

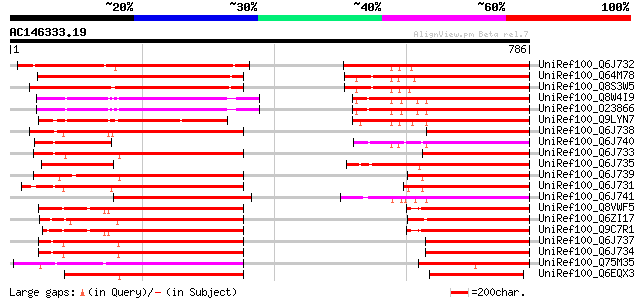
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146333.19 - phase: 0 /pseudo
(786 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6J732 AML1 [Lycopersicon esculentum] 358 4e-97
UniRef100_Q64M78 Putative AML1 [Oryza sativa] 311 6e-83
UniRef100_Q8S3W5 Mei2-like protein [Hordeum vulgare var. distichum] 293 1e-77
UniRef100_Q8W4I9 Mei2-like protein [Arabidopsis thaliana] 288 4e-76
UniRef100_O23866 Mei2-like protein [Arabidopsis thaliana] 288 4e-76
UniRef100_Q9LYN7 Mei2-like protein [Arabidopsis thaliana] 285 5e-75
UniRef100_Q6J738 AML5 [Medicago truncatula] 261 4e-68
UniRef100_Q6J740 AML1 [Beta vulgaris] 259 3e-67
UniRef100_Q6J733 AML1 [Citrus unshiu] 258 4e-67
UniRef100_Q6J735 AML1 [Sorghum bicolor] 257 8e-67
UniRef100_Q6J739 AML1 [Medicago truncatula] 254 7e-66
UniRef100_Q6J731 AML1 [Solanum tuberosum] 252 3e-65
UniRef100_Q6J741 Mei2-like protein [Pinus taeda] 249 2e-64
UniRef100_Q8VWF5 RNA-binding protein MEI2, putative [Arabidopsis... 249 2e-64
UniRef100_Q6ZI17 Putative meiosis protein mei2 [Oryza sativa] 241 5e-62
UniRef100_Q9C7R1 RNA-binding protein MEI2, putative; 36123-32976... 241 6e-62
UniRef100_Q6J737 AML1 [Aegilops speltoides] 238 5e-61
UniRef100_Q6J734 AML15 [Triticum aestivum] 238 5e-61
UniRef100_Q75M35 Hypothetical protein P0668H12.3 [Oryza sativa] 234 6e-60
UniRef100_Q6EQX3 Putative AML1 [Oryza sativa] 230 1e-58
>UniRef100_Q6J732 AML1 [Lycopersicon esculentum]
Length = 971
Score = 358 bits (918), Expect = 4e-97
Identities = 190/324 (58%), Positives = 227/324 (69%), Gaps = 44/324 (13%)
Query: 506 NISSSFPLPVHHEQWSNSYP--PPRTIWPNSPSYFDGIYAASTLQRLNQLPMSPSHMITT 563
N +++ P P H WSNS+ P +WPNSP+Y G+ AS Q+L+ +P +PSHM+
Sbjct: 608 NGTANCPSPGHQYMWSNSHQSQPQGMMWPNSPTYVGGV-CASRPQQLHSVPRAPSHMLNA 666
Query: 564 VLPTNNHHIQSPP-------FWDRRYTYAAEPI------------------TPHCVDFVP 598
++P NNHH+ S P WDRR+ YA E +PH ++F+P
Sbjct: 667 LVPINNHHVGSAPSVNPSLSLWDRRHAYAGESPDASGFHPGSLGSMRISGNSPHPLEFIP 726
Query: 599 HNMFPHFG----------LNV-HNQRGMVFPGRNH---MINSFDT-YKRVRSRRNVGASN 643
HN+F G NV H QR ++FPGR MI+SFD+ +R+RSRRN G S+
Sbjct: 727 HNVFSRTGGSCIDLPMSSSNVGHQQRNLMFPGRAQIIPMISSFDSPNERMRSRRNEGNSS 786
Query: 644 LADMKR-YELDIDCIKRGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHKGAYDFVYLPIDF 702
D K+ +ELDI+ I RG+D RTTLMIKNIPNKYTSKMLLAAIDE H+G YDF+YLPIDF
Sbjct: 787 QTDNKKQFELDIERIARGDDKRTTLMIKNIPNKYTSKMLLAAIDERHRGTYDFIYLPIDF 846
Query: 703 RNKCNVGYAFINMTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQNS 762
+NKCNVGYAFINMT PSLIVPFY FNGKKWEKFNSEKVASLAYARIQGK+AL+AHFQNS
Sbjct: 847 KNKCNVGYAFINMTEPSLIVPFYHAFNGKKWEKFNSEKVASLAYARIQGKSALIAHFQNS 906
Query: 763 SLMNEDKRCRPILIDTDGPNAGDQ 786
SLMNEDKRCRPIL TDGPNAGDQ
Sbjct: 907 SLMNEDKRCRPILFHTDGPNAGDQ 930
Score = 320 bits (819), Expect = 1e-85
Identities = 179/359 (49%), Positives = 237/359 (65%), Gaps = 11/359 (3%)
Query: 12 NYSLTLLCHVDSDQGIKPFEDSKEDDIVASRHESSLFSSSLSDLFSKKLRLSANNAFYGH 71
+Y++ + V +D P ED K ++ + E+ LFSSSLS+LFS+KLRL N + +GH
Sbjct: 105 DYNVGVRSIVSTDLASYPTEDDKIS-VLGGQCENGLFSSSLSELFSRKLRLPTNYSPHGH 163
Query: 72 SVDTIASNYEEEKLSDSLEELEAQIIGNLLPDEDDLLSGVTDGNNYIICDSNGDDIDELD 131
SV S+YEEE+ +SL+ELEA IGNLLPD+DDLL+GVTDG +Y+ GD+ ++LD
Sbjct: 164 SVGAADSHYEEERF-ESLKELEAHAIGNLLPDDDDLLAGVTDGLDYVGQPYAGDETEDLD 222
Query: 132 LFSSNGGFDLGDVENPSSIERNSEIIS------GVRNSSIAGENSYGEHPSRTLFVRNID 185
LFSS GG DLG+ + S+ ++NSE G N++I + + E+PSRTLFVRN++
Sbjct: 223 LFSSVGGMDLGE-DGSSTGQQNSEYAGNYTLPLGDSNAAIGSQKPFEENPSRTLFVRNVN 281
Query: 186 SDVKDSVLKALFEQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALNNRLFGRKKFDI 245
S V+DS L+ LFEQ+GDI T CKH+G MISYYDIRA+QNAM+AL N R+K DI
Sbjct: 282 SSVEDSELQTLFEQYGDIRTLYTACKHRGFVMISYYDIRASQNAMKALQNNPLRRRKLDI 341
Query: 246 HYPIPKDSPSRNGVNQGTLEVFLYDSSISNTELQHILNVYGGIKEIHENPRSQRHKLIEF 305
H+ IPKD+PS NQGTL VF DSS+SN EL+ I VYG IKEI E HK IEF
Sbjct: 342 HFSIPKDNPSEKNANQGTLLVFNLDSSVSNDELRQIFGVYGEIKEIRETQHRSHHKYIEF 401
Query: 306 YDFRAADAALHGINRNDTTMKRLKVDQMQSTNSESNIIQPMHPEFKQ-ECDLCLHQKSP 363
YD RAA+AAL +NR+D K++ ++ + + + Q E +Q E L LHQ SP
Sbjct: 402 YDVRAAEAALRALNRSDVAGKQIMIEAIHPGGTR-RLSQQFPSELEQDEPGLYLHQNSP 459
>UniRef100_Q64M78 Putative AML1 [Oryza sativa]
Length = 1001
Score = 311 bits (796), Expect = 6e-83
Identities = 175/325 (53%), Positives = 215/325 (65%), Gaps = 47/325 (14%)
Query: 508 SSSFPLPVHHEQWSNS-----YPPPRTIWPNSPSYFDGIYAASTLQRLNQLPMSPS-HMI 561
S S + H W+NS +P +WP+ S+ + + + S Q ++ +P +PS HMI
Sbjct: 628 SGSSSVRGHQLMWNNSSNFHHHPNSPVLWPSPGSFVNNVPSRSPAQ-MHGVPRAPSSHMI 686
Query: 562 TTVLPTNNHHIQSPP-----FWDRRYTYAAE-----------------PITPHCVDFVPH 599
VLP ++ H+ S P WDRR+ YA E P +P +
Sbjct: 687 DNVLPMHHLHVGSAPAINPSLWDRRHGYAGELTEAPNFHPGSVGSMGFPGSPQLHSMELN 746
Query: 600 NMFPHFGLNVHN--------------QRGMVFPGRNHMIN--SFDTY-KRVRSRRNVGAS 642
N++P G N + QRG +F GRN M+ SFD+ +R+RSRRN
Sbjct: 747 NIYPQTGGNCMDPTVSPAQIGGPSPQQRGSMFHGRNPMVPLPSFDSPGERMRSRRNDSNG 806
Query: 643 NLADMKR-YELDIDCIKRGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHKGAYDFVYLPID 701
N +D K+ YELD+D I RG+D+RTTLMIKNIPNKYTSKMLLAAIDE+HKG YDF+YLPID
Sbjct: 807 NQSDNKKQYELDVDRIVRGDDSRTTLMIKNIPNKYTSKMLLAAIDENHKGTYDFIYLPID 866
Query: 702 FRNKCNVGYAFINMTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQN 761
F+NKCNVGYAFINMT+P I+PFYQ FNGKKWEKFNSEKVASLAYARIQGK+AL+AHFQN
Sbjct: 867 FKNKCNVGYAFINMTNPQHIIPFYQTFNGKKWEKFNSEKVASLAYARIQGKSALIAHFQN 926
Query: 762 SSLMNEDKRCRPILIDTDGPNAGDQ 786
SSLMNEDKRCRPIL +DGPNAGDQ
Sbjct: 927 SSLMNEDKRCRPILFHSDGPNAGDQ 951
Score = 284 bits (726), Expect = 8e-75
Identities = 152/313 (48%), Positives = 202/313 (63%), Gaps = 4/313 (1%)
Query: 43 HESSLFSSSLSDLFSKKLRLSANNAFYGHSVDTIASNYEEEKLSDSLEELEAQIIGNLLP 102
+E+ LFSSS+SD+F KKLRL++ N G S++ + N+ +++ + EE+EAQIIGNLLP
Sbjct: 140 NENGLFSSSVSDIFDKKLRLTSKNGLVGQSIEKVDLNHVDDEPFELTEEIEAQIIGNLLP 199
Query: 103 DEDDLLSGVTDGNNYIICDSNGDDIDELDLFSSNGGFDLGDVENPSSIERNSEIISGVR- 161
