

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146342.2 + phase: 0
(340 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
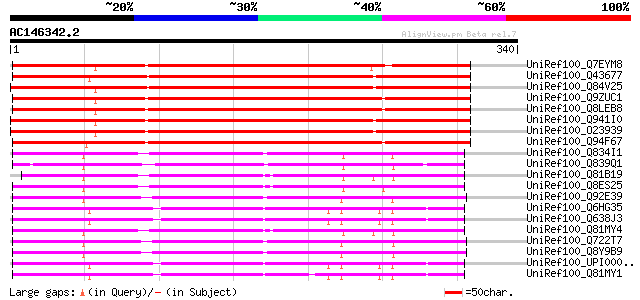
UniRef100_Q7EYM8 Putative oxidoreductase, zinc-binding [Oryza sa... 286 4e-76
UniRef100_Q43677 Auxin-induced protein [Vigna radiata] 283 6e-75
UniRef100_Q84V25 Quinone oxidoreductase [Fragaria ananassa] 278 2e-73
UniRef100_Q9ZUC1 Quinone oxidoreductase-like protein At1g23740, ... 276 6e-73
UniRef100_Q8LEB8 Quinone oxidoreductase-like protein [Arabidopsi... 276 8e-73
UniRef100_Q941I0 Putative quinone oxidoreductase [Fragaria anana... 273 6e-72
UniRef100_O23939 Ripening-induced protein [Fragaria vesca] 272 8e-72
UniRef100_Q94F67 Quinone oxidoreductase-like protein [Helianthus... 263 7e-69
UniRef100_Q834I1 Oxidoreductase, zinc-binding [Enterococcus faec... 170 4e-41
UniRef100_Q839Q1 Oxidoreductase, zinc-binding [Enterococcus faec... 169 1e-40
UniRef100_Q81B19 Quinone oxidoreductase [Bacillus cereus] 166 1e-39
UniRef100_Q8ES25 Zinc-binding oxidoreductase [Oceanobacillus ihe... 164 2e-39
UniRef100_Q92E39 Lin0622 protein [Listeria innocua] 164 3e-39
UniRef100_Q6HG35 Quinone oxidoreductase [Bacillus thuringiensis] 163 7e-39
UniRef100_Q638J3 Quinone oxidoreductase [Bacillus cereus] 163 7e-39
UniRef100_Q81MY4 Alcohol dehydrogenase, zinc-containing [Bacillu... 162 2e-38
UniRef100_Q722T7 Alcohol dehydrogenase, zinc-dependent [Listeria... 161 2e-38
UniRef100_Q8Y9B9 Lmo0613 protein [Listeria monocytogenes] 160 4e-38
UniRef100_UPI00003CB31E UPI00003CB31E UniRef100 entry 160 5e-38
UniRef100_Q81MY1 Alcohol dehydrogenase, zinc-containing [Bacillu... 159 1e-37
>UniRef100_Q7EYM8 Putative oxidoreductase, zinc-binding [Oryza sativa]
Length = 390
Score = 286 bits (733), Expect = 4e-76
Identities = 151/313 (48%), Positives = 210/313 (66%), Gaps = 11/313 (3%)
Query: 3 KAWFYEEYGPKEVLKLGDFP-IPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSD--FPA 59
KAW Y++YG VLKL D +P ++Q+LV+V AAALNP+D+KRR +D P
Sbjct: 83 KAWAYDDYGDGSVLKLNDAAAVPDIADDQVLVRVAAAALNPVDAKRRAGKFKATDSPLPT 142
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKP 119
VPG D+AGVV+ G VK GDEVYGNI + ++E PKQ G+LA++ VEE L+A KP
Sbjct: 143 VPGYDVAGVVVKAGRKVKGLKEGDEVYGNISE-KALEGPKQSGSLAEYTAVEEKLLALKP 201
Query: 120 KRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVV 179
K L F +AA LPLA++TA EG + F G+++ ++GGAGGVG+L +QLAK ++GAS V
Sbjct: 202 KSLGFAQAAGLPLAIETAHEGLERAGFSAGKSILILGGAGGVGSLAIQLAKHVYGASKVA 261
Query: 180 SSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVAKKDGAIVDI 239
++ STPKL+ +K GAD +DYTK +ED+ +K+D + D VG +K+ V K+ G++V +
Sbjct: 262 ATASTPKLELLKSLGADVAIDYTKENFEDLPDKYDVVLDAVGQGEKAVKVVKEGGSVVVL 321
Query: 240 TW---PASHERAVYSSLTVCGEILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIET 296
T P V S G +LEKL PYLE G++K ++DPKG + F V++AF Y+ET
Sbjct: 322 TGAVVPPGFRFVVTSD----GSVLEKLNPYLESGKVKPLVDPKGPFAFSQVVEAFSYLET 377
Query: 297 GRAWGKVVVTCFP 309
GRA GKVV++ P
Sbjct: 378 GRATGKVVISPIP 390
>UniRef100_Q43677 Auxin-induced protein [Vigna radiata]
Length = 317
Score = 283 bits (723), Expect = 6e-75
Identities = 146/311 (46%), Positives = 210/311 (66%), Gaps = 6/311 (1%)
Query: 3 KAWFYEEYGP-KEVLKLG-DFPIPSPLENQLLVQVYAAALNPIDSKRRMRPI--FPSDFP 58
+AW Y EYG +EVLK + P P +ENQ+L++V AAA+NP+D KR + S P
Sbjct: 9 RAWIYSEYGNIEEVLKFDPNVPTPHLMENQVLIKVVAAAINPVDYKRALGHFKDIDSPLP 68
Query: 59 AVPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARK 118
PG D+AGVV+ G VKKF +GDEVYG+I + +++ PK +G+LA++ VEE ++A K
Sbjct: 69 TAPGYDVAGVVVKVGNQVKKFQVGDEVYGDINEV-TLDHPKTIGSLAEYTAVEEKVLAHK 127
Query: 119 PKRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYV 178
