

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146341.6 + phase: 0
(540 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
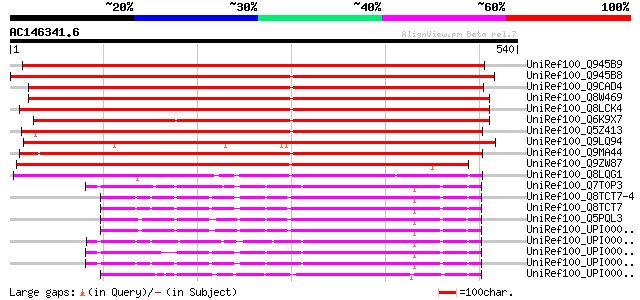
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q945B9 Growth-on protein GRO11 [Euphorbia esula] 785 0.0
UniRef100_Q945B8 Growth-on protein GRO10 [Euphorbia esula] 745 0.0
UniRef100_Q9CAD4 Hypothetical protein F24D7.12 [Arabidopsis thal... 744 0.0
UniRef100_Q8W469 Hypothetical protein At1g63690; F24D7.12 [Arabi... 743 0.0
UniRef100_Q8LCK4 Growth-on protein GRO10 [Arabidopsis thaliana] 740 0.0
UniRef100_Q6K9X7 Putative growth-on protein GRO10 [Oryza sativa] 727 0.0
UniRef100_Q5Z413 Putative growth-on protein GRO10 [Oryza sativa] 724 0.0
UniRef100_Q9LQ94 T1N6.3 protein [Arabidopsis thaliana] 647 0.0
UniRef100_Q9MA44 T20M3.9 protein [Arabidopsis thaliana] 422 e-116
UniRef100_Q9ZW87 Hypothetical protein At2g43105 [Arabidopsis tha... 419 e-115
UniRef100_Q8LQG1 Vacuolar sorting receptor-like protein [Oryza s... 385 e-105
UniRef100_Q7T0P3 MGC69113 protein [Xenopus laevis] 206 2e-51
UniRef100_Q8TCT7-4 Splice isoform 4 of Q8TCT7 [Homo sapiens] 206 2e-51
UniRef100_Q8TCT7 Signal peptide peptidase-like 2B [Homo sapiens] 206 2e-51
UniRef100_Q5PQL3 Hypothetical protein [Rattus norvegicus] 203 9e-51
UniRef100_UPI00001CFC14 UPI00001CFC14 UniRef100 entry 201 3e-50
UniRef100_UPI0000362B74 UPI0000362B74 UniRef100 entry 199 2e-49
UniRef100_UPI00003AC9AF UPI00003AC9AF UniRef100 entry 198 4e-49
UniRef100_UPI00003AC9AE UPI00003AC9AE UniRef100 entry 197 6e-49
UniRef100_UPI00001CF2D7 UPI00001CF2D7 UniRef100 entry 196 1e-48
>UniRef100_Q945B9 Growth-on protein GRO11 [Euphorbia esula]
Length = 538
Score = 785 bits (2026), Expect = 0.0
Identities = 383/495 (77%), Positives = 433/495 (87%), Gaps = 3/495 (0%)
Query: 14 YVFLFSVSLVLA--GDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENAEYVGVGARFGP 71
Y+ L + SL A GDIVH DD +P RPGC+NNFVLVKVPTW+DGVE+ EYVGVGARFG
Sbjct: 10 YLALLASSLCFASAGDIVHQDDKSPKRPGCDNNFVLVKVPTWVDGVEDIEYVGVGARFGR 69
Query: 72 TLESKEKHANHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANIADEAGASAIL 131
TLE+KEK AN T++V+ADPPD C PK KL ++ILVHRG CSFTTKA IA+ A ASAIL
Sbjct: 70 TLEAKEKDANKTKLVLADPPDLCQPPKFKLNRDVILVHRGNCSFTTKAKIAELANASAIL 129
Query: 132 IINYRTELFKMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVD 191
IIN TEL KMVCE NETDV I IPAVMLPQDAG +L ++QN+S VS+QLYSP RPLVD
Sbjct: 130 IINTETELLKMVCEANETDVHIQIPAVMLPQDAGGSLRDYMQNSSQVSVQLYSPERPLVD 189
Query: 192 VAEVFLWLMAVGTILCASYWSAWTAREAAIEQEKLLKDASDEYV-AESVGSRGYVEISTT 250
VAEVFLWLMAVGTIL ASYWSAW+ARE AIEQ+KLLKD SD++ E V S G V I+TT
Sbjct: 190 VAEVFLWLMAVGTILGASYWSAWSAREIAIEQDKLLKDGSDDFTHGEGVPSTGVVNINTT 249
Query: 251 AAILFVVLASCFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQT 310
+AILFVV+ASCFLVMLYKLMS WF++VLVVLFCIGG+EGLQTCL ALLSCFRWFQ+P ++
Sbjct: 250 SAILFVVIASCFLVMLYKLMSVWFMDVLVVLFCIGGVEGLQTCLVALLSCFRWFQHPGES 309
Query: 311 YVKIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILGIALIITVLQIVRIPN 370
++K+P GA+ +LTLAV+PFCIVFAV+WA+ R+ S+AWIGQDILGIALIITVLQIV +PN
Sbjct: 310 FIKLPVVGAISHLTLAVSPFCIVFAVIWAIHRRDSFAWIGQDILGIALIITVLQIVHVPN 369
Query: 371 LKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPMLLKLPRLFDPWGG 430
LKVGTVLLSCAFLYDI WVFVSK W +SVMIVVARGDKSGEDGIPMLLK+PR+FDPWGG
Sbjct: 370 LKVGTVLLSCAFLYDIFWVFVSKLWLKDSVMIVVARGDKSGEDGIPMLLKIPRMFDPWGG 429
Query: 431 YSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHG 490
YSIIGFGDIILPGL+V F+LRYDWL KKNLRAGYF+WAMTAYGLGLL+TYVALNLMDGHG
Sbjct: 430 YSIIGFGDIILPGLLVTFALRYDWLTKKNLRAGYFLWAMTAYGLGLLVTYVALNLMDGHG 489
Query: 491 QPALLYIVPFTLGKY 505
QPALLYIVPFTLG +
Sbjct: 490 QPALLYIVPFTLGTF 504
>UniRef100_Q945B8 Growth-on protein GRO10 [Euphorbia esula]
Length = 537
Score = 745 bits (1923), Expect = 0.0
Identities = 366/516 (70%), Positives = 432/516 (82%), Gaps = 3/516 (0%)
Query: 2 VSLGVIYSNIVAYVFLFSVSLVLAGDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENAE 61
+ LG + S +V + V+AGDIVH D AP +PGCEN+FVLVKV TW++G E+AE
Sbjct: 1 MDLGFVISVVVVISLVCYPYYVIAGDIVHDDASAPKKPGCENDFVLVKVQTWVNGEEDAE 60
Query: 62 YVGVGARFGPTLESKEKHANHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANI 121
+VGVGARFG T+ SKEK+AN TR+ ++DP DCCS PK K EII+V RG C FT KAN
Sbjct: 61 FVGVGARFGTTIVSKEKNANQTRLTLSDPRDCCSPPKRKFAGEIIMVDRGNCKFTAKANY 120
Query: 122 ADEAGASAILIINYRTELFKMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQ 181
A+ AGA+A+LIIN + EL+KMVC+ +ETD+DI IPAVMLPQDAG +LE+ + +N+ VS+Q
Sbjct: 121 AEAAGATAVLIINNQKELYKMVCDPDETDLDIKIPAVMLPQDAGASLEKMLLSNASVSVQ 180
Query: 182 LYSPLRPLVDVAEVFLWLMAVGTILCASYWSAWTAREAAIEQEKLLKDASDEYVAES-VG 240
LYSP RPLVD+AEVFLWLMAV TILCASYWSAW+ REA IE +KLLKDA DE + VG
Sbjct: 181 LYSPRRPLVDIAEVFLWLMAVITILCASYWSAWSTREAVIEHDKLLKDALDEIPNDKGVG 240
Query: 241 SRGYVEISTTAAILFVVLASCFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSC 300
V+I+T++A+LFVV+ASCFLVMLYKLMS+WF+E+LVVLFCIGG+EGLQTCL ALLS
Sbjct: 241 FSSVVDINTSSAVLFVVVASCFLVMLYKLMSYWFVELLVVLFCIGGVEGLQTCLVALLS- 299
Query: 301 FRWFQYPAQTYVKIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILGIALII 360
RWF++ ++YVK+PFFGA+ +LTLAV+PFCI FAVVWAV R S+AWIGQDILGIALII
Sbjct: 300 -RWFKHAGESYVKVPFFGALSHLTLAVSPFCITFAVVWAVYRNVSFAWIGQDILGIALII 358
Query: 361 TVLQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPMLLK 420
TVLQIV +PNLKVGTVLLSCAFLYDI WVFVSK F ESVMIVVARGD+SGEDGIPMLLK
Sbjct: 359 TVLQIVHVPNLKVGTVLLSCAFLYDIFWVFVSKKLFKESVMIVVARGDRSGEDGIPMLLK 418
Query: 421 LPRLFDPWGGYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITY 480
+PR+FDPWGGYSIIGFGDI+LPGL++AF+LRYDWLA K+LRAGYF+WAM AYGLGLLITY
Sbjct: 419 IPRMFDPWGGYSIIGFGDILLPGLLIAFALRYDWLANKSLRAGYFLWAMIAYGLGLLITY 478
Query: 481 VALNLMDGHGQPALLYIVPFTLGKYNHLTHKFESVN 516
VALNLMDGH QPALLYIVPFTLG + L K +N
Sbjct: 479 VALNLMDGHDQPALLYIVPFTLGTFLALGKKNGDLN 514
>UniRef100_Q9CAD4 Hypothetical protein F24D7.12 [Arabidopsis thaliana]
Length = 519
Score = 744 bits (1921), Expect = 0.0
Identities = 364/484 (75%), Positives = 417/484 (85%), Gaps = 2/484 (0%)
Query: 21 SLVLAGDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENAEYVGVGARFGPTLESKEKHA 80
S V AGDIVH D++AP +PGCEN+FVLVKV TWIDGVEN E+VGVGARFG + SKEK+A
Sbjct: 23 STVTAGDIVHQDNLAPKKPGCENDFVLVKVQTWIDGVENEEFVGVGARFGKRIVSKEKNA 82
Query: 81 NHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTELF 140
N T +V A+P D C+ KNKL+ ++++V RG C FT KAN A+ AGASA+LIIN + EL+
Sbjct: 83 NQTHLVFANPRDSCTPLKNKLSGDVVIVERGNCRFTAKANNAEAAGASALLIINNQKELY 142
Query: 141 KMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVDVAEVFLWLM 200
KMVCE +ETD+DI IPAVMLPQDAG +L++ + N+S VS QLYSP RP VDVAEVFLWLM
Sbjct: 143 KMVCEPDETDLDIQIPAVMLPQDAGASLQKMLANSSKVSAQLYSPRRPAVDVAEVFLWLM 202
Query: 201 AVGTILCASYWSAWTAREAAIEQEKLLKDASDEYVAESVGSRGYVEISTTAAILFVVLAS 260
A+GTILCASYWSAW+AREAAIE +KLLKDA DE + G G VEI++ +AI FVVLAS
Sbjct: 203 AIGTILCASYWSAWSAREAAIEHDKLLKDAIDEIPNTNDGGSGVVEINSISAIFFVVLAS 262
Query: 261 CFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYVKIPFFGAV 320
FLV+LYKLMS+WF+E+LVV+FCIGG+EGLQTCL ALLS RWFQ A TYVK+PF G +
Sbjct: 263 GFLVILYKLMSYWFVELLVVVFCIGGVEGLQTCLVALLS--RWFQRAADTYVKVPFLGPI 320
Query: 321 PYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILGIALIITVLQIVRIPNLKVGTVLLSC 380
YLTLAV+PFCIVFAV+WAV R S+AWIGQD+LGIALIITVLQIV +PNLKVGTVLLSC
Sbjct: 321 SYLTLAVSPFCIVFAVLWAVYRVHSFAWIGQDVLGIALIITVLQIVHVPNLKVGTVLLSC 380
Query: 381 AFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPMLLKLPRLFDPWGGYSIIGFGDII 440
AFLYDI WVFVSK FHESVMIVVARGDKSGEDGIPMLLK+PR+FDPWGGYSIIGFGDI+
Sbjct: 381 AFLYDIFWVFVSKKLFHESVMIVVARGDKSGEDGIPMLLKIPRMFDPWGGYSIIGFGDIL 440
Query: 441 LPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVPF 500
LPGL++AF+LRYDWLA K LR GYF+WAM AYGLGLLITYVALNLMDGHGQPALLYIVPF
Sbjct: 441 LPGLLIAFALRYDWLANKTLRTGYFIWAMVAYGLGLLITYVALNLMDGHGQPALLYIVPF 500
Query: 501 TLGK 504
TLGK
Sbjct: 501 TLGK 504
>UniRef100_Q8W469 Hypothetical protein At1g63690; F24D7.12 [Arabidopsis thaliana]
Length = 540
Score = 743 bits (1918), Expect = 0.0
Identities = 365/491 (74%), Positives = 418/491 (84%), Gaps = 2/491 (0%)
Query: 21 SLVLAGDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENAEYVGVGARFGPTLESKEKHA 80
S V AGDIVH D++AP +PGCEN+FVLVKV TWIDGVEN E+VGVGARFG + SKEK+A
Sbjct: 23 STVTAGDIVHQDNLAPKKPGCENDFVLVKVQTWIDGVENEEFVGVGARFGKRIVSKEKNA 82
Query: 81 NHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTELF 140
N T +V A+P D C+ KNKL+ ++++V RG C FT KAN A+ AGASA+LIIN + EL+
Sbjct: 83 NQTHLVFANPRDSCTPLKNKLSGDVVIVERGNCRFTAKANNAEAAGASALLIINNQKELY 142
Query: 141 KMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVDVAEVFLWLM 200
KMVCE +ETD+DI IPAVMLPQDAG +L++ + N+S VS QLYSP RP VDVAEVFLWLM
Sbjct: 143 KMVCEPDETDLDIQIPAVMLPQDAGASLQKMLANSSKVSAQLYSPRRPAVDVAEVFLWLM 202
Query: 201 AVGTILCASYWSAWTAREAAIEQEKLLKDASDEYVAESVGSRGYVEISTTAAILFVVLAS 260
A+GTILCASYWSAW+AREAAIE +KLLKDA DE + G G VEI++ +AI FVVLAS
Sbjct: 203 AIGTILCASYWSAWSAREAAIEHDKLLKDAIDEIPNTNDGGSGVVEINSISAIFFVVLAS 262
Query: 261 CFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYVKIPFFGAV 320
FLV+LYKLMS+WF+E+LVV+FCIGG+EGLQTCL ALLS RWFQ A TYVK+PF G +
Sbjct: 263 GFLVILYKLMSYWFVELLVVVFCIGGVEGLQTCLVALLS--RWFQRAADTYVKVPFLGPI 320
Query: 321 PYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILGIALIITVLQIVRIPNLKVGTVLLSC 380
YLTLAV+PFCIVFAV+WAV R S+AWIGQD+LGIALIITVLQIV +PNLKVGTVLLSC
Sbjct: 321 SYLTLAVSPFCIVFAVLWAVYRVHSFAWIGQDVLGIALIITVLQIVHVPNLKVGTVLLSC 380
Query: 381 AFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPMLLKLPRLFDPWGGYSIIGFGDII 440
AFLYDI WVFVSK FHESVMIVVARGDKSGEDGIPMLLK+PR+FDPWGGYSIIGFGDI+
Sbjct: 381 AFLYDIFWVFVSKKLFHESVMIVVARGDKSGEDGIPMLLKIPRMFDPWGGYSIIGFGDIL 440
Query: 441 LPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVPF 500
LPGL++AF+LRYDWLA K LR GYF+WAM AYGLGLLITYVALNLMDGHGQPALLYIVPF
Sbjct: 441 LPGLLIAFALRYDWLANKTLRTGYFIWAMVAYGLGLLITYVALNLMDGHGQPALLYIVPF 500
Query: 501 TLGKYNHLTHK 511
TLG L K
Sbjct: 501 TLGTMLTLARK 511
>UniRef100_Q8LCK4 Growth-on protein GRO10 [Arabidopsis thaliana]
Length = 540
