

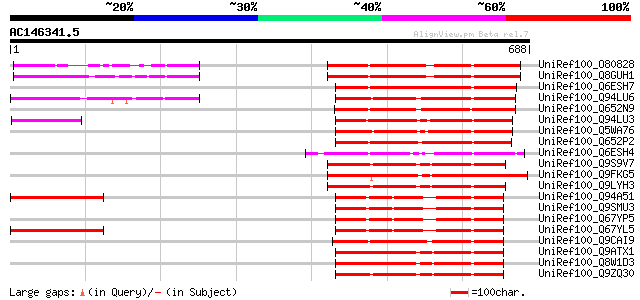
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146341.5 + phase: 0 /pseudo
(688 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O80828 Hypothetical protein At2g45910 [Arabidopsis tha... 293 2e-77
UniRef100_Q8GUH1 Hypothetical protein At2g45910 [Arabidopsis tha... 292 2e-77
UniRef100_Q6ESH7 Protein kinase-like [Oryza sativa] 276 2e-72
UniRef100_Q94LU6 Putative receptor kinase [Oryza sativa] 274 6e-72
UniRef100_Q652N9 Putative serine/threonine protein kinase [Oryza... 259 2e-67
UniRef100_Q94LU3 Putative serine/threonine protein kinase [Oryza... 255 3e-66
UniRef100_Q5WA76 Putative stress-induced protein sti1 [Oryza sat... 253 1e-65
UniRef100_Q652P2 Putative serine/threonine protein kinase [Oryza... 242 3e-62
UniRef100_Q6ESH4 Putative serine/threonine-specific protein kina... 219 2e-55
UniRef100_Q9S9V7 T1N24.15 protein [Arabidopsis thaliana] 218 6e-55
UniRef100_Q9FKG5 Similarity to receptor-like protein kinase [Ara... 212 3e-53
UniRef100_Q9LYH3 Putative receptor-like kinase [Arabidopsis thal... 212 3e-53
UniRef100_Q94A51 AT3g49060/T2J13_100 [Arabidopsis thaliana] 211 4e-53
UniRef100_Q9SMU3 Hypothetical protein T2J13.100 [Arabidopsis tha... 211 4e-53
UniRef100_Q67YP5 Hypothetical protein At3g49065 [Arabidopsis tha... 211 4e-53
UniRef100_Q67YL5 Hypothetical protein At3g49065 [Arabidopsis tha... 211 4e-53
UniRef100_Q9CAI9 Hypothetical protein F28P22.5 [Arabidopsis thal... 207 8e-52
UniRef100_Q9ATX1 Serine threonine kinase 1 [Zea mays] 207 1e-51
UniRef100_Q8W1D3 Serine threonine kinase [Zea mays] 207 1e-51
UniRef100_Q9ZQ30 Hypothetical protein At2g24370 [Arabidopsis tha... 205 4e-51
>UniRef100_O80828 Hypothetical protein At2g45910 [Arabidopsis thaliana]
Length = 799
Score = 293 bits (749), Expect = 2e-77
Identities = 148/258 (57%), Positives = 189/258 (72%), Gaps = 15/258 (5%)
Query: 422 DPQNFNRSLSYQ--VEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNN 479
+P + + YQ V+VLSK++HPN+ITLIG E +L+YEYLP GSLED L+ +N
Sbjct: 476 NPNSSQGPVEYQQEVDVLSKMRHPNIITLIGACPEGWSLVYEYLPGGSLEDRLTCK--DN 533
Query: 480 APPLTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVL 539
+PPL+WQ R+RIATE+C+AL+FLHSNK HS+VHGDLKP+NILLD+NLV+KLSDFG C +L
Sbjct: 534 SPPLSWQNRVRIATEICAALVFLHSNKAHSLVHGDLKPANILLDSNLVSKLSDFGTCSLL 593
Query: 540 SCQNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPA 599
S T GT AY+DPE +GELT KSDVYSFGI+LLRL+TG+PA
Sbjct: 594 HPNGSKSVRTDVT---------GTVAYLDPEASSSGELTPKSDVYSFGIILLRLLTGRPA 644
Query: 600 LGIKNEVLYALNNAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSEGW 659
L I NEV YAL+N G + +LDPLAGDWP V+AE+L ALRCC+ ++RP+L +E W
Sbjct: 645 LRISNEVKYALDN--GTLNDLLDPLAGDWPFVQAEQLARLALRCCETVSENRPDLGTEVW 702
Query: 660 RVLEPMKVSCSGTNNFGL 677
RVLEPM+ S G+++F L
Sbjct: 703 RVLEPMRASSGGSSSFHL 720
Score = 92.0 bits (227), Expect = 5e-17
Identities = 78/247 (31%), Positives = 117/247 (46%), Gaps = 50/247 (20%)
Query: 5 VDVYLAICQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSRRMTDLRS 64
+D YL ICQ+ GV A K+ IEM+ IE GI++LIS I+ LVMGAA+D+++SR T +
Sbjct: 118 LDDYLRICQQRGVRAEKMFIEMESIENGIVQLISELGIRKLVMGAAADRHYSREAT-MDD 176
Query: 65 KKAIYVCEQSPASCHIQFICKGYLIQTRDCSLENFRVEATSPLVQQMQNSEVGHSPHLIS 124
++ Y + S S L+Q E H IS
Sbjct: 177 TESEYASPRPSIS--------------------------ASDLLQTFSTPESEHQH--IS 208
Query: 125 RSISHDQELHHHRVRSISSASESGRSMASSVSSSERFSSFETVLTPNLTSDGNESLLDLK 184
R S D V + SS +SGR S+++ E SDG+E
Sbjct: 209 RVQSTDSV--QQLVSNGSSTEQSGRVSDGSLNTDEE----------ERESDGSE------ 250
Query: 185 MSYLSSIKEENLCHSSPPGVLDRGMDESVYDQLEQAIAEAVKARWDAYQETVKRRKAEKD 244
S + HSSP D G+D+S ++ +A +EA ++ +A+ ET++R+KAEK+
Sbjct: 251 --VTGSATVMSSGHSSPSSFPD-GVDDSFNVKIRKATSEAHSSKQEAFAETLRRQKAEKN 307
Query: 245 VIDTIRK 251
+D IR+
Sbjct: 308 ALDAIRR 314