P LSF EAASLPLA+ TA +G +T F G+++ V+GGAGGVG+LV+Q+AK +FGAS +
Sbjct: 128 PSNLSFVEAASLPLAIITAYQGLETAQFSVGKSILVLGGAGGVGSLVIQIAKHIFGASKI 187
Query: 179 VSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVAKKDGAIVD 238
++ ST KL+ + G D +DYTK +E++ EKFD +YD VG +++ K+ G +V
Sbjct: 188 AATGSTGKLELFENLGTDLPIDYTKENFEELAEKFDVVYDAVGQSERALKSVKEGGKVVT 247
Query: 239 ITWPASHERAVYSSLTVCGEILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIETGR 298
I PA+ A+ LT G++LEKLRPYLE G++K ++DPK + F ++AF Y++T R
Sbjct: 248 IVAPAT-PPAIPFLLTSDGDLLEKLRPYLENGQVKPILDPKSPFPFSQTVEAFSYLKTNR 306
Query: 299 AWGKVVVTCFP 309
A GKVV+ P
Sbjct: 307 ATGKVVIYPIP 317
>UniRef100_Q84V25 Quinone oxidoreductase [Fragaria ananassa]
Length = 322
Score = 278 bits (711), Expect = 2e-73
Identities = 146/313 (46%), Positives = 207/313 (65%), Gaps = 6/313 (1%)
Query: 1 MQKAWFYEEYGP-KEVLKLG-DFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSD-- 56
+ KAW Y EYG +VLK +P E+Q+L++V AA+LNP+D KR + +D
Sbjct: 12 VNKAWVYSEYGKTSDVLKFDPSVAVPEVKEDQVLIKVVAASLNPVDFKRALGYFKDTDSP 71
Query: 57 FPAVPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVA 116
P VPG D+AGVV+ G V KF +GDEVYG++ + ++ P + G+LA++ +E ++A
Sbjct: 72 LPTVPGYDVAGVVVKVGSQVTKFKVGDEVYGDLNEA-ALVNPTRFGSLAEYTAADERVLA 130
Query: 117 RKPKRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGAS 176
KPK LSF EAASLPLA++TA EG + + G+++ V+GGAGGVGT ++QLAK +FGAS
Sbjct: 131 HKPKDLSFIEAASLPLAIETAYEGLERAELSAGKSILVLGGAGGVGTHIIQLAKHVFGAS 190
Query: 177 YVVSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVAKKDGAI 236
V ++ ST KL F++ G D +DYTK ED+ EKFD +YD VG+ K+ K+ G +
Sbjct: 191 KVAATASTKKLDFLRTLGVDLAIDYTKENIEDLPEKFDVVYDAVGETDKAVKAVKEGGKV 250
Query: 237 VDITWPASHERAVYSSLTVCGEILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIET 296
V I PA+ A++ LT G +LEKL+PYLE G++K V+DP Y F +++AFGY+E+
Sbjct: 251 VTIVGPAT-PPAIHFVLTSKGSVLEKLKPYLESGKVKPVLDPTSPYPFTKLVEAFGYLES 309
Query: 297 GRAWGKVVVTCFP 309
RA GKVVV P
Sbjct: 310 SRATGKVVVYPIP 322
>UniRef100_Q9ZUC1 Quinone oxidoreductase-like protein At1g23740, chloroplast
precursor [Arabidopsis thaliana]
Length = 386
Score = 276 bits (706), Expect = 6e-73
Identities = 144/310 (46%), Positives = 205/310 (65%), Gaps = 5/310 (1%)
Query: 3 KAWFYEEYGPKEVLKL-GDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSD--FPA 59
KAW Y +YG +VLKL + +P E+Q+L++V AAALNP+D+KRR +D P
Sbjct: 79 KAWVYSDYGGVDVLKLESNIVVPEIKEDQVLIKVVAAALNPVDAKRRQGKFKATDSPLPT 138
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKP 119
VPG D+AGVV+ G VK GDEVY N+ + ++E PKQ G+LA++ VEE L+A KP
Sbjct: 139 VPGYDVAGVVVKVGSAVKDLKEGDEVYANVSE-KALEGPKQFGSLAEYTAVEEKLLALKP 197
Query: 120 KRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVV 179
K + F +AA LPLA++TA EG +F G+++ V+ GAGGVG+LV+QLAK ++GAS V
Sbjct: 198 KNIDFAQAAGLPLAIETADEGLVRTEFSAGKSILVLNGAGGVGSLVIQLAKHVYGASKVA 257
Query: 180 SSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVAKKDGAIVDI 239
++ ST KL+ V+ GAD +DYTK ED+ +K+D ++D +G C K+ V K+ G +V +
Sbjct: 258 ATASTEKLELVRSLGADLAIDYTKENIEDLPDKYDVVFDAIGMCDKAVKVIKEGGKVVAL 317
Query: 240 TWPASHERAVYSSLTVCGEILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIETGRA 299
T + + +T G++L+KL PY+E G++K V+DPKG + F V DAF Y+ET A
Sbjct: 318 TGAVTPPGFRF-VVTSNGDVLKKLNPYIESGKVKPVVDPKGPFPFSRVADAFSYLETNHA 376
Query: 300 WGKVVVTCFP 309
GKVVV P
Sbjct: 377 TGKVVVYPIP 386
>UniRef100_Q8LEB8 Quinone oxidoreductase-like protein [Arabidopsis thaliana]
Length = 309
Score = 276 bits (705), Expect = 8e-73
Identities = 143/310 (46%), Positives = 205/310 (66%), Gaps = 5/310 (1%)
Query: 3 KAWFYEEYGPKEVLKL-GDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSD--FPA 59
