFACSTSKLHGGRVGFNHREGKTSFLKFGVDESVVGGGGNGDVGER- 90
F L+R + G + ACST+ G RVG + R+G S L F G DV ER
Sbjct: 57 FRLRRRNSGL-VPVVACSTTPFMG-RVGLHWRDGNMSLLSFC---------GGTDVTERA 105
Query: 91 DWSQILSALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAF 150
D SQ SALLPFVVA+TAVAALS P +FTWVSK+LYAPALGGIMLSIGI LS++DFALAF
Sbjct: 106 DSSQFWSALLPFVVALTAVAALSYPPSFTWVSKDLYAPALGGIMLSIGIQLSVDDFALAF 165
Query: 151 KRPLPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTACVSGAQLSSYASFISKGD 210
KRP+PLS+GF+AQYVLKP+LGVL+A AFG+PR FYAGF+LT CV+GAQLSSYAS +SK D
Sbjct: 166 KRPVPLSVGFVAQYVLKPLLGVLVANAFGMPRTFYAGFILTCCVAGAQLSSYASSLSKAD 225
Query: 211 VALCIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYA 270
VA+ I+LTS TTIASVI TPLL+GLLIGSVVPVDAVAMSKSILQVVL P+TLGL+LNTYA
Sbjct: 226 VAMSILLTSSTTIASVIFTPLLSGLLIGSVVPVDAVAMSKSILQVVLVPITLGLVLNTYA 285
Query: 271 KPVVSILRPVMPFVAMICTSLCIGSPLAINRSQILSGEGLRLVAPVLIFHAAAFTLGYWF 330
KPVV++L+PVMPFVAM+CTSLCIGSPL+INRSQILS EGL L+ P++ FHA AF LGYWF
Sbjct: 286 KPVVTLLQPVMPFVAMVCTSLCIGSPLSINRSQILSAEGLGLIVPIVTFHAVAFALGYWF 345
Query: 331 SNLPSLR 337
S +P LR
Sbjct: 346 SKIPGLR 352
>UniRef100_Q8RXE8 Hypothetical protein At3g25410 [Arabidopsis thaliana]
Length = 431
Score = 409 bits (1052), Expect = e-113
Identities = 217/307 (70%), Positives = 252/307 (81%), Gaps = 12/307 (3%)
Query: 32 FSLQRVSKGCSISSFACSTSKLHGGRVGFNHREGKTSFLKFGVDESVVGGGGNGDVGER- 90
F L+R + G + ACST+ G RVG + R+G S L F G DV E+
Sbjct: 57 FRLRRRNSGL-VPVVACSTTPFMG-RVGLHWRDGNMSLLSFC---------GGTDVTEKA 105
Query: 91 DWSQILSALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAF 150
D SQ SALLPFVVA+TAVAALS P +FTWVSK+LYAPALGGIMLSIGI LS++DFALAF
Sbjct: 106 DSSQFWSALLPFVVALTAVAALSYPPSFTWVSKDLYAPALGGIMLSIGIQLSVDDFALAF 165
Query: 151 KRPLPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTACVSGAQLSSYASFISKGD 210
KRP+PLS+GF+AQYVLKP+LGVL+A AFG+PR FYAGF+LT CV+GAQLSSYAS +SK D
Sbjct: 166 KRPVPLSVGFVAQYVLKPLLGVLVANAFGMPRTFYAGFILTCCVAGAQLSSYASSLSKAD 225
Query: 211 VALCIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYA 270
VA+ I+LTS TTIASVI TPLL+GLLIGSVVPVDAVAMSKSILQVVL P+TLGL+LNTYA
Sbjct: 226 VAMSILLTSSTTIASVIFTPLLSGLLIGSVVPVDAVAMSKSILQVVLVPITLGLVLNTYA 285
Query: 271 KPVVSILRPVMPFVAMICTSLCIGSPLAINRSQILSGEGLRLVAPVLIFHAAAFTLGYWF 330
KPVV++L+PVMPFVAM+CTSLCIGSPL+INRSQILS EGL L+ P++ FHA AF LGYWF
Sbjct: 286 KPVVTLLQPVMPFVAMVCTSLCIGSPLSINRSQILSAEGLGLIVPIVTFHAVAFALGYWF 345
Query: 331 SNLPSLR 337
S +P LR
Sbjct: 346 SKIPGLR 352
>UniRef100_Q9LSV9 Similarity to transporter protein [Arabidopsis thaliana]
Length = 454
Score = 409 bits (1052), Expect = e-113
Identities = 217/307 (70%), Positives = 252/307 (81%), Gaps = 12/307 (3%)
Query: 32 FSLQRVSKGCSISSFACSTSKLHGGRVGFNHREGKTSFLKFGVDESVVGGGGNGDVGER- 90
F L+R + G + ACST+ G RVG + R+G S L F G DV E+
Sbjct: 57 FRLRRRNSGL-VPVVACSTTPFMG-RVGLHWRDGNMSLLSFC---------GGTDVTEKA 105
