

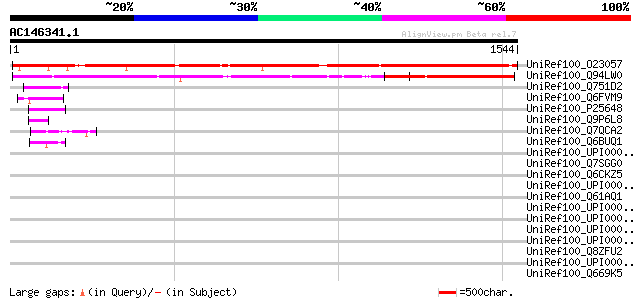
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146341.1 - phase: 0 /pseudo
(1544 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O23057 A_IG005I10.24 protein [Arabidopsis thaliana] 1421 0.0
UniRef100_Q94LW0 Hypothetical protein OSJNBa0082M15.26 [Oryza sa... 804 0.0
UniRef100_Q751D2 AGL226Cp [Ashbya gossypii] 62 1e-07
UniRef100_Q6FVM9 Similar to sp|P25648 Saccharomyces cerevisiae Y... 61 2e-07
UniRef100_P25648 Suppressor of RNA polymerase B SRB8 [Saccharomy... 52 1e-04
UniRef100_Q9P6L8 Suppressor of RNA polymerase B srb8 [Schizosacc... 52 1e-04
UniRef100_Q7QCA2 ENSANGP00000003011 [Anopheles gambiae str. PEST] 50 4e-04
UniRef100_Q6BUQ1 Debaryomyces hansenii chromosome C of strain CB... 50 4e-04
UniRef100_UPI000023E8E2 UPI000023E8E2 UniRef100 entry 48 0.002
UniRef100_Q7SGG0 Hypothetical protein B20J13.130 [Neurospora cra... 48 0.003
UniRef100_Q6CKZ5 Kluyveromyces lactis strain NRRL Y-1140 chromos... 47 0.006
UniRef100_UPI00003AEF48 UPI00003AEF48 UniRef100 entry 45 0.013
UniRef100_Q61AQ1 Hypothetical protein CBG13669 [Caenorhabditis b... 45 0.013
UniRef100_UPI000023601E UPI000023601E UniRef100 entry 45 0.023
UniRef100_UPI000042BF15 UPI000042BF15 UniRef100 entry 44 0.030
UniRef100_UPI000021B3B3 UPI000021B3B3 UniRef100 entry 43 0.066
UniRef100_UPI000042BF10 UPI000042BF10 UniRef100 entry 43 0.087
UniRef100_Q8ZFU2 Ribonuclease E [Yersinia pestis] 43 0.087
UniRef100_UPI000021AE56 UPI000021AE56 UniRef100 entry 42 0.11
UniRef100_Q669K5 Ribonuclease E [Yersinia pseudotuberculosis] 42 0.15
>UniRef100_O23057 A_IG005I10.24 protein [Arabidopsis thaliana]
Length = 2124
Score = 1421 bits (3679), Expect = 0.0
Identities = 813/1635 (49%), Positives = 1072/1635 (64%), Gaps = 174/1635 (10%)
Query: 8 SFQAIKKRLRAINESRAQKRK--------------------AGQVYGVPLSGLQLAKPGI 47
S A++K LRAINESRA KRK AGQVYGVPLSG L KPG
Sbjct: 9 SIGAVRKCLRAINESRALKRKVIKRLSDSNPFQFIFCAEFRAGQVYGVPLSGSLLCKPG- 67
Query: 48 FPELRPCGEDFRKKWIEGLSQPHKRLCTLADHVPHGYKRSSLLEVLIRNNVPLLRATWFV 107
FPE R CGE+ +K+WIE LSQ HKRL +LAD++P GY+R +L EVLIRNNVPLLRATWF+
Sbjct: 68 FPEQRSCGEETKKRWIESLSQQHKRLRSLADNIP-GYRRKTLFEVLIRNNVPLLRATWFI 126
Query: 108 KVTYLNQV---------------------RPGSVGISSGTADKIQPSRTEIWTK---DVI 143
KVTYLNQV R +G+S+G + T W +
Sbjct: 127 KVTYLNQVHCWAINWCDLVLLLFLQEHLTRHKLLGVSNGQKMLLNICNTS-WMNFCHGIA 185
Query: 144 HYLQTLLD----EFFSKNNSHSTLQNRERSPQMP-------------YAGTLLHKSDPLS 186
H+L + L+ F ++ +Q+++ Y+GT+L L
Sbjct: 186 HFLLSKLEIGHHRCFIQDQCKRIVQHQQAFTARKHLYILNGGIWCVFYSGTMLKGFFFLI 245
Query: 187 SFSAGEEPSSHFRWWYIVRLLQWHHAEGLILPSLVIDWVLNQLQEKDLLEVWELLLPIIY 246
S G S F + +H A LQEK++ E+ +LLLPI+Y
Sbjct: 246 SLLIGFSSSYSF--------VNFHFA---------------LLQEKEIFEILQLLLPIVY 282
Query: 247 GFLEIIVLSQSYVRALAGLTLRVIRDPAPGGSDLVDNSRRAYTTYALIEMLQYLILAVPD 306
G LE IVLSQ+YV++L + +R I++PAPGGSDLVDNSRRAYT ALIEM++YL+LA PD
Sbjct: 283 GVLESIVLSQTYVQSLVAIAVRFIQEPAPGGSDLVDNSRRAYTLSALIEMVRYLVLAAPD 342
Query: 307 TFVALDCFPLPSSVVSHTMNDGSFVLKSSEAAGKIKNSS-----DYFGRIVLC------- 354
TFVA D FPLP SV + ND S+ K+ E K++++S + GR VL
Sbjct: 343 TFVASDFFPLPPSVAACGPNDVSYTSKAYENLEKLRSNSAEISAQFQGRGVLSRFEFLSF 402
Query: 355 ------IQKRAEDLAKAASPGNPSHCLAKVAKALDNSLVLGDLREAYKFLFEDFCDGTVS 408
IQ+ A+DLAK AS G P H +AK +ALD +L GD+R AY +LFED C+G V
Sbjct: 403 DYTISTIQRSADDLAKIASAGYPQHNVAKAVQALDKALSDGDIRAAYSYLFEDLCNGAVD 462
Query: 409 ESWIVKVSPCLRLSLKWFGTVKTSLIHSVFFLCEWATCDFRDFCTTPPCDIKFTGRKDLS 468