Score = 740 bits (1911), Expect = 0.0
Identities = 367/502 (73%), Positives = 424/502 (84%), Gaps = 3/502 (0%)
Query: 11 IVAYVFLFSV-SLVLAGDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENAEYVGVGARF 69
+++ + L S+ S V AGDIVH D++AP +PGCEN+FVLVKV TWIDGVEN E+VGVGARF
Sbjct: 12 LLSSILLLSLRSTVTAGDIVHQDNLAPKKPGCENDFVLVKVQTWIDGVENEEFVGVGARF 71
Query: 70 GPTLESKEKHANHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANIADEAGASA 129
G + SKEK+AN T +V A+P D C+ KNKL+ ++++V RG C FT KAN A+ AGASA
Sbjct: 72 GKRIVSKEKNANQTHLVFANPRDSCTPLKNKLSGDVVIVERGNCRFTAKANNAEAAGASA 131
Query: 130 ILIINYRTELFKMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPL 189
+LIIN + EL+KMVCE +ETD+DI IPAVMLPQDAG +L++ + N+S VS QLYSP RP
Sbjct: 132 LLIINNQKELYKMVCEPDETDLDIQIPAVMLPQDAGASLQKMLANSSKVSAQLYSPRRPA 191
Query: 190 VDVAEVFLWLMAVGTILCASYWSAWTAREAAIEQEKLLKDASDEYVAESVGSRGYVEIST 249
VDVAEVFL LMA+GTILCASYWSAW+AREAAIE +KLLKDA DE + G G VEI+T
Sbjct: 192 VDVAEVFLCLMAIGTILCASYWSAWSAREAAIEHDKLLKDAIDEIPNTNDGGSGVVEINT 251
Query: 250 TAAILFVVLASCFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQ 309
+AI FVVLAS FLV+LYKLMS+WF+E+LVV+FCIGG+EGLQTCL ALLS RWFQ A
Sbjct: 252 ISAIFFVVLASGFLVILYKLMSYWFVELLVVVFCIGGVEGLQTCLVALLS--RWFQRAAD 309
Query: 310 TYVKIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILGIALIITVLQIVRIP 369
TYVK+PF G + YLTLAV+PFCIVFAV+WAV R S+AWIGQD+LGIALIITVLQIV +P
Sbjct: 310 TYVKVPFLGPISYLTLAVSPFCIVFAVLWAVYRVHSFAWIGQDVLGIALIITVLQIVHVP 369
Query: 370 NLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPMLLKLPRLFDPWG 429
NLKVGTVLLSCAFLYDI WVFVSK FHESVMIVVARGDKSGEDGIPMLLK+PR+FDPWG
Sbjct: 370 NLKVGTVLLSCAFLYDIFWVFVSKKLFHESVMIVVARGDKSGEDGIPMLLKIPRMFDPWG 429
Query: 430 GYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGH 489
GYSIIGFGDI+LPGL++AF+LRYDWLA K LR GYF+WAM AYGLGLLITYVALNLMDGH
Sbjct: 430 GYSIIGFGDILLPGLLIAFALRYDWLANKTLRTGYFIWAMVAYGLGLLITYVALNLMDGH 489
Query: 490 GQPALLYIVPFTLGKYNHLTHK 511
GQPALLYIVPFTLG L K
Sbjct: 490 GQPALLYIVPFTLGTMLTLARK 511
>UniRef100_Q6K9X7 Putative growth-on protein GRO10 [Oryza sativa]
Length = 538
Score = 727 bits (1876), Expect = 0.0
Identities = 348/487 (71%), Positives = 416/487 (84%), Gaps = 4/487 (0%)
Query: 26 GDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENAEYVGVGARFGPTLESKEKHANHTRV 85
GDIVH DD AP PGC N+FVLVKV TW++ E+ E+VGVGARFGPT+ESKEKHAN T +
Sbjct: 19 GDIVHQDDEAPKIPGCSNDFVLVKVQTWVNNREDGEFVGVGARFGPTIESKEKHANRTGL 78
Query: 86 VMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTELFKMVCE 145
++ADP DCC P K+ +++LV RG C FT KA A+ AGASAI+IIN+ EL+KMVC+
Sbjct: 79 LLADPIDCCDPPTQKVAGDVLLVQRGNCKFTKKAKNAEAAGASAIIIINHVHELYKMVCD 138
Query: 146 ENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVDVAEVFLWLMAVGTI 205
NETD+DI IPAV+LP+DAG +L++ + VS+QLYSP RPLVD AEVFLWLMAVGTI
Sbjct: 139 RNETDLDINIPAVLLPKDAGNDLQKLLTRGK-VSVQLYSPDRPLVDTAEVFLWLMAVGTI 197
Query: 206 LCASYWSAWTAREAAIEQEKLLKDASDEYV-AESVGSRGYVEISTTAAILFVVLASCFLV 264
LCASYWSAW+AREA IEQEKLLKD + + E+ GS G V+I+ T+AILFVV+ASCFL+
Sbjct: 198 LCASYWSAWSAREAVIEQEKLLKDGHESSLNLEAGGSSGMVDINMTSAILFVVIASCFLI 257
Query: 265 MLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYVKIPFFGAVPYLT 324
MLYKLMS WF+E+LVV+FCIGG+EGLQTCL ALLS RWF+ A+++VK+PFFGAV YLT
Sbjct: 258 MLYKLMSHWFVELLVVIFCIGGVEGLQTCLVALLS--RWFKPAAESFVKVPFFGAVSYLT 315
Query: 325 LAVTPFCIVFAVVWAVKRQASYAWIGQDILGIALIITVLQIVRIPNLKVGTVLLSCAFLY 384
+AV PFCIVFAV+WAV R+ +YAWIGQDILGIALI+TV+QIVRIPNLKVG+VLLSC+FLY
Sbjct: 316 IAVCPFCIVFAVIWAVYRRMTYAWIGQDILGIALIVTVIQIVRIPNLKVGSVLLSCSFLY 375
Query: 385 DILWVFVSKWWFHESVMIVVARGDKSGEDGIPMLLKLPRLFDPWGGYSIIGFGDIILPGL 444
DI WVF+SK WFHESVMIVVARGDK+ EDG+PMLLK+PR+FDPWGG+SIIGFGDI+LPGL
Sbjct: 376 DIFWVFISKMWFHESVMIVVARGDKTDEDGVPMLLKIPRMFDPWGGFSIIGFGDILLPGL 435
Query: 445 VVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVPFTLGK 504
++AF+LRYDW AKK L++GYF+W+M AYG GL+ITYVALNLMDGHGQPALLYIVPFTLG
Sbjct: 436 LIAFALRYDWAAKKTLQSGYFLWSMVAYGSGLMITYVALNLMDGHGQPALLYIVPFTLGT 495
Query: 505 YNHLTHK 511
+ L K
Sbjct: 496 FIALGRK 502
>UniRef100_Q5Z413 Putative growth-on protein GRO10 [Oryza sativa]
Length = 542
Score = 724 bits (1869), Expect = 0.0
Identities = 347/497 (69%), Positives = 418/497 (83%), Gaps = 8/497 (1%)
Query: 13 AYVFLFSVSLVLAG-----DIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENAEYVGVGA 67
A VF ++ LAG DIVHHDD AP PGC N+F+LVKV +W++G E+ EYVGVGA
Sbjct: 6 AAVFALLMASALAGAAAGGDIVHHDDEAPKIPGCSNDFILVKVQSWVNGKEDDEYVGVGA 65
Query: 68 RFGPTLESKEKHANHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANIADEAGA 127
RFGP + SKEKHAN TR+++ADP DCC+ PK K++ +I+LV RGKC FT KA A+ AGA
Sbjct: 66 RFGPQIVSKEKHANRTRLMLADPIDCCTSPKEKVSGDILLVQRGKCKFTKKAKFAEAAGA 125
Query: 128 SAILIINYRTELFKMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPLR 187
S I+IIN+ EL+KMVCE+NETD+DI IPAV+LP+DAG L + + + VS+Q YSP R
Sbjct: 126 SGIIIINHVHELYKMVCEKNETDLDINIPAVLLPRDAGFALHTVLTSGNSVSVQQYSPDR 185
Query: 188 PLVDVAEVFLWLMAVGTILCASYWSAWTAREAAIEQEKLLKDASDEYV-AESVGSRGYVE 246
P+VD AEVFLWLMAVGT+LCASYWSAW+AREA EQEKLLKD + + E+ S G ++