KAW Y +YG +VLKL + +P E+Q+L++V AA LNP+D+KRR +D P
Sbjct: 2 KAWVYSDYGGVDVLKLESNIAVPEIKEDQVLIKVVAAGLNPVDAKRRQGKFKATDSPLPT 61
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKP 119
VPG D+AGVV+ G VK F GDEVY N+ + ++E PKQ G+LA++ VEE L+A KP
Sbjct: 62 VPGYDVAGVVVKVGSAVKDFKEGDEVYANVSE-KALEGPKQFGSLAEYTAVEEKLLALKP 120
Query: 120 KRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVV 179
K + F +AA LPLA++TA EG +F G+++ V+ GAGGVG+L++QLAK ++GAS V
Sbjct: 121 KNIDFAQAAGLPLAIETADEGLVRTEFSAGKSILVLNGAGGVGSLMIQLAKHVYGASKVA 180
Query: 180 SSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVAKKDGAIVDI 239
++ ST KL+ V+ GAD +DYTK ED+ +K+D ++D +G C K+ V K+ G +V +
Sbjct: 181 ATASTGKLELVRSLGADLAIDYTKENIEDLPDKYDVVFDAIGMCDKAVKVIKEGGKVVAL 240
Query: 240 TWPASHERAVYSSLTVCGEILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIETGRA 299
T + + +T G++L+KL PY+E G++K V+DPKG + F V DAF Y+ET A
Sbjct: 241 TGAVTPPGFRF-VVTSNGDVLKKLNPYIESGKVKPVVDPKGPFPFSRVADAFSYLETNHA 299
Query: 300 WGKVVVTCFP 309
GKVVV P
Sbjct: 300 TGKVVVYPIP 309
>UniRef100_Q941I0 Putative quinone oxidoreductase [Fragaria ananassa]
Length = 339
Score = 273 bits (697), Expect = 6e-72
Identities = 146/314 (46%), Positives = 206/314 (65%), Gaps = 7/314 (2%)
Query: 1 MQKAWFYEEYGP-KEVLKLG-DFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSD-- 56
+ KAW Y EYG +VLK +P E+Q+L++V AA+LNP+D KR + +D
Sbjct: 28 VNKAWVYSEYGKTSDVLKFDPSVAVPEIKEDQVLIKVVAASLNPVDFKRALGYFKDTDSP 87
Query: 57 FPAVPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVA 116
P +PG D+AGVV+ G V KF +GDEVYG++ + ++ P + G+LA++ E ++A
Sbjct: 88 LPTIPGYDVAGVVVKVGSQVTKFKVGDEVYGDLNE-TALVNPTRFGSLAEYTAAYERVLA 146
Query: 117 RKPKRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGAS 176
KPK LSF EAASLPLA++TA EG + + G+++ V+GGAGGVGT ++QLAK +FGAS
Sbjct: 147 HKPKNLSFIEAASLPLAIETAHEGLERAELSAGKSVLVLGGAGGVGTHIIQLAKHVFGAS 206
Query: 177 YVVSSCSTPKLKFVKQFG-ADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVAKKDGA 235
V ++ ST KL ++ G AD +DYTK +ED+ EKFD +YD VG+ K+ K+ G
Sbjct: 207 KVAATASTKKLDLLRTLGAADLAIDYTKENFEDLPEKFDVVYDAVGETDKAVKAVKEGGK 266
Query: 236 IVDITWPASHERAVYSSLTVCGEILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIE 295
+V I PA+ A+ LT G +LEKL+PYLE G++K V+DP Y F V++AFGY+E
Sbjct: 267 VVTIVGPAT-PPAILFVLTSKGSVLEKLKPYLESGKVKPVLDPTSPYPFTKVVEAFGYLE 325
Query: 296 TGRAWGKVVVTCFP 309
+ RA GKVVV P
Sbjct: 326 SSRATGKVVVYPIP 339
>UniRef100_O23939 Ripening-induced protein [Fragaria vesca]
Length = 337
Score = 272 bits (696), Expect = 8e-72
Identities = 144/313 (46%), Positives = 205/313 (65%), Gaps = 6/313 (1%)
Query: 1 MQKAWFYEEYGP-KEVLKLG-DFPIPSPLENQLLVQVYAAALNPIDSKRRMRPIFPSD-- 56
+ KAW EYG +VLK +P E+Q+L++V AA+LNP+D KR + +D
Sbjct: 27 VNKAWVXSEYGKTSDVLKFDPSVAVPEIKEDQVLIKVVAASLNPVDFKRALGYFKDTDSP 86
Query: 57 FPAVPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVA 116
P +PG +AGVV+ G V KF +GDEVYG++ + ++ P + G+LA++ +E ++A
Sbjct: 87 LPTIPGYYVAGVVVKVGSQVTKFKVGDEVYGDLNE-TALVNPTRFGSLAEYTAADERVLA 145
Query: 117 RKPKRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGAS 176
KPK LSF EAASLPLA++TA EG + + G+++ V+GGAGGVGT ++QLAK +FGAS
Sbjct: 146 HKPKNLSFIEAASLPLAIETAHEGLERAELSAGKSVLVLGGAGGVGTHIIQLAKHVFGAS 205
Query: 177 YVVSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVAKKDGAI 236
V ++ ST KL ++ GAD +DYTK +ED+ EKFD +YD VG+ K+ K+ G +
Sbjct: 206 KVAATASTKKLDLLRTLGADLAIDYTKENFEDLPEKFDVVYDAVGETDKAVKAVKEGGKV 265
Query: 237 VDITWPASHERAVYSSLTVCGEILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIET 296
V I PA+ A+ LT G +LEKL+PYLE G++K V+DP Y F V++AFGY+E+