E+WI VSPCLR SL+W G + TS + SVFFL EWATCDFRDF P DIKF+GRKD S
Sbjct: 463 EAWITDVSPCLRSSLRWIGAISTSFVCSVFFLIEWATCDFRDFRAGVPKDIKFSGRKDCS 522
Query: 469 QVHIAVRLLKMKLRDVKSSLRRAKRSIHRASYAAKHESQRHNQNYITTGSSVISESPGPL 528
QV++ ++LLK K+ + + R+ K + +K +GS ESPGPL
Sbjct: 523 QVYLVIQLLKQKILGGEFTARKGKNCRNNFLGVSK-----------PSGSMDAFESPGPL 571
Query: 529 HDIIVCWIDQHVVHKGEGLKRLHLFIVELIRAGIFYPLAYVRQLIVSGIMNMDVNVVDVE 588
HDIIVCWIDQH VHKG G KRL L + ELIR+GIF P+AYVRQLIVSG++++ VD E
Sbjct: 572 HDIIVCWIDQHEVHKG-GAKRLQLLVFELIRSGIFNPIAYVRQLIVSGMIDVIQPAVDPE 630
Query: 589 RRKRHSHILKQLPGHFMRDALTESGIAEGPQLIEALQTYLTERRLILRNSPSDPRDDANA 648
RR RH ILKQLPG F+ + L E+ + G +L EA++TY ERRL+LR +
Sbjct: 631 RRMRHHRILKQLPGCFVHETLEEAQLFGGDKLSEAVRTYSNERRLLLRELLVEK--GKYW 688
Query: 649 NNAKLPTLKRKRNPASSKDGTSTVSTDQWKTVQSTASS--KSAKDGASMEELKETISVLL 706
NN L K K+ S S+V + +S + K K + ELKE IS LL
Sbjct: 689 NNLVLSDQKSKKISTS----LSSVIFPRACNAKSNSKGPRKHTKSSVDIRELKERISALL 744
Query: 707 QLPNSLSNLNSTGCDESESSVRKPTWPHYGKTEPVEGTPGCEECRRAKRQKLSEERSSSV 766
Q P + + DE ++SV++ + Y K + E TPGCE+CRRAKR K+++E+SS
Sbjct: 745 QFPGMSCGVETPVRDEFQNSVKRSSGSVYSKMDQPEATPGCEDCRRAKRPKMNDEKSSCY 804
Query: 767 PA-------DDDTWWVKKGLKSTEP-LKVDQPQKTTKQVTKSRQKNVRK-MSLAQLAASR 817
++D WW+KKG K+ E LKVD + TKQV + RQK RK SLAQL A+R
Sbjct: 805 QGNSPIASDEEDNWWIKKGSKTVESSLKVDPQIEITKQVPRGRQKMARKTQSLAQLQAAR 864
Query: 818 IEGSQGASTSHMCENKVSCPHHRTPVDGDALRSVDSMRTCDSRDIVFIGKALKKLRFVEK 877
IEGSQGASTSH+C+NKVSCPHH V+G+ + VD RT D+V +G +LK+L+FV+K
Sbjct: 865 IEGSQGASTSHVCDNKVSCPHHGPGVEGENQKVVDVFRTSTPVDMVSVGNSLKQLQFVDK 924
Query: 878 RAVAAWLLTVVKQVIEETEKNIGKVGQFGRAYSMVDDRNSIRWKLGEDELSAILYLIDIS 937
R++A WL T V+Q++EE +K+ +VGQF R + V+++N+IRWKLG DEL +IL+L+DIS
Sbjct: 925 RSIAVWLTTAVRQLVEEPQKSSVRVGQFNRG-APVEEKNTIRWKLGADELYSILFLLDIS 983
Query: 938 DDLVSAVKFLLWLMPKVLSSPNSTIHSGRNVLMLPRNAENQVCDVGEAFLLSSLRRYENI 997
DLVSA GRN++ +PRN EN +C++GEA L+SSLRRYENI
Sbjct: 984 LDLVSA--------------------GGRNLVTVPRNVENNMCEIGEAILVSSLRRYENI 1023
Query: 998 LVAADLIPEALSSTMHRAATLIASNGRVSNSGATAFTRYLLKKYSNVASVIEWEKTFKST 1057
L++ADL+PEA+++ M+RAA+L++SNG++S S A +TRY+LK+Y ++ SV+EW FK+T
Sbjct: 1024 LLSADLVPEAMTALMNRAASLMSSNGKISGSAALVYTRYILKRYGSLPSVVEWHNNFKAT 1083
Query: 1058 CDARLSAELESVRSVDGELGLPFGVPAGVEDPDDFFRQKIS-GSRLPSRVGAGMRDIVQR 1116
+ +L +EL+ RS +GE G P GVPAGV++PDD+ R+KIS G PSRVG MRD++QR
Sbjct: 1084 SEKKLLSELDHTRSGNGEYGNPLGVPAGVDNPDDYLRKKISIGGARPSRVGLSMRDVLQR 1143
Query: 1117 NVEEAFQYLFGKDRKLYAAGIPKGHALEKWDNGYQIAQQIVMGLIDSIKQTGGAAQEGDP 1176
+VEEA YL +KL G K EK D+GYQ+AQQIV+GL+D I+QTGGAAQEGDP
Sbjct: 1144 HVEEATHYL----KKLIGTGTMKASLAEKNDDGYQVAQQIVVGLMDCIRQTGGAAQEGDP 1199
Query: 1177 SLVLSAVSAIVGSVGPTLAKMPDFSSSN----NQSNIMSLNYARCILRMHITCLRLLKEA 1232
SLV SAVSAI+ SVG ++A++ DFS N + S + S N AR ILR+HITCL LLKEA
Sbjct: 1200 SLVSSAVSAIINSVGLSVARITDFSLGNIYQNHPSGVDSSNIARYILRIHITCLCLLKEA 1259
Query: 1233 LGERQSRVFDIALATEASNVFAGVFAPSKASRAQFQMSSAEVHDTSATNSN-ELGNNSIK 1291
LGERQSRVF+IALATE+S GVFAP K SR Q Q+S E +D++A NS ++ N + K
Sbjct: 1260 LGERQSRVFEIALATESSTALTGVFAPVKGSRGQHQLSP-ESYDSNANNSTIDMSNGTGK 1318
Query: 1292 TVVTKATKIAAAVSALVVGAVIYGVTSLERMVTILRLKEGLDVIQCIRTTRSNSNGNVRS 1351
+++ATKI AAVSALV+G++ +GV +LER+V +LRLK+ LD +Q +R T+S+SNG+ RS
Sbjct: 1319 MALSRATKITAAVSALVIGSITHGVITLERIVGLLRLKDYLDFVQFVRRTKSSSNGSARS 1378
Query: 1352 VGAFKADSSIEVHVHWFRLLIGNCRSLCEGLAVDLLGEPSISALSRMQRMLPLSLIFPPA 1411
+GA K +S IEV+VHWFRLL+GNC+++ EGL ++L+GE S+ A+SRMQRMLPL L+FPPA