Sbjct: 186 PVVDTAEVFLWLMAVGTVLCASYWSAWSAREALCEQEKLLKDGREVLLNVENGSSSGMID 245
Query: 247 ISTTAAILFVVLASCFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSCFRWFQY 306
I+ +AI+FVV+ASCFL+MLYK+MS WF+E+LVV+FC+GG+EGLQTCL ALLS RWF+
Sbjct: 246 INVASAIMFVVVASCFLIMLYKMMSSWFVELLVVIFCVGGVEGLQTCLVALLS--RWFRA 303
Query: 307 PAQTYVKIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILGIALIITVLQIV 366
++++ K+PFFGAV YLTLAV+PFCIVFAV+WAV R +YAWIGQDILGIALIITV+QIV
Sbjct: 304 ASESFFKVPFFGAVSYLTLAVSPFCIVFAVLWAVHRHFTYAWIGQDILGIALIITVIQIV 363
Query: 367 RIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPMLLKLPRLFD 426
R+PNLKVG+VLLSCAF YDI WVFVSK WFHESVMIVVARGDK+ EDG+PMLLK+PR+FD
Sbjct: 364 RVPNLKVGSVLLSCAFFYDIFWVFVSKRWFHESVMIVVARGDKTDEDGVPMLLKIPRMFD 423
Query: 427 PWGGYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLM 486
PWGGYSIIGFGDI+LPGL+VAF+LRYDW AKK+L+ GYF+W+M AYG GLLITYVALNLM
Sbjct: 424 PWGGYSIIGFGDILLPGLLVAFALRYDWAAKKSLQTGYFLWSMVAYGSGLLITYVALNLM 483
Query: 487 DGHGQPALLYIVPFTLG 503
DGHGQPALLYIVPFTLG
Sbjct: 484 DGHGQPALLYIVPFTLG 500
>UniRef100_Q9LQ94 T1N6.3 protein [Arabidopsis thaliana]
Length = 613
Score = 647 bits (1670), Expect = 0.0
Identities = 342/577 (59%), Positives = 399/577 (68%), Gaps = 74/577 (12%)
Query: 15 VFLFSVSLVLAGDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENAEYVGVGARFGPTLE 74
+ L+S S V AGDIVHHDD P RPGC NNFVLVKVPT ++G E EYVGVGARFGPTLE
Sbjct: 16 LLLYSASFVCAGDIVHHDDSLPQRPGCNNNFVLVKVPTRVNGSEYTEYVGVGARFGPTLE 75
Query: 75 SKEKHANHTRVVMADPPDCCSKPKNKLTNEIILVHR----GKCSFTTKANIADEAGASAI 130
SKEKHA ++ +ADPPDCCS PKNK+ LV C + S +
Sbjct: 76 SKEKHATLIKLAIADPPDCCSTPKNKVKFPFWLVSLVLIFPSCVIPVSLSFGSLQERSFL 135
Query: 131 LIINYRT--ELFKMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPLRP 188
I+ +++ + E +DI IP VMLP DAG +LE +++N+IV++QLYSP RP
Sbjct: 136 FIVVNAVLPPKLRLLKQLGENVLDITIPVVMLPVDAGRSLENIVKSNAIVTLQLYSPKRP 195
Query: 189 LVDVAEVFLWLMAVGTILCASYWSAWTAREAAIEQEKLLK-------------------- 228
VDVAEVFLWLMAVGTILCASYWSAWT RE AIEQ+KLLK
Sbjct: 196 AVDVAEVFLWLMAVGTILCASYWSAWTVREEAIEQDKLLKVIAGSKFRTILYSGHPPCSN 255
Query: 229 -------------------DASDEYVAESV-GSRGYVEISTTAAILFVVLASCFLVMLYK 268
D SDE + S SRG VE++ +AILFVV+ASCFL+MLYK
Sbjct: 256 LLTAKEKCPHSFVIYLKLQDGSDELLQLSTTSSRGVVEVTVISAILFVVVASCFLIMLYK 315
Query: 269 LMSFWFLEVLVVLFCIGGIE----------------GLQTC------------LTALLSC 300
LMSFWF+EVLVVLFCIGG+E ++ C ++L++
Sbjct: 316 LMSFWFIEVLVVLFCIGGVEQYLIEPSCTDVESFVGAIRGCKLVWSLYSHGMYSSSLINI 375
Query: 301 FRWFQYPAQTYVKIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILGIALII 360
F WF+ ++YVK+PF GAV YLTLA+ PFCI FAV WAVKRQ SYAWIGQDILGI+LII
Sbjct: 376 FLWFRRFGESYVKVPFLGAVSYLTLAICPFCIAFAVFWAVKRQYSYAWIGQDILGISLII 435
Query: 361 TVLQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPMLLK 420
TVLQIVR+PNLKVG VLLSCAF+YDI WVFVSKWWF ESVMIVVARGD+SGEDGIPMLLK
Sbjct: 436 TVLQIVRVPNLKVGFVLLSCAFMYDIFWVFVSKWWFRESVMIVVARGDRSGEDGIPMLLK 495
Query: 421 LPRLFDPWGGYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITY 480
+PR+FDPWGGYSIIGFGDIILPGL+V F+LRYDWLA K L++GYF+ M+AYGLGLLITY
Sbjct: 496 IPRMFDPWGGYSIIGFGDIILPGLLVTFALRYDWLANKRLKSGYFLGTMSAYGLGLLITY 555
Query: 481 VALNLMDGHGQPALLYIVPFTLGKYNHLTHKFESVNT 517
+ALNLMDGHGQPALLYIVPF LG L HK + T
Sbjct: 556 IALNLMDGHGQPALLYIVPFILGTLFVLGHKRGDLKT 592
>UniRef100_Q9MA44 T20M3.9 protein [Arabidopsis thaliana]
Length = 536
Score = 422 bits (1085), Expect = e-116
Identities = 217/495 (43%), Positives = 306/495 (60%), Gaps = 5/495 (1%)
Query: 11 IVAYVFLFSVSLVLAGDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENAEYVGVGARFG 70
+ A L+ + L+ G D AP PGC N F +VKV W++G + + A+FG
Sbjct: 11 LAAAAALYLIGLLCVGADTK-DVTAPKIPGCSNEFQMVKVENWVNGENGETFTAMTAQFG 69
Query: 71 PTLESKEKHANHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANIADEAGASAI 130
L S + A V + P D CS +KL+ I L RG+C+FT KA +A GA+A+
Sbjct: 70 TMLPSDKDKAVKLPVALTTPLDSCSNLTSKLSWSIALSVRGECAFTVKAQVAQAGGAAAL 129
Query: 131 LIINYRTELFKMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLV 190
++IN + EL +MVC E +T +++ IP +M+ +G L++ I N V + LY+P P+V
Sbjct: 130 VLINDKEELDEMVCGEKDTSLNVSIPILMITTSSGDALKKSIMQNKKVELLLYAPKSPIV 189
Query: 191 DVAEVFLWLMAVGTILCASYWSAWTAREAAIEQEKLL--KDASDEYVAESVGSRGYVEIS 248
D A VFLWLM+VGT+ AS WS T+ + EQ L K +S+ + ++IS
Sbjct: 190 DYAVVFLWLMSVGTVFVASVWSHVTSPKKNDEQYDELSPKKSSNVDATKGGAEEETLDIS 249
Query: 249 TTAAILFVVLASCFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSCFRWFQYPA 308
A++FV+ AS FLV+L+ MS WF+ +L + F IGG++G+ L++ R
Sbjct: 250 AMGAVIFVISASTFLVLLFFFMSSWFILILTIFFVIGGMQGMHNINVTLIT--RRCSKCG 307
Query: 309 QTYVKIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILGIALIITVLQIVRI 368
Q +K+P G L+L V FC V A++W + R+ S+AW GQDI GI ++I VLQ+ R+
Sbjct: 308 QKNLKLPLLGNTSILSLVVLLFCFVVAILWFMNRKTSHAWAGQDIFGICMMINVLQVARL 367
Query: 369 PNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPMLLKLPRLFDPW 428
PN++V T+LL CAF YDI WVF+S F +SVMI VARG K + IPMLL++PRL DPW
Sbjct: 368 PNIRVATILLCCAFFYDIFWVFISPLIFKQSVMIAVARGSKDTGESIPMLLRIPRLSDPW 427