Sbjct: 266 VTIVGPAT-PPAILFVLTSKGSVLEKLKPYLESGKVKPVLDPTSPYPFTKVVEAFGYLES 324
Query: 297 GRAWGKVVVTCFP 309
RA GKVVV P
Sbjct: 325 SRATGKVVVYPIP 337
>UniRef100_Q94F67 Quinone oxidoreductase-like protein [Helianthus annuus]
Length = 309
Score = 263 bits (671), Expect = 7e-69
Identities = 140/310 (45%), Positives = 198/310 (63%), Gaps = 5/310 (1%)
Query: 3 KAWFYEEYGPKEVLKLG-DFPIPSPLENQLLVQVYAAALNPIDSKRRMR--PIFPSDFPA 59
KAW Y+EYG +VLKL D +P ++Q+LV+V AAA+NP+D KRR+ S P
Sbjct: 2 KAWKYDEYGSVDVLKLATDVAVPEIKDDQVLVKVVAAAVNPVDYKRRLGYFKAIDSPLPI 61
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKP 119
+PG D+ GVV+ G VK GDEVYG+I D +E P Q GTLA++ VEE L+A KP
Sbjct: 62 IPGFDVLGVVLKVGSQVKDLKEGDEVYGDIND-KGLEGPTQFGTLAEYTAVEERLLALKP 120
Query: 120 KRLSFEEAASLPLAVQTAIEGFKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVV 179
K L F +AA+LPLA++TA EG + F +G+T+ V+ GAGGVG+ ++QLAK ++GAS V
Sbjct: 121 KNLDFIQAAALPLAIETAYEGLERAKFSEGKTILVLNGAGGVGSFIIQLAKHVYGASKVA 180
Query: 180 SSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGDCKKSFVVAKKDGAIVDI 239
++ ST KL+ +K GAD +DYTK +ED+ +K+D +YD +G +K+ + G V I
Sbjct: 181 ATSSTGKLELLKSLGADVAIDYTKENFEDLPDKYDVVYDAIGQPEKALKAVNETGVAVSI 240
Query: 240 TWPASHERAVYSSLTVCGEILEKLRPYLERGELKAVIDPKGEYDFENVIDAFGYIETGRA 299
T P + LT G IL KL PYLE G++K+ DPK + F+ + +A+ Y+E+
Sbjct: 241 TGPIPPPGFSF-ILTSDGSILTKLNPYLESGKIKSDNDPKSPFPFDKLNEAYAYLESTGP 299
Query: 300 WGKVVVTCFP 309
GKVV+ P
Sbjct: 300 TGKVVIYPIP 309
>UniRef100_Q834I1 Oxidoreductase, zinc-binding [Enterococcus faecalis]
Length = 313
Score = 170 bits (431), Expect = 4e-41
Identities = 108/317 (34%), Positives = 177/317 (55%), Gaps = 23/317 (7%)
Query: 3 KAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRR---MRPIFPSDFPA 59
KA +YG KEVL+ + +P E+Q+LV+ YA ++NPID K R ++ +F FP
Sbjct: 2 KAVVINQYGSKEVLEEAEVTLPELSEHQVLVKEYATSINPIDWKLREGYLKQMFDWSFPI 61
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKP 119
+ G D+AGV+ G V + +GD+V+ S + + GT A+ +V+++L+A+ P
Sbjct: 62 ILGWDVAGVITEVGSQVTDWQVGDKVF-------SRPETTRFGTYAEVTIVDDHLLAKIP 114
Query: 120 KRLSFEEAASLPLAVQTAIEG-FKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYV 178
+ +SFEEAA++PLA TA + F G K+GE + + GAGGVGT +QLAK ++V
Sbjct: 115 ETISFEEAAAVPLAGLTAWQALFDHGHLKEGEIVLIHAGAGGVGTYAIQLAK--EAGAHV 172
Query: 179 VSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGD--CKKSFVVAKKD-GA 235
+++ S +K+ GAD+V+DY T + ++ D ++DT+G K SF V K + G
Sbjct: 173 ITTASAKNHTLLKKIGADEVIDYHTTNFAEVLTDVDLVFDTMGGEVQKNSFAVLKPNTGR 232
Query: 236 IVDITWPASHERAVYSSLTV-------CGEILEKLRPYLERGELKAVIDPKGEYDFENVI 288
+V I + A +++ GE L+K+ + G++K++I + + +
Sbjct: 233 LVSIVGIEDKQLAAEKNISAESIWLQPNGEQLQKIADLMAAGKVKSIIGEVFPFSRQGIY 292
Query: 289 DAFGYIETGRAWGKVVV 305
DA ET A GK+VV
Sbjct: 293 DAHALSETHHAVGKIVV 309
>UniRef100_Q839Q1 Oxidoreductase, zinc-binding [Enterococcus faecalis]
Length = 339
Score = 169 bits (428), Expect = 1e-40
Identities = 109/341 (31%), Positives = 183/341 (52%), Gaps = 51/341 (14%)
Query: 3 KAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRR---MRPIFPSDFPA 59
KA +YG KE L + + P+P+ +N +LV++ AA++NPID K + ++ + P
Sbjct: 2 KAALIHKYGQKE-LAIEEVPLPTIHDNDVLVKIIAASINPIDLKTKDGKVKMLLDYQMPL 60
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPK-QLGTLAQFIVVEENLVARK 118
+ G D AG+V+ G NV+ F +GD VYG + PK ++GT A++I V++ VA K
Sbjct: 61 ILGSDFAGIVVSVGKNVQNFRLGDAVYGRV--------PKNRVGTFAEYIAVDQAAVAMK 112
Query: 119 PKRLSFEEAASLPLAVQTAIEGFKT-GDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASY 177
PK L+FEEAA++PL T+ + + + G+ + + G+GG+GT+ +QLAKL +Y
Sbjct: 113 PKNLTFEEAAAIPLVGLTSYQALHDIMNVQPGQKVLIQAGSGGIGTIAIQLAKL--AGAY 170
Query: 178 VVSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGD--CKKSFVVAKKDGA 235
