

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146332.8 - phase: 0 /pseudo
(199 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
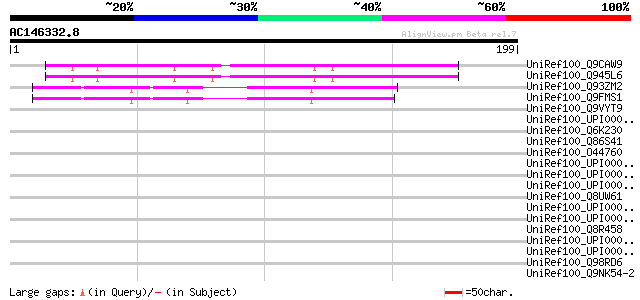
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9CAW9 Hypothetical protein F24K9.26 [Arabidopsis thal... 96 5e-19
UniRef100_Q945L6 AT3g11590/F24K9_26 [Arabidopsis thaliana] 96 5e-19
UniRef100_Q93ZM2 AT5g22310/MWD9_9 [Arabidopsis thaliana] 68 2e-10
UniRef100_Q9FMS1 Gb|AAF00675.1 [Arabidopsis thaliana] 66 6e-10
UniRef100_Q9VYT9 CG15741-PA [Drosophila melanogaster] 36 0.66
UniRef100_UPI0000467C78 UPI0000467C78 UniRef100 entry 35 0.86
UniRef100_Q6K230 Intracellular protein transport protein USO1-li... 35 1.1
UniRef100_Q86S41 Prion-like-(Q/n-rich)-domain-bearing protein pr... 35 1.5
UniRef100_O44760 Prion-like-(Q/n-rich)-domain-bearing protein pr... 35 1.5
UniRef100_UPI00004999D2 UPI00004999D2 UniRef100 entry 34 1.9
UniRef100_UPI00002C3039 UPI00002C3039 UniRef100 entry 34 1.9
UniRef100_UPI000042FC2C UPI000042FC2C UniRef100 entry 34 2.5
UniRef100_Q8UW61 Vitellogenin II [Oryzias latipes] 34 2.5
UniRef100_UPI0000432B10 UPI0000432B10 UniRef100 entry 33 3.3
UniRef100_UPI0000432B0F UPI0000432B0F UniRef100 entry 33 3.3
UniRef100_Q8R458 Protein kinase for splicing component [Rattus n... 33 3.3
UniRef100_UPI0000244851 UPI0000244851 UniRef100 entry 33 4.3
UniRef100_UPI0000318C1C UPI0000318C1C UniRef100 entry 33 4.3
UniRef100_Q98RD6 Hypothetical protein MYPU_0730 [Mycoplasma pulm... 33 4.3
UniRef100_Q9NK54-2 Splice isoform B of Q9NK54 [Drosophila melano... 33 4.3
>UniRef100_Q9CAW9 Hypothetical protein F24K9.26 [Arabidopsis thaliana]
Length = 622
Score = 95.9 bits (237), Expect = 5e-19
Identities = 72/174 (41%), Positives = 90/174 (51%), Gaps = 15/174 (8%)
Query: 15 KIRKRGCSS-TSSSSPFSRR-YRFKRAILMGKKSGYNTPAPMWKTSSKSPS-MATHHHHA 71
KIRKRGCSS TSS+S R YRFKRAI++GK+ G TP P W+ +SPS A+ HA
Sbjct: 16 KIRKRGCSSPTSSTSSILREGYRFKRAIVVGKRGGSTTPVPTWRLMGRSPSPRASGALHA 75
Query: 72 MKKTALH-SSRTGLPFKEKKEISVSARKLAATLWEINDLTPSRIKKES----MKSNKER- 125
+ H S+TG K VSARKLAATLWE+N++ R+ +E+ KS KER
Sbjct: 76 AASPSSHCGSKTG---KVSAPAPVSARKLAATLWEMNEMPSPRVVEEAAPMIRKSRKERI 132
Query: 126 ---DKVERLCRSVLLGPQKLDPLVSPFSERTKGIEVDGCKRNVSDLSHQFHFVD 176
S L P DP SP SER + +R S + D
Sbjct: 133 APLPPPRSSVHSGSLPPHLSDPSHSPVSERMERSGTGSRQRRASSTVQKLRLGD 186