D+DDLLSGV D Y +N DD D+ D+F + GG +L EN E N G+
Sbjct: 200 DDDDLLSGVVDEVGYPTNANNRDDADD-DIFYTGGGMELETDENKKLQEFNGSANDGIGL 258
Query: 162 -NSSIAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSAMISY 220
N + GE+ Y E PSRTLFVRNI+S+V+DS LK LFE FGDI CKH+G MISY
Sbjct: 259 LNGVLNGEHLYREQPSRTLFVRNINSNVEDSELKLLFEHFGDIRALYTACKHRGFVMISY 318
Query: 221 YDIRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYDSSISNTELQH 280
YDIR+A NA L N+ R+K DIHY IPKD+PS +NQGT+ +F D S++N +L
Sbjct: 319 YDIRSALNAKMELQNKALRRRKLDIHYSIPKDNPSEKDINQGTIVLFNVDLSLTNDDLHK 378
Query: 281 ILNVYGGIKEIHENPRSQRHKLIEFYDFRAADAALHGINRNDTTMKRLKVDQMQSTNSES 340
I YG IKEI + P+ HK+IEFYD RAA+AAL +NRND K++K++ + +
Sbjct: 379 IFGDYGEIKEIRDTPQKGHHKIIEFYDVRAAEAALRALNRNDIAGKKIKLETSR-LGAAR 437
Query: 341 NIIQPMHPEFKQE 353
+ Q M E QE
Sbjct: 438 RLSQHMSSELCQE 450
>UniRef100_Q8S3W5 Mei2-like protein [Hordeum vulgare var. distichum]
Length = 961
Score = 293 bits (750), Expect = 1e-77
Identities = 171/321 (53%), Positives = 205/321 (63%), Gaps = 45/321 (14%)
Query: 508 SSSFPLPVHHEQWSNS-----YPPPRTIWPNSPSYFDGIYAASTLQRLNQLPMSPSHMIT 562
+ S P H W+NS + IWPNS S+ + I + Q ++ + + S M+
Sbjct: 587 TGSCPFHGHQVAWNNSNNSHHHSSSPMIWPNSGSFINNIPSRPPTQ-VHGISRT-SRMLE 644
Query: 563 TVLPTNNHHIQSPP-----FWDRRYTYAAEPI-----------------TPHCVDFVPHN 600
LP N HH+ S P DRR YA EP+ +PH +
Sbjct: 645 NALPAN-HHVGSAPAVNPSILDRRTGYAGEPMEAPSFHPGSAGSMGFSGSPHLHQLELTS 703
Query: 601 MFP-----------HFGLNVHNQRGMVFPGRNHM--INSFDTY-KRVRSRRNVGASNLAD 646
MFP H G QRG +F GR H+ +SFD+ +R RSRRN +N +D
Sbjct: 704 MFPQSGGNPAMSPAHIGARSPQQRGHMFHGRGHIGPPSSFDSLGERARSRRNESCANQSD 763
Query: 647 MKR-YELDIDCIKRGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHKGAYDFVYLPIDFRNK 705
KR YELDI+ I GED+RTTLMIKNIPNKYTSKMLL AIDE+HKG YDFVYLPIDF+NK
Sbjct: 764 NKRQYELDIERIVCGEDSRTTLMIKNIPNKYTSKMLLTAIDENHKGTYDFVYLPIDFKNK 823
Query: 706 CNVGYAFINMTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQNSSLM 765
CNVGYAFINM SP IVPFY+ F+GK+WEKFNSEKVASLAYARIQG+++L+AHFQNSSLM
Sbjct: 824 CNVGYAFINMISPEHIVPFYKIFHGKRWEKFNSEKVASLAYARIQGRSSLIAHFQNSSLM 883
Query: 766 NEDKRCRPILIDTDGPNAGDQ 786
NEDKRCRPIL +DGPNAGDQ
Sbjct: 884 NEDKRCRPILFHSDGPNAGDQ 904
Score = 256 bits (654), Expect = 2e-66
Identities = 139/326 (42%), Positives = 208/326 (63%), Gaps = 10/326 (3%)
Query: 30 FEDSKEDDIVASRHESSLFSSSLSDLFSKKLRLSANNAFYGHSVDTIASNYEEEKLSDSL 89
F ++++ + +++E+ LFSSSL D+F KKLRL+ N G + ++ ++E +
Sbjct: 92 FSNARKGHLNITQYENGLFSSSLPDIFDKKLRLTPKNGLVGQPAEKELNHADDEPF-ELT 150
Query: 90 EELEAQIIGNLLPDEDDLLSGVTDGNNYIICDSNGDDIDELDLFSSNGGFDLGDVENPSS 149
+E+EAQ+IGNLLPD+DDLLSGV + +N DDID+ D+FS+ GG +L EN
Sbjct: 151 QEIEAQVIGNLLPDDDDLLSGVLYNVGHPARANNIDDIDD-DIFSTGGGMELEADENNKL 209
Query: 150 IERNSEIISGVRNSSIAGENS--YGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFD 207
++ N G N+ G N YGE+PSRTLF+RNI+++V+D+ LK LFEQ+GDI T
Sbjct: 210 LKHN-----GGANTGQTGLNGLPYGENPSRTLFIRNINANVEDTELKLLFEQYGDIQTLY 264
Query: 208 RTCKHQGSAMISYYDIRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQGTLEVF 267
KH G +ISYYDIR+A+ AM+AL ++ F + K +IHY IPK++ N NQGTL V
Sbjct: 265 TAYKHHGLVIISYYDIRSAERAMKALQSKPFRQWKLEIHYSIPKENLLENDNNQGTLAVI 324
Query: 268 LYDSSISNTELQHILNVYGGIKEIHENPRSQRHKLIEFYDFRAADAALHGINRNDTTMKR 327
D S++N +L+HI YG IK IHE + HK +EF+D RAA+AAL+ +N + K+
Sbjct: 325 NLDQSVTNDDLRHIFGGYGEIKAIHETTQKGYHKSVEFFDIRAAEAALYALNMREIAGKK 384
Query: 328 LKVDQMQSTNSESNIIQPMHPEFKQE 353
+++++ +++ HPE +QE
Sbjct: 385 IRLERC-CPGDGKRLMRHRHPELEQE 409
>UniRef100_Q8W4I9 Mei2-like protein [Arabidopsis thaliana]
Length = 915
Score = 288 bits (737), Expect = 4e-76
Identities = 163/314 (51%), Positives = 195/314 (61%), Gaps = 49/314 (15%)
Query: 520 WSNSY-----PPPRTIWPNSPSYFDGIYAASTLQRLNQLPMSPSHMITTVLPTNNHHIQS 574
WSNS P +WPNSPS+ + I + + +P M+ +HHI S
Sbjct: 554 WSNSNTQQQNPSSGMMWPNSPSHINSI--PTQRPPVTVFSRAPPIMVNMASSPVHHHIGS 611
Query: 575 PP-----FWDRRYTYAAEPITP-------------------HCVDFVPHNMFPHFG--LN 608
P FWDRR Y AE + H +D H F G ++
Sbjct: 612 APVLNSPFWDRRQAYVAESLESSGFHIGSHGSMGIPGSSPSHPMDIGSHKTFSVGGNRMD 671
Query: 609 VHNQRGMV---------FPGRN---HMINSFDT----YKRVRSRRNVGASNLADMKRYEL 652
V++Q ++ FPGR+ M SFD+ Y+ + RR+ +S+ AD K YEL
Sbjct: 672 VNSQNAVLRSPQQLSHLFPGRSPMGSMPGSFDSPNERYRNLSHRRSESSSSNADKKLYEL 731
Query: 653 DIDCIKRGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHKGAYDFVYLPIDFRNKCNVGYAF 712
D+D I RGED RTTLMIKNIPNKYTSKMLL+AIDEH KG YDF+YLPIDF+NKCNVGYAF
Sbjct: 732 DVDRILRGEDRRTTLMIKNIPNKYTSKMLLSAIDEHCKGTYDFLYLPIDFKNKCNVGYAF 791
Query: 713 INMTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQNSSLMNEDKRCR 772
IN+ P IVPF++ FNGKKWEKFNSEKVA+L YARIQGK AL+AHFQNSSLMNEDKRCR
Sbjct: 792 INLIEPEKIVPFFKAFNGKKWEKFNSEKVATLTYARIQGKTALIAHFQNSSLMNEDKRCR 851
Query: 773 PILIDTDGPNAGDQ 786
PIL TDGPNAGDQ
Sbjct: 852 PILFHTDGPNAGDQ 865
Score = 244 bits (624), Expect = 6e-63
Identities = 155/339 (45%), Positives = 198/339 (57%), Gaps = 20/339 (5%)
Query: 41 SRHESSLFSSSLSDLFSKKLRLSANNAFYGHSVDTIASNYEEEKLSDSLEELEAQIIGNL 100
S+ ESSLFSSS+SDLFS+KLRL ++ S +T+ ++ EEE S+SLEE+EAQ IGNL
Sbjct: 86 SQWESSLFSSSMSDLFSRKLRLQGSDMLSTMSANTVVTHREEEP-SESLEEIEAQTIGNL 144
Query: 101 LPDEDDLLSGVTDGNNYIICDSNGDDIDELDLFSSNGGFDL-GDVENPSSIERNSEIISG 159
LPDEDDL + VT + GD++DE DLFSS GG +L GD+ + S RN E G
Sbjct: 145 LPDEDDLFAEVTGEVGRKSRANTGDELDEFDLFSSVGGMELDGDIFSSVS-HRNGE--RG 201
Query: 160 VRNSSIAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSAMIS 219
NS GE + GE PSRTL V NI S+V+D LK LFEQFGDI CK++G M+S
Sbjct: 202 GNNS--VGELNRGEIPSRTLLVGNISSNVEDYELKVLFEQFGDIQALHTACKNRGFIMVS 259
Query: 220 YYDIRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYDSSISNTELQ 279
Y DIRAAQNA RAL N+L K DI Y I K++PS+ ++G L V DSSISN EL
Sbjct: 260 YCDIRAAQNAARALQNKLLRGTKLDIRYSISKENPSQKDTSKGALLVNNLDSSISNQELN 319
Query: 280 HILNVYGGIKEIHENPRSQRHKLIEFYDFRAADAALHGINRNDTTMKRLKVDQMQSTNSE 339
++ YG +KEI IEF+D RAA AAL G+N + K+L+
Sbjct: 320 RLVKSYGEVKEIRRTMHDNSQIYIEFFDVRAAAAALGGLNGLEVAGKKLQ---------- 369