V ++ S+ ++V+ GAD+V+DY +E++ +D+++DT+G +K+F V K G
Sbjct: 171 VATTTSSKNKEWVQALGADEVIDYRTQNFEEVLSDYDYVFDTMGGTILEKAFSVVKPQGK 230
Query: 236 IVDITWPASHERAVYSSLTVC-------------------------------GEILEKLR 264
+V ++ + A L + GE L L
Sbjct: 231 VVTLSGIPNERFAKEYGLPLWKQWAFKIATRKIHRLEQATDVSYHFLFMRPDGEQLALLT 290
Query: 265 PYLERGELKAVIDPKGEYDFENVIDAFGYIETGRAWGKVVV 305
++E+G+L+ +ID F + +A Y TGRA GK+VV
Sbjct: 291 EFIEQGKLQPIID--RVVPFSQIQEAVDYSLTGRAQGKIVV 329
>UniRef100_Q81B19 Quinone oxidoreductase [Bacillus cereus]
Length = 317
Score = 166 bits (419), Expect = 1e-39
Identities = 112/315 (35%), Positives = 168/315 (52%), Gaps = 27/315 (8%)
Query: 9 EYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRR---MRPIFPSDFPAVPGCDM 65
+YG K VL+ + P +N +L++VYAA +NP+D K R ++ + DFP V G D+
Sbjct: 8 QYGDKSVLQEIEMQTPLLGDNDVLIEVYAAGVNPVDWKIREGLLQDVISYDFPLVLGWDV 67
Query: 66 AGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPKRLSFE 125
AGVV G NV F +GDEVY S ++ GT A+++ V+E VA+KP+ LSFE
Sbjct: 68 AGVVAAIGKNVTAFKVGDEVY-------SRPDIERNGTYAEYVAVDEKYVAKKPRNLSFE 120
Query: 126 EAASLPLAVQTAIEGF-KTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVVSSCST 184
EAAS+PL T+ + K + +KG + + G+GG+GT +QLAK FGA YV ++ ST
Sbjct: 121 EAASIPLVGLTSWQSLVKFANVQKGNKVLIHAGSGGIGTFAIQLAK-SFGA-YVATTTST 178
Query: 185 PKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGD--CKKSFVVAKKDGAIVDITWP 242
++FVK GAD VVDY + + ++ ++D +G K S+ V +G + I P
Sbjct: 179 KNMQFVKDLGADTVVDYKTEDFSLLLHNYNIVFDVLGGDVLKNSYKVLAPNGKLASIYGP 238
Query: 243 ---------ASHERAVYSSLTVC---GEILEKLRPYLERGELKAVIDPKGEYDFENVIDA 290
S E+ + S G L + +E G++K V+ E V A
Sbjct: 239 KGMKIPQTEISREKNIESDHIFAEPNGYALSLIPELIESGKIKPVVTHVLPLHVEGVKKA 298
Query: 291 FGYIETGRAWGKVVV 305
E+ RA GK+V+
Sbjct: 299 HHISESERALGKIVL 313
>UniRef100_Q8ES25 Zinc-binding oxidoreductase [Oceanobacillus iheyensis]
Length = 311
Score = 164 bits (416), Expect = 2e-39
Identities = 112/316 (35%), Positives = 171/316 (53%), Gaps = 22/316 (6%)
Query: 3 KAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRR---MRPIFPSDFPA 59
KA E YG + L D P P+ +NQ+L++ YA ++NPID K R ++ F +FP
Sbjct: 2 KAIVIENYGHADELHEQDVPKPTINDNQILIEQYATSINPIDWKLREGYLKDGFNFEFPI 61
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKP 119
+ G D AGVV G NV +F +GD V+ S GT A+F+ VEE+ VA+ P
Sbjct: 62 ILGWDSAGVVAEVGKNVSQFKVGDRVF-------SRPATTAQGTYAEFVAVEESRVAKIP 114
Query: 120 KRLSFEEAASLPLAVQTAIEGF-KTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYV 178
+ +SFEEAAS+PLA TA + KKG+ + + G+GGVG+ +Q AK FGA YV
Sbjct: 115 ENVSFEEAASVPLAGLTAWQCLVDFSAIKKGDKVLIHAGSGGVGSFAIQFAK-HFGA-YV 172
Query: 179 VSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGD--CKKSFVVAKKDGAI 236
++ S VK+ GAD+ ++Y + + ++ E +D + DT+G ++SF V K+ G +
Sbjct: 173 ATTASGKNESLVKELGADRFINYKEENFNEVLEDYDIVVDTLGGDILEQSFEVLKEGGKL 232
Query: 237 VDITWPASHERAV-------YSSLTVCGEILEKLRPYLERGELKAVIDPKGEYDFENVID 289
V I E+A + L G+ L ++ LE G++K+VI + + +
Sbjct: 233 VSIAGNPDEEKAKEKGIKAGFLWLEENGKQLSEVADLLENGKVKSVIGHTFPLTEQGLRE 292
Query: 290 AFGYIETGRAWGKVVV 305
A ET A GK+V+
Sbjct: 293 AHQLSETHHARGKIVI 308
>UniRef100_Q92E39 Lin0622 protein [Listeria innocua]
Length = 313
Score = 164 bits (415), Expect = 3e-39
Identities = 105/318 (33%), Positives = 173/318 (54%), Gaps = 23/318 (7%)
Query: 3 KAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRR---MRPIFPSDFPA 59
KA E YG KE LK + +P +NQ++V+ A ++NPID K R ++ + +FP
Sbjct: 2 KAVVIENYGGKEELKEKEVAMPKAGKNQVIVKEVATSINPIDWKLREGYLKQMMDWEFPI 61
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKP 119
+ G D+AGV+ G V + +GDEV+ + + GT A++ V+++L+A P