Query: 429 GGYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDG 488
GGY++IGFGDI+ PGL++ F R+D K + GYF W M YGLGL +TY+ L +M+G
Sbjct: 428 GGYNMIGFGDILFPGLLICFIFRFDKENNKGVSNGYFPWLMFGYGLGLFLTYLGLYVMNG 487
Query: 489 HGQPALLYIVPFTLG 503
HGQPALLY+VP TLG
Sbjct: 488 HGQPALLYLVPCTLG 502
>UniRef100_Q9ZW87 Hypothetical protein At2g43105 [Arabidopsis thaliana]
Length = 543
Score = 419 bits (1076), Expect = e-115
Identities = 219/506 (43%), Positives = 308/506 (60%), Gaps = 27/506 (5%)
Query: 8 YSNIVAYVFLFSVSLVLAGDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENAEYVGVGA 67
YS +V + L S+ A D+ +D + PGC N F +VKV W+DGVE G+ A
Sbjct: 11 YSALVLILLLLGFSVAAADDVSWTEDSSLESPGCTNKFQMVKVLNWVDGVEGDFLTGLTA 70
Query: 68 RFGPTLESKEKHANHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANIADEAGA 127
+FG L S A DP D CS ++L I L RG C+FT KA A+ AGA
Sbjct: 71 QFGAALPSVPDQALRFPAAFVDPLDSCSHLSSRLDGHIALSIRGNCAFTEKAKHAEAAGA 130
Query: 128 SAILIINYRTELFKMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPLR 187
SA+L+IN + +L +M C E +T +++ IP +M+ + +G L + + +N V + LY+P R
Sbjct: 131 SALLVINDKEDLDEMGCMEKDTSLNVSIPVLMISKSSGDALNKSMVDNKNVELLLYAPKR 190
Query: 188 PLVDVAEVFLWLMAVGTILCASYWSAWTAREAAIEQEKLL-KDASDEYVAESVGSRGYVE 246
P VD+ L LMAVGT++ AS WS T + A E +L KD S + + ++
Sbjct: 191 PAVDLTAGLLLLMAVGTVVVASLWSELTDPDQANESYSILAKDVSSAGTRKDDPEKEILD 250
Query: 247 ISTTAAILFVVLASCFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSCFRWFQY 306
IS T A+ F+V AS FL++L+ MS WF+ VL + FCIGG++G+ + A++ R ++
Sbjct: 251 ISVTGAVFFIVTASIFLLLLFYFMSSWFVWVLTIFFCIGGMQGMHNIIMAVI--LRKCRH 308
Query: 307 PAQTYVKIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILGIALIITVLQIV 366
A+ VK+P G + L+L V C+ FAV W +KR SY+W+GQDILGI L+IT LQ+V
Sbjct: 309 LARKSVKLPLLGTMSVLSLLVNIVCLAFAVFWFIKRHTSYSWVGQDILGICLMITALQVV 368
Query: 367 RIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPMLLKLPRLFD 426
R+PN+KV TVLL CAF+YDI WVF+S FHESVMIVVA+GD S + IPMLL++PR FD
Sbjct: 369 RLPNIKVATVLLCCAFVYDIFWVFISPLIFHESVMIVVAQGDSSTGESIPMLLRIPRFFD 428
Query: 427 PWGGYSIIGFGDIILPGLVVAFS------------------------LRYDWLAKKNLRA 462
PWGGY +IGFGDI+ PGL+++F+ LRYD + K+ +
Sbjct: 429 PWGGYDMIGFGDILFPGLLISFASRVSFSTILSSNPLPRLILAPLFDLRYDKIKKRVISN 488
Query: 463 GYFVWAMTAYGLGLLITYVALNLMDG 488
GYF+W YG+GLL+TY+ L ++ G
Sbjct: 489 GYFLWLTIGYGIGLLLTYLGLAVILG 514
>UniRef100_Q8LQG1 Vacuolar sorting receptor-like protein [Oryza sativa]
Length = 569
Score = 385 bits (989), Expect = e-105
Identities = 212/505 (41%), Positives = 293/505 (57%), Gaps = 18/505 (3%)
Query: 5 GVIYSNIVAYVFLFSVSLVLAGDIVHHDDVAPTRPGCENNFVLVKVPTWIDGVENAEYVG 64
G+ Y + + V D D V+P PGC+N F VKV W+DG E + G
Sbjct: 17 GLAYLLVSVLLLASRVPGAAGADSEFEDGVSPKFPGCDNPFQKVKVTYWVDGDERSSLTG 76
Query: 65 VGARFGPTLESKEKHANHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANIADE 124
+ ARFG L + + + V+ P C+K L + I + RG+C+F KA A+
Sbjct: 77 ITARFGEVLPATGSDGDKRKAVVPAPKTGCAKSSAPLASSIAVAERGECTFLEKAKTAES 136
Query: 125 AGASAILIIN------YRTELFKMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIV 178
GA+A+L+IN R +L KMVC +N+T +IGIP VM+ Q AG + + + V
Sbjct: 137 GGAAALLLINDEDGQVLRVDLQKMVCTQNDTVPNIGIPVVMVSQSAGRKILSGMDGGAKV 196
Query: 179 SIQLYSPLRPLVDVAEVFLWLMAVGTILCASYWSAWTAREAAIEQEKLLKDASDEYVAES 238
I +Y+P +P D A FLWLMAVG++ CAS WS + ++K E A+S
Sbjct: 197 DILMYAPEKPSFDGAIPFLWLMAVGSVACASVWSFVVVGD----EDKNAPTLGGEEAADS 252
Query: 239 VGSRGYVEISTTAAILFVVLASCFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALL 298
VE+ T A++F+V AS L+ L+ S W +LVVLFC+ G++GL + L+
Sbjct: 253 E----IVELQTKTALVFIVTASLVLLFLFFFKSTWSAWLLVVLFCLSGLQGLHYVASTLI 308
Query: 299 SCFRWFQYPAQTYVKIPFFGAVPYLTLAVTPFCIVFAVVWAVKRQASYAWIGQDILGIAL 358
R + V +P G V +TL + P ++F VVWAV + + +AW+GQD++GI +
Sbjct: 309 --VRTCDRCREAKVALPVLGNVTVVTLVILPLALIFVVVWAVHQNSPFAWVGQDLMGICM 366
Query: 359 IITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWWFHESVMIVVARGDKSGEDGIPML 418
+I VLQ+V +PN+KV T LL AF+YDI WVF+S + F +SVMI VARG G +PM+
Sbjct: 367 MILVLQVVHLPNIKVATALLVSAFMYDIFWVFISPFIFKKSVMITVARGSDEG-PSLPMV 425
Query: 419 LKLPRLFDPWGGYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLI 478
LK+P+ FD W GY +IGFGDI+ PGL+VAFS RYD K+L GYF+ M Y GL
Sbjct: 426 LKMPKEFDTWNGYDMIGFGDILFPGLLVAFSFRYDRANGKDLTDGYFLCLMIGYAFGLSC 485
Query: 479 TYVALNLMDGHGQPALLYIVPFTLG 503
TYV L LM GQPALLY+VP TLG
Sbjct: 486 TYVGLYLMKS-GQPALLYLVPSTLG 509
>UniRef100_Q7T0P3 MGC69113 protein [Xenopus laevis]
Length = 606
Score = 206 bits (523), Expect = 2e-51
Identities = 144/433 (33%), Positives = 234/433 (53%), Gaps = 29/433 (6%)
Query: 81 NHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTELF 140
N T + + P D P+ N I +V RG C+F K +A GA +LI++ T +
Sbjct: 41 NETEIALCLPSDV---PEGGFINRIPMVMRGNCTFYEKVRLAQINGARGLLIVSRETLVP 97
Query: 141 KMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVDVAEVFLWLM 200
+ ++DI P +L L++ + + V + +Y+P P++D V ++LM
Sbjct: 98 PGGNQSQFEEIDI--PVALLSYSDMLDIGKTFGKS--VKVAMYAPNEPVLDYNMVIIFLM 153
Query: 201 AVGTILCASYWSAWTAREAAIEQEKLLKDASDEYVAESVGSRGYVEISTTAAILFVVLAS 260
AVGT+ YW+ + + K D SD+ + V+++ +FVV+