Query: 340 SNIIQPMHPEFKQECDLCLHQKSPLLKPTTSFQGREHVH 378
+ P +PE + C + P TS+ H
Sbjct: 370 ---LVPTYPEGTRYTSQCAANDTEGCLPKTSYSNTSSGH 405
>UniRef100_O23866 Mei2-like protein [Arabidopsis thaliana]
Length = 884
Score = 288 bits (737), Expect = 4e-76
Identities = 163/314 (51%), Positives = 195/314 (61%), Gaps = 49/314 (15%)
Query: 520 WSNSY-----PPPRTIWPNSPSYFDGIYAASTLQRLNQLPMSPSHMITTVLPTNNHHIQS 574
WSNS P +WPNSPS+ + I + + +P M+ +HHI S
Sbjct: 523 WSNSNTQQQNPSSGMMWPNSPSHINSI--PTQRPPVTVFSRAPPIMVNMASSPVHHHIGS 580
Query: 575 PP-----FWDRRYTYAAEPITP-------------------HCVDFVPHNMFPHFG--LN 608
P FWDRR Y AE + H +D H F G ++
Sbjct: 581 APVLNSPFWDRRQAYVAESLESSGFHIGSHGSMGIPGSSPSHPMDIGSHKTFSVGGNRMD 640
Query: 609 VHNQRGMV---------FPGRN---HMINSFDT----YKRVRSRRNVGASNLADMKRYEL 652
V++Q ++ FPGR+ M SFD+ Y+ + RR+ +S+ AD K YEL
Sbjct: 641 VNSQNAVLRSPQQLSHLFPGRSPMGSMPGSFDSPNERYRNLSHRRSESSSSNADKKLYEL 700
Query: 653 DIDCIKRGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHKGAYDFVYLPIDFRNKCNVGYAF 712
D+D I RGED RTTLMIKNIPNKYTSKMLL+AIDEH KG YDF+YLPIDF+NKCNVGYAF
Sbjct: 701 DVDRILRGEDRRTTLMIKNIPNKYTSKMLLSAIDEHCKGTYDFLYLPIDFKNKCNVGYAF 760
Query: 713 INMTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQNSSLMNEDKRCR 772
IN+ P IVPF++ FNGKKWEKFNSEKVA+L YARIQGK AL+AHFQNSSLMNEDKRCR
Sbjct: 761 INLIEPEKIVPFFKAFNGKKWEKFNSEKVATLTYARIQGKTALIAHFQNSSLMNEDKRCR 820
Query: 773 PILIDTDGPNAGDQ 786
PIL TDGPNAGDQ
Sbjct: 821 PILFHTDGPNAGDQ 834
Score = 244 bits (624), Expect = 6e-63
Identities = 155/339 (45%), Positives = 198/339 (57%), Gaps = 20/339 (5%)
Query: 41 SRHESSLFSSSLSDLFSKKLRLSANNAFYGHSVDTIASNYEEEKLSDSLEELEAQIIGNL 100
S+ ESSLFSSS+SDLFS+KLRL ++ S +T+ ++ EEE S+SLEE+EAQ IGNL
Sbjct: 55 SQWESSLFSSSMSDLFSRKLRLQGSDMLSTMSANTVVTHREEEP-SESLEEIEAQTIGNL 113
Query: 101 LPDEDDLLSGVTDGNNYIICDSNGDDIDELDLFSSNGGFDL-GDVENPSSIERNSEIISG 159
LPDEDDL + VT + GD++DE DLFSS GG +L GD+ + S RN E G
Sbjct: 114 LPDEDDLFAEVTGEVGRKSRANTGDELDEFDLFSSVGGMELDGDIFSSVS-HRNGE--RG 170
Query: 160 VRNSSIAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSAMIS 219
NS GE + GE PSRTL V NI S+V+D LK LFEQFGDI CK++G M+S
Sbjct: 171 GNNS--VGELNRGEIPSRTLLVGNISSNVEDYELKVLFEQFGDIQALHTACKNRGFIMVS 228
Query: 220 YYDIRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYDSSISNTELQ 279
Y DIRAAQNA RAL N+L K DI Y I K++PS+ ++G L V DSSISN EL
Sbjct: 229 YCDIRAAQNAARALQNKLLRGTKLDIRYSISKENPSQKDTSKGALLVNNLDSSISNQELN 288
Query: 280 HILNVYGGIKEIHENPRSQRHKLIEFYDFRAADAALHGINRNDTTMKRLKVDQMQSTNSE 339
++ YG +KEI IEF+D RAA AAL G+N + K+L+
Sbjct: 289 RLVKSYGEVKEIRRTMHDNSQIYIEFFDVRAAAAALGGLNGLEVAGKKLQ---------- 338
Query: 340 SNIIQPMHPEFKQECDLCLHQKSPLLKPTTSFQGREHVH 378
+ P +PE + C + P TS+ H
Sbjct: 339 ---LVPTYPEGTRYTSQCAANDTEGCLPKTSYSNTSSGH 374
>UniRef100_Q9LYN7 Mei2-like protein [Arabidopsis thaliana]
Length = 907
Score = 285 bits (728), Expect = 5e-75
Identities = 163/315 (51%), Positives = 194/315 (60%), Gaps = 49/315 (15%)
Query: 520 WSNSYPPPRT-----IWPNSPSYFDGIYAASTLQRLNQLPMSPSHMITTVLPTNNHHIQS 574
WSNS +WPNSPS +G+ + + + + M+ +HHI S
Sbjct: 550 WSNSNSQQHNQSSGMMWPNSPSRVNGV-PSQRIPPVTAFSRASPLMVNMASSPVHHHIGS 608
Query: 575 PP-----FWDRRYTYAAEP-------------------ITPHCVDFVPHNMFPHFGLN-- 608
P FWDRR Y AE H +DF H +F H G N
Sbjct: 609 APVLNSPFWDRRQAYVAESPESSGFHLGSPGSMGFPGSSPSHPMDFGSHKVFSHVGGNRM 668
Query: 609 --------VHNQRGM--VFPGRNHMIN---SFDT----YKRVRSRRNVGASNLADMKRYE 651
+ + R M +F GR+ M++ SFD Y+ + RR+ S+ A+ K YE
Sbjct: 669 EANSKNAVLRSSRQMPHLFTGRSPMLSVSGSFDLPNERYRNLSHRRSESNSSNAEKKLYE 728
Query: 652 LDIDCIKRGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHKGAYDFVYLPIDFRNKCNVGYA 711
LD+D I RGED+RTTLMIKNIPNKYTSKMLLAAIDE+ KG YDF+YLPIDF+NKCNVGYA
Sbjct: 729 LDVDRILRGEDSRTTLMIKNIPNKYTSKMLLAAIDEYCKGTYDFLYLPIDFKNKCNVGYA 788
Query: 712 FINMTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQNSSLMNEDKRC 771
FIN+ P IVPFY+ FNGKKWEKFNSEKVASLAY RIQGK+AL+AHFQNSSLMNEDKRC
Sbjct: 789 FINLIEPENIVPFYKAFNGKKWEKFNSEKVASLAYGRIQGKSALIAHFQNSSLMNEDKRC 848
Query: 772 RPILIDTDGPNAGDQ 786
RPIL T GPNAGDQ
Sbjct: 849 RPILFHTAGPNAGDQ 863
Score = 221 bits (563), Expect = 7e-56
Identities = 137/288 (47%), Positives = 180/288 (61%), Gaps = 12/288 (4%)
Query: 44 ESSLFSSSLSDLFSKKLRLSANNAFYGHSVDTIASNYEEEKLSDSLEELEAQIIGNLLPD 103
ESSLFSSSLSDLFS+KLRL ++ + +++N EEE S+SLEE+EAQ IGNLLPD
Sbjct: 87 ESSLFSSSLSDLFSRKLRLPRSD-----KLAFMSANREEEP-SESLEEMEAQTIGNLLPD 140
Query: 104 EDDLLSGVTDGNNYIICDSNGDDIDELDLFSSNGGFDL-GDVENPSSIERNSEIISGVRN 162
EDDL + V + + GDD+D+ DLFSS GG +L GDV + S +R+ + S V
Sbjct: 141 EDDLFAEVVGEGVHKSRANGGDDLDDCDLFSSVGGMELDGDVFSSVS-QRDGKRGSNV-- 197
Query: 163 SSIAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSAMISYYD 222
S E+ GE SR LFVRN+DS ++D L LF+QFGD+ K++G M+SYYD
Sbjct: 198 -STVAEHPQGEILSRILFVRNVDSSIEDCELGVLFKQFGDVRALHTAGKNRGFIMVSYYD 256
Query: 223 IRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYDSSISNTELQHIL 282
IRAAQ A RAL+ RL +K DI Y IPK++P N ++G L V DSSISN EL I
Sbjct: 257 IRAAQKAARALHGRLLRGRKLDIRYSIPKENPKENS-SEGALWVNNLDSSISNEELHGIF 315
Query: 283 NVYGGIKEIHENPRSQRHKLIEFYDFRAADAALHGINRNDTTMKRLKV 330
+ YG I+E+ IEF+D R A AL G+N + ++LK+
Sbjct: 316 SSYGEIREVRRTMHENSQVYIEFFDVRKAKVALQGLNGLEVAGRQLKL 363
>UniRef100_Q6J738 AML5 [Medicago truncatula]
Length = 865
Score = 261 bits (668), Expect = 4e-68
Identities = 150/345 (43%), Positives = 213/345 (61%), Gaps = 24/345 (6%)
Query: 31 EDSKEDDIVASRHESSLFSSSLSDLFSKKLRLS-ANNAFYGHSVDTIASNY----EEEKL 85
E S D + ++SLFSSSL L +KL L+ N F SVD I++N+ +E +L
Sbjct: 30 EVSSGSDSYHASSDASLFSSSLPVLPHEKLNLNETENGF--QSVDDISTNFKKHHQEAEL 87
Query: 86 SDSLEELEAQIIGNLLPDEDD-LLSGVTDGNNYIICDSNGDDIDELDLFSSNGGFDLGDV 144
+ SL+ IG +LPD+D+ LL+G+ D + + +D++E DLF S+GG +L
Sbjct: 88 NGSLDNGNNHAIGTMLPDDDEELLAGIMDDFDLRGLPGSLEDLEEYDLFDSSGGLELETD 147
Query: 145 ENPS--------SIERNS--------EIISGVRNSSIAGENSYGEHPSRTLFVRNIDSDV 188
S S+ +S + +GV ++AGE+ YGEHPSRTLFVRNI+S+V
Sbjct: 148 PQESLSVGISKLSLSDSSVGNSMPPYSLPNGVGGGAVAGEHPYGEHPSRTLFVRNINSNV 207
