

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146331.8 + phase: 2 /pseudo
(279 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
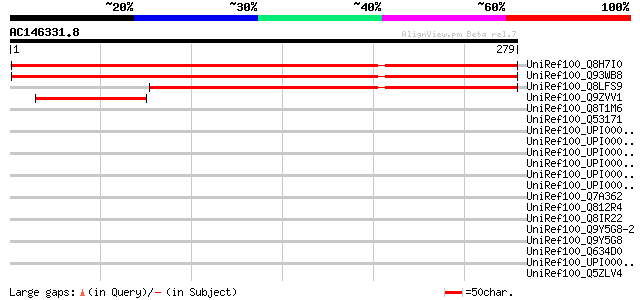
UniRef100_Q8H7I0 Hypothetical protein [Arabidopsis thaliana] 277 2e-73
UniRef100_Q93WB8 Hypothetical protein At3g13410 [Arabidopsis tha... 276 4e-73
UniRef100_Q8LFS9 Hypothetical protein [Arabidopsis thaliana] 201 2e-50
UniRef100_Q9ZVV1 T5A14.4 protein [Arabidopsis thaliana] 69 2e-10
UniRef100_Q8T1M6 Hypothetical protein [Dictyostelium discoideum] 38 0.25
UniRef100_Q53171 Methyl accepting chemotaxis protein [Rhodobacte... 36 1.3
UniRef100_UPI00002B8934 UPI00002B8934 UniRef100 entry 35 1.6
UniRef100_UPI00001D12C3 Protocadherin-T1 [Rattus norvegicus] 35 2.2
UniRef100_UPI00004314D3 UPI00004314D3 UniRef100 entry 35 2.2
UniRef100_UPI0000366374 UPI0000366374 UniRef100 entry 35 2.2
UniRef100_UPI0000366373 UPI0000366373 UniRef100 entry 35 2.2
UniRef100_UPI00000135A6 UPI00000135A6 UniRef100 entry 35 2.2
UniRef100_Q7A362 Surface protein SasA [Staphylococcus aureus] 35 2.2
UniRef100_Q812R4 Hypothetical Membrane Associated Protein [Bacil... 35 2.2
UniRef100_Q8IR22 CG32580-PA [Drosophila melanogaster] 35 2.2
UniRef100_Q9Y5G8-2 Splice isoform 2 of Q9Y5G8 [Homo sapiens] 35 2.2
UniRef100_Q9Y5G8 Protocadherin gamma A5 precursor [Homo sapiens] 35 2.2
UniRef100_Q634D0 Hypothetical protein [Bacillus cereus] 35 2.8
UniRef100_UPI00003CB97D UPI00003CB97D UniRef100 entry 34 3.7
UniRef100_Q5ZLV4 Hypothetical protein [Gallus gallus] 34 4.8
>UniRef100_Q8H7I0 Hypothetical protein [Arabidopsis thaliana]
Length = 321
Score = 277 bits (708), Expect = 2e-73
Identities = 144/281 (51%), Positives = 198/281 (70%), Gaps = 6/281 (2%)
Query: 2 SENG-LKESVNYQVISLKDLAKSVLSEAGWSNFLCKGKKFQDPLDLALLFVGGELQSSDL 60
S NG L E+VNYQV+S KDL SV ++ GWSNFLC KK + P+D+AL+F+G EL SSD+
Sbjct: 42 SANGELDEAVNYQVMSAKDLVGSVFTQGGWSNFLCSEKKLEQPVDVALVFIGRELLSSDV 101
Query: 61 SLNKHADSALSYLLKDSFVRSNTSMAFPYVSASEDVNLEDSLVSGFAEACGDDLGIGNVA 120
S +++D AL L + F SN S+AFPY++A E+ +E+ L+SG EAC +++G+ N+
Sbjct: 102 SSKRNSDPALVNTLNNLFTASNFSLAFPYIAAPEEERMENLLLSGLKEACPNNVGVSNIV 161
Query: 121 FLGSCSMGNGNREETAALQSVQAYLTKRKEESHKGKTDLVVFCN-GPQASKNVDSTKSEG 179
F SC + +G ++ + LQS + +L R+E +G+TDLVV C+ G +++ + SE
Sbjct: 162 FSDSCFVEDGTIQKLSDLQSFKDHLLARRETRKEGETDLVVLCSEGSESNSQAGQSHSER 221