Sbjct: 154 AVGTVAVGGYWAGSRDVKKRYMKHKR-DDGSDKKHDDET-----VDVTPIMICVFVVMCC 207
Query: 261 CFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYVKIPFFGAV 320
LV+LY + V++ +FC GL +CL+ + F + + K+P+F
Sbjct: 208 SMLVLLYYFYDH-LVYVIIGIFCFAASIGLYSCLSPFVRRFPYGKCRVPDN-KLPYFHKR 265
Query: 321 PYL-TLAVTPFCIVFAVVWAVKRQAS-YAWIGQDILGIALIITVLQIVRIPNLKVGTVLL 378
P + L + FCI+ +V+W V R +AW+ QDILGIA + +L+ +R+P K T+LL
Sbjct: 266 PPVWKLLLAAFCIMVSVIWGVYRNKDQWAWVLQDILGIAFCLYMLKTIRMPTFKGCTLLL 325
Query: 379 SCAFLYDILWVFVSKWWFH--ESVMIVVARG--DKSGEDGIPMLLKLPRL-FDPWG---- 429
F+YD+ +VF++ + ES+M+ VA G + + ++ +PM+LK+PRL P
Sbjct: 326 FVLFVYDVFFVFITPYLTKRGESIMVEVASGPSNSTTQEKLPMVLKVPRLNSSPLALCDR 385
Query: 430 GYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGH 489
+S++GFGDI++PGL+VA+ R+D + + YFV AYG+GLL+T+VAL LM
Sbjct: 386 PFSLLGFGDILVPGLLVAYCHRFDIQVQSS--RIYFVACTIAYGIGLLLTFVALALMQ-K 442
Query: 490 GQPALLYIVPFTL 502
GQPALLY+VP TL
Sbjct: 443 GQPALLYLVPCTL 455
>UniRef100_Q8TCT7-4 Splice isoform 4 of Q8TCT7 [Homo sapiens]
Length = 483
Score = 206 bits (523), Expect = 2e-51
Identities = 145/419 (34%), Positives = 223/419 (52%), Gaps = 31/419 (7%)
Query: 97 PKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTELFKMVCEENETDVDIGIP 156
P +N+I LV RG C+F K +A +GA +LI++ R L + + D +IGIP
Sbjct: 54 PARGFSNQIPLVARGNCTFYEKVRLAQGSGARGLLIVS-RERLVPPGGNKTQYD-EIGIP 111
Query: 157 AVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVDVAEVFLWLMAVGTILCASYWSAWTA 216
+L L++ V LY+P P++D V +++MAVGT+ YW+ +
Sbjct: 112 VALLSYKDMLDIFTRFGRT--VRAALYAPKEPVLDYNMVIIFIMAVGTVAIGGYWAG--S 167
Query: 217 REAAIEQEKLLKDASDEYVAESVGSRGYVEISTTAAILFVVLASCFLVMLYKLMSFWFLE 276
R+ K +D E + V+++ +FVV+ LV+LY +
Sbjct: 168 RDVKKRYMKHKRDDGPEKQEDEA-----VDVTPVMTCVFVVMCCSMLVLLYYFYDL-LVY 221
Query: 277 VLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYVK--IPFFGAVPYLT-LAVTPFCIV 333
V++ +FC+ GL +CL C R + +P+F P L + FC+
Sbjct: 222 VVIGIFCLASATGLYSCLAP---CVRRLPFGKCRIPNNSLPYFHKRPQARMLLLALFCVA 278
Query: 334 FAVVWAVKR-QASYAWIGQDILGIALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVS 392
+VVW V R + +AW+ QD LGIA + +L+ +R+P K T+LL FLYDI +VF++
Sbjct: 279 VSVVWGVFRNEDQWAWVLQDALGIAFCLYMLKTIRLPTFKACTLLLLVLFLYDIFFVFIT 338
Query: 393 KWWFH--ESVMIVVARG--DKSGEDGIPMLLKLPRL-FDPWG----GYSIIGFGDIILPG 443
+ S+M+ VA G D + + +PM+LK+PRL P +S++GFGDI++PG
Sbjct: 339 PFLTKSGSSIMVEVATGPSDSATREKLPMVLKVPRLNSSPLALCDRPFSLLGFGDILVPG 398
Query: 444 LVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVPFTL 502
L+VA+ R+D + + YFV AYG+GLL+T+VAL LM GQPALLY+VP TL
Sbjct: 399 LLVAYCHRFDIQVQSS--RVYFVACTIAYGVGLLVTFVALALMQ-RGQPALLYLVPCTL 454
>UniRef100_Q8TCT7 Signal peptide peptidase-like 2B [Homo sapiens]
Length = 564
Score = 206 bits (523), Expect = 2e-51
Identities = 145/419 (34%), Positives = 223/419 (52%), Gaps = 31/419 (7%)
Query: 97 PKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTELFKMVCEENETDVDIGIP 156
P +N+I LV RG C+F K +A +GA +LI++ R L + + D +IGIP
Sbjct: 54 PARGFSNQIPLVARGNCTFYEKVRLAQGSGARGLLIVS-RERLVPPGGNKTQYD-EIGIP 111
Query: 157 AVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVDVAEVFLWLMAVGTILCASYWSAWTA 216
+L L++ V LY+P P++D V +++MAVGT+ YW+ +
Sbjct: 112 VALLSYKDMLDIFTRFGRT--VRAALYAPKEPVLDYNMVIIFIMAVGTVAIGGYWAG--S 167
Query: 217 REAAIEQEKLLKDASDEYVAESVGSRGYVEISTTAAILFVVLASCFLVMLYKLMSFWFLE 276
R+ K +D E + V+++ +FVV+ LV+LY +
Sbjct: 168 RDVKKRYMKHKRDDGPEKQEDEA-----VDVTPVMTCVFVVMCCSMLVLLYYFYDL-LVY 221
Query: 277 VLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYVK--IPFFGAVPYLT-LAVTPFCIV 333
V++ +FC+ GL +CL C R + +P+F P L + FC+
Sbjct: 222 VVIGIFCLASATGLYSCLAP---CVRRLPFGKCRIPNNSLPYFHKRPQARMLLLALFCVA 278
Query: 334 FAVVWAVKR-QASYAWIGQDILGIALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVS 392
+VVW V R + +AW+ QD LGIA + +L+ +R+P K T+LL FLYDI +VF++
Sbjct: 279 VSVVWGVFRNEDQWAWVLQDALGIAFCLYMLKTIRLPTFKACTLLLLVLFLYDIFFVFIT 338
Query: 393 KWWFH--ESVMIVVARG--DKSGEDGIPMLLKLPRL-FDPWG----GYSIIGFGDIILPG 443
+ S+M+ VA G D + + +PM+LK+PRL P +S++GFGDI++PG
Sbjct: 339 PFLTKSGSSIMVEVATGPSDSATREKLPMVLKVPRLNSSPLALCDRPFSLLGFGDILVPG 398
Query: 444 LVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVPFTL 502
L+VA+ R+D + + YFV AYG+GLL+T+VAL LM GQPALLY+VP TL
Sbjct: 399 LLVAYCHRFDIQVQSS--RVYFVACTIAYGVGLLVTFVALALMQ-RGQPALLYLVPCTL 454
>UniRef100_Q5PQL3 Hypothetical protein [Rattus norvegicus]
Length = 577
Score = 203 bits (517), Expect = 9e-51
Identities = 144/420 (34%), Positives = 225/420 (53%), Gaps = 34/420 (8%)
Query: 97 PKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTELFKMVCEENETDVD-IGI 155
P TN+I LV RG C+F K +A +GA +LI++ + N+T + I I
Sbjct: 76 PVEDFTNQIALVARGNCTFYEKVRLAQGSGAHGLLIVSKER---LVPPRGNKTQYEEISI 132
Query: 156 PAVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVDVAEVFLWLMAVGTILCASYWSAWT 215
P +L ++ R + V + LY+P P++D V +++MAVGT+ YW+
Sbjct: 133 PVALLSHRDLQDIFRRFGHE--VMVALYAPSEPVMDYNMVIIFIMAVGTVALGGYWAG-- 188
Query: 216 AREAAIEQEKLLKDASDEYVAESVGSRGYVEISTTAAILFVVLASCFLVMLYKLMSFWFL 275