Query: 189 KDSVLKALFEQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALNNRLFGRKKFDIHYP 248
+D+ L+ LFEQ+GDI T CKH+G MISYYDIRAA+ AMRAL N+ R+K DIH+
Sbjct: 208 EDTELRTLFEQYGDIRTLYTACKHRGFVMISYYDIRAARTAMRALQNKPLRRRKLDIHFS 267
Query: 249 IPKDSPSRNGVNQGTLEVFLYDSSISNTELQHILNVYGGIKEIHENPRSQRHKLIEFYDF 308
IPKD+PS +NQGTL VF D S+SN +L+ I YG +KEI E P + HK IE+YD
Sbjct: 268 IPKDNPSDKDINQGTLVVFNLDPSVSNEDLRQIFGAYGEVKEIRETPHKRHHKFIEYYDV 327
Query: 309 RAADAALHGINRNDTTMKRLKVDQMQSTNSESNIIQPMHPEFKQE 353
RAA+AAL +NR+D KR+K++ + + N++ ++ E Q+
Sbjct: 328 RAAEAALKSLNRSDIAGKRIKLEPSRPGGARRNLMLQLNQELDQD 372
Score = 240 bits (612), Expect = 1e-61
Identities = 118/156 (75%), Positives = 133/156 (84%), Gaps = 1/156 (0%)
Query: 632 RVRSRR-NVGASNLADMKRYELDIDCIKRGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHK 690
RVRSR + S + + K ++LD+D I+ GED RTTLMIKNIPNKYTSKMLLAAIDE+HK
Sbjct: 666 RVRSRWIDNNGSQVDNKKLFQLDLDKIRSGEDTRTTLMIKNIPNKYTSKMLLAAIDENHK 725
Query: 691 GAYDFVYLPIDFRNKCNVGYAFINMTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARIQ 750
G YDF+YLPIDF+NKCNVGYAFINM SPSLI+PFY+ F+GKKWEKFNSEKVASLAYARIQ
Sbjct: 726 GTYDFLYLPIDFKNKCNVGYAFINMLSPSLIIPFYETFHGKKWEKFNSEKVASLAYARIQ 785
Query: 751 GKAALVAHFQNSSLMNEDKRCRPILIDTDGPNAGDQ 786
GK ALV HFQNSSLMNEDKRCRPI+ +DG DQ
Sbjct: 786 GKNALVNHFQNSSLMNEDKRCRPIVFHSDGSEVADQ 821
>UniRef100_Q6J740 AML1 [Beta vulgaris]
Length = 617
Score = 259 bits (661), Expect = 3e-67
Identities = 158/312 (50%), Positives = 186/312 (58%), Gaps = 50/312 (16%)
Query: 521 SNSYPPPRT---IWPNSPSYFDGIYAASTLQRLNQLPMSPSHMITTVLPTNNHHIQSPP- 576
SNSY + + PNSPS+ + +A + L RL P +P M+ V P ++ + S P
Sbjct: 285 SNSYQQHQLRSLLMPNSPSFLNNNHA-NHLGRLPGSPGAPHLMMNMVSPVHHLQVGSAPV 343
Query: 577 -------FWDRRYTYAAE------------------------------PITPHCVDFVPH 599
WDR+ YA E PI+ V
Sbjct: 344 APVMKASLWDRQQIYAGESHEASGYQMGAIRNVGLPGYSASPAREISSPISFSHVHGT-- 401
Query: 600 NMFPHFGLNVHNQRGMVFPGRNHMINSFDTY-----KRVRSRRNVGASNLADMKRYELDI 654
M + G+ Q +F RN M NS T +RVR RN + AD K YEL+I
Sbjct: 402 EMSKNAGIQSPQQLSHMFHERNTM-NSLPTSFGSPSERVRRSRNETNLSHADRKHYELNI 460
Query: 655 DCIKRGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHKGAYDFVYLPIDFRNKCNVGYAFIN 714
+ I RG+D RTTLMIKNIPNKYTSKMLLA IDE H+G YDF+YLPIDF+NKCN+GYAFIN
Sbjct: 461 ERILRGDDIRTTLMIKNIPNKYTSKMLLATIDEQHRGKYDFIYLPIDFKNKCNMGYAFIN 520
Query: 715 MTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQNSSLMNEDKRCRPI 774
M P IV F+Q F G+KWEKFNSEKVASLAYARIQGK AL+AHFQNSSLMNEDKRCRPI
Sbjct: 521 MIDPLQIVSFHQTFEGRKWEKFNSEKVASLAYARIQGKGALIAHFQNSSLMNEDKRCRPI 580
Query: 775 LIDTDGPNAGDQ 786
L TDGPNAGDQ
Sbjct: 581 LFHTDGPNAGDQ 592
Score = 92.4 bits (228), Expect = 5e-17
Identities = 50/116 (43%), Positives = 74/116 (63%), Gaps = 1/116 (0%)
Query: 38 IVASRHESSLFSSSLSDLFSKKLRLSANNAFYGHSVDTIASNYEEEKLSDSLEELEAQII 97
+V +++ESSLFSSSLS+L + KLRLS+N F +A +E + SLEE E Q I
Sbjct: 11 VVGAQNESSLFSSSLSELVNHKLRLSSNK-FLSRPSSAVAPQIDEVEPFKSLEETEGQTI 69
Query: 98 GNLLPDEDDLLSGVTDGNNYIICDSNGDDIDELDLFSSNGGFDLGDVENPSSIERN 153
NLLPDE++L SG+ D ++ + GDD+D+ D+F+ +GG +L E+ + RN
Sbjct: 70 NNLLPDEEELFSGIVDEMGHLNNANGGDDLDDFDVFTYSGGIELEAEEHLQAGRRN 125
>UniRef100_Q6J733 AML1 [Citrus unshiu]
Length = 858
Score = 258 bits (660), Expect = 4e-67
Identities = 149/336 (44%), Positives = 207/336 (61%), Gaps = 20/336 (5%)
Query: 37 DIVASRHESSLFSSSLSDLFSKKLRLSANNAFYGHSVDTIASNYEEE----KLSDSLEEL 92
D + ++SLFSSSL L +KL L+A SVD I+S + D LE +
Sbjct: 35 DSYLASDDASLFSSSLPVLPHEKLNLNAMG-LGRQSVDNISSGLSKVHHGVSSDDPLEGI 93
Query: 93 EAQIIGNLLPD-EDDLLSGVTDGNNYIICDSNGDDIDELDLFSSNGGFDL-GDVENPSSI 150
IGN LPD ED+LL+G+ D + S+ +D+++ D+F S GG +L G+ + S+
Sbjct: 94 MNPAIGNSLPDDEDELLAGIMDDFDLRGLPSSLEDLEDYDIFGSGGGMELEGEPQESLSM 153
Query: 151 ERNSEIISGVRNSS-------------IAGENSYGEHPSRTLFVRNIDSDVKDSVLKALF 197
+ IS + + +AGE+ YGEHPSRTLFVRNI+S+V+DS L+ALF
Sbjct: 154 SMSKISISDSASGNGLLHYSVPNGAGTVAGEHPYGEHPSRTLFVRNINSNVEDSELRALF 213
Query: 198 EQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRN 257
EQ+GDI T CKH+G MISYYDIRAA+ AMRAL N+ R+K DIH+ IPKD+PS
Sbjct: 214 EQYGDIRTLYTACKHRGFVMISYYDIRAARTAMRALQNKPLRRRKLDIHFSIPKDNPSDK 273
Query: 258 GVNQGTLEVFLYDSSISNTELQHILNVYGGIKEIHENPRSQRHKLIEFYDFRAADAALHG 317
+NQGTL VF D S+SN +L+ I YG +KEI E P + HK IEFYD RAA+AAL
Sbjct: 274 DLNQGTLVVFNLDPSVSNEDLRQIFGAYGEVKEIRETPHKRHHKFIEFYDVRAAEAALKS 333
Query: 318 INRNDTTMKRLKVDQMQSTNSESNIIQPMHPEFKQE 353
+NR+D KR+K++ + + N++ ++ E +Q+
Sbjct: 334 LNRSDIAGKRIKLEPSRPGGARRNLMLQLNQELEQD 369
Score = 251 bits (642), Expect = 5e-65
Identities = 122/164 (74%), Positives = 140/164 (84%), Gaps = 2/164 (1%)
Query: 625 NSFDTYKRVRSRR--NVGASNLADMKRYELDIDCIKRGEDNRTTLMIKNIPNKYTSKMLL 682
NS Y+R RSRR N + + K+++L+++ I+ GED RTTLMIKNIPNKYTSKMLL
Sbjct: 653 NSEGLYERGRSRRIENNNGNQIDSKKQFQLELEKIRSGEDTRTTLMIKNIPNKYTSKMLL 712
Query: 683 AAIDEHHKGAYDFVYLPIDFRNKCNVGYAFINMTSPSLIVPFYQGFNGKKWEKFNSEKVA 742
AAIDE+HKG YDF+YLPIDF+NKCNVGYAFINM SP I+PFY+ FNGKKWEKFNSEKVA
Sbjct: 713 AAIDENHKGTYDFLYLPIDFKNKCNVGYAFINMLSPLHIIPFYEAFNGKKWEKFNSEKVA 772
Query: 743 SLAYARIQGKAALVAHFQNSSLMNEDKRCRPILIDTDGPNAGDQ 786
SLAYARIQG+AALVAHFQNSSLMNEDKRCRPIL ++GP AGDQ
Sbjct: 773 SLAYARIQGRAALVAHFQNSSLMNEDKRCRPILFHSEGPEAGDQ 816
>UniRef100_Q6J735 AML1 [Sorghum bicolor]
Length = 818
Score = 257 bits (657), Expect = 8e-67
Identities = 151/306 (49%), Positives = 192/306 (62%), Gaps = 34/306 (11%)
Query: 510 SFPLPVHHEQWSNSYPPPRTIWPNSPSYFDGIYAASTLQRLNQLPMSPSHMITTVL-PTN 568
S PL HH WSNS P++ P++P + +N P+ P HM T P +
Sbjct: 483 SCPLHGHHYTWSNSNGFPQS--PSAPMLWSNFQQPV---HVNCYPVMPPHMRRTAAHPMD 537
Query: 569 NHHIQSPPFWDRRYT--YAAEPITPHCVDFVPH-NMFPHFGLNVH---NQRGMVFP---- 618
HH+ S P + ++ P + V F ++P + N R +F
Sbjct: 538 QHHLGSAPNSVGGFANAHSFHPGSLESVGFPGSPQLYPDLSVFASARGNYRETMFSPVSA 597
Query: 619 -------------GRNHMIN---SFD-TYKRVRSRRNVG-ASNLADMKRYELDIDCIKRG 660
GRN M++ S+D T R+RSRR+ G A+ + K++ELD+D I +G
Sbjct: 598 GFPSIQQIFHATNGRNPMVHVSTSYDATNDRIRSRRHDGNAAQSENKKQFELDLDRIAKG 657