>UniRef100_Q8GUH1 Hypothetical protein At2g45910 [Arabidopsis thaliana]
Length = 834
Score = 292 bits (748), Expect = 2e-77
Identities = 148/258 (57%), Positives = 189/258 (72%), Gaps = 15/258 (5%)
Query: 422 DPQNFNRSLSYQ--VEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNN 479
+P + + YQ V+VLSK++HPN+ITLIG E +L+YEYLP GSLED L+ +N
Sbjct: 511 NPNSSQGPVEYQQEVDVLSKMRHPNIITLIGACPEGWSLVYEYLPGGSLEDRLTCK--DN 568
Query: 480 APPLTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVL 539
+PPL+WQ R+RIATE+C+AL+FLHSNK HS+VHGDLKP+NILLD+NLV+KLSDFG C +L
Sbjct: 569 SPPLSWQNRVRIATEICAALVFLHSNKAHSLVHGDLKPANILLDSNLVSKLSDFGTCSLL 628
Query: 540 SCQNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPA 599
S T GT AY+DPE +GELT KSDVYSFGI+LLRL+TG+PA
Sbjct: 629 HPNGSKSVRTDIT---------GTVAYLDPEASSSGELTPKSDVYSFGIILLRLLTGRPA 679
Query: 600 LGIKNEVLYALNNAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSEGW 659
L I NEV YAL+N G + +LDPLAGDWP V+AE+L ALRCC+ ++RP+L +E W
Sbjct: 680 LRISNEVKYALDN--GTLNDLLDPLAGDWPFVQAEQLARLALRCCETVSENRPDLGTEVW 737
Query: 660 RVLEPMKVSCSGTNNFGL 677
RVLEPM+ S G+++F L
Sbjct: 738 RVLEPMRASSGGSSSFHL 755
Score = 151 bits (382), Expect = 5e-35
Identities = 95/247 (38%), Positives = 146/247 (58%), Gaps = 15/247 (6%)
Query: 5 VDVYLAICQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSRRMTDLRS 64
+D YL ICQ+ GV A K+ IEM+ IE GI++LIS I+ LVMGAA+D+++SRRMTDL+S
Sbjct: 118 LDDYLRICQQRGVRAEKMFIEMESIENGIVQLISELGIRKLVMGAAADRHYSRRMTDLKS 177
Query: 65 KKAIYVCEQSPASCHIQFICKGYLIQTRDCSLENFRVEATSPLVQQMQNSEVGHSPHLIS 124
+KAI+V ++P C I F CKGYLI TR+ ++++ E SP + S L +
Sbjct: 178 RKAIFVRREAPTLCQIWFTCKGYLIHTREATMDDTESEYASP------RPSISASDLLQT 231
Query: 125 RSISHDQELHHHRVRSISSASESGRSMASSVSSSERFSSFETVLTPNLTSDGNESLLDLK 184
S + H RV+S ++S + + S+ SS+E+ V +L +D E D
Sbjct: 232 FSTPESEHQHISRVQS----TDSVQQLVSNGSSTEQSG---RVSDGSLNTDEEERESD-G 283
Query: 185 MSYLSSIKEENLCHSSPPGVLDRGMDESVYDQLEQAIAEAVKARWDAYQETVKRRKAEKD 244
S + HSSP D G+D+S ++ +A +EA ++ +A+ ET++R+KAEK+
Sbjct: 284 SEVTGSATVMSSGHSSPSSFPD-GVDDSFNVKIRKATSEAHSSKQEAFAETLRRQKAEKN 342
Query: 245 VIDTIRK 251
+D IR+
Sbjct: 343 ALDAIRR 349
>UniRef100_Q6ESH7 Protein kinase-like [Oryza sativa]
Length = 398
Score = 276 bits (705), Expect = 2e-72
Identities = 136/240 (56%), Positives = 178/240 (73%), Gaps = 4/240 (1%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIA 492
++ VLS+++HPNL+TLIG +E+ L+YE+LP GSLED L+ NN PPLTWQ R RI
Sbjct: 65 EIAVLSRVRHPNLVTLIGSCREAFGLVYEFLPKGSLEDRLACL--NNTPPLTWQVRTRII 122
Query: 493 TELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTT 552
E+CSAL FLHSNKPH IVHGDLKP+NILLDAN V+KL DFGICR+L N ++ +TT
Sbjct: 123 YEMCSALSFLHSNKPHPIVHGDLKPANILLDANFVSKLGDFGICRLLIQTNTGAAAAATT 182
Query: 553 QFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNN 612
+ + T+ KGTFAYMDPEFL TGELT +SDVYS GI++LRL+TGKP I V A+ +
Sbjct: 183 RLYRTTTPKGTFAYMDPEFLTTGELTPRSDVYSLGIIILRLLTGKPPQKIAEVVEDAIES 242
Query: 613 AGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSEGWRVLEPMKVSCSGT 672
G + S+LDP AG WP V+A +L H LRC +M+++ RP+L ++ W+V+EP+ + S T
Sbjct: 243 --GGLHSILDPSAGSWPFVQANQLAHLGLRCAEMSRRRRPDLATDVWKVVEPLMKAASLT 300
>UniRef100_Q94LU6 Putative receptor kinase [Oryza sativa]
Length = 881
Score = 274 bits (701), Expect = 6e-72
Identities = 134/238 (56%), Positives = 182/238 (76%), Gaps = 9/238 (3%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIA 492
+V VLS+++HPNL+TL+G E+ L+YE+LPNGSLEDHL+ N + PLTWQ R RI
Sbjct: 560 EVAVLSRVRHPNLVTLVGYCSEASGLVYEFLPNGSLEDHLACESNTS--PLTWQIRTRII 617
Query: 493 TELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTT 552
E+CSALIFLHS+KPH+++HGDLKP+NILLDANLV+KL DFGI R+L + S+ +T
Sbjct: 618 GEICSALIFLHSDKPHAVIHGDLKPANILLDANLVSKLGDFGISRLL-----NRSSTVST 672
Query: 553 QFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNN 612