+ + +K +K D+ + V+++ +FVV+ LV+LY +
Sbjct: 189 ----SHDVKKYMKHKRDDVPEKQEDEA--VDVTPVMICVFVVMCCFMLVLLYYFYDR-LV 241
Query: 276 EVLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYV--KIPFFGAVPYLT-LAVTPFCI 332
V++ +FC+ GL +CL C R + +P+F P L + FC+
Sbjct: 242 YVIIGIFCLASSTGLYSCLAP---CVRKLPFCTCRVPDNNLPYFHKRPQARMLLLALFCV 298
Query: 333 VFAVVWAVKR-QASYAWIGQDILGIALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFV 391
+VVW V R + +AW+ QD LGIA + +L+ +R+P K T+LL F+YDI +VF+
Sbjct: 299 TVSVVWGVFRNEDQWAWVLQDTLGIAFCLYMLRTIRLPTFKACTLLLLVLFVYDIFFVFI 358
Query: 392 SKWWFH--ESVMIVVARG--DKSGEDGIPMLLKLPRL-FDPWG----GYSIIGFGDIILP 442
+ + S+M+ VA G + S + +PM+LK+PRL P +S++GFGDI++P
Sbjct: 359 TPYLTKSGNSIMVEVATGPSNSSTHEKLPMVLKVPRLNTSPLSLCDRPFSLLGFGDILVP 418
Query: 443 GLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVPFTL 502
GL+VA+ R+D + + YFV AYGLGLL+T+VAL LM HGQPALLY+VP TL
Sbjct: 419 GLLVAYCHRFDIQVQSS--RIYFVACTIAYGLGLLVTFVALVLM-RHGQPALLYLVPCTL 475
>UniRef100_UPI00001CFC14 UPI00001CFC14 UniRef100 entry
Length = 579
Score = 201 bits (512), Expect = 3e-50
Identities = 143/420 (34%), Positives = 225/420 (53%), Gaps = 33/420 (7%)
Query: 97 PKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTELFKMVCEENETDVD-IGI 155
P TN+I LV RG C+F K +A +GA +LI++ + N+T + I I
Sbjct: 76 PVEDFTNQIALVARGNCTFYEKVRLAQGSGAHGLLIVSKER---LVPPRGNKTQYEEISI 132
Query: 156 PAVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVDVAEVFLWLMAVGTILCASYWSAWT 215
P +L ++ R + V + LY+P P++D V +++MAVGT+ YW+
Sbjct: 133 PVALLSHRDLQDIFRRFGHE--VMVALYAPSEPVMDYNMVIIFIMAVGTVALGGYWAG-- 188
Query: 216 AREAAIEQEKLLKDASDEYVAESVGSRGYVEISTTAAILFVVLASCFLVMLYKLMSFWFL 275
+ +++ +K D+ + V+++ +FVV+ LV+LY +
Sbjct: 189 ---SHDVKKRYMKHKRDDVPEKQEDEA--VDVTPVMICVFVVMCCFMLVLLYYFYDR-LV 242
Query: 276 EVLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYV--KIPFFGAVPYLT-LAVTPFCI 332
V++ +FC+ GL +CL C R + +P+F P L + FC+
Sbjct: 243 YVIIGIFCLASSTGLYSCLAP---CVRKLPFCTCRVPDNNLPYFHKRPQARMLLLALFCV 299
Query: 333 VFAVVWAVKR-QASYAWIGQDILGIALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFV 391
+VVW V R + +AW+ QD LGIA + +L+ +R+P K T+LL F+YDI +VF+
Sbjct: 300 TVSVVWGVFRNEDQWAWVLQDTLGIAFCLYMLRTIRLPTFKACTLLLLVLFVYDIFFVFI 359
Query: 392 SKWWFH--ESVMIVVARG--DKSGEDGIPMLLKLPRL-FDPWG----GYSIIGFGDIILP 442
+ + S+M+ VA G + S + +PM+LK+PRL P +S++GFGDI++P
Sbjct: 360 TPYLTKSGNSIMVEVATGPSNSSTHEKLPMVLKVPRLNTSPLSLCDRPFSLLGFGDILVP 419
Query: 443 GLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVPFTL 502
GL+VA+ R+D + + YFV AYGLGLL+T+VAL LM HGQPALLY+VP TL
Sbjct: 420 GLLVAYCHRFDIQVQSS--RIYFVACTIAYGLGLLVTFVALVLM-RHGQPALLYLVPCTL 476
>UniRef100_UPI0000362B74 UPI0000362B74 UniRef100 entry
Length = 481
Score = 199 bits (506), Expect = 2e-49
Identities = 143/431 (33%), Positives = 234/431 (54%), Gaps = 32/431 (7%)
Query: 83 TRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTELFKM 142
T V+ P + P+ N I +V RG C+F K +A GA +LI++ + L
Sbjct: 49 TTSVLCSPSEV---PEGGFPNRIPMVLRGNCTFYEKVRLAQINGAKGLLIVS-KDRLTPP 104
Query: 143 VCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVDVAEVFLWLMAV 202
+ + + +I IP +L L++ + +V+ +Y+P P++D V ++LMAV
Sbjct: 105 AGNKTQYE-EIDIPVALLSYSDMLDISKTFGKGRLVA--MYAPNEPVLDYNMVIIFLMAV 161
Query: 203 GTILCASYWSAWTAREAAIEQEKLLKDASDEYVAESVGSRGYVEISTTAAILFVVLASCF 262
GT+ YW+ R+ E A+++ E+V T +FVV+
Sbjct: 162 GTVAVGGYWAGSRDRKKRTLSEA--GRAAEKQDEETVDV-------TPMICVFVVMCCSM 212
Query: 263 LVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYVKIPFFGAVPY 322
LV+LY + + V+V+ FC+ GL +CL + + + +P+ P+
Sbjct: 213 LVLLYFFYDYLAIWVIVI-FCLASSVGLHSCLWPFVRRLPFCKCRVPEN-NLPYLQKRPH 270
Query: 323 LT-LAVTPFCIVFAVVWAVKR-QASYAWIGQDILGIALIITVLQIVRIPNLKVGTVLLSC 380
++ L ++ FC+ +V W V R + ++AW+ QD LGIA + +L+ VR+P K T+LLS
Sbjct: 271 VSMLLLSAFCVGVSVTWMVFRNEDAWAWVLQDTLGIAFCLYMLKTVRLPTFKACTLLLSV 330
Query: 381 AFLYDILWVFVSKWWFH--ESVMIVVARG--DKSGEDGIPMLLKLPRL-FDPWG----GY 431
F+YD+ +VF++ + + ES+M+ VA G D + + +PM+LK+PRL P +
Sbjct: 331 LFVYDVFFVFITPFLTNSGESIMVEVAAGPSDSTTHEKLPMVLKVPRLNSSPLALCDRPF 390
Query: 432 SIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQ 491
S++GFGDI++PGL+V + R+D L + + YFV AYG+GLLIT+VAL +M GQ
Sbjct: 391 SLLGFGDILVPGLLVVYCHRFDILIQSS--RIYFVACTIAYGIGLLITFVALAVMQ-MGQ 447
Query: 492 PALLYIVPFTL 502
PALLY+VP TL
Sbjct: 448 PALLYLVPCTL 458
>UniRef100_UPI00003AC9AF UPI00003AC9AF UniRef100 entry
Length = 468
Score = 198 bits (503), Expect = 4e-49
Identities = 144/435 (33%), Positives = 227/435 (52%), Gaps = 44/435 (10%)
Query: 81 NHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIINYR--TE 138
+ T V+ P D P N I +V RG C+F K +A GA +LI++
Sbjct: 41 DQTASVLCSPSDV---PDGGFNNRIPMVMRGNCTFYEKVRLAQINGARGLLIVSRERLVS 97
Query: 139 LFKMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVDVAEVFLW 198
LF C + +DIG+ +V++ V +Y+P P++D V ++
Sbjct: 98 LFLSFCITDALMLDIGLFSVVVA----------------VKGAMYAPNEPVLDYNMVIIF 141
Query: 199 LMAVGTILCASYWSAWTAREAAIEQEKLLKDASDEYVAESVGSRGYVEISTTAAILFVVL 258
+MAVGT+ YW+ +R+ K +D E + V+++ +FVV+
Sbjct: 142 VMAVGTVAIGGYWAG--SRDVKKRYMKHKRDDGAEKHEDET-----VDVTPIMICVFVVM 194