>UniRef100_Q945L6 AT3g11590/F24K9_26 [Arabidopsis thaliana]
Length = 622
Score = 95.9 bits (237), Expect = 5e-19
Identities = 72/174 (41%), Positives = 90/174 (51%), Gaps = 15/174 (8%)
Query: 15 KIRKRGCSS-TSSSSPFSRR-YRFKRAILMGKKSGYNTPAPMWKTSSKSPS-MATHHHHA 71
KIRKRGCSS TSS+S R YRFKRAI++GK+ G TP P W+ +SPS A+ HA
Sbjct: 16 KIRKRGCSSPTSSTSSILREGYRFKRAIVVGKRGGSTTPVPTWRLMGRSPSPRASGALHA 75
Query: 72 MKKTALH-SSRTGLPFKEKKEISVSARKLAATLWEINDLTPSRIKKES----MKSNKER- 125
+ H S+TG K VSARKLAATLWE+N++ R+ +E+ KS KER
Sbjct: 76 AASPSSHCGSKTG---KVSAPAPVSARKLAATLWEMNEMPSPRVVEEAAPMIRKSRKERI 132
Query: 126 ---DKVERLCRSVLLGPQKLDPLVSPFSERTKGIEVDGCKRNVSDLSHQFHFVD 176
S L P DP SP SER + +R S + D
Sbjct: 133 APLPPPRSSVHSGSLPPHLSDPSHSPVSERMERSGTGSRQRRASSTVQKLRLGD 186
>UniRef100_Q93ZM2 AT5g22310/MWD9_9 [Arabidopsis thaliana]
Length = 481
Score = 67.8 bits (164), Expect = 2e-10
Identities = 58/160 (36%), Positives = 79/160 (49%), Gaps = 36/160 (22%)
Query: 10 KQKHNKIRKRGCSSTSSSSPFSRRYRFKRAILMGKKS-----GYNTPAPMWKTSSKSPSM 64
++K KIRKRG SS+SSSS +RR RFKRAI GK++ G TP T++K+P +
Sbjct: 4 RKKGCKIRKRGGSSSSSSS-LARRNRFKRAIFAGKRAAQDDGGSGTPVKSI-TAAKTPVL 61
Query: 65 ATHH-------HHAMKKTALHSSRTGLPFKEKKEISVSARKLAATLWEINDLTPSRIKKE 117
+ HH ++K+ VSARKLAATLWEIND + +
Sbjct: 62 LSFSPENLPIDHHQLQKSC-----------------VSARKLAATLWEINDDADPPVNSD 104
Query: 118 -----SMKSNKERDKVERLCRSVLLGPQKLDPLVSPFSER 152
S K ++ R K S+ P+ DP+ SER
Sbjct: 105 KDCLRSKKPSRYRAKKSTEFSSIDFPPRSSDPISRLSSER 144
>UniRef100_Q9FMS1 Gb|AAF00675.1 [Arabidopsis thaliana]
Length = 450
Score = 65.9 bits (159), Expect = 6e-10
Identities = 57/159 (35%), Positives = 78/159 (48%), Gaps = 36/159 (22%)
Query: 10 KQKHNKIRKRGCSSTSSSSPFSRRYRFKRAILMGKKS-----GYNTPAPMWKTSSKSPSM 64
++K KIRKRG SS+SSSS +RR RFKRAI GK++ G TP T++K+P +
Sbjct: 4 RKKGCKIRKRGGSSSSSSS-LARRNRFKRAIFAGKRAAQDDGGSGTPVKSI-TAAKTPVL 61
Query: 65 ATHH-------HHAMKKTALHSSRTGLPFKEKKEISVSARKLAATLWEINDLTPSRIKKE 117
+ HH ++K+ VSARKLAATLWEIND + +
Sbjct: 62 LSFSPENLPIDHHQLQKSC-----------------VSARKLAATLWEINDDADPPVNSD 104
Query: 118 -----SMKSNKERDKVERLCRSVLLGPQKLDPLVSPFSE 151
S K ++ R K S+ P+ DP+ SE
Sbjct: 105 KDCLRSKKPSRYRAKKSTEFSSIDFPPRSSDPISRLSSE 143
>UniRef100_Q9VYT9 CG15741-PA [Drosophila melanogaster]
Length = 135
Score = 35.8 bits (81), Expect = 0.66
Identities = 33/118 (27%), Positives = 49/118 (40%), Gaps = 5/118 (4%)