F+ T+ +GTFAYMDPEFL TGELT++SD+YSFGI++LRL+TGKPALGI EV AL+
Sbjct: 673 SFYQTTNPRGTFAYMDPEFLTTGELTARSDIYSFGIIILRLVTGKPALGIAREVEVALDK 732
Query: 613 AGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSEGWRVLEPMKVSCS 670
G ++ ++D AGDWP V+AEKL+ L+C +++++ RP+ + W V+EP+ S S
Sbjct: 733 --GELELLVDRSAGDWPFVQAEKLMLLGLQCAELSRRKRPDRMNHVWSVVEPLVKSAS 788
Score = 105 bits (262), Expect = 4e-21
Identities = 87/272 (31%), Positives = 137/272 (49%), Gaps = 35/272 (12%)
Query: 1 MQKTVDVYLAICQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSRRMT 60
++K +D YL C +M V K+ IE + I GI ELI + + LVMGAA+DK +SR+M
Sbjct: 120 VEKNLDEYLEQCTKMKVKCEKIVIENEDIANGITELILLHGVSKLVMGAAADKQYSRKMK 179
Query: 61 DLRSKKAIYVCEQSPASCHIQFICKGYLIQTRDCSLENFRVEATSPLVQQMQNSEV--GH 118
+SK A+ V ++ SC I F+CK +LI TRD +P+ Q+ + G
Sbjct: 180 LPKSKTALSVTVKANPSCKIWFVCKEHLIYTRD---------FVAPISPNSQSPDTIRGS 230
Query: 119 SPHLISRSISHDQELHH-----------HRVRSISSASESGRSMAS-------SVSSSER 160
+L +R + +Q ++ V S+S + G S+ S S SS R
Sbjct: 231 ISNLAARGGTTNQYANNAVNGYVQRSMSEMVESLSRLNMEGTSVDSWDSFRRGSFPSSYR 290
Query: 161 FSSFETVLTPNLTSDGNESLLDLK-MSYLSSIKEENLCHSSPPGVLDRGMDESVYDQLEQ 219
SS +T + SD + S + +S L+ N G D G + +++D+LE+
Sbjct: 291 ASS---TVTEEVLSDSSSSGIPRDGISTLAGCDFPNSALHHEQG--DAGSNANLFDKLEE 345
Query: 220 AIAEAVKARWDAYQETVKRRKAEKDVIDTIRK 251
A AEA K R AY E+++R+K E+++I +K
Sbjct: 346 AFAEAEKYRKQAYDESLRRQKTEEELISYHQK 377
>UniRef100_Q652N9 Putative serine/threonine protein kinase [Oryza sativa]
Length = 407
Score = 259 bits (661), Expect = 2e-67
Identities = 128/240 (53%), Positives = 177/240 (73%), Gaps = 9/240 (3%)
Query: 431 SYQVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIR 490
S++VE+LS+++HPNL+TLIG ++++ L+YEY+PNGSL+D L+ +N+ PL+WQ R R
Sbjct: 87 SHEVEILSRVRHPNLVTLIGACKDARALVYEYMPNGSLDDRLACK--DNSKPLSWQLRTR 144
Query: 491 IATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNS 550
IA+ +CSALIFLHSNKPHSIVH DLK SNILLD N V KLS FG+CR+L + +
Sbjct: 145 IASNICSALIFLHSNKPHSIVHSDLKASNILLDGNNVAKLSGFGVCRML-----TDEFKA 199
Query: 551 TTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYAL 610
TT + + KGTF Y+DPE+ +G+LT SDVYSFGI+LLRL+TG+ G+ +V A+
Sbjct: 200 TTTLYRHTHPKGTFVYIDPEYAISGDLTPLSDVYSFGIILLRLLTGRSGFGLLKDVQRAV 259
Query: 611 NNAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSEGWRVLEPMKVSCS 670
A G ++++LD AGDWP++ AE+L LRCC++ +K+RP+L +E W VLEPM S S
Sbjct: 260 --AKGCLQAILDSSAGDWPLMHAEQLSRVGLRCCEIRRKNRPDLQTEVWTVLEPMLRSAS 317
>UniRef100_Q94LU3 Putative serine/threonine protein kinase [Oryza sativa]
Length = 675
Score = 255 bits (652), Expect = 3e-66
Identities = 131/234 (55%), Positives = 170/234 (71%), Gaps = 9/234 (3%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIA 492
+V +LS+++HPNL+TLIG E+ L+YE LPNGSLED L N +N PPLTWQ RI+I
Sbjct: 340 EVAILSRVRHPNLVTLIGACTEASALVYELLPNGSLEDRL--NCVDNTPPLTWQVRIQII 397
Query: 493 TELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTT 552
TE+CSALIFLH ++PH +VHGDLKP NILLDANL +KLSDFGI R+L +SS S
Sbjct: 398 TEICSALIFLHKHRPHPVVHGDLKPGNILLDANLQSKLSDFGISRLLL---ESSVTGSDA 454
Query: 553 QFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNN 612
+ TS GT AYMDPEF TGELT +SD YSFG+ ++RL+TG+ L + V ALN+
Sbjct: 455 HY--TSRPMGTPAYMDPEFFATGELTPQSDTYSFGVTIMRLLTGRAPLRLIRTVREALND 512
Query: 613 AGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSEGWRVLEPMK 666
+++SVLD AGDWP+V E+L H AL+C +++K+ RP+L + W V+EPMK
Sbjct: 513 Y--DLQSVLDHSAGDWPLVHVEQLAHIALQCTELSKQRRPDLEHDVWEVIEPMK 564
Score = 67.8 bits (164), Expect = 1e-09
Identities = 37/93 (39%), Positives = 55/93 (58%)
Query: 3 KTVDVYLAICQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSRRMTDL 62
+ VD YL C R V A K I++G++ LI Y + LVMGAASD+++ R+M
Sbjct: 26 QNVDNYLDQCSRKKVKAEKEVFFFTKIDEGLLHLIEIYGVTKLVMGAASDRHYKRKMKAP 85