Sbjct: 1379 MGASKVESPIEVYVHWFRLLVGNCKTVSEGLVLELVGESSVVAISRMQRMLPLKLVFPPA 1438
Query: 1412 YSIFAFVRWRPFI--LNANVAVRDDMNQLYQSLTMAIADAIKHSPFRDVCFRDCQGLYDL 1469
YSI AFV WRPF+ N+N +V +D ++LYQSLTMA D IKH PFRDVCFRD QGLY+L
Sbjct: 1439 YSIIAFVLWRPFVSNSNSNSSVHEDTHRLYQSLTMAFHDVIKHLPFRDVCFRDTQGLYEL 1498
Query: 1470 MVADGSDAEFAALLELNGSDMHLKSMAFVPLRSRLFLDAMIDCKMPPSIFTKDDVNRVSG 1529
+VAD +DAEFA++ E +G DMHLKS+AF PLR+RLFL+++IDCK+P S ++ + G
Sbjct: 1499 IVADSTDAEFASVFESHGLDMHLKSVAFAPLRARLFLNSLIDCKVPSSGYSHE------G 1552
Query: 1530 PSESKTKLANGDSKL 1544
SE+K + +KL
Sbjct: 1553 VSEAKNRHQGNGTKL 1567
>UniRef100_Q94LW0 Hypothetical protein OSJNBa0082M15.26 [Oryza sativa]
Length = 2073
Score = 804 bits (2077), Expect = 0.0
Identities = 513/1253 (40%), Positives = 721/1253 (56%), Gaps = 88/1253 (7%)
Query: 10 QAIKKRLRAINESRAQKRKAGQVYGVPLSGLQLAKPGIFPELRPCGEDFRKKWIEGLSQP 69
+A+KKR RAINESRAQKRKAGQVYGVPLSG L KPG++PE ED R+KWIE L QP
Sbjct: 68 EALKKRFRAINESRAQKRKAGQVYGVPLSGSLLTKPGMYPEQMHSNEDTRRKWIEALVQP 127
Query: 70 HKRLCTLADHVPHGYKRSSLLEVLIRNNVPLLRATWFVKVTYLNQVRPGSVGISSGTADK 129
++RL +LA+ VP G++R L LIR NVPLLRA+W VKVTYLNQV+ S +SS D
Sbjct: 128 NRRLWSLAEQVPRGFRRKLLFNYLIRYNVPLLRASWLVKVTYLNQVQTSSNNVSSAAPDS 187
Query: 130 IQPSRTEIWTKDVIHYLQTLLDEFFSKNNSHSTLQNRERSPQMPYAG--TLLHKSDPLSS 187
+ R++ WTKDVI YLQ LLDE SKN ++E+S AG + K+ +
Sbjct: 188 L---RSQHWTKDVIEYLQLLLDELCSKNGFFGLPSSQEQSLPCLVAGDSPIKLKTGASPA 244
Query: 188 FSAGEEPSSHFRWWYIVRLLQWHHAEGLILPSLVIDWVLNQLQEKDLLEVWELLLPIIYG 247
+ EEPS HF+W Y++R++Q H E L++PSL+I+WV NQLQE+D EV ELLLPI+
Sbjct: 245 SADVEEPSLHFKWSYMIRIVQCHLMEQLLVPSLLIEWVFNQLQERDSTEVLELLLPIVLS 304
Query: 248 FLEIIVLSQSYVRALAGLTLRVIRDPAPGGSDLVDNSRRAYTTYALIEMLQYLILAVPDT 307
++ I LSQ+Y+ L + ++ + D +PG + +N +R+ T AL+E+LQYLILAVPDT
Sbjct: 305 LVDTITLSQTYIHMLVEILIQRLSDASPGSLSVKNNPKRSSITSALVELLQYLILAVPDT 364
Query: 308 FVALDCFPLPSSVVSHTMNDGSFVLKSSE---AAGKIKNSSDYF--GRIVLCIQKRAEDL 362
FV+LDCFPLPS V G+ + + A+ + +N+S + G + +Q+RA DL
Sbjct: 365 FVSLDCFPLPSVVAPDVYGKGALLKIAGGGKIASSRRQNASRHLSCGYAICSVQRRASDL 424
Query: 363 AKAASPGNPSHCLAKVAKALDNSLVLGDLREAYKFLFEDFCDGTVSESWIVKVSPCLRLS 422
+ A+P A V +ALD +LV G+L AY +F D + E+WI +VSPCLR S
Sbjct: 425 SLVANPNLQVRGAANVVQALDKALVTGNLTAAYTSVFNYLSDTLMEETWIKEVSPCLRSS 484
Query: 423 LKWFGTVKTSLIHSVFFLCEWATCDFRDFCTTPPC-DIKFTGRKDLSQVHIAVRLLKMKL 481
L W G V+ SL+ SVFF+CEWATC FRD C T C ++KF+G KD SQV++AV LLK K+
Sbjct: 485 LMWMGAVELSLVCSVFFICEWATCTFRD-CRTSQCQNVKFSGSKDFSQVYMAVSLLKDKM 543
Query: 482 RDV------KSSLRRAKRSIHRASYAAKHESQRHNQNYITTG------------SSVISE 523
++ KSS + A + H S H S + +G I
Sbjct: 544 NEINNLSSSKSSSQLAMKD-HLKSATLNHSSIKVTAMETASGFRDSTGSIDENNKKDIFS 602
Query: 524 SPGPLHDIIVCWIDQHVVHKGEGLKRLHLFIVELIRAGIFYPLAYVRQLIVSGIMNMDVN 583
SPGPLHDIIVCW+DQH + G K + +F+ ELIR+GIFYP YVRQLIVSGI + +
Sbjct: 603 SPGPLHDIIVCWLDQHEISDASGFKSVDVFMTELIRSGIFYPQTYVRQLIVSGITIWNDS 662
Query: 584 VVDVERRKRHSHILKQLPGHFMRDALTESGIAEGPQLIEALQTYLTERRLILRNSPSDPR 643
+ D+E++ RH ILK LPG + + L E+ IAE L E + TY +ERRL+L S
Sbjct: 663 LFDLEKKTRHYKILKHLPGFCLFNILEEAKIAEDQVLYEIVSTYSSERRLVLSELSSGLA 722
Query: 644 DDANANNAKLPTLKRKRNPASSKDGTSTVSTDQWKTVQSTASSKSAKDGASMEELKETIS 703
DAN R P SS Q + + + +EE+K IS
Sbjct: 723 TDANVEG---------RVPLSS------CLQKQPDLLMDSTDDNHGRVAVQVEEVKLMIS 767
Query: 704 VLLQLPNSLSNLNSTGCDESESSVRKPTWPHYGKTEPVEGTPGCEECRRAKRQKLSE--- 760
LL L S + L +G +E++ + + T + + GC++ R KRQKL +
Sbjct: 768 GLLNLGYS-TLLAESGREETKKTKKGQTDLVDSEEDVGHAKTGCKDSSRTKRQKLDKNVF 826