Query: 661 EDNRTTLMIKNIPNKYTSKMLLAAIDEHHKGAYDFVYLPIDFRNKCNVGYAFINMTSPSL 720
ED+RTTLMIKNIPNKY K+LLA IDE+H+G YDF+YLPIDF+NKCNVGYAFINMT P
Sbjct: 658 EDSRTTLMIKNIPNKYNCKLLLAVIDENHRGTYDFIYLPIDFKNKCNVGYAFINMTDPQH 717
Query: 721 IVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQNSSLMNEDKRCRPILIDTDG 780
I+PFY+ FNGKKWEKFNSEKVASLAYARIQG+ AL+AHFQNSSLMNE+K CRP+L DG
Sbjct: 718 IIPFYKTFNGKKWEKFNSEKVASLAYARIQGRNALIAHFQNSSLMNEEKWCRPMLFHKDG 777
Query: 781 PNAGDQ 786
P+AGDQ
Sbjct: 778 PHAGDQ 783
Score = 75.9 bits (185), Expect = 4e-12
Identities = 44/108 (40%), Positives = 68/108 (62%), Gaps = 3/108 (2%)
Query: 49 SSSLSDLFSKKLRLSANNAFYGHSVDTIASNYEEEKLSDSLEELEAQIIGNLLPDEDDLL 108
SSS +++FS K + YG S D S E ++ S++ELEAQ IG+LLPD+DDL+
Sbjct: 150 SSSYTEVFSSKSARLVASGVYGQSADANGSGCEGDEPLGSMKELEAQTIGDLLPDDDDLI 209
Query: 109 SGVTDGNNYIICDSNGDDIDELDLFSSNGGFDLGDVENPSSIERNSEI 156
SG+ DG Y SN DD DE D+F + GG +L + ++ +++++ E+
Sbjct: 210 SGIIDGFEYTGL-SNKDDADE-DIFYTGGGLEL-EHDDSNNVDKFREV 254
Score = 44.7 bits (104), Expect = 0.011
Identities = 25/79 (31%), Positives = 40/79 (49%)
Query: 286 GGIKEIHENPRSQRHKLIEFYDFRAADAALHGINRNDTTMKRLKVDQMQSTNSESNIIQP 345
G + EI++ P S K +EFY+ RAA L+ +N+ D + ++KV+ S + S +
Sbjct: 262 GDVNEIYKAPTSCNKKFVEFYNTRAAQETLNDLNKGDMSCSQIKVEHSYSGGAGSCFTEQ 321
Query: 346 MHPEFKQECDLCLHQKSPL 364
E KQ + SPL
Sbjct: 322 CSGEQKQNAVPHQLKNSPL 340
>UniRef100_Q6J739 AML1 [Medicago truncatula]
Length = 856
Score = 254 bits (649), Expect = 7e-66
Identities = 146/339 (43%), Positives = 205/339 (60%), Gaps = 27/339 (7%)
Query: 37 DIVASRHESSLFSSSLSDLFSKKLRLS---ANNAFYGHSVD----TIASNYEEEKLSDSL 89
DI ++ SLFS+SL L +KL L+ ++ G VD T+ ++E++ +D
Sbjct: 41 DIFHESNDVSLFSTSLPVLPHEKLNLTDSEQDSEQSGQPVDDNLLTLGKVHKEDEGNDLF 100
Query: 90 EELEAQIIGNLLPDEDDLLSGVTDGNNYIICDSNGDDIDELDLFSSNGGFDLGDVENPSS 149
++ E L DED+LL+G+ D + + +D+DE DLF + GGF++ D E
Sbjct: 101 DDFETM----LPDDEDELLAGIMDDFDLRRLPNQLEDLDENDLFVNGGGFEM-DFEPQEG 155
Query: 150 IE---RNSEIISGVRNSSI------------AGENSYGEHPSRTLFVRNIDSDVKDSVLK 194
+ I G+ ++ I AGE+ YGEHPSRTLFVRNI+S+V+DS L+
Sbjct: 156 LSFGISKMSISDGIASNGIGPYAIPNGVGTVAGEHPYGEHPSRTLFVRNINSNVEDSELR 215
Query: 195 ALFEQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALNNRLFGRKKFDIHYPIPKDSP 254
LFEQ+GDI T CKH+G MISYYDIRAA+ AMRAL N+ R+K DIH+ IPKD+P
Sbjct: 216 TLFEQYGDIRTLYTACKHRGFVMISYYDIRAARTAMRALQNKPLRRRKLDIHFSIPKDNP 275
Query: 255 SRNGVNQGTLEVFLYDSSISNTELQHILNVYGGIKEIHENPRSQRHKLIEFYDFRAADAA 314
S +NQGTL VF D S+SN +L+ I YG +KEI E P + HK IEFYD RAADAA
Sbjct: 276 SEKDINQGTLVVFNLDPSVSNDDLRQIFGAYGEVKEIRETPHKRHHKFIEFYDVRAADAA 335
Query: 315 LHGINRNDTTMKRLKVDQMQSTNSESNIIQPMHPEFKQE 353
L +NR+D KR+K++ + + N++Q + E +Q+
Sbjct: 336 LKALNRSDIAGKRIKLEPSRPGGARRNLMQQLSQELEQD 374
Score = 241 bits (614), Expect = 8e-62
Identities = 125/190 (65%), Positives = 147/190 (76%), Gaps = 6/190 (3%)
Query: 603 PHFGLNVHNQRGMVFPGRNHM----INSFDTY-KRVRSRRNVGASNLADMKR-YELDIDC 656
P+FG+ G +F G + ++S + + +R RSRR N + K+ Y+LD+D
Sbjct: 629 PNFGMMSLPGHGSLFLGNSLYAGPGVSSIEGFGERGRSRRPDNIVNQVESKKLYQLDLDK 688
Query: 657 IKRGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHKGAYDFVYLPIDFRNKCNVGYAFINMT 716
I GED RTTLMIKNIPNKYTSKMLLAAIDE+H+G YDF+YLPIDF+NKCNVGYAFINM
Sbjct: 689 IVNGEDTRTTLMIKNIPNKYTSKMLLAAIDENHQGTYDFLYLPIDFKNKCNVGYAFINMV 748
Query: 717 SPSLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQNSSLMNEDKRCRPILI 776
SPS IV F++ FNGKKWEKFNSEKVASLAYARIQGKAALV HFQNSSLMNEDKRCRPIL
Sbjct: 749 SPSHIVAFFKAFNGKKWEKFNSEKVASLAYARIQGKAALVMHFQNSSLMNEDKRCRPILF 808
Query: 777 DTDGPNAGDQ 786
++G + DQ
Sbjct: 809 HSEGQDTSDQ 818
>UniRef100_Q6J731 AML1 [Solanum tuberosum]
Length = 843
Score = 252 bits (643), Expect = 3e-65
Identities = 151/355 (42%), Positives = 219/355 (61%), Gaps = 26/355 (7%)
Query: 18 LCHVDSDQGIKPFEDSKEDDIVASRHESSLFSSSLSDLFSKKLRLSANNAFYGH-SVDTI 76
+ +V ++G P+E+S V +++SLFSSS+ L +KL++S + +GH SVD
Sbjct: 9 IINVSKEKGGTPWENS-----VLITNDASLFSSSVPVLQHEKLKVSDGD--HGHQSVDDA 61
Query: 77 ASN----YEEEKLSDSLEELEAQIIGNLLPD-EDDLLSGVTDGNNYIICDSNGDDIDELD 131
+ + + ++ L++ E + IG+LLPD ED+LL+G+ DG + ++ DD++E D
Sbjct: 62 SPSLKIIHPGVEVDVLLDDGENRAIGSLLPDDEDELLAGIMDGFDPSQFPNHTDDLEEYD 121
Query: 132 LFSSNGGFDL---GDVENPSSIER--------NSEIISGVRNS--SIAGENSYGEHPSRT 178
LF S GG +L G I R N I G+ N ++ GE+ GEHPSRT
Sbjct: 122 LFGSGGGLELEFDGQEHLNLGISRVSLVDPDSNGAAIYGLSNGGGAVTGEHPLGEHPSRT 181
Query: 179 LFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALNNRLF 238
LFVRNI+S+V+DS L+ LFEQ+GDI T CKH+G MISY+DIRAA+ AMRAL N+
Sbjct: 182 LFVRNINSNVEDSELRTLFEQYGDIRTLYTACKHRGFVMISYFDIRAARTAMRALQNKPL 241
Query: 239 GRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYDSSISNTELQHILNVYGGIKEIHENPRSQ 298
R+K DIH+ IPKD+PS VNQGTL VF D S+SN +L+ + YG IKEI E P +
Sbjct: 242 RRRKLDIHFSIPKDNPSDKDVNQGTLVVFNLDPSVSNDDLRKVFGPYGEIKEIRETPHKR 301
Query: 299 RHKLIEFYDFRAADAALHGINRNDTTMKRLKVDQMQSTNSESNIIQPMHPEFKQE 353
HK IE+YD RAA+AAL +N++ KR+K++ + + N++ E +Q+
Sbjct: 302 HHKFIEYYDVRAAEAALRSLNKSAIAGKRIKLEPSRPGGARRNLVLQSSQEPEQD 356
Score = 249 bits (636), Expect = 2e-64
Identities = 129/201 (64%), Positives = 150/201 (74%), Gaps = 11/201 (5%)
Query: 597 VPHNMF----PHFGLNVHNQRGMVFPGRNHM----INSFDTY-KRVRSRR--NVGASNLA 645
VP NM P FG+ + +F G H SF+ +R R+RR N + +
Sbjct: 602 VPRNMSDNASPRFGMMSSQKHSPLFLGNGHFPGHAATSFEGLTERSRTRRVDNNNGNQID 661
Query: 646 DMKRYELDIDCIKRGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHKGAYDFVYLPIDFRNK 705
+ K ++LD+D I+ GED RTTLMIKNIPNKYTSKMLLAAIDE HKG +DF+YLPIDF+NK
Sbjct: 662 NKKLFQLDLDKIRCGEDTRTTLMIKNIPNKYTSKMLLAAIDEQHKGTFDFLYLPIDFKNK 721
Query: 706 CNVGYAFINMTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQNSSLM 765
CNVGYAFINM SPSLI+PFY+ FNGKKWEKFNSEKVA+LAYARIQGK ALVAHFQNSSLM
Sbjct: 722 CNVGYAFINMLSPSLIIPFYEAFNGKKWEKFNSEKVAALAYARIQGKTALVAHFQNSSLM 781
Query: 766 NEDKRCRPILIDTDGPNAGDQ 786
NEDKRCRPIL ++ GDQ
Sbjct: 782 NEDKRCRPILFHSESSELGDQ 802
>UniRef100_Q6J741 Mei2-like protein [Pinus taeda]