Query: 91 DWSQILSALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAF 150
D SQ SALLPFVVA+TAVAALS P +FTWVSK+LYAPALGGIMLSIGI LS++DFALAF
Sbjct: 106 DSSQFWSALLPFVVALTAVAALSYPPSFTWVSKDLYAPALGGIMLSIGIQLSVDDFALAF 165
Query: 151 KRPLPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTACVSGAQLSSYASFISKGD 210
KRP+PLS+GF+AQYVLKP+LGVL+A AFG+PR FYAGF+LT CV+GAQLSSYAS +SK D
Sbjct: 166 KRPVPLSVGFVAQYVLKPLLGVLVANAFGMPRTFYAGFILTCCVAGAQLSSYASSLSKAD 225
Query: 211 VALCIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYA 270
VA+ I+LTS TTIASVI TPLL+GLLIGSVVPVDAVAMSKSILQVVL P+TLGL+LNTYA
Sbjct: 226 VAMSILLTSSTTIASVIFTPLLSGLLIGSVVPVDAVAMSKSILQVVLVPITLGLVLNTYA 285
Query: 271 KPVVSILRPVMPFVAMICTSLCIGSPLAINRSQILSGEGLRLVAPVLIFHAAAFTLGYWF 330
KPVV++L+PVMPFVAM+CTSLCIGSPL+INRSQILS EGL L+ P++ FHA AF LGYWF
Sbjct: 286 KPVVTLLQPVMPFVAMVCTSLCIGSPLSINRSQILSAEGLGLIVPIVTFHAVAFALGYWF 345
Query: 331 SNLPSLR 337
S +P LR
Sbjct: 346 SKIPGLR 352
>UniRef100_Q6K739 Bile acid:sodium symporter-like [Oryza sativa]
Length = 423
Score = 386 bits (991), Expect = e-106
Identities = 202/290 (69%), Positives = 237/290 (81%), Gaps = 6/290 (2%)
Query: 48 CSTSKLHGGRVGFNHREGKTSFLKFGVDESVVGGGGNGDVGERDWSQILSALLPFVVAVT 107
CS L GRVG+ REG L F V + G D SQ LSALLP VVA T
Sbjct: 62 CSAPSL--GRVGWPRREGAAWLLSFRAGP--VSSPSSAAAG--DPSQALSALLPLVVAAT 115
Query: 108 AVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRPLPLSIGFIAQYVLK 167
AVAAL P+TF+WVSKE YAPALGGIMLSIGI LS++DFALAFKRP+PL+IG++AQY++K
Sbjct: 116 AVAALGNPATFSWVSKEYYAPALGGIMLSIGIKLSIDDFALAFKRPVPLTIGYMAQYIVK 175
Query: 168 PVLGVLIAKAFGLPRMFYAGFVLTACVSGAQLSSYASFISKGDVALCIVLTSYTTIASVI 227
P++GVLIA+AFG+P F+AGFVLT CVSGAQLSSYASF+SKGDVAL I+LTS +TI+SV+
Sbjct: 176 PLMGVLIARAFGMPSAFFAGFVLTCCVSGAQLSSYASFLSKGDVALSILLTSCSTISSVV 235
Query: 228 VTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYAKPVVSILRPVMPFVAMI 287
VTP+LTGLLIGSVVPVD +AM+KSILQVVL PVTLGLLLNTYAK VV++++PVMPFVAM+
Sbjct: 236 VTPVLTGLLIGSVVPVDGIAMAKSILQVVLVPVTLGLLLNTYAKAVVNVIQPVMPFVAML 295
Query: 288 CTSLCIGSPLAINRSQILSGEGLRLVAPVLIFHAAAFTLGYWFSNLPSLR 337
CTSLCIGSPLAINRS+ILS EG L+ P++ FH AAF +GYW S LP LR
Sbjct: 296 CTSLCIGSPLAINRSKILSSEGFLLLLPIVTFHIAAFIVGYWISKLPMLR 345
>UniRef100_Q8VYY4 Hypothetical protein At4g22840 [Arabidopsis thaliana]
Length = 409
Score = 201 bits (511), Expect = 3e-50
Identities = 106/251 (42%), Positives = 162/251 (64%), Gaps = 10/251 (3%)
Query: 97 SALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRPLPL 156
+++LP VV + + AL P +FTW + + PALG +M ++GI + +DF AFKRP +
Sbjct: 98 NSILPHVVLASTILALIYPPSFTWFTSRYFVPALGFLMFAVGINSNEKDFLEAFKRPKAI 157
Query: 157 SIGFIAQYVLKPVLGVLIAKA----FGLPRMFYAGFVLTACVSGAQLSSYASFISKGDVA 212
+G++ QY++KPVLG + A F LP AG +L +CVSGAQLS+YA+F++ +A
Sbjct: 158 LLGYVGQYLVKPVLGFIFGLAAVSLFQLPTPIGAGIMLVSCVSGAQLSNYATFLTDPALA 217
Query: 213 -LCIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYAK 271