Query: 63 RSKKAIYVCEQSPASCHIQFICKGYLIQTRDCS 95
+S+ AI V +++ + C+I FIC G L R+ S
Sbjct: 86 QSQTAISVMQRAHSYCNIWFICNGKLTCVREAS 118
>UniRef100_Q5WA76 Putative stress-induced protein sti1 [Oryza sativa]
Length = 805
Score = 253 bits (646), Expect = 1e-65
Identities = 129/234 (55%), Positives = 170/234 (72%), Gaps = 9/234 (3%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIA 492
+V +LS+++HP+L+TL+G ES TL+YE+LPNGSLED L + LTWQ RIRI
Sbjct: 488 EVSILSRVRHPHLVTLLGACSESSTLVYEFLPNGSLEDFLMCSDKRQT--LTWQARIRII 545
Query: 493 TELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTT 552
E+CSALIFLH NKPH +VHGDLKP+NILL NLV+KLSDFGI R+L SS+NN+T
Sbjct: 546 AEICSALIFLHKNKPHPVVHGDLKPANILLGVNLVSKLSDFGISRLLI---QSSTNNTT- 601
Query: 553 QFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNN 612
+ T GT YMDPEFL TGELT +SDVYSFGIV+LRL+TGKP +GIKN V A+
Sbjct: 602 -LYRTMHPVGTPLYMDPEFLSTGELTPQSDVYSFGIVVLRLLTGKPPVGIKNIVEDAMEK 660
Query: 613 AGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSEGWRVLEPMK 666
G++ SV+D G+WP + E+L + ALRC +++++ RP+L E W ++E ++
Sbjct: 661 --GDLNSVIDTSVGEWPHLHIEQLAYLALRCTELSRRCRPDLSGEVWAIVEAIR 712
>UniRef100_Q652P2 Putative serine/threonine protein kinase [Oryza sativa]
Length = 326
Score = 242 bits (617), Expect = 3e-62
Identities = 123/233 (52%), Positives = 163/233 (69%), Gaps = 9/233 (3%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIA 492
+VE+L +++HPNL+TLIGV +E+K L+YE+LPNGSLED L + PL W+ RI+IA
Sbjct: 76 EVEILGRMRHPNLVTLIGVCREAKALVYEFLPNGSLEDRLQCK--HQTDPLPWRMRIKIA 133
Query: 493 TELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTT 552
++C+ALIFLHSNKP I HGDLKP NILL N V KL DFGI R L+ +N + T
Sbjct: 134 ADICTALIFLHSNKPKGIAHGDLKPDNILLGDNFVGKLGDFGISRSLNL-----TNTTIT 188
Query: 553 QFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNN 612
+ T+ KGT YMDP ++ +GELT++ DVYSFG+VLLRL+TGK LG+ +EV ALNN
Sbjct: 189 PYHQTNQIKGTLGYMDPGYIASGELTAQYDVYSFGVVLLRLLTGKSPLGLPSEVEAALNN 248
Query: 613 AGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSEGWRVLEPM 665
++ V+D AG+WP ++KL ALRCC ++K RP+L E W VL+ M
Sbjct: 249 E--MLQQVVDASAGEWPPEYSKKLAILALRCCRYDRKERPDLAKEAWGVLQAM 299
>UniRef100_Q6ESH4 Putative serine/threonine-specific protein kinase [Oryza sativa]
Length = 721
Score = 219 bits (559), Expect = 2e-55
Identities = 129/295 (43%), Positives = 177/295 (59%), Gaps = 32/295 (10%)
Query: 393 VKVDTERYLKV--SFVTLTWR*KC*APTV-LKDPQNFNRSLSY--QVEVLSKLKHPNLIT 447
+K+ T R++ V F+ T A TV L PQ L + +V VLS+L+HPN++
Sbjct: 350 LKIGTGRFMNVYKGFIRNT------AITVMLLHPQGLQGQLEFHQEVVVLSRLRHPNVMM 403
Query: 448 LIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIATELCSALIFLHSNKP 507
LIG E+ ++YE+LPNGSLED LS N PPLTW+ R RI E+CSAL F+HS KP
Sbjct: 404 LIGACPEAFGMVYEFLPNGSLEDQLSCK--KNTPPLTWKMRTRIIGEICSALTFIHSQKP 461
Query: 508 HSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTTQFWITSFAKGTFAYM 567
H +VHG+L P NILLDAN V+KL +C++L N + NN++ GT +Y+
Sbjct: 462 HPVVHGNLNPMNILLDANFVSKLH---VCQLLRKYN--TGNNTS----------GTSSYI 506
Query: 568 DPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNNAGGNVKSVLDPLAGD 627
DPEFL TGEL + DVYSFGI++L L+TGK I V A+ + S++D AG
Sbjct: 507 DPEFLSTGELAPRCDVYSFGIIILHLLTGKSPQNITTIVEDAMEKR--QLHSIMDTSAGS 564
Query: 628 WPIVEAEKLVHFALRCCDMNKKSRPELCSEGWRVLEPMKVSCSGTNNFGLKSCLD 682
WP V+A +L H LRC +++ + RP+L E W V++P+ S NFG K +
Sbjct: 565 WPFVQANQLAHLGLRCANLSGRHRPDLTGEVWGVIKPLLKDAS--QNFGCKQAFE 617
>UniRef100_Q9S9V7 T1N24.15 protein [Arabidopsis thaliana]
Length = 675
Score = 218 bits (554), Expect = 6e-55
Identities = 110/236 (46%), Positives = 158/236 (66%), Gaps = 9/236 (3%)
Query: 422 DPQNFNRSLSYQVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAP 481
D + +VEVL ++HP+++ L+G E L+YE++ NGSLED L R GN+ P
Sbjct: 426 DAAQGKKQFQQEVEVLCSIRHPHMVLLLGACPEYGCLVYEFMENGSLEDRLFRTGNS--P 483