Query: 761 --ERSSSVPAD-DDTWWVKKGLKSTEPLKVDQPQKTTKQVTKSRQKNVRK-MSLAQLAAS 816
+ S V +D +D WWV+K K E V+ ++ +Q + + V+K ++AQLAA+
Sbjct: 827 PFQGSPLVQSDEEDFWWVRKEQKQ-ELFTVETIHQSIEQTSGGKATVVQKTQNIAQLAAA 885
Query: 817 RIEGSQGASTSHMCENKVSCPHHRTPVDGDALRSVDSMRTCDSRDIVFIGKALKKLRFVE 876
RI+GSQGASTSH+C+N +SCPHH+ + D L+ D M + +GK+LK+LR +E
Sbjct: 886 RIDGSQGASTSHVCDNNLSCPHHKPGTNSDILKDADHM---SMLTLAEVGKSLKRLRLLE 942
Query: 877 KRAVAAWLLTVVKQVIEETEKNIGKVGQFGRAYSMVDDRNSIR--WKLGEDELSAILYLI 934
+R+++ WLL +KQ+IE E K A S V I W+ GEDEL ++LY++
Sbjct: 943 RRSISIWLLKSIKQLIEGDEVKHSKANNSISA-STVQHSGKIASGWRFGEDELLSVLYIM 1001
Query: 935 DISDDLVSAVKFLLWLMPKVLSSPNSTIHSGRNVLMLPRNAENQVCDVGEAFLLSSLRRY 994
D DL+S+V+ L+WL+ K+ ++ GR V M P++ ENQV V EAFL SSL RY
Sbjct: 1002 DTCCDLLSSVRLLIWLLSKIYIGRTTSGQVGRGV-MHPKHKENQVFQVAEAFLFSSLLRY 1060
Query: 995 ENILVAADLIPEALSSTMHRAATLIASNGRVSNSGATAFTRYLLKKYSNVASVIEWEKTF 1054
ENIL+A DL+PE LS +M+R T+ S R S S A A+ RY L+KY +V SV WE+ F
Sbjct: 1061 ENILIAMDLLPEVLSVSMNR--TVHKSGERQSTSVAFAYARYFLRKYRDVTSVARWERNF 1118
Query: 1055 KSTCDARLSAELESVRSVDGELGLPFGVPAGVEDPDDFFRQKISGSRLPSRVGAGMRDIV 1114
+ST D RL AEL+S +S+ G+ + +G+ D RQ +G P + + V
Sbjct: 1119 RSTSDKRLLAELDSGKSITGD-----SIISGIS--SDCIRQ--NGGANPDGDHSLVASAV 1169
Query: 1115 QRNVEEAFQYLFGKDRKLYAAGIPKGHALEKWDNGYQIAQQIVMGLIDS---IKQTGG-- 1169
V+ A + K + P ++ N + Q I+ I+S +++T G
Sbjct: 1170 SAIVDNA-GHAIAKHLDISGGNNPGVTSI----NSLNLIQHILDIHINSLALLRETLGDR 1224
Query: 1170 AAQEGDPSLVLSAVSAIVGSVGPTLAKMPDFSS-----SNNQSNIMSLNYARC 1217
++ + SL + A SA+ S P A SS S N +N + N ++C
Sbjct: 1225 FSRIFEISLAVEASSAVAASFAPPKAHRSQQSSETHDESGNHANEVPSNPSKC 1277
Score = 359 bits (922), Expect = 3e-97
Identities = 195/398 (48%), Positives = 274/398 (67%), Gaps = 10/398 (2%)
Query: 1143 LEKWDNGYQIA-QQIVMGLI-DSIKQTGGAAQEGDPSLVLSAVSAIVGSVGPTLAKMPDF 1200
L + D+G I I+ G+ D I+Q GGA +GD SLV SAVSAIV + G +AK D
Sbjct: 1127 LAELDSGKSITGDSIISGISSDCIRQNGGANPDGDHSLVASAVSAIVDNAGHAIAKHLDI 1186
Query: 1201 SSSNNQ--SNIMSLNYARCILRMHITCLRLLKEALGERQSRVFDIALATEASNVFAGVFA 1258
S NN ++I SLN + IL +HI L LL+E LG+R SR+F+I+LA EAS+ A FA
Sbjct: 1187 SGGNNPGVTSINSLNLIQHILDIHINSLALLRETLGDRFSRIFEISLAVEASSAVAASFA 1246
Query: 1259 PSKASRAQFQMSSAEVHDTSATNSNELGNNSIKTVVTKATKIAAAVSALVVGAVIYGVTS 1318
P KA R+Q S+E HD S ++NE+ +N K KA K++AAVSALVVGA+IYGV S
Sbjct: 1247 PPKAHRSQ---QSSETHDESGNHANEVPSNPSKCFNVKAVKVSAAVSALVVGAIIYGVVS 1303
Query: 1319 LERMVTILRLKEGLDVIQCIRTTRSNSNGNVRSVGAFKADSSIEVHVHWFRLLIGNCRSL 1378
LERM+ +LRLKEGLD++Q +R +++++NG S+G FK DSS EV VHWF++LIGNC+++
Sbjct: 1304 LERMMVVLRLKEGLDILQFLRISKASTNGVTHSIGNFKIDSSTEVLVHWFKILIGNCKTV 1363
Query: 1379 CEGLAVDLLGEPSISALSRMQRMLPLSLIFPPAYSIFAFVRWRPFILNANVAVRDDMNQL 1438
G+ ++LG+ + A SR+QR LPL ++ PPAYSIFA V WRP++ + + + +D+ QL
Sbjct: 1364 YNGVIAEILGDSYVLAFSRLQRTLPLGIVLPPAYSIFAMVLWRPYLYDTSTSNHEDI-QL 1422
Query: 1439 YQSLTMAIADAIKHSPFRDVCFRDCQGLYDLMVADGSDAEFAALLELNGSDMHLKSMAFV 1498
YQSL AI+D +H PFRDVCFR+ YDL+ AD D+EFAA++EL D LK+++
Sbjct: 1423 YQSLLGAISDITRHQPFRDVCFRNMHLFYDLLAADVGDSEFAAIVELRSPDECLKALS-- 1480
Query: 1499 PLRSRLFLDAMIDCKMPPSIFTKDDVNRVSGPSESKTK 1536
PLR+R+FL+A++DC++P ++ G +E+ TK
Sbjct: 1481 PLRARVFLNALLDCEIPVTMRDDGTYALEPGCAEASTK 1518
>UniRef100_Q751D2 AGL226Cp [Ashbya gossypii]
Length = 1358
Score = 62.0 bits (149), Expect = 1e-07
Identities = 41/140 (29%), Positives = 66/140 (46%), Gaps = 4/140 (2%)
Query: 42 LAKPGI-FPELRPCGEDFRKKWIEGLSQPHKRLCTLADHVPHGYKRSSLLEVLIRNNVPL 100