L IV+TS +T +V+VTP+L+ LLIG +PVD M SILQVV+AP+ GLLLN
Sbjct: 218 PLSIVMTSLSTATAVLVTPMLSLLLIGKKLPVDVKGMISSILQVVIAPIAAGLLLNKLFP 277
Query: 272 PVVSILRPVMPFVAMICTSLCIGSPLAINRSQILSGEGLRLVAPVLIFHAAAFTLGYW-- 329
V + +RP +P ++++ T+ C+G+PLA+N + ++S G ++ V +FH +AF GY+
Sbjct: 278 KVSNAIRPFLPILSVLDTACCVGAPLALNINSVMSPFGATILLLVTMFHLSAFLAGYFLT 337
Query: 330 ---FSNLPSLR 337
F N P +
Sbjct: 338 GSVFRNAPDAK 348
>UniRef100_Q650U0 Hypothetical protein P0669G04.12 [Oryza sativa]
Length = 401
Score = 196 bits (498), Expect = 9e-49
Identities = 104/253 (41%), Positives = 156/253 (61%), Gaps = 10/253 (3%)
Query: 97 SALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRPLPL 156
+ ++P VV + + AL P +FTW + YAPALG +M ++G+ S++DF A +RP +
Sbjct: 92 NTIIPHVVLGSTILALVYPPSFTWFTTRYYAPALGFLMFAVGVNSSVKDFIEAIQRPDAI 151
Query: 157 SIGFIAQYVLKPVLGVLIAKA----FGLPRMFYAGFVLTACVSGAQLSSYASFISKGDVA 212
+ G++ Q+++KP LG L F LP AG +L +CVSGAQLS+YA+F++ +A
Sbjct: 152 AAGYVGQFIIKPFLGFLFGTLAVTIFNLPTALGAGIMLVSCVSGAQLSNYATFLTDPHMA 211
Query: 213 -LCIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYAK 271
L IV+TS +T +V VTP L+ LIG +PVD M SI+Q+V+AP+ GLLLN Y
Sbjct: 212 PLSIVMTSLSTATAVFVTPTLSYFLIGKKLPVDVKGMMSSIVQIVVAPIAAGLLLNRYLP 271
Query: 272 PVVSILRPVMPFVAMICTSLCIGSPLAINRSQILSGEGLRLVAPVLIFHAAAFTLGY--- 328
+ S ++P +P +++ T+LC+GSPLAIN +LS GL V + FH ++F GY
Sbjct: 272 RLCSAIQPFLPPLSVFVTALCVGSPLAINIKAVLSPFGLATVLLLFAFHTSSFIAGYHLA 331
Query: 329 --WFSNLPSLRFL 339
WF ++ L
Sbjct: 332 GTWFRESADVKAL 344
>UniRef100_Q8S2V7 Putative sodium-dependent bile acid symporter [Arabidopsis
thaliana]
Length = 409
Score = 194 bits (493), Expect = 3e-48
Identities = 100/252 (39%), Positives = 155/252 (60%), Gaps = 2/252 (0%)
Query: 94 QILSALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRP 153
++L+ L P V + + + +PS TW+ +L++ LG +MLS+G+ L+ EDF + P
Sbjct: 99 ELLTTLFPLWVILGTLVGIFKPSLVTWLETDLFSLGLGFLMLSMGLTLTFEDFRRCLRNP 158
Query: 154 LPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTACVSGAQLSSYASFISKGDVAL 213
+ +GF+AQY++KP+LG LIA L G +L +C G Q S+ A++ISKG+VAL
Sbjct: 159 WTVGVGFLAQYMIKPILGFLIAMTLKLSAPLATGLILVSCCPGGQASNVATYISKGNVAL 218
Query: 214 CIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYAKPV 273
+++T+ +TI ++I+TPLLT LL G +VPVDA ++ S QVVL P +G+L N +
Sbjct: 219 SVLMTTCSTIGAIIMTPLLTKLLAGQLVPVDAAGLALSTFQVVLVPTIIGVLANEFFPKF 278
Query: 274 VSILRPVMPFVAMICTSLCIGSPLAINRSQILSGEGLRLVAPVLIFHAAAFTLGYWFSNL 333
S + V P + +I T+L SP+ + +L +G +L+ PV + HAAAF +GYW S
Sbjct: 279 TSKIITVTPLIGVILTTLLCASPIG-QVADVLKTQGAQLILPVALLHAAAFAIGYWISKF 337
Query: 334 PSLRFLTSITIN 345
S TS TI+
Sbjct: 338 -SFGESTSRTIS 348
>UniRef100_O49665 Predicted protein [Arabidopsis thaliana]
Length = 460
Score = 193 bits (490), Expect = 8e-48
Identities = 103/251 (41%), Positives = 159/251 (63%), Gaps = 10/251 (3%)