Query: 482 PLTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSC 541
PL+W+ R IA E+ +AL FLH KP +VH DLKP+NILLD N V+K+SD G+ R++
Sbjct: 484 PLSWRKRFEIAAEIATALSFLHQAKPEPLVHRDLKPANILLDKNYVSKISDVGLARLV-- 541
Query: 542 QNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALG 601
+S +S TQF +TS A GTF Y+DPE+ TG LT+KSDVYS GI+LL++ITG+P +G
Sbjct: 542 --PASIADSVTQFHMTS-AAGTFCYIDPEYQQTGMLTTKSDVYSLGILLLQIITGRPPMG 598
Query: 602 IKNEVLYALNNAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSE 657
+ ++V A++ G K +LDP+ DWP+ EA+ AL+C ++ K+ RP+L E
Sbjct: 599 LAHQVSRAISK--GTFKEMLDPVVPDWPVQEAQSFATLALKCAELRKRDRPDLGKE 652
>UniRef100_Q9FKG5 Similarity to receptor-like protein kinase [Arabidopsis thaliana]
Length = 763
Score = 212 bits (540), Expect = 3e-53
Identities = 111/269 (41%), Positives = 167/269 (61%), Gaps = 9/269 (3%)
Query: 422 DPQNFNRSLSYQVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNN--- 478
D + + ++E+LSK++HP+L+ L+G E +L+YEY+ NGSLE+ L + N
Sbjct: 428 DKSSLTKQFHQELEILSKIRHPHLLLLLGACPERGSLVYEYMHNGSLEERLMKRRPNVDT 487
Query: 479 -NAPPLTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICR 537
PPL W R RIA E+ SAL FLH+N+P IVH DLKP+NILLD N V+K+ D G+ +
Sbjct: 488 PQPPPLRWFERFRIAWEIASALYFLHTNEPRPIVHRDLKPANILLDRNNVSKIGDVGLSK 547
Query: 538 VLSCQNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGK 597
+++ ++++T F T GTF Y+DPE+ TG +T +SD+Y+FGI+LL+L+T +
Sbjct: 548 MVNLD----PSHASTVFNETG-PVGTFFYIDPEYQRTGVVTPESDIYAFGIILLQLVTAR 602
Query: 598 PALGIKNEVLYALNNAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSE 657
A+G+ + + AL + G +LD AGDWP+ EA+++V LRC +M K+ RP+L E
Sbjct: 603 SAMGLAHSIEKALRDQTGKFTEILDKTAGDWPVKEAKEMVMIGLRCAEMRKRDRPDLGKE 662
Query: 658 GWRVLEPMKVSCSGTNNFGLKSCLDKQLN 686
VLE +K S N + +D N
Sbjct: 663 ILPVLERLKEVASIARNMFADNLIDHHHN 691
Score = 38.1 bits (87), Expect = 0.88
Identities = 45/200 (22%), Positives = 72/200 (35%), Gaps = 44/200 (22%)
Query: 22 LHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSRRMTDLRSKKAIYVCEQSPASCHIQ 81
LH++ + I I + + + I LV+GA+S S ++ RS + + + +P C +
Sbjct: 59 LHVQPNDIADAISKAVQDHGISELVIGASSSIIFSWKLK--RSNLSSRIADATPRFCSVH 116
Query: 82 FICKGYLIQTR------------DCSLENFRVEATSPLVQQMQNSEVGHSPHLISR---- 125
I KG L+ R D S F ++ S V + + +P L R
Sbjct: 117 VISKGKLLNVRKSDMDTETSIADDRSESRFSSDSHSGTVSSTSSHQFSSTPLLFQRIQAL 176
Query: 126 -----------SISHDQELHHHRVRSIS---------------SASESGRSMASSVSSSE 159
+++ HHH R+ S S SG S SS
Sbjct: 177 TTVNQKVGTNIGKQNNEPHHHHHNRAGSLDVDESKLLNQKGFYRTSSSGIGYGGSDISSW 236
Query: 160 RFSSFETVLTPNLTSDGNES 179
R S E + + SD S
Sbjct: 237 RSSQMEEASSSSTYSDPTSS 256
>UniRef100_Q9LYH3 Putative receptor-like kinase [Arabidopsis thaliana]
Length = 703
Score = 212 bits (539), Expect = 3e-53
Identities = 107/236 (45%), Positives = 156/236 (65%), Gaps = 9/236 (3%)
Query: 422 DPQNFNRSLSYQVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAP 481
D + +VEVLS ++HP+++ L+G E L+YE++ NGSLED L R GN+ P
Sbjct: 454 DAAQGKKQFQQEVEVLSSIRHPHMVLLLGACPEYGCLVYEFMDNGSLEDRLFRRGNS--P 511
Query: 482 PLTWQTRIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSC 541
PL+W+ R +IA E+ +AL FLH KP +VH DLKP+NILLD N V+K+SD G+ R++
Sbjct: 512 PLSWRKRFQIAAEIATALSFLHQAKPEPLVHRDLKPANILLDKNYVSKISDVGLARLV-- 569
Query: 542 QNDSSSNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALG 601
+S N+ TQ+ +TS A GTF Y+DPE+ TG+LT+KSD++S GI+LL++IT K +G
Sbjct: 570 --PASVANTVTQYHMTS-AAGTFCYIDPEYQQTGKLTTKSDIFSLGIMLLQIITAKSPMG 626
Query: 602 IKNEVLYALNNAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPELCSE 657
+ + V A++ G K +LDP+ DWP+ EA LRC ++ K+ RP+L E
Sbjct: 627 LAHHVSRAIDK--GTFKDMLDPVVPDWPVEEALNFAKLCLRCAELRKRDRPDLGKE 680
Score = 37.7 bits (86), Expect = 1.1
Identities = 20/86 (23%), Positives = 44/86 (50%), Gaps = 1/86 (1%)