LA PG P + R +W++ LS P+ L LA +PHG+KR +LE +PL
Sbjct: 115 LAGPGFSLPNRVTLTDQRRTQWLQELSSPNVSLSKLAKSIPHGFKRKQILEQCYMRQLPL 174
Query: 101 LRATWFVKVTYLNQVRPGSVGISSG-TADKIQPSRTEIWTKDVIHYLQTLLDEFFSKNNS 159
RA W +K +Y + + + + G T + + +WT ++ ++ LL E N
Sbjct: 175 QRAIWLIKSSYSIEWKSLTSKLKPGQTNEGVVSQLYNLWTNSMVSIMERLLFEMPKYYN- 233
Query: 160 HSTLQNRERSPQMPYAGTLL 179
T Q + P++ Y LL
Sbjct: 234 -DTAQAKIWKPRVSYYINLL 252
>UniRef100_Q6FVM9 Similar to sp|P25648 Saccharomyces cerevisiae YCR081w DNA-directed
RNA polymerase II [Candida glabrata]
Length = 1358
Score = 61.2 bits (147), Expect = 2e-07
Identities = 42/156 (26%), Positives = 67/156 (42%), Gaps = 19/156 (12%)
Query: 23 RAQKRKAGQVYGVPLSGLQLAKPGIFPELRPCGEDFR------------KKWIEGLSQPH 70
R ++R+ + G G+ +K F +L CG DF+ + W++ LS P+
Sbjct: 89 RIREREINTIKGNSKDGMAASK---FTDL--CGPDFKLPSRLTLTDHRKELWLQELSSPY 143
Query: 71 KRLCTLADHVPHGYKRSSLLEVLIRNNVPLLRATWFVKVTYLNQVRPG--SVGISSGTAD 128
L + +PHG KR +LE +PL RA W +K Y + + G S D
Sbjct: 144 TPLHKITSFIPHGLKRRQVLEQCYIKQIPLKRAVWLIKCCYSIEWKTGIAKYANSPQEKD 203
Query: 129 KIQPSRTEIWTKDVIHYLQTLLDEFFSKNNSHSTLQ 164
K + WT + +H L+ L+ E N L+
Sbjct: 204 KFNAHLLKEWTDNFVHILEKLIFEMTQHYNDSVKLE 239
>UniRef100_P25648 Suppressor of RNA polymerase B SRB8 [Saccharomyces cerevisiae]
Length = 1427
Score = 52.4 bits (124), Expect = 1e-04
Identities = 31/116 (26%), Positives = 52/116 (44%), Gaps = 4/116 (3%)
Query: 57 DFRKK-WIEGLSQPHKRLCTLADHVPHGYKRSSLLEVLIRNNVPLLRATWFVKVTYLNQV 115
D RK+ W+ LS H L + +PHG KR ++E +PL RA W +K Y +
Sbjct: 123 DHRKETWLHELSSSHTSLVKIGKFIPHGLKRRQVIEQCYLKFIPLKRAIWLIKCCYFIEW 182
Query: 116 RPG---SVGISSGTADKIQPSRTEIWTKDVIHYLQTLLDEFFSKNNSHSTLQNRER 168
+ ++G D I + WT ++ L+ L+ + + N L+ +R
Sbjct: 183 KSNHKKKRSNAAGADDAISMHLLKDWTDTFVYILEKLIFDMTNHYNDSQQLRTWKR 238
>UniRef100_Q9P6L8 Suppressor of RNA polymerase B srb8 [Schizosaccharomyces pombe]
Length = 1233
Score = 52.0 bits (123), Expect = 1e-04
Identities = 22/61 (36%), Positives = 35/61 (57%)
Query: 56 EDFRKKWIEGLSQPHKRLCTLADHVPHGYKRSSLLEVLIRNNVPLLRATWFVKVTYLNQV 115
E R +W++GLS P L +L +PHG +L +L++ VP RA WF++ +N+
Sbjct: 131 ESRRDRWLQGLSDPMVPLSSLIKTIPHGLWGEDILRMLVKFRVPFTRAIWFIRCAGVNEA 190
Query: 116 R 116
R
Sbjct: 191 R 191
>UniRef100_Q7QCA2 ENSANGP00000003011 [Anopheles gambiae str. PEST]
Length = 1651
Score = 50.4 bits (119), Expect = 4e-04
Identities = 52/212 (24%), Positives = 93/212 (43%), Gaps = 41/212 (19%)
Query: 62 WIEGLSQPHKRLCTLADHVPHGYKRSSLLEVLIRNNVPLLRATWFVKVTYLNQVRPGSVG 121
W + L+ K L +LA P K+ + +L N V + RA WF+K++ +V
Sbjct: 21 WFKDLAGT-KPLSSLAKKAPSFNKKEEIFAMLCENQVTMQRAAWFIKLS-----SAYTVA 74
Query: 122 ISSGTADKIQ-PSRTEIWTKDVIHYLQTLLDEFFSKNNSHSTLQNRERSPQMPYAGTLLH 180
+S K Q P WT +I +++ L+ K + H + G L
Sbjct: 75 VSEAKIKKRQMPDPATEWTGTMIKFMKDLI----PKLHEHY------------HQGPLQE 118
Query: 181 KSDPLSSFSAGEEPS-SHFRWWYIVRLLQWHHAEGLILPSLVIDWVLNQLQ--------E 231
K S+GEE + +W Y +L ++ + EGL+ ++W+++ L+ +
Sbjct: 119 KP------SSGEEQRIAQKQWNYSTQLCKYMYEEGLLDKQEFLNWIIDLLEKMKSSPNAD 172
Query: 232 KDLLEVWELLLPIIYGFLEIIVLSQSYVRALA 263
LL ++ LP+ +L +V S+ + R LA
Sbjct: 173 DGLLRIY---LPLAMQYLYDLVQSERFCRRLA 201
>UniRef100_Q6BUQ1 Debaryomyces hansenii chromosome C of strain CBS767 of Debaryomyces
hansenii [Debaryomyces hansenii]
Length = 1661
Score = 50.4 bits (119), Expect = 4e-04
Identities = 26/121 (21%), Positives = 56/121 (45%), Gaps = 16/121 (13%)
Query: 59 RKKWIEGLSQPHKRLCTLADHVPHGYKRSSLLEVLIRNNVPLLRATWFVKVT-------- 110