Query: 22 SSTSSSSPFSRRYRFKRAILMGKKSGYNTPAPMWKTSSKSPSMATHHHHAMKKTALHSSR 81
SSTSS+SP S S ++ +P TS S S ++ A T +
Sbjct: 17 SSTSSTSPASSTSPASSTSPTSSTSPTSSTSPTSSTSPTSSSSSSSSSTATTTTTTVAPT 76
Query: 82 TGLPFKEKKEISVSARKLAATLWEIN-----DLTPSRIKKESMKSNKERDKVERLCRS 134
T + S S +K+ T +N D ++ KK++ K NK+ KV R RS
Sbjct: 77 TTTTTEASSSSSSSDKKVRHTRHYVNRRHRKDKKTTKSKKKNSKKNKKTKKVRRTRRS 134
>UniRef100_UPI0000467C78 UPI0000467C78 UniRef100 entry
Length = 303
Score = 35.4 bits (80), Expect = 0.86
Identities = 28/122 (22%), Positives = 57/122 (45%), Gaps = 19/122 (15%)
Query: 5 QTKHLKQKHNKIR-KRGCSSTSSSSPFSRRYRFKRAILMGKKSGYNTPAPMWKTSSKSPS 63
+T + N+ + K S SS +P+ R+RFK++ + KK+ + KT K+ +
Sbjct: 2 ETNEFENTDNETKSKENSSDESSDTPYKNRFRFKKSSIQIKKNS------LLKTKIKNAN 55
Query: 64 MATHHHHAMKKTALHSSRTGLPFK-EKKEISVSARKLAATLWEINDLTPSRIKKESMKSN 122
++ T + S G P + K+ +S+++ ND+ ++K+S+K N
Sbjct: 56 --------LEGTTQNDSALGTPVQLNNKDDQISSKENNVI---TNDIPNEELEKDSIKKN 104
Query: 123 KE 124
+
Sbjct: 105 SQ 106
>UniRef100_Q6K230 Intracellular protein transport protein USO1-like [Oryza sativa]
Length = 711
Score = 35.0 bits (79), Expect = 1.1
Identities = 25/92 (27%), Positives = 39/92 (42%), Gaps = 8/92 (8%)
Query: 18 KRGCSSTSSSSPFSRRYRFKRAILMGKKSGYNTPAPMWKTSSKSPSMATHHHHAMKKTAL 77
+RG R R RA +++G TP+P WK + +A +
Sbjct: 46 RRGEGDCRGEEEVPLRVRLGRAAR--RRAGPCTPSPSWKLEGEEVEVAAGELAPVHPAVA 103
Query: 78 HSSRTGLPFKEKKEISVSARKLAATLWEINDL 109
+ R+ S SAR+L A+LWEI+D+
Sbjct: 104 PARRSSA------SASASARQLGASLWEIHDV 129
>UniRef100_Q86S41 Prion-like-(Q/n-rich)-domain-bearing protein protein 64, isoform b
[Caenorhabditis elegans]
Length = 662
Score = 34.7 bits (78), Expect = 1.5
Identities = 22/58 (37%), Positives = 31/58 (52%), Gaps = 3/58 (5%)
Query: 5 QTKHLKQKHNKIRKRGCSSTSSSSPFSRRYRFKRAILMGKKSGYNTPAPMWKTSSKSP 62
QT+ KQ+HN R S+SSSS S +Y A+ SG +TP +TS ++P
Sbjct: 451 QTRQRKQQHNGYNNRPTVSSSSSSSTSSKYFSPDAVETPTNSGSSTPP---QTSVRNP 505
>UniRef100_O44760 Prion-like-(Q/n-rich)-domain-bearing protein protein 64, isoform a
[Caenorhabditis elegans]
Length = 760
Score = 34.7 bits (78), Expect = 1.5
Identities = 22/58 (37%), Positives = 31/58 (52%), Gaps = 3/58 (5%)
Query: 5 QTKHLKQKHNKIRKRGCSSTSSSSPFSRRYRFKRAILMGKKSGYNTPAPMWKTSSKSP 62
QT+ KQ+HN R S+SSSS S +Y A+ SG +TP +TS ++P
Sbjct: 549 QTRQRKQQHNGYNNRPTVSSSSSSSTSSKYFSPDAVETPTNSGSSTPP---QTSVRNP 603