Length = 632
Score = 249 bits (637), Expect = 2e-64
Identities = 152/330 (46%), Positives = 191/330 (57%), Gaps = 52/330 (15%)
Query: 502 NVSANISSSFPLPVHHEQWSNSYPPPRTI-WPNSPSYFDGIYAASTLQRLNQLPMS--PS 558
++S++ + L H W + P I WP P ++ P S S
Sbjct: 248 HLSSSSGVNGTLSGHQYLWGSPSPYSHHITWPGPP-------LGHSVNANGSQPYSGRQS 300
Query: 559 HMITTVLPTNNHHIQSPPFW----DRRYTYAAE-PITPH----------CVDFVP----- 598
+++ + ++HH+ S P DR ++Y E P P C + P
Sbjct: 301 PYVSSAIAPHHHHVGSAPSGEPSLDRHFSYLTETPDMPFVNPSSLGSMSCANGSPVISIG 360
Query: 599 -HNMFPHFGLNVHNQR--------GMVFPGRNHMINSFDTY------------KRVRSRR 637
H + G+++ N G++ P R + S + +R RSRR
Sbjct: 361 AHAVLNAGGVSISNNSNIECGSPIGVLSPQRKSRMFSGGGFTGSIANFSEGLNERGRSRR 420
Query: 638 NVGASNLADMKR-YELDIDCIKRGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHKGAYDFV 696
++ AD K+ Y+LD+D I RGED RTT+MIKNIPNKYTSKMLLA IDEHH+G YDF+
Sbjct: 421 GDNNTSQADNKKQYQLDLDKIMRGEDARTTIMIKNIPNKYTSKMLLATIDEHHRGTYDFL 480
Query: 697 YLPIDFRNKCNVGYAFINMTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALV 756
YLPIDF+NKCNVGYAFINMTSP+ I+PFYQ FNGKKWEKFNSEKVASLAYARIQGKAAL+
Sbjct: 481 YLPIDFKNKCNVGYAFINMTSPTHIIPFYQAFNGKKWEKFNSEKVASLAYARIQGKAALI 540
Query: 757 AHFQNSSLMNEDKRCRPILIDTDGPNAGDQ 786
AHFQNSSLMNEDKRCRPIL GDQ
Sbjct: 541 AHFQNSSLMNEDKRCRPILFQPGAAGTGDQ 570
Score = 214 bits (545), Expect = 8e-54
Identities = 106/210 (50%), Positives = 142/210 (67%), Gaps = 2/210 (0%)
Query: 158 SGVRN--SSIAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGS 215
SGV N ++I GE+ YGEHPSRTLFVRNI+S+V+DS L+ FE +GDI T CKH+G
Sbjct: 29 SGVSNVPATIVGEHPYGEHPSRTLFVRNINSNVEDSELRTYFEHYGDIRTLYTACKHRGF 88
Query: 216 AMISYYDIRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYDSSISN 275
MISYYDIRAA+NAMRAL N+ R+K DIH+ IPKD+PS +NQGTL VF D S+SN
Sbjct: 89 VMISYYDIRAARNAMRALQNKPLRRRKLDIHFSIPKDNPSDKDINQGTLVVFNLDPSVSN 148
Query: 276 TELQHILNVYGGIKEIHENPRSQRHKLIEFYDFRAADAALHGINRNDTTMKRLKVDQMQS 335
+L+ I YG +KEI E P + HK IEFYD RAA+ AL +N++D KR+K++ +
Sbjct: 149 DDLRRIFGAYGEVKEIRETPHKRHHKFIEFYDVRAAEEALRALNKSDIAGKRIKLEPSRP 208
Query: 336 TNSESNIIQPMHPEFKQECDLCLHQKSPLL 365
+ +++Q + E +Q+ L P +
Sbjct: 209 GGARRSLMQHLSQELEQDVGSPLMNSPPAI 238
>UniRef100_Q8VWF5 RNA-binding protein MEI2, putative [Arabidopsis thaliana]
Length = 800
Score = 249 bits (636), Expect = 2e-64
Identities = 143/324 (44%), Positives = 198/324 (60%), Gaps = 20/324 (6%)
Query: 44 ESSLFSSSLSDLFSKKLRLSANNAFYGHSVDTIASNYEEEKLSDSLEELEAQIIGNLLPD 103
+++LFSSSL KL+LS N + DT S K ++S ++ E+ IGNLLPD
Sbjct: 27 DATLFSSSLPVFPRGKLQLSDNRDGFSLIDDTAVSR--TNKFNESADDFESHSIGNLLPD 84
Query: 104 EDDLLSGVTDGNNYIICDSNGDDIDELDLFSSNGGFDLG-------DVENP-----SSIE 151
E+DLL+G+ D + D D+ DLF S GG +L + P SS+
Sbjct: 85 EEDLLTGMMDDLDL----GELPDADDYDLFGSGGGMELDADFRDNLSMSGPPRLSLSSLG 140
Query: 152 RNSEIISGVRNSS--IAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRT 209
N+ + N + +AGE+ YGEHPSRTLFVRNI+S+V+DS L ALFEQ+GDI T T
Sbjct: 141 GNAIPQFNIPNGAGTVAGEHPYGEHPSRTLFVRNINSNVEDSELTALFEQYGDIRTLYTT 200
Query: 210 CKHQGSAMISYYDIRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLY 269
CKH+G MISYYDIR+A+ AMR+L N+ R+K DIH+ IPKD+PS +NQGTL VF
Sbjct: 201 CKHRGFVMISYYDIRSARMAMRSLQNKPLRRRKLDIHFSIPKDNPSEKDMNQGTLVVFNL 260
Query: 270 DSSISNTELQHILNVYGGIKEIHENPRSQRHKLIEFYDFRAADAALHGINRNDTTMKRLK 329
D SISN +L I +G IKEI E P + HK +EFYD R A+AAL +NR + KR+K
Sbjct: 261 DPSISNDDLHGIFGAHGEIKEIRETPHKRHHKFVEFYDVRGAEAALKALNRCEIAGKRIK 320
Query: 330 VDQMQSTNSESNIIQPMHPEFKQE 353
V+ + + +++ ++ + + +
Sbjct: 321 VEPSRPGGARRSLMLQLNQDLEND 344
Score = 232 bits (591), Expect = 4e-59
Identities = 122/189 (64%), Positives = 146/189 (76%), Gaps = 14/189 (7%)
Query: 601 MFPHFGLNVHNQRGMVFPGRNHMINSFDT-YKRVRSRRNVGASNLADM-KRYELDIDCIK 658
MF GLN PGR + FD+ Y+ R RR SN + K+++LD++ I
Sbjct: 598 MFLSSGLN---------PGR--FASGFDSLYENGRPRRVENNSNQVESRKQFQLDLEKIL 646
Query: 659 RGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHKGAYDFVYLPIDFRNKCNVGYAFINMTSP 718
GED+RTTLMIKNIPNKYTSKMLLAAIDE ++G Y+F+YLPIDF+NKCNVGYAFINM +P
Sbjct: 647 NGEDSRTTLMIKNIPNKYTSKMLLAAIDEKNQGTYNFLYLPIDFKNKCNVGYAFINMLNP 706
Query: 719 SLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQNSSLMNEDKRCRPILIDT 778
LI+PFY+ FNGKKWEKFNSEKVASLAYARIQGK+AL+AHFQNSSLMNED RCRPI+ DT
Sbjct: 707 ELIIPFYEAFNGKKWEKFNSEKVASLAYARIQGKSALIAHFQNSSLMNEDMRCRPIIFDT 766
Query: 779 -DGPNAGDQ 786
+ P + +Q
Sbjct: 767 PNNPESVEQ 775
>UniRef100_Q6ZI17 Putative meiosis protein mei2 [Oryza sativa]
Length = 848
Score = 241 bits (616), Expect = 5e-62
Identities = 123/185 (66%), Positives = 144/185 (77%), Gaps = 4/185 (2%)
Query: 603 PHFGLNVHNQRGMVFPGRNHMINSFDTYKRVRSRRNVGASNLADMKR-YELDIDCIKRGE 661
P G + + PG + NS + R R+RR + AD K+ Y+LD++ I++G+
Sbjct: 625 PRLGQSFYGNPTYQGPGSFGLDNSIE---RGRNRRVDSSVFQADSKKQYQLDLEKIRKGD 681
Query: 662 DNRTTLMIKNIPNKYTSKMLLAAIDEHHKGAYDFVYLPIDFRNKCNVGYAFINMTSPSLI 721
D RTTLMIKNIPNKYTSKMLLAAIDE HKG YDF YLPIDF+NKCNVGYAFINM SP I
Sbjct: 682 DTRTTLMIKNIPNKYTSKMLLAAIDEFHKGTYDFFYLPIDFKNKCNVGYAFINMISPVHI 741
Query: 722 VPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQNSSLMNEDKRCRPILIDTDGP 781
V FYQ FNGKKWEKFNSEKVASLAYARIQG+ AL++HFQNSSLMNEDKRCRPIL ++GP
Sbjct: 742 VSFYQAFNGKKWEKFNSEKVASLAYARIQGRTALISHFQNSSLMNEDKRCRPILFHSNGP 801
Query: 782 NAGDQ 786
+AG+Q
Sbjct: 802 DAGNQ 806
Score = 234 bits (596), Expect = 1e-59
Identities = 133/329 (40%), Positives = 203/329 (61%), Gaps = 23/329 (6%)
Query: 45 SSLFSSSLSDLFSKKLRLSANNAFYGHSVDTIASNYEEEKLSDSLE------ELEAQIIG 98
+SLFS+SL L +K+ ++A +D ++ +E L D E + + + I
Sbjct: 48 ASLFSTSLPVLPHEKINF-LDSARGTPLMDDASAKLKE--LDDDPEGKDYKFDFDLRQID 104
Query: 99 NLLPDEDDLLSGVTDGNNYIICDSNGDDIDELDLFSSNGGFDLGD--VENPSSIERNSEI 156
+LLP+EDDL +G+T+ ++ ++++E D+F S GG +L VE+ ++ N+ I
Sbjct: 105 DLLPNEDDLFAGITNEIEPAGQTNSMEELEEFDVFGSGGGMELDTDPVESITAGLGNTSI 164
Query: 157 ISGVRN------------SSIAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIH 204
G+R S++AGE+ YGEHPSRTLFVRNI+S+V D+ L++LFEQ+GDI