++ W+ L+ P + L +++ +PHG K L+++L N+P RA W K
Sbjct: 178 KEAWLRDLANPDEPLLNISNKLPHGIKNKMLVDILCSKNIPTSRALWLTKCVLYSELLVL 237
Query: 111 ---YLNQVRPGSVGISSGTADKIQPSRTEIWTKDVIHYLQTLLDEFFSKNNSHSTLQNRE 167
Y +++ + + T++ + + WT ++ Y FSK+ + T+Q ++
Sbjct: 238 KKKYQSRLPNNPHPVENTTSETFETQWLQEWTHQLVDYFYK-----FSKDMCNITIQEKK 292
Query: 168 R 168
+
Sbjct: 293 Q 293
>UniRef100_UPI000023E8E2 UPI000023E8E2 UniRef100 entry
Length = 1529
Score = 48.1 bits (113), Expect = 0.002
Identities = 38/157 (24%), Positives = 68/157 (43%), Gaps = 10/157 (6%)
Query: 59 RKKWIEGLSQPHKRLCTLADHVPHGYKRSSLLEVLIRNNVPLLRATWFVKVTYLNQVRPG 118
R+ W++ L+ P L L+ +PHG + +LL+ + NVP RA W K N++R
Sbjct: 291 REVWLKDLANPVISLRRLSRTIPHGIRGRTLLDQCLNKNVPAERAVWLAKCVGANEIR-- 348
Query: 119 SVGISSGTADKIQPSRTEI-WTKDVIHYLQTLLDEF---FSKNNSHSTLQNRERSPQMPY 174
G+ + E+ W +D +++ +D FS+ + + + R Y
Sbjct: 349 --GLKRKGVNGAFVMGGELKWARDWTVFVEQFVDAVVTGFSEKDWKNRVTYAIRLATNLY 406
Query: 175 AGTLLHKSDPLSSFSAGEEPS--SHFRWWYIVRLLQW 209
+ LL + L +G E S S W ++ + W
Sbjct: 407 SEQLLDRDHYLDWIVSGIENSLQSRIPMWLLIAQIYW 443
>UniRef100_Q7SGG0 Hypothetical protein B20J13.130 [Neurospora crassa]
Length = 1789
Score = 47.8 bits (112), Expect = 0.003
Identities = 48/199 (24%), Positives = 82/199 (41%), Gaps = 21/199 (10%)
Query: 41 QLAKPGIF-PELRPCGEDFRKK-WIEGLSQPHKRLCTLADHVPHGYKRSSLLEVLIRNNV 98
Q+ P F P R D +K+ W++ L+ L L+ +PHG + LL+ + NV
Sbjct: 367 QVTAPSTFKPPPRVTLTDTKKEVWLKDLANSAIPLRKLSRSIPHGVRNKVLLDQCLNKNV 426
Query: 99 PLLRATWFVKVTYLNQVRPGSVGISSGTADKIQPSRTEIWTKDVIHYLQTLLDEFFSKNN 158
P+ RA W K N++R A ++ R W KD ++ ++ F
Sbjct: 427 PIDRAVWLAKCVGTNELRTFRRLGKGNNASVLEGERK--WLKDWTLVIENFIERVF---- 480
Query: 159 SHSTLQNRERSPQMPYAGTLLHKSDPLSSFSAGEEPSSHFRWWYIVRLLQWHHAEGLILP 218
+ + ++ YA L ++ G H+ W + L HH++ LP
Sbjct: 481 --FSFGEQNWKAKVNYAIQL-----AAHLYAEGLMDREHYLDWTVSSLESSHHSQ---LP 530
Query: 219 SLVIDWVLNQLQEKDLLEV 237
+I + Q+ KDLL +
Sbjct: 531 MWII---IAQIYWKDLLRL 546
>UniRef100_Q6CKZ5 Kluyveromyces lactis strain NRRL Y-1140 chromosome F of strain NRRL
Y- 1140 of Kluyveromyces lactis [Kluyveromyces lactis]
Length = 1322
Score = 46.6 bits (109), Expect = 0.006
Identities = 28/108 (25%), Positives = 52/108 (47%), Gaps = 3/108 (2%)
Query: 59 RKKWIEGLSQPHKRLCTLADHVPHGYKRSSLLEVLIRNNVPLLRATWFVKVTYLNQVRPG 118
R W++ LS + L L VPHG+++ +LE VP+ R W +K Y + + G
Sbjct: 127 RNLWLQELSSSNTSLRKLTKSVPHGFRKRHILEQCYTQFVPIYRVIWLIKSCYAIEWK-G 185
Query: 119 SVGISSG--TADKIQPSRTEIWTKDVIHYLQTLLDEFFSKNNSHSTLQ 164
+ + G T +++ + + WT +++ L+ L+ E N L+
Sbjct: 186 MISKAKGETTHEELLSTLYKDWTDNMVLILEKLVFEVSKYYNDPGQLK 233
>UniRef100_UPI00003AEF48 UPI00003AEF48 UniRef100 entry
Length = 660
Score = 45.4 bits (106), Expect = 0.013
Identities = 52/203 (25%), Positives = 82/203 (39%), Gaps = 41/203 (20%)
Query: 71 KRLCTLADHVPHGYKRSSLLEVLIRNNVPLLRATWFVKVT--YLNQVRPGSVGISSGTAD 128
K L LA VP K+ + L + +VPLLRA W +K+T Y + + T
Sbjct: 92 KPLTILAKKVPILSKKEDVFAYLAKYSVPLLRAAWLIKMTCAYYAAISEAKIKKRQATDP 151
Query: 129 KIQPSRTEIWTKDVIHYLQTLL---DEFFSKNNSHSTLQNRERSPQMPYAGTLLHKSDPL 185
I+ WT+ + YL+ L EF+ H T S MP
Sbjct: 152 NIE------WTQIITRYLREQLAKVAEFY-----HVTSSQGNSSVVMPQ----------- 189
Query: 186 SSFSAGEEPSSHFRWWYIVRLLQWHHAEGLILPSLVIDWVLNQLQ-----EKDLLEVWEL 240
E + +W Y +L + EG++ + W+L+ L+ + D+L +L
Sbjct: 190 ------EMEQALKQWEYNEKLSFYMFQEGMLERHEYLTWILDVLEKIRPTDDDIL---KL 240
Query: 241 LLPIIYGFLEIIVLSQSYVRALA 263
+LP++ + E V S R LA
Sbjct: 241 VLPLMLQYSEEFVQSAYLSRRLA 263