Query: 259 ASCFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYVKIPFFG 318
LV+LY + V++ +FC+ GL +CL+ + F + +P+F
Sbjct: 195 CCSMLVLLYFFYDH-LVYVIIGIFCLAASIGLYSCLSPFVRRFPLGKCRIPDN-NLPYFH 252
Query: 319 AVPYLT-LAVTPFCIVFAVVWAVKR-QASYAWIGQDILGIALIITVLQIVRIPNLKVGTV 376
P + L + FCI +VVW V R + +AW+ QD LGIA + +L+ +R+P K T+
Sbjct: 253 KRPQVRMLLLAVFCISVSVVWGVFRNEDQWAWVLQDALGIAFCLYMLKTIRLPTFKGCTL 312
Query: 377 LLSCAFLYDILWVFVSKWWFH--ESVMIVVARG--DKSGEDGIPMLLKLPRL-FDPWG-- 429
LL F+YD+ +VF++ + ES+M+ VA G D + + +PM+LK+PRL P
Sbjct: 313 LLLVLFVYDVFFVFITPFLTKTGESIMVEVAAGPSDSATHEKLPMVLKVPRLNSSPLALC 372
Query: 430 --GYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMD 487
+S++GFGDI++PGL+VA+ R+D + + YFV AYG+GLL+T+VAL LM
Sbjct: 373 DRPFSLLGFGDILVPGLLVAYCHRFDIQVQSS--RVYFVACTIAYGIGLLVTFVALALMQ 430
Query: 488 GHGQPALLYIVPFTL 502
GQPALLY+VP TL
Sbjct: 431 -MGQPALLYLVPCTL 444
>UniRef100_UPI00003AC9AE UPI00003AC9AE UniRef100 entry
Length = 483
Score = 197 bits (501), Expect = 6e-49
Identities = 144/433 (33%), Positives = 230/433 (52%), Gaps = 30/433 (6%)
Query: 81 NHTRVVMADPPDCCSKPKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTELF 140
+ T V+ P D P N I +V RG C+F K +A GA +LI++ R L
Sbjct: 48 DQTASVLCSPSDV---PDGGFNNRIPMVMRGNCTFYEKVRLAQINGARGLLIVS-RERLV 103
Query: 141 KMVCEENETDVDIGIPAVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVDVAEVFLWLM 200
++ + +I IP +L L++ + + V +Y+P P++D V +++M
Sbjct: 104 PPGGNRSQYE-EIDIPVALLSYSDMLDIVKSFGRS--VKGAMYAPNEPVLDYNMVIIFVM 160
Query: 201 AVGTILCASYWSAWTAREAAIEQEKLLKDASDEYVAESVGSRGYVEISTTAAILFVVLAS 260
AVGT+ YW+ +R+ K +D E + V+++ +FVV+
Sbjct: 161 AVGTVAIGGYWAG--SRDVKKRYMKHKRDDGAEKHEDET-----VDVTPIMICVFVVMCC 213
Query: 261 CFLVMLYKLMSFWFLEVLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYVKIPFFGAV 320
LV+LY + V++ +FC+ GL +CL+ + F + +P+F
Sbjct: 214 SMLVLLYFFYDH-LVYVIIGIFCLAASIGLYSCLSPFVRRFPLGKCRIPDN-NLPYFHKR 271
Query: 321 PYLT-LAVTPFCIVFAVVWAVKR-QASYAWIGQDILGIALIITVLQIVRIPNLKVGTVLL 378
P + L + FCI +VVW V R + +AW+ QD LGIA + +L+ +R+P K T+LL
Sbjct: 272 PQVRMLLLAVFCISVSVVWGVFRNEDQWAWVLQDALGIAFCLYMLKTIRLPTFKGCTLLL 331
Query: 379 SCAFLYDILWVFVSKWWFH--ESVMIVVARG--DKSGEDGIPMLLKLPRL-FDPWG---- 429
F+YD+ +VF++ + ES+M+ VA G D + + +PM+LK+PRL P
Sbjct: 332 LVLFVYDVFFVFITPFLTKTGESIMVEVAAGPSDSATHEKLPMVLKVPRLNSSPLALCDR 391
Query: 430 GYSIIGFGDIILPGLVVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGH 489
+S++GFGDI++PGL+VA+ R+D + + YFV AYG+GLL+T+VAL LM
Sbjct: 392 PFSLLGFGDILVPGLLVAYCHRFDIQVQSS--RVYFVACTIAYGIGLLVTFVALALMQ-M 448
Query: 490 GQPALLYIVPFTL 502
GQPALLY+VP TL
Sbjct: 449 GQPALLYLVPCTL 461
>UniRef100_UPI00001CF2D7 UPI00001CF2D7 UniRef100 entry
Length = 523
Score = 196 bits (498), Expect = 1e-48
Identities = 131/418 (31%), Positives = 223/418 (53%), Gaps = 35/418 (8%)
Query: 97 PKNKLTNEIILVHRGKCSFTTKANIADEAGASAILIINYRTELFKMVCEENETDVDIGIP 156
P + + N+ ++VH G C F KA IA E GA+A+LI N + + +V I I
Sbjct: 82 PPDGIKNKAVVVHWGPCHFLEKAKIAQEGGAAALLIANNSVLIPSSRNKSAFQNVTILI- 140
Query: 157 AVMLPQDAGLNLERHIQNNSIVSIQLYSPLRPLVDVAEVFLWLMAVGTILCASYWSAWTA 216
AV+ +D +++ + ++ +++++YSP P D V ++++AV T+ YWS
Sbjct: 141 AVITQKDFN-DMKETLGDD--ITVKMYSPSWPNFDYTLVVIFVIAVFTVALGGYWSGLIE 197
Query: 217 REAAIEQEKLLKDASDEYVAESVGSRGYVEISTTAAILFVVLASCFLVMLYKLMSFWFLE 276
+E K ++DA D + Y+ S +LFVV+ +V+LY W +
Sbjct: 198 ----LESMKAVEDAEDREARKK--KEDYLTFSPLTVVLFVVICCVMIVLLYFFYK-WLVY 250
Query: 277 VLVVLFCIGGIEGLQTCLTALLSCFRWFQYPAQTYVKIPFFGAVPYLTLAVTPFCIVFAV 336
V++ +FCI L CL AL+ + P + + + ++ CI AV
Sbjct: 251 VMIAIFCIASATSLYNCLAALIH-----RMPCGQCTILCCGKNIKVSLIFLSGLCISVAV 305
Query: 337 VWAVKRQAS-YAWIGQDILGIALIITVLQIVRIPNLKVGTVLLSCAFLYDILWVFVSKWW 395
VWAV R +AWI QDILGIA + +++ +++PN +LL +YD+ +VF++ +
Sbjct: 306 VWAVFRNEDRWAWILQDILGIAFCLNLIKTMKLPNFMSCVILLGLLLIYDVFFVFITPFI 365
Query: 396 FH--ESVMIVVARGDKSGEDGIPMLLKLPRLFD---------PWGGYSIIGFGDIILPGL 444
ES+M+ +A G + +P+++++P+L D P S++GFGDII+PGL
Sbjct: 366 TKNGESIMVELAAGPFENAEKLPVVIRVPKLMDYSVMSVCSVP---VSVLGFGDIIVPGL 422
Query: 445 VVAFSLRYDWLAKKNLRAGYFVWAMTAYGLGLLITYVALNLMDGHGQPALLYIVPFTL 502
++A+ R+D ++ Y++ + AY +G++IT+V L +M GQPALLY+VP TL
Sbjct: 423 LIAYCRRFDVQTGSSI---YYISSTIAYAVGMIITFVVLMVMK-TGQPALLYLVPCTL 476
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.327 0.142 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 898,152,406
Number of Sequences: 2790947
Number of extensions: 37616743
Number of successful extensions: 108502
Number of sequences better than 10.0: 280
Number of HSP's better than 10.0 without gapping: 168
Number of HSP's successfully gapped in prelim test: 112
Number of HSP's that attempted gapping in prelim test: 107851
Number of HSP's gapped (non-prelim): 310
length of query: 540
length of database: 848,049,833
effective HSP length: 132
effective length of query: 408
effective length of database: 479,644,829
effective search space: 195695090232
effective search space used: 195695090232
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC146341.6