Sbjct: 165 ADGLRGNGVNHFGPSNSASTVAGEHPYGEHPSRTLFVRNINSNVDDTELRSLFEQYGDIR 224
Query: 205 TFDRTCKHQGSAMISYYDIRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQGTL 264
T KH+G MISY+DIRAA+ AMR L N+ R+K DIH+ IPK++PS +NQGTL
Sbjct: 225 TLYTATKHRGFVMISYFDIRAARGAMRGLQNKPLRRRKLDIHFSIPKENPSDKDLNQGTL 284
Query: 265 EVFLYDSSISNTELQHILNVYGGIKEIHENPRSQRHKLIEFYDFRAADAALHGINRNDTT 324
+F D S+SN E++ I YG +KEI E P + HK IEFYD RAA+AAL +N+++
Sbjct: 285 VIFNLDPSVSNEEVRQIFGTYGEVKEIRETPNKKHHKFIEFYDVRAAEAALRSLNKSEIA 344
Query: 325 MKRLKVDQMQSTNSESNIIQPMHPEFKQE 353
KR+K++ + + N++Q + + Q+
Sbjct: 345 GKRIKLEPSRPGGTRRNLMQQLGHDIDQD 373
>UniRef100_Q9C7R1 RNA-binding protein MEI2, putative; 36123-32976 [Arabidopsis
thaliana]
Length = 779
Score = 241 bits (615), Expect = 6e-62
Identities = 140/318 (44%), Positives = 194/318 (60%), Gaps = 22/318 (6%)
Query: 50 SSLSDLFSKKLRLSANNAFYGHSVDTIASNYEEEKLSDSLEELEAQIIGNLLPDEDDLLS 109
SS + LFS ++LS N + DT S K ++S ++ E+ IGNLLPDE+DLL+
Sbjct: 25 SSDATLFS--MQLSDNRDGFSLIDDTAVSR--TNKFNESADDFESHSIGNLLPDEEDLLT 80
Query: 110 GVTDGNNYIICDSNGDDIDELDLFSSNGGFDLG-------DVENP-----SSIERNSEII 157
G+ D + D D+ DLF S GG +L + P SS+ N+
Sbjct: 81 GMMDDLDL----GELPDADDYDLFGSGGGMELDADFRDNLSMSGPPRLSLSSLGGNAIPQ 136
Query: 158 SGVRNSS--IAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGS 215
+ N + +AGE+ YGEHPSRTLFVRNI+S+V+DS L ALFEQ+GDI T TCKH+G
Sbjct: 137 FNIPNGAGTVAGEHPYGEHPSRTLFVRNINSNVEDSELTALFEQYGDIRTLYTTCKHRGF 196
Query: 216 AMISYYDIRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYDSSISN 275
MISYYDIR+A+ AMR+L N+ R+K DIH+ IPKD+PS +NQGTL VF D SISN
Sbjct: 197 VMISYYDIRSARMAMRSLQNKPLRRRKLDIHFSIPKDNPSEKDMNQGTLVVFNLDPSISN 256
Query: 276 TELQHILNVYGGIKEIHENPRSQRHKLIEFYDFRAADAALHGINRNDTTMKRLKVDQMQS 335
+L I +G IKEI E P + HK +EFYD R A+AAL +NR + KR+KV+ +
Sbjct: 257 DDLHGIFGAHGEIKEIRETPHKRHHKFVEFYDVRGAEAALKALNRCEIAGKRIKVEPSRP 316
Query: 336 TNSESNIIQPMHPEFKQE 353
+ +++ ++ + + +
Sbjct: 317 GGARRSLMLQLNQDLEND 334
Score = 232 bits (591), Expect = 4e-59
Identities = 122/189 (64%), Positives = 146/189 (76%), Gaps = 14/189 (7%)
Query: 601 MFPHFGLNVHNQRGMVFPGRNHMINSFDT-YKRVRSRRNVGASNLADM-KRYELDIDCIK 658
MF GLN PGR + FD+ Y+ R RR SN + K+++LD++ I
Sbjct: 586 MFLSSGLN---------PGR--FASGFDSLYENGRPRRVENNSNQVESRKQFQLDLEKIL 634
Query: 659 RGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHKGAYDFVYLPIDFRNKCNVGYAFINMTSP 718
GED+RTTLMIKNIPNKYTSKMLLAAIDE ++G Y+F+YLPIDF+NKCNVGYAFINM +P
Sbjct: 635 NGEDSRTTLMIKNIPNKYTSKMLLAAIDEKNQGTYNFLYLPIDFKNKCNVGYAFINMLNP 694
Query: 719 SLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQNSSLMNEDKRCRPILIDT 778
LI+PFY+ FNGKKWEKFNSEKVASLAYARIQGK+AL+AHFQNSSLMNED RCRPI+ DT
Sbjct: 695 ELIIPFYEAFNGKKWEKFNSEKVASLAYARIQGKSALIAHFQNSSLMNEDMRCRPIIFDT 754
Query: 779 -DGPNAGDQ 786
+ P + +Q
Sbjct: 755 PNNPESVEQ 763
>UniRef100_Q6J737 AML1 [Aegilops speltoides]
Length = 869
Score = 238 bits (607), Expect = 5e-61
Identities = 117/157 (74%), Positives = 133/157 (84%), Gaps = 1/157 (0%)
Query: 631 KRVRSRRNVGASNLADMKR-YELDIDCIKRGEDNRTTLMIKNIPNKYTSKMLLAAIDEHH 689
+R R+RR ++ AD K+ Y+LD+D I++GED RTTLMIKNIPNKYTSKMLLAAIDE H
Sbjct: 666 ERGRNRRVDSSAFQADSKKQYQLDLDKIRKGEDTRTTLMIKNIPNKYTSKMLLAAIDELH 725
Query: 690 KGAYDFVYLPIDFRNKCNVGYAFINMTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARI 749
KG YDF YLPIDF+NKCNVGYAFINM SP IV FYQ FNGKKWEKFNSEKVASLAY RI
Sbjct: 726 KGTYDFFYLPIDFKNKCNVGYAFINMISPVHIVSFYQAFNGKKWEKFNSEKVASLAYGRI 785
Query: 750 QGKAALVAHFQNSSLMNEDKRCRPILIDTDGPNAGDQ 786
QG+ AL++HFQNSSLMNEDKRCRPIL ++GP G+Q
Sbjct: 786 QGRNALISHFQNSSLMNEDKRCRPILFHSNGPETGNQ 822
Score = 233 bits (595), Expect = 1e-59
Identities = 133/330 (40%), Positives = 205/330 (61%), Gaps = 22/330 (6%)
Query: 44 ESSLFSSSLSDLFSKKLRLSANNAFYGHSVDTIASNY----EEEKLSDSLEELEAQIIGN 99
++S+FS+SL L +KL S ++A S+D ++ ++ + +D + + + I +
Sbjct: 60 DTSIFSTSLPVLPHEKLNFS-DSAHGTPSMDDASAKLKDFDDDPQGNDYPFDFDLRQIDD 118
Query: 100 LLPDEDDLLSGVTDGNNYIICDSNGDDIDELDLFSSNGGFDLGDVENPSSIER---NSEI 156
LLPDED+L +G+T+ + ++++E D+F S GG +L D + SI N+ I
Sbjct: 119 LLPDEDELFAGITNEIEPSSQANPAEELEEFDVFGSGGGMEL-DSDPLDSITAGLGNASI 177
Query: 157 ISGVRNS-------------SIAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDI 203
G+R + ++AGE+ GEHPSRTLFVRNI+S+V+DS L++LFEQFGDI
Sbjct: 178 GDGLRANGVNNNFGLSNSPGAVAGEHPLGEHPSRTLFVRNINSNVEDSELRSLFEQFGDI 237
Query: 204 HTFDRTCKHQGSAMISYYDIRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQGT 263
T KH+G MISY+DIRAA+ AMR+L N+ R+K DIH+ IPK++PS +NQGT
Sbjct: 238 RTLYTATKHRGFVMISYFDIRAARGAMRSLQNKPLRRRKLDIHFSIPKENPSDKDLNQGT 297
Query: 264 LEVFLYDSSISNTELQHILNVYGGIKEIHENPRSQRHKLIEFYDFRAADAALHGINRNDT 323
L +F D S+SN E++ I YG +KEI E P + HK IEFYD RAA+AAL +N+++
Sbjct: 298 LVIFNLDPSVSNEEVRQIFGTYGEVKEIRETPNKKHHKFIEFYDVRAAEAALRSLNKSEI 357
Query: 324 TMKRLKVDQMQSTNSESNIIQPMHPEFKQE 353
KR+K++ + + +++Q + E + E
Sbjct: 358 AGKRIKLEPSRPGGTRRSLVQHLGHELEDE 387
>UniRef100_Q6J734 AML15 [Triticum aestivum]
Length = 870
Score = 238 bits (607), Expect = 5e-61
Identities = 117/157 (74%), Positives = 133/157 (84%), Gaps = 1/157 (0%)
Query: 631 KRVRSRRNVGASNLADMKR-YELDIDCIKRGEDNRTTLMIKNIPNKYTSKMLLAAIDEHH 689
+R R+RR ++ AD K+ Y+LD+D I++GED RTTLMIKNIPNKYTSKMLLAAIDE H
Sbjct: 667 ERGRNRRVDSSAFQADSKKHYQLDLDKIRKGEDTRTTLMIKNIPNKYTSKMLLAAIDELH 726
Query: 690 KGAYDFVYLPIDFRNKCNVGYAFINMTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARI 749
KG YDF YLPIDF+NKCNVGYAFINM SP IV FYQ FNGKKWEKFNSEKVASLAY RI
Sbjct: 727 KGTYDFFYLPIDFKNKCNVGYAFINMISPVHIVSFYQAFNGKKWEKFNSEKVASLAYGRI 786
Query: 750 QGKAALVAHFQNSSLMNEDKRCRPILIDTDGPNAGDQ 786
QG+ AL++HFQNSSLMNEDKRCRPIL ++GP G+Q
Sbjct: 787 QGRNALISHFQNSSLMNEDKRCRPILFHSNGPETGNQ 823
Score = 232 bits (592), Expect = 3e-59
Identities = 132/330 (40%), Positives = 204/330 (61%), Gaps = 22/330 (6%)
Query: 44 ESSLFSSSLSDLFSKKLRLSANNAFYGHSVDTIASNY----EEEKLSDSLEELEAQIIGN 99
++S+FS+SL L +KL ++A S+D ++ ++ + +D + + + I +