Query: 97 SALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRPLPL 156
+++LP VV + + AL P +FTW + + G +M ++GI + +DF AFKRP +
Sbjct: 149 NSILPHVVLASTILALIYPPSFTWFTSRFFPLEQGFLMFAVGINSNEKDFLEAFKRPKAI 208
Query: 157 SIGFIAQYVLKPVLGVLIAKA----FGLPRMFYAGFVLTACVSGAQLSSYASFISKGDVA 212
+G++ QY++KPVLG + A F LP AG +L +CVSGAQLS+YA+F++ +A
Sbjct: 209 LLGYVGQYLVKPVLGFIFGLAAVSLFQLPTPIGAGIMLVSCVSGAQLSNYATFLTDPALA 268
Query: 213 -LCIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYAK 271
L IV+TS +T +V+VTP+L+ LLIG +PVD M SILQVV+AP+ GLLLN
Sbjct: 269 PLSIVMTSLSTATAVLVTPMLSLLLIGKKLPVDVKGMISSILQVVIAPIAAGLLLNKLFP 328
Query: 272 PVVSILRPVMPFVAMICTSLCIGSPLAINRSQILSGEGLRLVAPVLIFHAAAFTLGYW-- 329
V + +RP +P ++++ T+ C+G+PLA+N + ++S G ++ V +FH +AF GY+
Sbjct: 329 KVSNAIRPFLPILSVLDTACCVGAPLALNINSVMSPFGATILLLVTMFHLSAFLAGYFLT 388
Query: 330 ---FSNLPSLR 337
F N P +
Sbjct: 389 GSVFRNAPDAK 399
>UniRef100_Q5VRB2 Na(+) dependent transporter-like [Oryza sativa]
Length = 419
Score = 192 bits (488), Expect = 1e-47
Identities = 96/252 (38%), Positives = 153/252 (60%), Gaps = 1/252 (0%)
Query: 94 QILSALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRP 153
++L+ L P V + + + +PS TW+ +L+ LG +MLS+G+ L+ EDF + P
Sbjct: 108 ELLTTLFPVWVILGTIIGIYKPSMVTWLETDLFTVGLGFLMLSMGLTLTFEDFRRCMRNP 167
Query: 154 LPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTACVSGAQLSSYASFISKGDVAL 213
+ +GF+AQY++KP+LG IA L G +L +C G Q S+ A++ISKG+VAL
Sbjct: 168 WTVGVGFLAQYLIKPMLGFAIAMTLKLSAPLATGLILVSCCPGGQASNVATYISKGNVAL 227
Query: 214 CIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYAKPV 273
+++T+ +TI ++++TPLLT LL G +VPVDA ++ S QVVL P +G+L + Y
Sbjct: 228 SVLMTTCSTIGAIVMTPLLTKLLAGQLVPVDAAGLAISTFQVVLLPTIVGVLAHEYFPKF 287
Query: 274 VSILRPVMPFVAMICTSLCIGSPLAINRSQILSGEGLRLVAPVLIFHAAAFTLGYWFSNL 333
+ + P + ++ T+L SP+ S++L +G +L+ PV + H AAF LGYW S +
Sbjct: 288 TERIISITPLIGVLLTTLLCASPIG-QVSEVLKAQGGQLIIPVALLHVAAFALGYWLSKV 346
Query: 334 PSLRFLTSITIN 345
S TS TI+
Sbjct: 347 SSFGESTSRTIS 358
>UniRef100_Q672Q4 Putative anion:sodium symporter precursor [Lycopersicon esculentum]
Length = 407
Score = 184 bits (467), Expect = 4e-45
Identities = 104/295 (35%), Positives = 165/295 (55%), Gaps = 12/295 (4%)
Query: 61 NHREGKTSFLKFGVDESVVG-------GGGNGDVGERDWSQI---LSALLPFVVAVTAVA 110
N + KTS ++ + +V G + G + +I L+ L P V + +
Sbjct: 54 NQNQSKTSLVQSPCNRKIVCCEAASNVSGESSSTGMTQYEKIIETLTTLFPLWVILGTII 113
Query: 111 ALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRPLPLSIGFIAQYVLKPVL 170
+ +PS TW+ +L+ LG +MLS+G+ L+ +DF + P + +GF+AQY +KP+L
Sbjct: 114 GIYKPSAVTWLETDLFTLGLGFLMLSMGLTLTFDDFRRCLRNPWTVGVGFLAQYFIKPLL 173
Query: 171 GVLIAKAFGLPRMFYAGFVLTACVSGAQLSSYASFISKGDVALCIVLTSYTTIASVIVTP 230
G IA A L G +L +C G Q S+ A++ISKG+VAL +++T+ +T+ ++++TP
Sbjct: 174 GFTIAMALKLSAPLATGLILVSCCPGGQASNVATYISKGNVALSVLMTTCSTVGAIVMTP 233