Sbjct: 62 ILGWDVAGVISEVGEGVTDWKVGDEVFAR-------PETTRFGTYAEYTAVDDHLLAPLP 114
Query: 120 KRLSFEEAASLPLAVQTAIEG-FKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYV 178
+ +SFEEAAS+PLA TA + F +KGE + + GAGGVGTL +QLAK + + V
Sbjct: 115 EGISFEEAASIPLAGLTAWQALFDHAKLQKGEKVLIHAGAGGVGTLAIQLAK--YAGAEV 172
Query: 179 VSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVG---DCKKSFVVAKKDGA 235
+++ S + +K GAD+V+DY + ++D+ D ++DT+G + V+ + G
Sbjct: 173 ITTASAKNHELLKSLGADQVIDYKEVNFKDVLSDIDVVFDTMGGQIETDSYDVLKEGTGR 232
Query: 236 IVDITWPASHERAVYSSLTV-------CGEILEKLRPYLERGELKAVIDPKGEYDFENVI 288
+V I ++ +RA ++T GE L+KL L +K ++ + + V
Sbjct: 233 LVSIVGISNEDRAKEKNVTANGIWLQPNGEQLKKLGELLANKTVKPIVGATFPFSEKGVF 292
Query: 289 DAFGYIETGRAWGKVVVT 306
DA ET A GK+V++
Sbjct: 293 DAHALSETHHAVGKIVIS 310
>UniRef100_Q6HG35 Quinone oxidoreductase [Bacillus thuringiensis]
Length = 331
Score = 163 bits (412), Expect = 7e-39
Identities = 108/333 (32%), Positives = 170/333 (50%), Gaps = 37/333 (11%)
Query: 3 KAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPI--FPSDFPAV 60
KA +G V +L + P L +L+ V A ++NPID+K R + +FPA+
Sbjct: 2 KAQIIHSFGDSSVFQLEEVAKPKLLPGHVLIHVKATSVNPIDTKMRSGAVSSVAPEFPAI 61
Query: 61 PGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPK 120
D+AG+VI G V KF GDEVYG F G LA+F++ + L+A KPK
Sbjct: 62 LHGDVAGIVIEVGEGVSKFKTGDEVYGCAGGFKETG-----GALAEFMLADARLIAHKPK 116
Query: 121 RLSFEEAASLPLAVQTAIEG-FKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVV 179
++ EEAA+LPL TA E F + K G+ + + G GGVG + +QLAK GA
Sbjct: 117 NITMEEAAALPLVAITAWESLFDRANIKSGQNVLIHGATGGVGHVAIQLAK-WAGAKVFT 175
Query: 180 SSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEK------FDFIYDTVG--DCKKSFVVAK 231
++ K++ + GAD ++Y + ++ +K F+ I+DTVG + SF A
Sbjct: 176 TASQQNKMEISHRLGADVAINYKEESVQEYVQKHTNGNGFEVIFDTVGGKNLDNSFEAAA 235
Query: 232 KDGAIVDITWPASHE-----------RAVYSSLTV--------CGEILEKLRPYLERGEL 272
+G +V I ++H+ + +L + CGEIL K+ +E G+L
Sbjct: 236 VNGTVVTIAARSTHDLSPLHAKGLSLHVTFMALKILHTDKRNDCGEILTKITQIVEEGKL 295
Query: 273 KAVIDPKGEYDFENVIDAFGYIETGRAWGKVVV 305
+ ++D K + F+ V A Y+E+ +A GK+V+
Sbjct: 296 RPLLDSK-SFTFDEVAQAHEYLESNKAIGKIVL 327
>UniRef100_Q638J3 Quinone oxidoreductase [Bacillus cereus]
Length = 331
Score = 163 bits (412), Expect = 7e-39
Identities = 108/333 (32%), Positives = 170/333 (50%), Gaps = 37/333 (11%)
Query: 3 KAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPI--FPSDFPAV 60
KA +G V +L + P L +L+ V A ++NPID+K R + +FPA+
Sbjct: 2 KAQIIHSFGDSSVFQLEEVAKPKLLPGHVLIHVKATSVNPIDTKMRSGAVSSVAPEFPAI 61
Query: 61 PGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPK 120
D+AG+VI G V KF GDEVYG F G LA+F++ + L+A KPK
Sbjct: 62 LHGDVAGIVIEVGEGVSKFKTGDEVYGCAGGFKETG-----GALAEFMLADARLIAHKPK 116
Query: 121 RLSFEEAASLPLAVQTAIEG-FKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVV 179
++ EEAA+LPL TA E F + K G+ + + G GGVG + +QLAK GA
Sbjct: 117 NVTMEEAAALPLVAITAWESLFDRANMKSGQNVLIHGATGGVGHVAIQLAK-WAGAKVFT 175
Query: 180 SSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEK------FDFIYDTVG--DCKKSFVVAK 231
++ K++ + GAD ++Y + ++ +K F+ I+DTVG + SF A
Sbjct: 176 TASQQNKMEIAHRLGADIAINYKEESVQEYVQKHTNGNGFEVIFDTVGGKNLDNSFEAAA 235
Query: 232 KDGAIVDITWPASHE-----------RAVYSSLTV--------CGEILEKLRPYLERGEL 272
+G +V I ++H+ + +L + CGEIL K+ +E G+L
Sbjct: 236 VNGTVVTIAARSTHDLSPLHAKGLSLHVTFMALKILHTDKRNDCGEILTKITQIVEEGKL 295
Query: 273 KAVIDPKGEYDFENVIDAFGYIETGRAWGKVVV 305
+ ++D K + F+ V A Y+E+ +A GK+V+
Sbjct: 296 RPLLDSK-SFTFDEVAQAHEYLESNKAIGKIVL 327
>UniRef100_Q81MY4 Alcohol dehydrogenase, zinc-containing [Bacillus anthracis]
Length = 317
Score = 162 bits (409), Expect = 2e-38