Query: 8 YLAICQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSRRMTDLRSKK- 66
Y C R G++ ++ ++ + K +++ ++ + NLV+G++S +R + +S
Sbjct: 75 YRGYCARKGISMMEVILDDTDVSKAVLDYVNNNLVTNLVLGSSSKSPFARSLKFTKSHDV 134
Query: 67 AIYVCEQSPASCHIQFICKGYLIQTR 92
A V + +P C + I KG + +R
Sbjct: 135 ASSVLKSTPEFCSVYVISKGKVHSSR 160
>UniRef100_Q94A51 AT3g49060/T2J13_100 [Arabidopsis thaliana]
Length = 805
Score = 211 bits (538), Expect = 4e-53
Identities = 106/222 (47%), Positives = 154/222 (68%), Gaps = 21/222 (9%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIA 492
+VE+LS+++HPNL+TL+G ES++LIY+Y+PNGSLED S NN P L+W++RIRIA
Sbjct: 503 RVEILSRVRHPNLVTLMGACPESRSLIYQYIPNGSLEDCFS--SENNVPALSWESRIRIA 560
Query: 493 TELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTT 552
+E+CSAL+FLHSN P I+HG+LKPS ILLD+NLVTK++D+GI +++ S+
Sbjct: 561 SEICSALLFLHSNIP-CIIHGNLKPSKILLDSNLVTKINDYGISQLIPIDGLDKSD---- 615
Query: 553 QFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNN 612
++DP + + E+T +SD+Y+FGI+LL+L+T +P GI +V AL N
Sbjct: 616 ------------PHVDPHYFVSREMTLESDIYAFGIILLQLLTRRPVSGILRDVKCALEN 663
Query: 613 AGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPEL 654
N+ +VLD AGDWP+ +KL + A+RCC N +RP+L
Sbjct: 664 --DNISAVLDNSAGDWPVARGKKLANVAIRCCKKNPMNRPDL 703
Score = 103 bits (256), Expect = 2e-20
Identities = 56/124 (45%), Positives = 82/124 (65%)
Query: 1 MQKTVDVYLAICQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSRRMT 60
+ + ++ YL + + KL I IE+ I+ELI+R+ I+ LVMGAASDK++S +MT
Sbjct: 84 VDELMNSYLQLLSETEIQTDKLCIAGQNIEECIVELIARHKIKWLVMGAASDKHYSWKMT 143
Query: 61 DLRSKKAIYVCEQSPASCHIQFICKGYLIQTRDCSLENFRVEATSPLVQQMQNSEVGHSP 120
DL+SKKAI+VC+++P SCHI F+CKGYLI TR + ++ + PLVQ ++E S
Sbjct: 144 DLKSKKAIFVCKKAPDSCHIWFLCKGYLIFTRASNDDSNNRQTMPPLVQLDSDNETRKSE 203
Query: 121 HLIS 124
L S
Sbjct: 204 KLES 207
>UniRef100_Q9SMU3 Hypothetical protein T2J13.100 [Arabidopsis thaliana]
Length = 1175
Score = 211 bits (538), Expect = 4e-53
Identities = 106/222 (47%), Positives = 154/222 (68%), Gaps = 21/222 (9%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIA 492
+VE+LS+++HPNL+TL+G ES++LIY+Y+PNGSLED S NN P L+W++RIRIA
Sbjct: 429 RVEILSRVRHPNLVTLMGACPESRSLIYQYIPNGSLEDCFS--SENNVPALSWESRIRIA 486
Query: 493 TELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTT 552
+E+CSAL+FLHSN P I+HG+LKPS ILLD+NLVTK++D+GI +++ S+
Sbjct: 487 SEICSALLFLHSNIP-CIIHGNLKPSKILLDSNLVTKINDYGISQLIPIDGLDKSD---- 541
Query: 553 QFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNN 612
++DP + + E+T +SD+Y+FGI+LL+L+T +P GI +V AL N
Sbjct: 542 ------------PHVDPHYFVSREMTLESDIYAFGIILLQLLTRRPVSGILRDVKCALEN 589
Query: 613 AGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPEL 654
N+ +VLD AGDWP+ +KL + A+RCC N +RP+L
Sbjct: 590 --DNISAVLDNSAGDWPVARGKKLANVAIRCCKKNPMNRPDL 629
Score = 42.7 bits (99), Expect = 0.036
Identities = 23/56 (41%), Positives = 36/56 (64%)
Query: 1 MQKTVDVYLAICQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHS 56
+ + ++ YL + + KL I IE+ I+ELI+R+ I+ LVMGAASDK++S
Sbjct: 84 VDELMNSYLQLLSETEIQTDKLCIAGQNIEECIVELIARHKIKWLVMGAASDKHYS 139
>UniRef100_Q67YP5 Hypothetical protein At3g49065 [Arabidopsis thaliana]
Length = 509
Score = 211 bits (538), Expect = 4e-53
Identities = 106/222 (47%), Positives = 154/222 (68%), Gaps = 21/222 (9%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIA 492
+VE+LS+++HPNL+TL+G ES++LIY+Y+PNGSLED S NN P L+W++RIRIA
Sbjct: 207 RVEILSRVRHPNLVTLMGACPESRSLIYQYIPNGSLEDCFS--SENNVPALSWESRIRIA 264
Query: 493 TELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTT 552
+E+CSAL+FLHSN P I+HG+LKPS ILLD+NLVTK++D+GI +++ S+
Sbjct: 265 SEICSALLFLHSNIP-CIIHGNLKPSKILLDSNLVTKINDYGISQLIPIDGLDKSD---- 319
Query: 553 QFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNN 612
++DP + + E+T +SD+Y+FGI+LL+L+T +P GI +V AL N