>UniRef100_Q61AQ1 Hypothetical protein CBG13669 [Caenorhabditis briggsae]
Length = 1511
Score = 45.4 bits (106), Expect = 0.013
Identities = 45/185 (24%), Positives = 76/185 (40%), Gaps = 21/185 (11%)
Query: 636 RNSPSDPRDDANANNAKLPT-LKRKRNPASSKDGTSTVSTDQWKTVQSTASSKSAKDGAS 694
R S S D + +A + RKR PA++ +ST +TD A + D +
Sbjct: 1159 RTSSSQADDPGPSTSAATASPANRKRAPAATSSSSSTSATD--------AKKPKSNDEYA 1210
Query: 695 MEELKETISVLLQLPNSLSNLNSTGCDESESSVRKPTW----PHYGKTEPVEGTPGCEEC 750
+ EL++ ++ LL+ + S + D S R+P W H + E
Sbjct: 1211 LSELRDKMTSLLETTTTNSTMTEYKIDPSFIEGRRPDWIKKMEHDMEKIKAERAAATATS 1270
Query: 751 RRAKRQKLSEERSSSVPADDDTWWVKKGLKSTEPLKVDQPQKTTKQVTKSRQKNVRKMSL 810
A +K EE+ S V VK+ K T + ++T K+V + +K V K +
Sbjct: 1271 SGASSKKKKEEKRSEV--------VKEEKKETAKAPKLEKKETPKKVIEKEKKEVEKKNK 1322
Query: 811 AQLAA 815
+ +A
Sbjct: 1323 DEQSA 1327
>UniRef100_UPI000023601E UPI000023601E UniRef100 entry
Length = 1428
Score = 44.7 bits (104), Expect = 0.023
Identities = 25/93 (26%), Positives = 42/93 (44%), Gaps = 3/93 (3%)
Query: 59 RKKWIEGLSQPHKRLCTLADHVPHGYKRSSLLEVLIRNNVPLLRATWFVKVTYLNQVRPG 118
R+ W+ L+ P L L+ +PHG + +LL+ + +P+ RA W VK N++R
Sbjct: 298 REAWLRDLANPSVPLRRLSRTIPHGIRGKNLLDQCLSKWIPVNRAVWLVKCVGANEIRAC 357
Query: 119 SVGISSGTADKIQPSRTEIWTKDVIHYLQTLLD 151
GT + W +D +Q L+
Sbjct: 358 K---RKGTGGTLVVGSEVKWVRDWTSGVQQFLE 387
>UniRef100_UPI000042BF15 UPI000042BF15 UniRef100 entry
Length = 1087
Score = 44.3 bits (103), Expect = 0.030
Identities = 48/209 (22%), Positives = 78/209 (36%), Gaps = 12/209 (5%)
Query: 638 SPSDPRDDANANNAKLPTLKRKRNPASSKDGTSTVSTDQWKTVQSTASSKSAKDGASMEE 697
S S P +++ + P+SS D ++ S+ + STA S SA+ +S E
Sbjct: 391 SSSTPEPSSSSETSSTQESSSTEGPSSSTDSSTEASSSE----SSTAPSSSAEASSSTES 446
Query: 698 LKETISVLLQLPNSLSNLNSTGCDESESSVRKPTWPHYGKTEPVEGTPGCEECRRAKRQK 757
E S + P+S +S+ ESS ++P+ ++ EG EE +
Sbjct: 447 STEEPSSSTEGPSSSQESSSS----EESSTQEPS-SSTKESSSTEGPSSTEESSSTEGPS 501
Query: 758 LSEERS---SSVPADDDTWWVKKGLKSTEPLKVDQPQKTTKQVTKSRQKNVRKMSLAQLA 814
S + S +S + D+ G STE +D + T T S +
Sbjct: 502 SSTDSSTDITSASSTDEQSSSGTGQSSTEDEPIDSTESDTSSATDSSTATDSSATNTDTN 561
Query: 815 ASRIEGSQGASTSHMCENKVSCPHHRTPV 843
+ + S TS N S T V
Sbjct: 562 SESTDSSTATDTSSTDSNTASSTETNTDV 590
>UniRef100_UPI000021B3B3 UPI000021B3B3 UniRef100 entry
Length = 1690
Score = 43.1 bits (100), Expect = 0.066
Identities = 33/115 (28%), Positives = 52/115 (44%), Gaps = 16/115 (13%)
Query: 9 FQAIKKR--LRAIN----ESRAQKRKAGQVYGVPLSGLQLAKPGIFPELRPCGEDFRKK- 61
FQA+K+ L+ ++ E +R GQV +A P P R D +++
Sbjct: 523 FQALKQNRGLQGLSLVYSEVMRHRRDIGQV---------VAPPSFKPPPRVTLTDTKREV 573
Query: 62 WIEGLSQPHKRLCTLADHVPHGYKRSSLLEVLIRNNVPLLRATWFVKVTYLNQVR 116
W+ L+ P L L+ +PHG + LL+ + VP RA W K N++R
Sbjct: 574 WLRDLANPAITLRRLSRTIPHGIRGRVLLDQCLNKKVPAERAVWLAKCVGANEIR 628
>UniRef100_UPI000042BF10 UPI000042BF10 UniRef100 entry
Length = 1086
Score = 42.7 bits (99), Expect = 0.087
Identities = 47/209 (22%), Positives = 77/209 (36%), Gaps = 12/209 (5%)
Query: 638 SPSDPRDDANANNAKLPTLKRKRNPASSKDGTSTVSTDQWKTVQSTASSKSAKDGASMEE 697
S S P +++ + P+SS D ++ S+ + STA S S + +S E
Sbjct: 392 SSSTPEPSSSSETSSTQESSSTEGPSSSTDSSTEASSSE----SSTAPSSSTEASSSTES 447
Query: 698 LKETISVLLQLPNSLSNLNSTGCDESESSVRKPTWPHYGKTEPVEGTPGCEECRRAKRQK 757
E S + P+S +S+ ESS ++P+ ++ EG EE +
Sbjct: 448 STEEPSSSTEGPSSSQESSSS----EESSTQEPS-SSTKESSSTEGPSSTEESSSTEGPS 502
Query: 758 LSEERS---SSVPADDDTWWVKKGLKSTEPLKVDQPQKTTKQVTKSRQKNVRKMSLAQLA 814
S + S +S + D+ G STE +D + T T S +