Identities = 113/321 (35%), Positives = 170/321 (52%), Gaps = 27/321 (8%)
Query: 3 KAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRR---MRPIFPSDFPA 59
KA +YG K VL+ + P +N +L++VYAA +NP+D K R ++ + DFP
Sbjct: 2 KAIGLTQYGDKSVLQEIEMQTPLLGDNDVLIEVYAAGVNPVDWKIREGLLQDVISYDFPL 61
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKP 119
V G D+AGVV G NV F +GDEVY S ++ GT A+++ V+E VA+KP
Sbjct: 62 VLGWDVAGVVAAIGKNVTVFKVGDEVY-------SRPDIERNGTYAEYVAVDEKYVAKKP 114
Query: 120 KRLSFEEAASLPLAVQTAIEGF-KTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYV 178
+ LSFEEAAS+PL T+ + K + +KG + + G+GG+GT +QLAK FGA +V
Sbjct: 115 RNLSFEEAASIPLVGLTSWQSLVKFANVQKGNKVLIHAGSGGIGTFAIQLAK-SFGA-HV 172
Query: 179 VSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVGD--CKKSFVVAKKDGAI 236
++ ST ++FVK GAD VVDY + + ++ ++D +G K S+ V +G +
Sbjct: 173 ATTTSTKNMQFVKDLGADTVVDYKTEDFSLLLHNYNIVFDVLGGDVLKDSYKVLAPNGKL 232
Query: 237 VDITWP---------ASHERAVYSSLTVC---GEILEKLRPYLERGELKAVIDPKGEYDF 284
I P S E+ + S G L + +E G++K V+
Sbjct: 233 ASIYGPKGMEIPQTEISREKNIESDHIFTEPNGYELSLITELIEGGKIKPVVTHVLPLHV 292
Query: 285 ENVIDAFGYIETGRAWGKVVV 305
E V A E+ RA GK+V+
Sbjct: 293 EGVKKAHHISESERARGKIVL 313
>UniRef100_Q722T7 Alcohol dehydrogenase, zinc-dependent [Listeria monocytogenes]
Length = 313
Score = 161 bits (408), Expect = 2e-38
Identities = 104/318 (32%), Positives = 172/318 (53%), Gaps = 23/318 (7%)
Query: 3 KAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRR---MRPIFPSDFPA 59
KA E YG KE LK + +P +NQ++V+ A ++NPID K R ++ + +FP
Sbjct: 2 KAVVIENYGGKEELKEKEVAMPKAGKNQVIVKEAATSINPIDWKLREGYLKQMMDWEFPI 61
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKP 119
+ G D+AGV+ G V + +GDEV+ + + GT A++ V+++L+A P
Sbjct: 62 ILGWDVAGVISEVGEGVTDWKVGDEVFAR-------PETTRFGTYAEYTAVDDHLLAPLP 114
Query: 120 KRLSFEEAASLPLAVQTAIEG-FKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYV 178
+ +SF+EAAS+PLA TA + F +KGE + + GAGGVGTL +QLAK + V
Sbjct: 115 EGISFDEAASIPLAGLTAWQALFDHAKLQKGEKVLIHAGAGGVGTLAIQLAK--HAGAEV 172
Query: 179 VSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVG---DCKKSFVVAKKDGA 235
+++ S + +K GAD+V+DY + ++D+ D ++DT+G + V+ + G
Sbjct: 173 ITTASAKNHELLKSLGADQVIDYKEVNFKDVLSDIDVVFDTMGGQIETDSYDVLKEGTGR 232
Query: 236 IVDITWPASHERAVYSSLTVC-------GEILEKLRPYLERGELKAVIDPKGEYDFENVI 288
+V I ++ ERA ++T GE L++L L +K ++ + + V
Sbjct: 233 LVSIVGISNEERAKEKNVTATGIWLQPNGEQLKELGKLLANKTVKPIVGATFPFSEKGVF 292
Query: 289 DAFGYIETGRAWGKVVVT 306
DA ET A GK+V++
Sbjct: 293 DAHALSETHHAVGKIVIS 310
>UniRef100_Q8Y9B9 Lmo0613 protein [Listeria monocytogenes]
Length = 313
Score = 160 bits (406), Expect = 4e-38
Identities = 103/318 (32%), Positives = 172/318 (53%), Gaps = 23/318 (7%)
Query: 3 KAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRR---MRPIFPSDFPA 59
KA E YG KE LK + +P +NQ++V+ A ++NPID K R ++ + +FP
Sbjct: 2 KAVVIENYGGKEELKEKEVAMPKAGKNQVIVKEAATSINPIDWKLREGYLKQMMDWEFPI 61
Query: 60 VPGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKP 119
+ G D+AGV+ G V + +GDEV+ + + GT A++ V+++L+A P
Sbjct: 62 ILGWDVAGVISEVGEGVTDWKVGDEVFAR-------PETTRFGTYAEYTAVDDHLLAPLP 114
Query: 120 KRLSFEEAASLPLAVQTAIEG-FKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYV 178
+ +SF+EAAS+PLA TA + F +KGE + + GAGGVGTL +QLAK + V
Sbjct: 115 EGISFDEAASIPLAGLTAWQALFDHAKLQKGEKVLIHAGAGGVGTLAIQLAK--HAGAEV 172
Query: 179 VSSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEKFDFIYDTVG---DCKKSFVVAKKDGA 235
+++ S + +K GAD+V+DY + ++D+ D ++DT+G + V+ + G
Sbjct: 173 ITTASAKNHELLKSLGADQVIDYKEVNFKDVLSDIDVVFDTMGGQIETDSYDVLKEGTGR 232
Query: 236 IVDITWPASHERAVYSSLTVC-------GEILEKLRPYLERGELKAVIDPKGEYDFENVI 288
+V I ++ +RA ++T GE L++L L +K ++ + + V