Query: 231 LLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYAKPVVSILRPVMPFVAMICTS 290
LLT LL G +VPVDA ++ S QVVL P +G+L N + S + + P + +I T+
Sbjct: 234 LLTKLLAGQLVPVDAAGLAISTFQVVLVPTVIGVLSNEFFPKFTSKIVTITPLIGVILTT 293
Query: 291 LCIGSPLAINRSQILSGEGLRLVAPVLIFHAAAFTLGYWFSNLPSLRFLTSITIN 345
L SP+ + +L +G +L+ PV HAAAF LGY S S TS TI+
Sbjct: 294 LLCASPIG-QVADVLKTQGAQLLLPVAALHAAAFFLGYQISKF-SFGESTSRTIS 346
>UniRef100_O81017 Putative Na+ dependent ileal bile acid transporter [Arabidopsis
thaliana]
Length = 338
Score = 183 bits (464), Expect = 8e-45
Identities = 95/231 (41%), Positives = 142/231 (61%), Gaps = 2/231 (0%)
Query: 115 PSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRPLPLSIGFIAQYVLKPVLGVLI 174
P TW+ +L+ LG +MLS+G+ L+ EDF + P + +GF+AQY++KP+LG LI
Sbjct: 59 PLWVTWLETDLFTLGLGFLMLSMGLTLTFEDFRRCLRNPWTVGVGFLAQYMIKPILGFLI 118
Query: 175 AKAFGLPRMFYAGFVLTACVSGAQLSSYASFISKGDVALCIVLTSYTTIASVIVTPLLTG 234
A L G +L +C G Q S+ A++ISKG+VAL +++T+ +TI ++I+TPLLT
Sbjct: 119 AMTLKLSAPLATGLILVSCCPGGQASNVATYISKGNVALSVLMTTCSTIGAIIMTPLLTK 178
Query: 235 LLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYAKPVVSILRPVMPFVAMICTSLCIG 294
LL G +VPVDA ++ S QVVL P +G+L N + S + V P + +I T+L
Sbjct: 179 LLAGQLVPVDAAGLALSTFQVVLVPTIIGVLANEFFPKFTSKIITVTPLIGVILTTLLCA 238
Query: 295 SPLAINRSQILSGEGLRLVAPVLIFHAAAFTLGYWFSNLPSLRFLTSITIN 345
SP+ + +L +G +L+ PV + HAAAF +GYW S S TS TI+
Sbjct: 239 SPIG-QVADVLKTQGAQLILPVALLHAAAFAIGYWISKF-SFGESTSRTIS 287
>UniRef100_Q7XVB3 OSJNBa0072D21.3 protein [Oryza sativa]
Length = 406
Score = 172 bits (437), Expect = 1e-41
Identities = 103/278 (37%), Positives = 154/278 (55%), Gaps = 10/278 (3%)
Query: 77 SVVGGGGNGD-----VGERDW----SQILSALLPFVVAVTAVAALSQPSTFTWVSKELYA 127
S + GGG+G VG RD ++LS P VA AL +P F WVS
Sbjct: 74 SKIPGGGSGALEAGVVGWRDLLLQVGEVLSLGFPVWVASACAVALWRPPAFLWVSPMAQI 133
Query: 128 PALGGIMLSIGIGLSMEDFALAFKRPLPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAG 187
+ ML +G+ L+++D A P L+ GF+ QY + P+ G LI+K LP + AG
Sbjct: 134 VGISFTMLGMGMTLTLDDLKTALLMPKELASGFLLQYSVMPLSGFLISKLLNLPSYYAAG 193
Query: 188 FVLTACVSGAQLSSYASFISKGDVALCIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVA 247
+L +C G S+ +++++G+VAL +++T+ +T A+ +TPLLT L G V VD +
Sbjct: 194 LILVSCCPGGTASNIVTYLARGNVALSVLMTAASTFAAAFLTPLLTSKLAGQYVAVDPMG 253
Query: 248 MSKSILQVVLAPVTLGLLLNTYAKPVVSILRPVMPFVAMICTSLCIGSPLAINRSQILSG 307
+ S QVVLAPV LG LLN Y +V ++ P+MPF+A+ ++ G+ +A N S ILS
Sbjct: 254 LFVSTSQVVLAPVLLGALLNQYCNGLVQLVSPLMPFIAVATVAVLCGNAIAQNASAILS- 312
Query: 308 EGLRLVAPVLIFHAAAFTLGYWFSNLPSLRFLTSITIN 345
GL++V V HA+ F GY S + +S TI+
Sbjct: 313 SGLQVVMSVCWLHASGFFFGYVLSRTIGIDISSSRTIS 350
>UniRef100_Q9SZ68 Putative transport protein [Arabidopsis thaliana]
Length = 379
Score = 170 bits (431), Expect = 5e-41
Identities = 96/249 (38%), Positives = 143/249 (56%), Gaps = 29/249 (11%)
Query: 98 ALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRPLPLS 157