>UniRef100_UPI00004999D2 UPI00004999D2 UniRef100 entry
Length = 1738
Score = 34.3 bits (77), Expect = 1.9
Identities = 32/126 (25%), Positives = 51/126 (40%), Gaps = 21/126 (16%)
Query: 7 KHLKQKHNKIRKRGC--------SSTSSSSPFSRRYRFKRAILMGKKSGYNTPAPMWKTS 58
+HLK+ N + C SST + SP S KS TP S
Sbjct: 1602 QHLKESLNPSKSTNCIISAPPSVSSTPTISPASTP----------SKSKEETPRKHTINS 1651
Query: 59 SKSPSMATHHHHAMKKTALHSSRTGLPFKEKKEISVSARKLAATLWEINDLTPSRIKKES 118
S S++ H++K++ + ++ + K IS S R EI+DL P + E
Sbjct: 1652 FTSESISETPKHSLKQSKIEKHKS---LRSAKTISFSKRDKKKDKKEIDDLVPETVDGEE 1708
Query: 119 MKSNKE 124
++ +E
Sbjct: 1709 VQLERE 1714
>UniRef100_UPI00002C3039 UPI00002C3039 UniRef100 entry
Length = 299
Score = 34.3 bits (77), Expect = 1.9
Identities = 27/126 (21%), Positives = 55/126 (43%), Gaps = 7/126 (5%)
Query: 6 TKHLKQKHNKIRKRGCSSTSSSSPFSRRYRFKRAILMGKKSGYNTPAPMWKTSSKSP--- 62
TK K K+ + + P + + + R + + K Y T ++ SSKS
Sbjct: 138 TKAFKNKNYEKAAKSFLEAEIDEPNNHIHSYNRGVSLYKSKNYKTAIEAFEQSSKSKDKD 197
Query: 63 -SMATHHHHAMKKTALHSSRTGLPFKEKKEISVSARKLAATLWEINDLTPSRIKKESMKS 121
S + ++ A AL + + + + EK S++ + + E + ++KK+ K
Sbjct: 198 LSFKSQYNLANTFAALKNFQESIKYYEK---SLTTKPESKKAKENLEWAKKQLKKQKNKQ 254
Query: 122 NKERDK 127
+K++DK
Sbjct: 255 DKDKDK 260
>UniRef100_UPI000042FC2C UPI000042FC2C UniRef100 entry
Length = 732
Score = 33.9 bits (76), Expect = 2.5
Identities = 27/98 (27%), Positives = 46/98 (46%), Gaps = 8/98 (8%)
Query: 8 HLKQKHNKIRKRGCSSTSSSSPFSRRYRFKRAILMGKKSGYNTPAPMWKTSSKSPSMATH 67
+ K ++KR S SSSS SRR+ F R + K+ P+P +SS S + T
Sbjct: 570 NFKNLDQLVQKRNDSRASSSSSNSRRFEFIRGL---KEENERVPSPS-SSSSSSSATKTS 625
Query: 68 HHHAMKKTALHSSRTGLPFKEKKEISVSARKLAATLWE 105
++ K + SRT +++++S + +WE
Sbjct: 626 QNNFEKSSESAISRT----DDQQDLSSTNTGSEGRMWE 659
>UniRef100_Q8UW61 Vitellogenin II [Oryzias latipes]
Length = 1725
Score = 33.9 bits (76), Expect = 2.5
Identities = 33/128 (25%), Positives = 55/128 (42%), Gaps = 7/128 (5%)
Query: 5 QTKHLKQKHNKIRKRGC-SSTSSSSPFSRRYRFKRAILMGKKSGYNTPAPMWKTSSKSPS 63
+ K++ K KI G +ST+SSS S R R+ L S ++ KTSS+S S
Sbjct: 1068 EDKNVLLKLKKILTPGLKNSTTSSSSSSSSSRRSRSSLSSATSSLSSSRTSSKTSSRSSS 1127
Query: 64 MATHH------HHAMKKTALHSSRTGLPFKEKKEISVSARKLAATLWEINDLTPSRIKKE 117
+ KT+ SS + ++ K I ++ + W+ + + SR +
Sbjct: 1128 KTSSRSSSKTSSRTSSKTSSSSSSSSSSRRKSKTIDLAEVLNRTSKWQSSSSSSSRSSRP 1187
Query: 118 SMKSNKER 125
S S+ R
Sbjct: 1188 SRSSSSVR 1195