Query: 180 EVLSELISSVEESGAKYAVLYVSDISRSIQYPSYRDLQRFLAESTTGNGSTN-STACDGV 238
E + EL+SSVE+SG+KY LYVSD Y SY+ LQRFLAE+ GN + +T CD +
Sbjct: 222 ESILELVSSVEQSGSKYTALYVSD---PYWYTSYKTLQRFLAETAKGNSTPEIATGCDEL 278
Query: 239 CQLKSSLLEGLLVGIVLLIILISGLCCMMGIDSPTRFEAPQ 279
C+ KSSLLEG+LVGIV L+ILISGLCCM GID+PTRFE PQ
Sbjct: 279 CKFKSSLLEGILVGIVFLLILISGLCCMAGIDTPTRFETPQ 319
>UniRef100_Q93WB8 Hypothetical protein At3g13410 [Arabidopsis thaliana]
Length = 321
Score = 276 bits (706), Expect = 4e-73
Identities = 144/281 (51%), Positives = 197/281 (69%), Gaps = 6/281 (2%)
Query: 2 SENG-LKESVNYQVISLKDLAKSVLSEAGWSNFLCKGKKFQDPLDLALLFVGGELQSSDL 60
S NG L E+VNYQV+S KDL SV ++ GWSNFLC KK + P+D+AL+F+G EL SSD+
Sbjct: 42 SANGELDEAVNYQVMSAKDLVGSVFTQGGWSNFLCSEKKLEQPVDVALVFIGRELLSSDV 101
Query: 61 SLNKHADSALSYLLKDSFVRSNTSMAFPYVSASEDVNLEDSLVSGFAEACGDDLGIGNVA 120
S +++D AL L + F SN S+AFPY++A E+ +E+ L+SG EAC +++G+ N+
Sbjct: 102 SSKRNSDPALVNTLNNLFTASNFSLAFPYIAAPEEERMENLLLSGLKEACPNNVGVSNIV 161
Query: 121 FLGSCSMGNGNREETAALQSVQAYLTKRKEESHKGKTDLVVFCN-GPQASKNVDSTKSEG 179
F SC + +G ++ + LQS + +L R+E +G+TDLVV C+ G +++ + SE
Sbjct: 162 FSDSCFVEDGTIQKLSDLQSFKDHLLARRETRKEGETDLVVLCSEGSESNSQAGQSHSER 221
Query: 180 EVLSELISSVEESGAKYAVLYVSDISRSIQYPSYRDLQRFLAESTTGNGSTN-STACDGV 238
E EL+SSVE+SG+KY LYVSD Y SY+ LQRFLAE+ GN + +T CD +
Sbjct: 222 ESFLELVSSVEQSGSKYTALYVSD---PYWYTSYKTLQRFLAETAKGNSTPEIATGCDEL 278
Query: 239 CQLKSSLLEGLLVGIVLLIILISGLCCMMGIDSPTRFEAPQ 279
C+ KSSLLEG+LVGIV L+ILISGLCCM GID+PTRFE PQ
Sbjct: 279 CKFKSSLLEGILVGIVFLLILISGLCCMAGIDTPTRFETPQ 319
>UniRef100_Q8LFS9 Hypothetical protein [Arabidopsis thaliana]
Length = 203
Score = 201 bits (511), Expect = 2e-50
Identities = 103/204 (50%), Positives = 144/204 (70%), Gaps = 5/204 (2%)
Query: 78 FVRSNTSMAFPYVSASEDVNLEDSLVSGFAEACGDDLGIGNVAFLGSCSMGNGNREETAA 137
F SN S+AFPY++A E+ +E+ L+SG EAC +++G+ N+ F SC + +G ++ +
Sbjct: 1 FTASNFSLAFPYIAAPEEERMENLLLSGLKEACPNNVGVSNIVFSDSCFVEDGTIQKLSD 60
Query: 138 LQSVQAYLTKRKEESHKGKTDLVVFCN-GPQASKNVDSTKSEGEVLSELISSVEESGAKY 196
LQS + +L R+E +G+TDLVV C+ G +++ ++SE E + EL+SSVE+SG+KY
Sbjct: 61 LQSFKDHLLARRETRKEGETDLVVLCSEGSESNSQSGQSRSERESILELVSSVEQSGSKY 120
Query: 197 AVLYVSDISRSIQYPSYRDLQRFLAESTTGNGSTN-STACDGVCQLKSSLLEGLLVGIVL 255
LYVSD Y SY+ LQRFLAE+ GN + +T CD +C+ KSSLLEG+LVGIV
Sbjct: 121 TALYVSD---PYWYTSYKTLQRFLAETAKGNSTPEIATGCDELCKFKSSLLEGILVGIVF 177
Query: 256 LIILISGLCCMMGIDSPTRFEAPQ 279
L+ILISGLCCM GID+PTRFE PQ