+ +P + ++ + AL P +FTW + P LG +M ++GI + DF A KRP +
Sbjct: 82 SFIPHGILLSTILALVYPPSFTWFKPRYFVPGLGFMMFAVGINSNERDFLEALKRPDAIF 141
Query: 158 IGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTACVSGAQLSSYASFISKGDVA-LCIV 216
G+I QY++KP+LG AG +L +CVSGAQLS+Y +F++ +A L IV
Sbjct: 142 AGYIGQYLIKPLLG--------------AGIMLVSCVSGAQLSNYTTFLTDPSLAALSIV 187
Query: 217 LTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYAKPVV-- 274
+TS +T +V+VTP+L+ LLIG +PVD M SILQVV+ P+ GLLLN K +
Sbjct: 188 MTSISTATAVLVTPMLSLLLIGKKLPVDVFGMISSILQVVITPIAAGLLLNRKNKKLTLR 247
Query: 275 ------------SILRPVMPFVAMICTSLCIGSPLAINRSQILSGEGLRLVAPVLIFHAA 322
+ ++P +P + +I S CIG+PLA+N ILS G ++ V+ FH
Sbjct: 248 YKCECRLFPRLSNAIKPFLPALTVIDMSCCIGAPLALNIDSILSPFGATILFLVITFHLL 307
Query: 323 AFTLGYWFS 331
AF GY+F+
Sbjct: 308 AFVAGYFFT 316
>UniRef100_Q84WD8 Putative Na+ dependent ileal bile acid transporter [Arabidopsis
thaliana]
Length = 271
Score = 170 bits (431), Expect = 5e-41
Identities = 89/212 (41%), Positives = 132/212 (61%), Gaps = 2/212 (0%)
Query: 134 MLSIGIGLSMEDFALAFKRPLPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTAC 193
MLS+G+ L+ EDF + P + +GF+AQY++KP+LG LIA L G +L +C
Sbjct: 1 MLSMGLTLTFEDFRRCLRNPWTVGVGFLAQYMIKPILGFLIAMTLKLSAPLATGLILVSC 60
Query: 194 VSGAQLSSYASFISKGDVALCIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMSKSIL 253
G Q S+ A++ISKG+VAL +++T+ +TI ++I+TPLLT LL G +VPVDA ++ S
Sbjct: 61 CPGGQASNVATYISKGNVALSVLMTTCSTIGAIIMTPLLTKLLAGQLVPVDAAGLALSTF 120
Query: 254 QVVLAPVTLGLLLNTYAKPVVSILRPVMPFVAMICTSLCIGSPLAINRSQILSGEGLRLV 313
QVVL P +G+L N + S + V P + +I T+L SP+ + +L +G +L+
Sbjct: 121 QVVLVPTIIGVLANEFFPKFTSKIITVTPLIGVILTTLLCASPIG-QVADVLKTQGAQLI 179
Query: 314 APVLIFHAAAFTLGYWFSNLPSLRFLTSITIN 345
PV + HAAAF +GYW S S TS TI+
Sbjct: 180 LPVALLHAAAFAIGYWISKF-SFGESTSRTIS 210
>UniRef100_Q94A17 AT4g12030/F16J13_100 [Arabidopsis thaliana]
Length = 273
Score = 164 bits (414), Expect = 5e-39
Identities = 89/204 (43%), Positives = 129/204 (62%), Gaps = 5/204 (2%)
Query: 133 IMLSIGIGLSMEDFALAFKRPLPLSIGFIAQYVLKPVLG----VLIAKAFGLPRMFYAGF 188
+M ++GI + DF A KRP + G+I QY++KP+LG V+ F LP AG
Sbjct: 1 MMFAVGINSNERDFLEALKRPDAIFAGYIGQYLIKPLLGYIFGVIAVSLFNLPTSIGAGI 60
Query: 189 VLTACVSGAQLSSYASFISKGDVA-LCIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVA 247
+L +CVSGAQLS+Y +F++ +A L IV+TS +T +V+VTP+L+ LLIG +PVD
Sbjct: 61 MLVSCVSGAQLSNYTTFLTDPSLAALSIVMTSISTATAVLVTPMLSLLLIGKKLPVDVFG 120
Query: 248 MSKSILQVVLAPVTLGLLLNTYAKPVVSILRPVMPFVAMICTSLCIGSPLAINRSQILSG 307
M SILQVV+ P+ GLLLN + + ++P +P + +I S CIG+PLA+N ILS
Sbjct: 121 MISSILQVVITPIAAGLLLNRLFPRLSNAIKPFLPALTVIDMSCCIGAPLALNIDSILSP 180
Query: 308 EGLRLVAPVLIFHAAAFTLGYWFS 331
G ++ V+ FH AF GY+F+
Sbjct: 181 FGATILFLVITFHLLAFVAGYFFT 204
>UniRef100_Q93YR2 Hypothetical protein T30F21.11 [Arabidopsis thaliana]
Length = 401