>UniRef100_UPI0000432B10 UPI0000432B10 UniRef100 entry
Length = 564
Score = 33.5 bits (75), Expect = 3.3
Identities = 37/126 (29%), Positives = 60/126 (47%), Gaps = 25/126 (19%)
Query: 13 HNKIRKRGCSSTSS--SSPFSRRYRFKRAILMGKKSGYNTPAPMWKTSSKSPSMATHHHH 70
+NK +++ S T+S SSP SR++ +P K+ S+SPS +
Sbjct: 319 NNKSKQKSVSPTASKKSSPVSRKH-----------------SPRDKSYSRSPSKSYTRSV 361
Query: 71 AMKKTALHSSR--TGLPFKEKKEISVSARKLAATLWEINDLTPSRIKKESMKSNKERDKV 128
+++K + S R + +KK SVS K +A+ + +PSR S KS K +
Sbjct: 362 SLEKRSSRSPRRHSRSISTDKKSRSVSRSKKSASPRHRSSTSPSR----SKKSPKSPVRS 417
Query: 129 ERLCRS 134
+RL RS
Sbjct: 418 KRLSRS 423
>UniRef100_UPI0000432B0F UPI0000432B0F UniRef100 entry
Length = 971
Score = 33.5 bits (75), Expect = 3.3
Identities = 37/126 (29%), Positives = 60/126 (47%), Gaps = 25/126 (19%)
Query: 13 HNKIRKRGCSSTSS--SSPFSRRYRFKRAILMGKKSGYNTPAPMWKTSSKSPSMATHHHH 70
+NK +++ S T+S SSP SR++ +P K+ S+SPS +
Sbjct: 697 NNKSKQKSVSPTASKKSSPVSRKH-----------------SPRDKSYSRSPSKSYTRSV 739
Query: 71 AMKKTALHSSR--TGLPFKEKKEISVSARKLAATLWEINDLTPSRIKKESMKSNKERDKV 128
+++K + S R + +KK SVS K +A+ + +PSR S KS K +
Sbjct: 740 SLEKRSSRSPRRHSRSISTDKKSRSVSRSKKSASPRHRSSTSPSR----SKKSPKSPVRS 795
Query: 129 ERLCRS 134
+RL RS
Sbjct: 796 KRLSRS 801
>UniRef100_Q8R458 Protein kinase for splicing component [Rattus norvegicus]
Length = 1258
Score = 33.5 bits (75), Expect = 3.3
Identities = 35/149 (23%), Positives = 65/149 (43%), Gaps = 21/149 (14%)
Query: 22 SSTSSSSPFSRRYRFKRAILMGKKSGY--------NTPAPMWKTSSKSPSMATHHHHAMK 73
SST S SP+SRR R + S Y +P+P + S SP ++ ++
Sbjct: 286 SSTRSPSPYSRRQRSVSPYSRRRSSSYERSGSYSGRSPSPYGRRRSSSPFLS---KRSLS 342
Query: 74 KTALHS-----SRTGLPFKEKKEISVSARKLAATLWEINDLTPSRIKKESM----KSNKE 124
++ + S SR+ P + S S +K + + + ++P R+ S S K+
Sbjct: 343 RSPISSRKSMKSRSRSPAYSRHSSSHSKKKRSGSRSRHSSISPVRLPLNSSLGAELSRKK 402
Query: 125 RDKVERLCRSVLLGPQ-KLDPLVSPFSER 152
+++ + L G + K P++ P E+
Sbjct: 403 KERAAAAAAAKLDGKESKGSPIILPKKEK 431
>UniRef100_UPI0000244851 UPI0000244851 UniRef100 entry
Length = 324
Score = 33.1 bits (74), Expect = 4.3
Identities = 23/62 (37%), Positives = 30/62 (48%), Gaps = 2/62 (3%)
Query: 79 SSRTGLPFKEKKEISVSARKLAAT--LWEINDLTPSRIKKESMKSNKERDKVERLCRSVL 136
S +T L F E+ EI+ K A WEI + +KKE + R K + CRSVL
Sbjct: 61 SMKTILAFLEQLEIAHCVEKEAEEEDFWEILHEDTAAVKKEKEEQKAFRKKQDNKCRSVL 120
Query: 137 LG 138
G
Sbjct: 121 NG 122