Sbjct: 178 LLILISGLCCMAGIDTPTRFETPQ 201
>UniRef100_Q9ZVV1 T5A14.4 protein [Arabidopsis thaliana]
Length = 76
Score = 68.6 bits (166), Expect = 2e-10
Identities = 33/61 (54%), Positives = 43/61 (70%)
Query: 15 ISLKDLAKSVLSEAGWSNFLCKGKKFQDPLDLALLFVGGELQSSDLSLNKHADSALSYLL 74
+S KDL SV + GWSNFLC KK Q P+D+AL+F+G EL SSD+S N+++D L L
Sbjct: 1 MSAKDLVDSVFTLGGWSNFLCSEKKLQQPVDVALVFIGRELLSSDVSSNQNSDPVLVNTL 60
Query: 75 K 75
K
Sbjct: 61 K 61
>UniRef100_Q8T1M6 Hypothetical protein [Dictyostelium discoideum]
Length = 295
Score = 38.1 bits (87), Expect = 0.25
Identities = 20/61 (32%), Positives = 35/61 (56%), Gaps = 4/61 (6%)
Query: 223 STTGNGSTNSTACDGVCQ----LKSSLLEGLLVGIVLLIILISGLCCMMGIDSPTRFEAP 278
S G+GS++S + + Q + +L L+ +LL IL +G+CC+ + P R+EAP
Sbjct: 231 SQQGSGSSSSGSNEPTKQHFTYVTGPVLSAYLIISILLAILFTGICCISDLQVPDRYEAP 290
Query: 279 Q 279
+
Sbjct: 291 K 291
>UniRef100_Q53171 Methyl accepting chemotaxis protein [Rhodobacter sphaeroides]
Length = 792
Score = 35.8 bits (81), Expect = 1.3
Identities = 27/102 (26%), Positives = 46/102 (44%), Gaps = 2/102 (1%)
Query: 128 GNGNREETAALQSVQAYLTKRKEESHKGKTDLVVFCNGPQASKNVDSTKSEGEVLSELIS 187
G+ R V+A L +R E+ + TDL++ +G Q + VD G+ L +++S
Sbjct: 631 GDAGRGFAVVASEVRA-LAQRSSEAAREITDLILK-SGNQVRRGVDLVGKTGDALKQIVS 688
Query: 188 SVEESGAKYAVLYVSDISRSIQYPSYRDLQRFLAESTTGNGS 229
SV E + + VS +S+ L +ST N +
Sbjct: 689 SVSEISTLVSDIAVSSRQQSVSLAEINCAVNNLDQSTQQNAA 730
>UniRef100_UPI00002B8934 UPI00002B8934 UniRef100 entry
Length = 265
Score = 35.4 bits (80), Expect = 1.6
Identities = 24/64 (37%), Positives = 35/64 (54%), Gaps = 3/64 (4%)
Query: 1 VSENGLKESVNYQVISLKDLAKSVLSEAGWSNFLCKGKKFQDPLDLALLFVGGELQSSDL 60
+S+ GLK +N +VIS+KD+ K V E +N K K D L+ VG + + L
Sbjct: 23 LSKQGLKFQLNSKVISVKDIGKQVQIEYQDNN---KNNKNMISADKVLIAVGRKPYTEGL 79
Query: 61 SLNK 64
+LNK
Sbjct: 80 NLNK 83
>UniRef100_UPI00001D12C3 Protocadherin-T1 [Rattus norvegicus]
Length = 3987
Score = 35.0 bits (79), Expect = 2.2
Identities = 29/99 (29%), Positives = 41/99 (41%), Gaps = 15/99 (15%)
Query: 143 AYLTKRKEESHKGKTDLVVFCNGPQASKNVDSTKS------EGEVLSELISSVEESGAKY 196
+YL E + +G + V P KN T S +G LS +S +G Y
Sbjct: 2086 SYLAYIPENNARGASIFSVTAQDPDTDKNAQITYSLAEDTLQGVPLSSFVSINSNTGVLY 2145
Query: 197 AVLYVSDISRSIQYPSYRDLQRFLAESTTGNG--STNST 233
A+ S Y +RDLQ + +GN S+NST
Sbjct: 2146 ALC-------SFDYEQFRDLQLHITARDSGNPPLSSNST 2177