Sbjct: 320 ------------PHVDPHYFVSREMTLESDIYAFGIILLQLLTRRPVSGILRDVKCALEN 367
Query: 613 AGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPEL 654
N+ +VLD AGDWP+ +KL + A+RCC N +RP+L
Sbjct: 368 --DNISAVLDNSAGDWPVARGKKLANVAIRCCKKNPMNRPDL 407
>UniRef100_Q67YL5 Hypothetical protein At3g49065 [Arabidopsis thaliana]
Length = 805
Score = 211 bits (538), Expect = 4e-53
Identities = 106/222 (47%), Positives = 154/222 (68%), Gaps = 21/222 (9%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIA 492
+VE+LS+++HPNL+TL+G ES++LIY+Y+PNGSLED S NN P L+W++RIRIA
Sbjct: 503 RVEILSRVRHPNLVTLMGACPESRSLIYQYIPNGSLEDCFS--SENNVPALSWESRIRIA 560
Query: 493 TELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTT 552
+E+CSAL+FLHSN P I+HG+LKPS ILLD+NLVTK++D+GI +++ S+
Sbjct: 561 SEICSALLFLHSNIP-CIIHGNLKPSKILLDSNLVTKINDYGISQLIPIDGLDKSD---- 615
Query: 553 QFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNN 612
++DP + + E+T +SD+Y+FGI+LL+L+T +P GI +V AL N
Sbjct: 616 ------------PHVDPHYFVSREMTLESDIYAFGIILLQLLTRRPVSGILRDVKCALEN 663
Query: 613 AGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPEL 654
N+ +VLD AGDWP+ +KL + A+RCC N +RP+L
Sbjct: 664 --DNISAVLDNSAGDWPVARGKKLANVAIRCCKKNPMNRPDL 703
Score = 103 bits (256), Expect = 2e-20
Identities = 56/124 (45%), Positives = 82/124 (65%)
Query: 1 MQKTVDVYLAICQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSRRMT 60
+ + ++ YL + + KL I IE+ I+ELI+R+ I+ LVMGAASDK++S +MT
Sbjct: 84 VDELMNSYLQLLSETEIQTDKLCIAGQNIEECIVELIARHKIKWLVMGAASDKHYSWKMT 143
Query: 61 DLRSKKAIYVCEQSPASCHIQFICKGYLIQTRDCSLENFRVEATSPLVQQMQNSEVGHSP 120
DL+SKKAI+VC+++P SCHI F+CKGYLI TR + ++ + PLVQ ++E S
Sbjct: 144 DLKSKKAIFVCKKAPDSCHIWFLCKGYLIFTRASNDDSNNRQTMPPLVQLDSDNETRKSE 203
Query: 121 HLIS 124
L S
Sbjct: 204 KLES 207
>UniRef100_Q9CAI9 Hypothetical protein F28P22.5 [Arabidopsis thaliana]
Length = 697
Score = 207 bits (527), Expect = 8e-52
Identities = 104/228 (45%), Positives = 150/228 (65%), Gaps = 11/228 (4%)
Query: 428 RSLSYQVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQT 487
+ ++EVLS ++HPN++ L+G E L+YEY+ NG+LED L NN PPL+W+
Sbjct: 420 KQFQQEIEVLSSMRHPNMVILLGACPEYGCLVYEYMENGTLEDRLFCK--NNTPPLSWRA 477
Query: 488 RIRIATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVL-SCQNDSS 546
R RIA+E+ + L+FLH KP +VH DLKP+NILLD +L K+SD G+ R++ D+
Sbjct: 478 RFRIASEIATGLLFLHQAKPEPLVHRDLKPANILLDKHLTCKISDVGLARLVPPAVADTY 537
Query: 547 SNNSTTQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEV 606
SN T A GTF Y+DPE+ TG L KSD+YSFG+VLL++IT +PA+G+ ++V
Sbjct: 538 SNYHMTS------AAGTFCYIDPEYQQTGMLGVKSDLYSFGVVLLQIITAQPAMGLGHKV 591
Query: 607 LYALNNAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPEL 654
A+ N N++ +LDP +WP E +L AL+CC++ KK RP+L
Sbjct: 592 EMAVEN--NNLREILDPTVSEWPEEETLELAKLALQCCELRKKDRPDL 637
>UniRef100_Q9ATX1 Serine threonine kinase 1 [Zea mays]
Length = 787
Score = 207 bits (526), Expect = 1e-51
Identities = 106/223 (47%), Positives = 149/223 (66%), Gaps = 8/223 (3%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHL-SRNGNNNAPPLTWQTRIRI 491
+VEVLS ++HPN++ L+G E L+YEY+ +GSL+D L R+G P + WQ R RI
Sbjct: 492 EVEVLSCIRHPNMVLLLGACPEYGCLVYEYMASGSLDDCLFRRSGGAGGPVIPWQHRFRI 551
Query: 492 ATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNST 551
E+ + L+FLH KP +VH DLKP NILLD N V+K+SD G+ R++ S ++
Sbjct: 552 CAEIATGLLFLHQTKPEPLVHRDLKPGNILLDRNYVSKISDVGLARLV----PPSVADTV 607
Query: 552 TQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALN 611
TQ+ +TS A GTF Y+DPE+ TG L KSDVYSFG++LL++IT KP +G+ + V AL
Sbjct: 608 TQYRMTSTA-GTFCYIDPEYQQTGMLGVKSDVYSFGVMLLQIITAKPPMGLSHHVGRALE 666
Query: 612 NAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPEL 654
G ++ +LDP DWP+ EA+ L ALRCC++ +K RP+L
Sbjct: 667 R--GELQDMLDPAVPDWPLEEAQCLAEMALRCCELRRKDRPDL 707