>UniRef100_UPI0000318C1C UPI0000318C1C UniRef100 entry
Length = 1004
Score = 33.1 bits (74), Expect = 4.3
Identities = 25/116 (21%), Positives = 44/116 (37%), Gaps = 22/116 (18%)
Query: 8 HLKQKHNKIRKRGCSSTSSSSPFSRRYRFKRAILMGKKSGYNTPAPMWKTSSKSPSMATH 67
+ K +NK++K+ S+ ++SS FS S SP H
Sbjct: 160 YCKHHYNKMQKKLRSAENASSAFSHS----------------------TGRSSSPPQDKH 197
Query: 68 HHHAMKKTALHSSRTGLPFKEKKEISVSARKLAATLWEINDLTPSRIKKESMKSNK 123
HHH +K + H ++ + KK A + ++ + + +K KS K
Sbjct: 198 HHHKQRKVSSHKDKSRQKERHKKSSDSLASSVTTSIIDKVSCSHHSMKDSEGKSKK 253
>UniRef100_Q98RD6 Hypothetical protein MYPU_0730 [Mycoplasma pulmonis]
Length = 231
Score = 33.1 bits (74), Expect = 4.3
Identities = 25/95 (26%), Positives = 46/95 (48%), Gaps = 2/95 (2%)
Query: 88 EKKEISVSARKLAATLWEINDLTPSRIKKESMKSNKERDKVERLCRSVLLGPQKLDPLVS 147
+KK+I + + L+ N T IK + + DK+ L LL KL+ ++S
Sbjct: 61 DKKDIELKIKNLSENNNSDNTTTMPNIKNDKVFIVHGHDKIALLELKDLLRDLKLNVVIS 120
Query: 148 PFSERTKGIEV-DGCKRNVSDLSHQFHFVDPYFRG 181
++ T+G+ + DG ++S +S F + P +G
Sbjct: 121 Q-NKTTRGLSILDGVLEDISSVSFAFDLLTPDDKG 154
>UniRef100_Q9NK54-2 Splice isoform B of Q9NK54 [Drosophila melanogaster]
Length = 1695
Score = 33.1 bits (74), Expect = 4.3
Identities = 26/107 (24%), Positives = 49/107 (45%), Gaps = 3/107 (2%)
Query: 23 STSSSSPFSRRYRFKRAILMGKKSGYNTPAPMWKTSSKSPSMATHHHHAMKKTALHSSRT 82
ST SSS S + RF A + ++ P P K K + + TA+ ++ T
Sbjct: 615 STGSSSSNSNQQRFPSAPIQPEEGPQPQPKPQLKIKIKQEQLVATRKSSRTATAIVTAAT 674
Query: 83 GLPFKEKKEISVSARKLAATLWE-INDLTPS--RIKKESMKSNKERD 126
++++ + RK+A L + + +L +IKKE ++ K ++
Sbjct: 675 ASSHQQQQLRQTTCRKMANKLEDRMGELVKPKIKIKKEVIEEQKVKE 721
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.128 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 314,900,082
Number of Sequences: 2790947
Number of extensions: 11846711
Number of successful extensions: 36654
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 50
Number of HSP's that attempted gapping in prelim test: 36588
Number of HSP's gapped (non-prelim): 83
length of query: 199
length of database: 848,049,833
effective HSP length: 121
effective length of query: 78
effective length of database: 510,345,246
effective search space: 39806929188
effective search space used: 39806929188
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 71 (32.0 bits)
Medicago: description of AC146332.8