Score = 154 bits (389), Expect = 4e-36
Identities = 86/252 (34%), Positives = 138/252 (54%), Gaps = 1/252 (0%)
Query: 94 QILSALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRP 153
+ +S P V++ + L +PSTF WV+ L ML +G+ L+++D A P
Sbjct: 95 EAVSTAFPIWVSLGCLLGLMRPSTFNWVTPNWTIVGLTITMLGMGMTLTLDDLRGALSMP 154
Query: 154 LPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTACVSGAQLSSYASFISKGDVAL 213
L GF+ QY + P+ ++K LP + AG +L C G S+ ++I++G+VAL
Sbjct: 155 KELFAGFLLQYSVMPLSAFFVSKLLNLPPHYAAGLILVGCCPGGTASNIVTYIARGNVAL 214
Query: 214 CIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYAKPV 273
+++T+ +T+++VI+TPLLT L + VDA+ + S LQVVL PV G LN Y K +
Sbjct: 215 SVLMTAASTVSAVIMTPLLTAKLAKQYITVDALGLLMSTLQVVLLPVLAGAFLNQYFKKL 274
Query: 274 VSILRPVMPFVAMICTSLCIGSPLAINRSQILSGEGLRLVAPVLIFHAAAFTLGYWFSNL 333
V + PVMP +A+ ++ G + N S IL G ++V + H + F GY FS +
Sbjct: 275 VKFVSPVMPPIAVGTVAILCGYAIGQNASAILM-SGKQVVLASCLLHISGFLFGYLFSRI 333
Query: 334 PSLRFLTSITIN 345
+ +S TI+
Sbjct: 334 LGIDVASSRTIS 345
>UniRef100_Q6E4A7 Hypothetical protein [Cynodon dactylon]
Length = 219
Score = 150 bits (380), Expect = 4e-35
Identities = 71/176 (40%), Positives = 115/176 (65%)
Query: 94 QILSALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRP 153
++L+ L P V + + + +PS TW+ +L+ LG +MLS+G+ L+ +DF + P
Sbjct: 44 ELLTTLFPVWVILGTIIGIYKPSMVTWLETDLFTLGLGFLMLSMGLTLTFDDFKRCLRNP 103
Query: 154 LPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTACVSGAQLSSYASFISKGDVAL 213
+ +GF+AQY++KP+LG IA + L G VL +C G Q S+ A++ISKG+VAL
Sbjct: 104 WTVGVGFVAQYLIKPLLGFAIATSLNLSAPLATGLVLVSCCPGGQASNVATYISKGNVAL 163
Query: 214 CIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTY 269
+++T+ +TI ++++TPLLT +L G +VPVDAV ++ S QVVL P +G+L + Y
Sbjct: 164 SVLMTTCSTIGAIVMTPLLTKILAGQLVPVDAVGLAMSTFQVVLMPTIVGVLAHEY 219
>UniRef100_Q8NR06 Predicted Na+-dependent transporter [Corynebacterium glutamicum]
Length = 335
Score = 150 bits (378), Expect = 7e-35
Identities = 83/210 (39%), Positives = 130/210 (61%), Gaps = 6/210 (2%)
Query: 119 TWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRPLPLSIGFIAQYVLKPVLGVLIAKAF 178
+WV+ P LG IM S+G+ L DFAL KRPLP+ IG IAQ+V+ P++ +L+
Sbjct: 47 SWVN-----PLLGIIMFSMGLTLKPVDFALVAKRPLPVLIGVIAQFVIMPLIALLVVWVL 101
Query: 179 GLPRMFYAGFVLTACVSGAQLSSYASFISKGDVALCIVLTSYTTIASVIVTPLLTGLLIG 238
LP AG +L C G S+ S++S+GDVAL + +TS +T+ + I TPLLT L G
Sbjct: 102 QLPAEIAAGVILVGCAPGGTSSNVVSYLSRGDVALSVTMTSISTLLAPIFTPLLTLWLAG 161
Query: 239 SVVPVDAVAMSKSILQVVLAPVTLGLLLNTYAKPVVSILRPVMPFVAMICTSLCIGSPLA 298
+P++A M+ SI+QVVL PV GL++ ++ + P++P++++I SL + +A
Sbjct: 162 QYMPLNAADMAVSIVQVVLIPVVGGLVVRLIFPTLIGKVLPLLPWISVIAISLIVAIVVA 221
Query: 299 INRSQILSGEGLRLVAPVLIFHAAAFTLGY 328
+R +IL GL ++A V+I + ++LGY
Sbjct: 222 GSRDKILEA-GLLVLAAVIIHNTLGYSLGY 250
>UniRef100_Q7U8I3 Sodium/bile acid cotransporter family [Synechococcus sp.]