Score = 37.0 bits (84), Expect = 2.0
Identities = 26/113 (23%), Positives = 52/113 (46%), Gaps = 7/113 (6%)
Query: 8 YLAICQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSRRMTDLRSKKA 67
+ C R + + ++ + K IIE ++ ++ LV+GA + R D+ +
Sbjct: 83 FRCFCTRKDINCKDVVLDEHDVAKSIIEFSAQAAVEKLVLGATTRGGFVRFKADIPTT-- 140
Query: 68 IYVCEQSPASCHIQFICKGYLIQTRDCSLENFRVEATSPLVQQMQNSEVGHSP 120
+ + +P C + + KG + R+ + RV SPL Q+Q+S++ P
Sbjct: 141 --ISKGAPDFCSVYIVNKGKVSSQRNSTRAAPRV---SPLRSQIQSSQIAAMP 188
>UniRef100_Q8W1D3 Serine threonine kinase [Zea mays]
Length = 787
Score = 207 bits (526), Expect = 1e-51
Identities = 106/223 (47%), Positives = 149/223 (66%), Gaps = 8/223 (3%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHL-SRNGNNNAPPLTWQTRIRI 491
+VEVLS ++HPN++ L+G E L+YEY+ +GSL+D L R+G P + WQ R RI
Sbjct: 493 EVEVLSCIRHPNMVLLLGACPEYGCLVYEYMASGSLDDCLFRRSGGAGGPVIPWQHRFRI 552
Query: 492 ATELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNST 551
E+ + L+FLH KP +VH DLKP NILLD N V+K+SD G+ R++ S ++
Sbjct: 553 CAEIATGLLFLHQTKPEPLVHRDLKPGNILLDRNYVSKISDVGLARLV----PPSVADTV 608
Query: 552 TQFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALN 611
TQ+ +TS A GTF Y+DPE+ TG L KSDVYSFG++LL++IT KP +G+ + V AL
Sbjct: 609 TQYRMTSTA-GTFCYIDPEYQQTGMLGVKSDVYSFGVMLLQIITAKPPMGLSHHVGRALE 667
Query: 612 NAGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPEL 654
G ++ +LDP DWP+ EA+ L ALRCC++ +K RP+L
Sbjct: 668 R--GELQDMLDPAVPDWPLEEAQCLAEMALRCCELRRKDRPDL 708
Score = 35.4 bits (80), Expect = 5.7
Identities = 25/109 (22%), Positives = 51/109 (45%), Gaps = 7/109 (6%)
Query: 8 YLAICQRMGVTATKLHIEMDCIEKGIIELISRYNIQNLVMGAASDKYHSRRMTDLRSKKA 67
+ C R + + ++ + K IIE ++ ++ LV+GA + R D+ +
Sbjct: 84 FRCFCTRKDINCKDVVLDEHDVAKSIIEFSAQAAVEKLVLGATTRGGFVRFKADIPTT-- 141
Query: 68 IYVCEQSPASCHIQFICKGYLIQTRDCSLENFRVEATSPLVQQMQNSEV 116
+ + +P C + + KG + R+ + RV SPL Q+Q+S++
Sbjct: 142 --ISKGAPDFCSVYIVNKGKVSSQRNSTRAAPRV---SPLRSQIQSSQI 185
>UniRef100_Q9ZQ30 Hypothetical protein At2g24370 [Arabidopsis thaliana]
Length = 816
Score = 205 bits (521), Expect = 4e-51
Identities = 105/222 (47%), Positives = 149/222 (66%), Gaps = 9/222 (4%)
Query: 433 QVEVLSKLKHPNLITLIGVNQESKTLIYEYLPNGSLEDHLSRNGNNNAPPLTWQTRIRIA 492
+VEVLS ++HPN++ L+G E L+YE++ NGSLED L R GN+ PPL+WQ R RIA
Sbjct: 523 EVEVLSCIRHPNMVLLLGACPECGCLVYEFMANGSLEDRLFRLGNS--PPLSWQMRFRIA 580
Query: 493 TELCSALIFLHSNKPHSIVHGDLKPSNILLDANLVTKLSDFGICRVLSCQNDSSSNNSTT 552
E+ + L+FLH KP +VH DLKP NILLD N V+K+SD G+ R++ + ++ T
Sbjct: 581 AEIGTGLLFLHQAKPEPLVHRDLKPGNILLDRNFVSKISDVGLARLV----PPTVADTVT 636
Query: 553 QFWITSFAKGTFAYMDPEFLGTGELTSKSDVYSFGIVLLRLITGKPALGIKNEVLYALNN 612
Q+ +TS A GTF Y+DPE+ TG L KSD+YS GI+ L+LIT KP +G+ + V AL
Sbjct: 637 QYRMTSTA-GTFCYIDPEYQQTGMLGVKSDIYSLGIMFLQLITAKPPMGLTHYVERALEK 695
Query: 613 AGGNVKSVLDPLAGDWPIVEAEKLVHFALRCCDMNKKSRPEL 654
G + +LDP+ DWP+ + E+ AL+C ++ +K RP+L
Sbjct: 696 --GTLVDLLDPVVSDWPMEDTEEFAKLALKCAELRRKDRPDL 735
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.331 0.141 0.432
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,038,838,354
Number of Sequences: 2790947
Number of extensions: 41516878
Number of successful extensions: 175909
Number of sequences better than 10.0: 17449
Number of HSP's better than 10.0 without gapping: 6244
Number of HSP's successfully gapped in prelim test: 11209
Number of HSP's that attempted gapping in prelim test: 144338
Number of HSP's gapped (non-prelim): 19956
length of query: 688
length of database: 848,049,833
effective HSP length: 134
effective length of query: 554
effective length of database: 474,062,935
effective search space: 262630865990
effective search space used: 262630865990
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 78 (34.7 bits)
Medicago: description of AC146341.5