Sbjct: 233 LVSIVGISNEDRAKEKNVTATGIWLQPNGEQLKELGKLLANKTIKPIVGATFPFSEKGVF 292
Query: 289 DAFGYIETGRAWGKVVVT 306
DA ET A GK+V++
Sbjct: 293 DAHALSETHHAVGKIVIS 310
>UniRef100_UPI00003CB31E UPI00003CB31E UniRef100 entry
Length = 331
Score = 160 bits (405), Expect = 5e-38
Identities = 108/333 (32%), Positives = 168/333 (50%), Gaps = 37/333 (11%)
Query: 3 KAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPI--FPSDFPAV 60
KA +G V +L + P L +L+ V A ++NPID+K R + +FPA+
Sbjct: 2 KAQIIHSFGDSSVFQLEEVSKPKLLPGHVLIHVKATSVNPIDTKMRSGAVSAVAPEFPAI 61
Query: 61 PGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPK 120
D+AG+VI G V KF GDEVYG F G LA+F++ + L+A KP
Sbjct: 62 LHGDVAGIVIEVGEGVSKFKSGDEVYGCAGGFKETG-----GALAEFMLADARLIAHKPN 116
Query: 121 RLSFEEAASLPLAVQTAIEG-FKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVV 179
L+ EEAA+LPL TA E F + K G+ + + G GGVG + +QLAK GA
Sbjct: 117 NLTMEEAAALPLVAITAWESLFDRANIKPGQNVLIHGATGGVGHVAIQLAK-WAGAKVFT 175
Query: 180 SSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEK------FDFIYDTVG--DCKKSFVVAK 231
++ K++ GAD ++Y + ++ +K F+ I+DTVG + SF A
Sbjct: 176 TASQQSKMEISHHLGADIAINYKEESVQEYVQKHTNGNGFEVIFDTVGGKNLDHSFEAAA 235
Query: 232 KDGAIVDITWPASHE-----------RAVYSSLTV--------CGEILEKLRPYLERGEL 272
+G +V I ++H+ + +L + CGEIL K+ +E G+L
Sbjct: 236 VNGTVVTIAARSTHDLSPLHTKGLTLHVTFMALKILHTDKRNHCGEILTKITQIIEEGKL 295
Query: 273 KAVIDPKGEYDFENVIDAFGYIETGRAWGKVVV 305
+ ++D K + F+ V A Y+E+ +A GK+V+
Sbjct: 296 RPLLDSK-PFTFDEVAQAHEYLESNKAIGKIVL 327
>UniRef100_Q81MY1 Alcohol dehydrogenase, zinc-containing [Bacillus anthracis]
Length = 335
Score = 159 bits (401), Expect = 1e-37
Identities = 109/340 (32%), Positives = 170/340 (49%), Gaps = 47/340 (13%)
Query: 3 KAWFYEEYGPKEVLKLGDFPIPSPLENQLLVQVYAAALNPIDSKRRMRPI--FPSDFPAV 60
KA +G V +L + P L +L+ V A ++NPID+K R + +FPA+
Sbjct: 2 KAQIIHSFGDSSVFQLEEVSKPKLLPGHVLIDVKATSVNPIDTKMRSGAVSAVAPEFPAI 61
Query: 61 PGCDMAGVVIGKGVNVKKFDIGDEVYGNIQDFNSMEKPKQLGTLAQFIVVEENLVARKPK 120
D+AG+VI G V KF GDEVYG F G LA+F++ + L+A KP
Sbjct: 62 LHGDVAGIVIEVGEGVSKFKCGDEVYGCAGGFKETG-----GALAEFMLADARLIAHKPN 116
Query: 121 RLSFEEAASLPLAVQTAIEG-FKTGDFKKGETMFVVGGAGGVGTLVLQLAKLMFGASYVV 179
++ EEAA+LPL TA E F + K G+ + + G GGVG + +QLAK GA+
Sbjct: 117 NITMEEAAALPLVAITAWESLFDRANIKSGQNVLIHGATGGVGHVAIQLAK-WAGANVFT 175
Query: 180 SSCSTPKLKFVKQFGADKVVDYTKTKYEDIEEK-------------FDFIYDTVG--DCK 224
++ K++ + GAD ++Y K E ++E F+ I+DTVG +
Sbjct: 176 TASQQNKMEIAHRLGADVAINY---KEESVQESVQEYVQKHTNGNGFEVIFDTVGGKNLD 232
Query: 225 KSFVVAKKDGAIVDITWPASHE-----------RAVYSSLTV--------CGEILEKLRP 265
SF A +G +V I ++H+ + +L + CGEIL K+
Sbjct: 233 NSFEAAAVNGTVVTIAARSTHDLSPLHAKGLSLHVTFMALKILHTDKRNDCGEILTKITK 292
Query: 266 YLERGELKAVIDPKGEYDFENVIDAFGYIETGRAWGKVVV 305
+E G+L+ ++D K + F+ V A Y+E+ +A GK+V+
Sbjct: 293 IVEEGKLRPLLDSK-SFTFDEVAQAHEYLESNKAIGKIVL 331
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.139 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 590,054,127
Number of Sequences: 2790947
Number of extensions: 25622602
Number of successful extensions: 73617
Number of sequences better than 10.0: 3476
Number of HSP's better than 10.0 without gapping: 1057
Number of HSP's successfully gapped in prelim test: 2420
Number of HSP's that attempted gapping in prelim test: 68013
Number of HSP's gapped (non-prelim): 4191
length of query: 340
length of database: 848,049,833
effective HSP length: 128
effective length of query: 212
effective length of database: 490,808,617
effective search space: 104051426804
effective search space used: 104051426804
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC146342.2