Length = 393
Score = 148 bits (373), Expect = 3e-34
Identities = 86/249 (34%), Positives = 129/249 (51%), Gaps = 1/249 (0%)
Query: 97 SALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKRPLPL 156
+ L P + ++ AL P F W L LG IML +G+GL+ DF ++P +
Sbjct: 85 TTLFPLWTLLGSLLALLHPPLFVWFRGPLITIGLGVIMLGMGLGLTPRDFLRVSRQPRAV 144
Query: 157 SIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTACVSGAQLSSYASFISKGDVALCIV 216
+G AQ+++ P L IA LP G +L C G S+ + I+KGDVAL +V
Sbjct: 145 ILGVAAQFLVMPSLAAAIAVLLQLPAPLAVGLILVGCCPGGTASNVVTLIAKGDVALSVV 204
Query: 217 LTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYAKPVVSI 276
+TS +T+A+V++TP LT LL VPVD + +LQVVL PV G+LL + +
Sbjct: 205 MTSISTMAAVVLTPRLTELLASQYVPVDGWLLLLRVLQVVLVPVACGVLLKQGVPRLANR 264
Query: 277 LRPVMPFVAMICTSLCIGSPLAINRSQILSGEGLRLVAPVLIFHAAAFTLGYWFSNLPSL 336
+ PVMP +A++ L + S + R Q+L +G L+ L+ H F LG+ + L
Sbjct: 265 VEPVMPPIAVVAIVLIVASVVGSQR-QLLLEQGALLLLACLLLHGGGFLLGFLMARLVGA 323
Query: 337 RFLTSITIN 345
TI+
Sbjct: 324 SSQAQCTIS 332
>UniRef100_Q9KEJ4 Sodium-dependent transporter [Bacillus halodurans]
Length = 323
Score = 145 bits (367), Expect = 1e-33
Identities = 77/236 (32%), Positives = 130/236 (54%), Gaps = 2/236 (0%)
Query: 93 SQILSALLPFVVAVTAVAALSQPSTFTWVSKELYAPALGGIMLSIGIGLSMEDFALAFKR 152
SQ S F V + + A P FTW++ + LG IM +G+ L + DF + ++
Sbjct: 8 SQFFSKYFAFFVIIISFVAFLSPDHFTWITPHITI-LLGVIMFGMGLTLKLSDFRIVLQK 66
Query: 153 PLPLSIGFIAQYVLKPVLGVLIAKAFGLPRMFYAGFVLTACVSGAQLSSYASFISKGDVA 212
P+P+ +G +AQ+V+ P++ +A AF LP AG VL G S+ +++KG+VA
Sbjct: 67 PIPVLVGVLAQFVIMPLVAFALAYAFNLPPELAAGLVLVGACPGGTASNVMVYLAKGNVA 126
Query: 213 LCIVLTSYTTIASVIVTPLLTGLLIGSVVPVDAVAMSKSILQVVLAPVTLGLLLNTYAKP 272
+ +TS +T+ + IVTP + LL G +P+DA AM SILQ+++ P+ LGL + A
Sbjct: 127 ASVAMTSVSTMLAPIVTPFILLLLAGQWLPIDAKAMFVSILQMIIVPIALGLFVRKMAPN 186
Query: 273 VVSILRPVMPFVAMICTSLCIGSPLAINRSQILSGEGLRLVAPVLIFHAAAFTLGY 328
V V+P V+++ + + + N++ ++SG L +A V++ + LGY
Sbjct: 187 AVDKSTAVLPLVSIVAIMAIVSAVVGANQANLMSGAALLFLA-VMLHNVFGLLLGY 241
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.324 0.138 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 591,957,834
Number of Sequences: 2790947
Number of extensions: 23898092
Number of successful extensions: 84641
Number of sequences better than 10.0: 534
Number of HSP's better than 10.0 without gapping: 363
Number of HSP's successfully gapped in prelim test: 172
Number of HSP's that attempted gapping in prelim test: 84050
Number of HSP's gapped (non-prelim): 598
length of query: 379
length of database: 848,049,833
effective HSP length: 129
effective length of query: 250
effective length of database: 488,017,670
effective search space: 122004417500
effective search space used: 122004417500
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 76 (33.9 bits)
Medicago: description of AC146343.8