Sbjct: 503 SSTDSSTDITSASSTDEQSSSGTGQSSTEDEPIDSTESDTSSATDSSTATDSSATNTDTN 562
Query: 815 ASRIEGSQGASTSHMCENKVSCPHHRTPV 843
+ + S TS N S T V
Sbjct: 563 SESTDSSTATDTSSTDSNTASSTETNTDV 591
>UniRef100_Q8ZFU2 Ribonuclease E [Yersinia pestis]
Length = 1221
Score = 42.7 bits (99), Expect = 0.087
Identities = 53/225 (23%), Positives = 91/225 (39%), Gaps = 32/225 (14%)
Query: 586 DVERRKRHSHILKQLPGHFMRDALTESGIAEGPQLIEALQTYLTERRLILRNSPSDPRDD 645
DVE+ S + L F A E Q E ++T +E R RN +PR
Sbjct: 562 DVEQPGFFSRLFSGLKNMFGASAEAEV------QPAEVVKTDTSENR---RNDRRNPRRQ 612
Query: 646 ANANNAKLPTLKRKRNPASSKDGTSTVSTDQWKTVQSTASSKSAKDGASMEELKETISVL 705
N + R+ SS+D T+ +T++ +T+ + +DGA+ + S
Sbjct: 613 NNGRKERNDRTPREGRDNSSRDNTNRDNTNR----DNTSRDNANRDGANRDNSNRDNS-- 666
Query: 706 LQLPNSLSNLNSTGCDESESSVRKPTWP---HYGKTEPVEGTPGCEE---CRRAKRQKLS 759
N++ G ++ + R+P P G+TE VE E RR RQ+
Sbjct: 667 -----GRDNVSREGREDQRRNNRRPAQPTTTSQGQTEVVEADKAQREEQPQRRGDRQRRR 721
Query: 760 EERSSSVPADDDTWWVKKGLKSTEPLKVDQP-QKTTKQVTKSRQK 803
++ P + +K + ++ QP Q+ +QV + RQ+
Sbjct: 722 QDEKRQAPQE-----IKADVAEAPVIEEVQPEQEERQQVMQRRQR 761
>UniRef100_UPI000021AE56 UPI000021AE56 UniRef100 entry
Length = 790
Score = 42.4 bits (98), Expect = 0.11
Identities = 40/148 (27%), Positives = 60/148 (40%), Gaps = 8/148 (5%)
Query: 667 DGTSTVSTDQWKTVQSTASSKSAKDGASMEELKETISVLLQLPNSLSNLNSTGCDESESS 726
D + TD WK Q+T SK + S +++ V P + S D+ S
Sbjct: 537 DEPAPTETD-WKVTQATEESKKTQAKVSSVNIQKPKVVSAPEPEAKSKKLGQLVDKENGS 595
Query: 727 VRKPTW-PHYGKTEPVEGTPGCEECRRAKRQKLSEERSSSVPADDDTWWVKKGLKSTEPL 785
+ P P T+P GTP AK+ KL + +P T V K + +
Sbjct: 596 IPTPNASPTLEATQPNTGTPTTLSLSDAKQTKLQFGQRKKLPGAPTTKTVNKRTRLPD-- 653
Query: 786 KVDQPQKTTKQ---VTKSRQKNVRKMSL 810
QP KTTK V K+ +K+ K ++
Sbjct: 654 -APQPSKTTKSKIPVKKAAKKDATKKAV 680
>UniRef100_Q669K5 Ribonuclease E [Yersinia pseudotuberculosis]
Length = 1230
Score = 42.0 bits (97), Expect = 0.15
Identities = 54/231 (23%), Positives = 93/231 (39%), Gaps = 34/231 (14%)
Query: 586 DVERRKRHSHILKQLPGHFMRDALTESGIAEGPQLIEALQTYLTERRLILRNSPSDPRDD 645
DVE+ S + L F A E Q E ++T +E R RN +PR
Sbjct: 562 DVEQPGFFSRLFSGLKNMFGASAEAEV------QPAEVVKTDASENR---RNDRRNPRRQ 612
Query: 646 ANANNAKLPTLKRKRNPASSKDGTSTVSTDQWKTVQ------STASSKSAKDGASMEELK 699
N + R+ SS+D T+ +T++ T + +T+ + +DGA+ +
Sbjct: 613 NNGRKERNDRTPREGRDNSSRDNTNRDNTNRDNTSRDNTNRDNTSRDNANRDGANRDNSN 672
Query: 700 ETISVLLQLPNSLSNLNSTGCDESESSVRKPTWP---HYGKTEPVEGTPGCEE---CRRA 753
S N++ G ++ + R+P P G+TE VE E RR
Sbjct: 673 RDNS-------GRDNVSREGREDQRRNNRRPAQPTTTSQGQTEVVEADKAQREEQPQRRG 725
Query: 754 KRQKLSEERSSSVPADDDTWWVKKGLKSTEPLKVDQP-QKTTKQVTKSRQK 803
RQ+ ++ P + +K + ++ QP Q+ +QV + RQ+
Sbjct: 726 DRQRRRQDEKRQAPQE-----IKADVAEAPVIEEVQPEQEERQQVMQRRQR 771
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,438,187,340
Number of Sequences: 2790947
Number of extensions: 100158160
Number of successful extensions: 255010
Number of sequences better than 10.0: 81
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 72
Number of HSP's that attempted gapping in prelim test: 254888
Number of HSP's gapped (non-prelim): 153
length of query: 1544
length of database: 848,049,833
effective HSP length: 141
effective length of query: 1403
effective length of database: 454,526,306
effective search space: 637700407318
effective search space used: 637700407318
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 82 (36.2 bits)
Medicago: description of AC146341.1