Sbjct: 60 DTSIFSTSLPVLPHEKLNFP-DSAHGTPSMDDASAKLKDFDDDPQGNDYPFDFDLRQIDD 118
Query: 100 LLPDEDDLLSGVTDGNNYIICDSNGDDIDELDLFSSNGGFDLGDVENPSSIER---NSEI 156
LLPDED+L +G+T+ + ++++E D+F S GG +L D + SI N+ I
Sbjct: 119 LLPDEDELFAGITNEMEPSSQANPVEELEEFDVFGSGGGMEL-DSDPLDSITAGLGNASI 177
Query: 157 ISGVRNS-------------SIAGENSYGEHPSRTLFVRNIDSDVKDSVLKALFEQFGDI 203
G+R + ++AGE+ GEHPSRTLFVRNI+S+V+DS L++LFEQFGDI
Sbjct: 178 SDGIRANGVNNNFGLSNSPGAVAGEHPLGEHPSRTLFVRNINSNVEDSELRSLFEQFGDI 237
Query: 204 HTFDRTCKHQGSAMISYYDIRAAQNAMRALNNRLFGRKKFDIHYPIPKDSPSRNGVNQGT 263
T KH+G MISY+DIRAA+ AMR+L N+ R+K DIH+ IPK++PS +NQGT
Sbjct: 238 RTLYTATKHRGFVMISYFDIRAARGAMRSLQNKPLRRRKLDIHFSIPKENPSDKDLNQGT 297
Query: 264 LEVFLYDSSISNTELQHILNVYGGIKEIHENPRSQRHKLIEFYDFRAADAALHGINRNDT 323
L +F D S+SN E++ I YG +KEI E P + HK IEFYD RAA+AAL +N+++
Sbjct: 298 LVIFNLDPSVSNEEVRQIFGTYGEVKEIRETPNKKHHKFIEFYDVRAAEAALRSLNKSEI 357
Query: 324 TMKRLKVDQMQSTNSESNIIQPMHPEFKQE 353
KR+K++ + + +++Q + E + E
Sbjct: 358 AGKRIKLEPSRPGGTRRSLVQHLGHELEDE 387
>UniRef100_Q75M35 Hypothetical protein P0668H12.3 [Oryza sativa]
Length = 932
Score = 234 bits (598), Expect = 6e-60
Identities = 125/201 (62%), Positives = 145/201 (71%), Gaps = 33/201 (16%)
Query: 619 GRNHMIN---SFD-TYKRVRSRRNVGASNLADMKR-YELDIDCIKRGEDNRTTLMIKNIP 673
GRN MI+ S+D T R+RSRR+ G ++ KR +ELDID I +GED+RTTLMIKNIP
Sbjct: 678 GRNPMIHVSTSYDATNDRMRSRRHDGNPAQSENKRQFELDIDRIAKGEDSRTTLMIKNIP 737
Query: 674 NKYTSKMLLAAIDEHHKGAYDFVYLPIDFR----------------------------NK 705
NKY K+LLA IDE+H+G YDF+YLPIDF+ NK
Sbjct: 738 NKYNCKLLLAVIDENHRGTYDFIYLPIDFKVYFFFLSVKDCNCEYNFRTDFCNVHFSQNK 797
Query: 706 CNVGYAFINMTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAALVAHFQNSSLM 765
CNVGYAFINMT P I+PFY+ FNGKKWEKFNSEKVASLAYARIQG++AL+AHFQNSSLM
Sbjct: 798 CNVGYAFINMTDPQHIIPFYKTFNGKKWEKFNSEKVASLAYARIQGRSALIAHFQNSSLM 857
Query: 766 NEDKRCRPILIDTDGPNAGDQ 786
NEDK CRP+L DGPNAGDQ
Sbjct: 858 NEDKWCRPMLFHKDGPNAGDQ 878
Score = 222 bits (565), Expect = 4e-56
Identities = 139/355 (39%), Positives = 207/355 (58%), Gaps = 14/355 (3%)
Query: 7 ENDTSNYSLT---LLCHVDSDQGIKPFEDSKEDDIVASRHE----SSLFSSSLSDLFSKK 59
EN + + SL+ +L + S G+ E + + A +H L SSSLS++FS K
Sbjct: 98 ENASGSPSLSWGEILTNPISRLGLSTRETAFVEPTTADQHVPGYGKGLSSSSLSEVFSGK 157
Query: 60 LRLSANNAFYGHSVDTIASNYEEEKLSDSLEELEAQIIGNLLPDEDD-LLSGVTDGNNYI 118
R + S T S Y+ ++ +S+E +EAQ IG+LLPD+DD L+SG+ DG +
Sbjct: 158 SREIVSGVLC-QSTGTHTSIYDSDEPLESMEAIEAQTIGDLLPDDDDDLISGIADGFEFT 216
Query: 119 ICDSNGDDIDELDLFSSNGGFDLGDVENPSSIERNSEIISGVRNSSIAGENSYGEHPSRT 178
+N DD DE D+F + GG +L EN S+ + ++ G S I+ +S + PSRT
Sbjct: 217 GMSTNQDDADE-DIFCTGGGMEL---ENNDSV-KGDKVQDGSFKSQISSGHSINKQPSRT 271
Query: 179 LFVRNIDSDVKDSVLKALFEQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALNNRLF 238
L VRNI ++++DS L LF+Q+GDI + KH G +SYYDIRAAQNAMRAL+++
Sbjct: 272 LVVRNITANIEDSDLTVLFQQYGDIRMLYTSFKHHGFVTVSYYDIRAAQNAMRALHSKPL 331
Query: 239 GRKKFDIHYPIPKDSPSRNGVNQGTLEVFLYDSSISNTELQHILNVYGGIKEIHENPRSQ 298
G K D+ + PK++ +++G L V DSSISN +L +L+VYG +KEI +P S
Sbjct: 332 GLMKLDVQFSFPKENVPGKDIDKGMLVVSNIDSSISNDDLLQMLSVYGDVKEISSSPISC 391
Query: 299 RHKLIEFYDFRAADAALHGINRNDTTMKRLKVDQMQSTNSESNIIQPMHPEFKQE 353
K +EFYD RAA+ ALH +N+ + + KV+ Q + S + Q E+KQ+
Sbjct: 392 TKKFVEFYDVRAAEEALHDLNKGGISGPKFKVELSQHGEAGSCLRQQHSREWKQD 446
>UniRef100_Q6EQX3 Putative AML1 [Oryza sativa]
Length = 811
Score = 230 bits (586), Expect = 1e-58
Identities = 115/284 (40%), Positives = 186/284 (65%), Gaps = 15/284 (5%)
Query: 84 KLSDSLEELEAQIIGNLLPDEDDLLSGVTDGNNYIICDSNGDDIDELDLFSSNGGFDLGD 143
+++D ++++ +IGNLLPD+++LL+GV + +++ + ++ +E D+F ++GG +L D
Sbjct: 87 RVADPMDDVAQHLIGNLLPDDEELLAGVIEDFDHVKLRTQVEESEEYDVFRNSGGMEL-D 145
Query: 144 VENPSSIE---RNSEIISGVRNSS-----------IAGENSYGEHPSRTLFVRNIDSDVK 189
++ SI + +++G +S+ + GE+ YGEHPSRTLFVRNI+S+V+
Sbjct: 146 IDPLESITFGTAKASLVNGTGSSTNQYSIQNGAGTVTGEHPYGEHPSRTLFVRNINSNVE 205
Query: 190 DSVLKALFEQFGDIHTFDRTCKHQGSAMISYYDIRAAQNAMRALNNRLFGRKKFDIHYPI 249
DS L++LFE FGDI + KH+G MISYYDIR A+NA AL ++ R+K DIHY I
Sbjct: 206 DSELRSLFEPFGDIRSMYTATKHRGFVMISYYDIRHARNAKTALQSKPLRRRKLDIHYSI 265
Query: 250 PKDSPSRNGVNQGTLEVFLYDSSISNTELQHILNVYGGIKEIHENPRSQRHKLIEFYDFR 309
PK++PS +NQGTL +F + ++SN EL I +G ++EI E P + H+ IEFYD R
Sbjct: 266 PKENPSDKDMNQGTLVIFNLEPAVSNEELLQIFGAFGEVREIRETPHKRHHRFIEFYDVR 325
Query: 310 AADAALHGINRNDTTMKRLKVDQMQSTNSESNIIQPMHPEFKQE 353
AA++AL +N++D KR+K++ + + + IQ + EF+Q+
Sbjct: 326 AAESALRSLNKSDIAGKRVKLEPSRPGGARRSFIQHFNHEFEQD 369
Score = 209 bits (532), Expect = 3e-52
Identities = 103/143 (72%), Positives = 115/143 (80%), Gaps = 2/143 (1%)
Query: 637 RNVGASNLADMKR--YELDIDCIKRGEDNRTTLMIKNIPNKYTSKMLLAAIDEHHKGAYD 694
+ VG S + R Y+LD++ I G+D RTTLMIKNIPNKYTS MLL IDE H+G YD
Sbjct: 628 QTVGNSGCQEDSRVQYQLDLEKIIAGKDTRTTLMIKNIPNKYTSNMLLEVIDETHEGTYD 687
Query: 695 FVYLPIDFRNKCNVGYAFINMTSPSLIVPFYQGFNGKKWEKFNSEKVASLAYARIQGKAA 754
F YLPIDF+NKCNVGYAFINM SP IV F++ F G+KWEKFNSEKV SLAYARIQGKAA
Sbjct: 688 FFYLPIDFKNKCNVGYAFINMASPGYIVSFFKAFAGRKWEKFNSEKVVSLAYARIQGKAA 747
Query: 755 LVAHFQNSSLMNEDKRCRPILID 777
LV HFQNSSLMNEDKRCRP+L D
Sbjct: 748 LVNHFQNSSLMNEDKRCRPMLFD 770
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.324 0.139 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,330,100,718
Number of Sequences: 2790947
Number of extensions: 57368796
Number of successful extensions: 178948
Number of sequences better than 10.0: 933
Number of HSP's better than 10.0 without gapping: 69
Number of HSP's successfully gapped in prelim test: 866
Number of HSP's that attempted gapping in prelim test: 177379
Number of HSP's gapped (non-prelim): 1967
length of query: 786
length of database: 848,049,833
effective HSP length: 135
effective length of query: 651
effective length of database: 471,271,988
effective search space: 306798064188
effective search space used: 306798064188
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 79 (35.0 bits)
Medicago: description of AC146333.19