>UniRef100_UPI00004314D3 UPI00004314D3 UniRef100 entry
Length = 516
Score = 35.0 bits (79), Expect = 2.2
Identities = 33/102 (32%), Positives = 48/102 (46%), Gaps = 11/102 (10%)
Query: 152 SHKGKTDLVVFCNGPQASKNVDSTKSEGEVLSELISSVEES--GAKYAVLYVSDISRSIQ 209
S +GK + V K T+SEG+ L E +++ +S Y + +SD R
Sbjct: 240 SRQGKINFVDLAGSEMTKK----TQSEGKTLEEA-NNINKSLMVLGYCIASLSDGKRKGG 294
Query: 210 YPSYRD--LQRFLAESTTGNGSTNSTACDGVCQLKSSLLEGL 249
+ YRD L + LA+S GNG T AC + KS+ E L
Sbjct: 295 HIPYRDSKLTKLLADSLAGNGVTLMIAC--ISPAKSNASETL 334
>UniRef100_UPI0000366374 UPI0000366374 UniRef100 entry
Length = 3221
Score = 35.0 bits (79), Expect = 2.2
Identities = 27/79 (34%), Positives = 37/79 (46%), Gaps = 1/79 (1%)
Query: 34 LCKGKKFQDPLDLALLFVGGELQSSDLSLNK-HADSALSYLLKDSFVRSNTSMAFPYVSA 92
L + K PLD ++ + +LQ SL + H +S LLK + S TS A P VSA
Sbjct: 1211 LSQNKPISPPLDPSIPKIDAKLQDLTSSLARYHLTGQISPLLKKDMLVSQTSTASPVVSA 1270
Query: 93 SEDVNLEDSLVSGFAEACG 111
V +D +G E G
Sbjct: 1271 VSPVINQDMYGTGPIEVKG 1289
>UniRef100_UPI0000366373 UPI0000366373 UniRef100 entry
Length = 3250
Score = 35.0 bits (79), Expect = 2.2
Identities = 27/79 (34%), Positives = 37/79 (46%), Gaps = 1/79 (1%)
Query: 34 LCKGKKFQDPLDLALLFVGGELQSSDLSLNK-HADSALSYLLKDSFVRSNTSMAFPYVSA 92
L + K PLD ++ + +LQ SL + H +S LLK + S TS A P VSA
Sbjct: 1211 LSQNKPISPPLDPSIPKIDAKLQDLTSSLARYHLTGQISPLLKKDMLVSQTSTASPVVSA 1270
Query: 93 SEDVNLEDSLVSGFAEACG 111
V +D +G E G
Sbjct: 1271 VSPVINQDMYGTGPIEVKG 1289
>UniRef100_UPI00000135A6 UPI00000135A6 UniRef100 entry
Length = 16223
Score = 35.0 bits (79), Expect = 2.2
Identities = 29/110 (26%), Positives = 49/110 (44%), Gaps = 5/110 (4%)
Query: 88 PYVSASEDVNLEDSLVSGFAEACGDDLGIGNVAFLGSCSMGNGNRE--ETAALQSVQAYL 145
P++S ++ E S G C L IG + + C++ E ++ S+ YL
Sbjct: 10452 PFLSTISNIKFEQSSFHGCDCTC--QLEIGKDSCICECNLKESTSERLKSVTASSLYIYL 10509
Query: 146 TKRKEESHKGKTDLVVFCNGPQASKNVDSTKSEGEVLSELISSVEESGAK 195
+ +H TDL N + + +KS ++ +E ISS +ES AK
Sbjct: 10510 PQSSTTAHSLLTDLSAE-NQTNQEETSELSKSLPQLTTEEISSFQESSAK 10558
>UniRef100_Q7A362 Surface protein SasA [Staphylococcus aureus]
Length = 2271
Score = 35.0 bits (79), Expect = 2.2
Identities = 39/194 (20%), Positives = 75/194 (38%), Gaps = 12/194 (6%)
Query: 57 SSDLSLNKHADSALSYLLKDSFVRSNTSMAFPYVSASEDVNLEDSLVSGFAEACGDDLGI 116
+S S+++ A ++ S + S S ++ S S+ +L S +E+ I
Sbjct: 1120 ASSESISQSASTSTSGSVSTSTSLSTSNSERTSTSVSDSTSLSTSESDSISESTSTSDSI 1179
Query: 117 GNVAFLG---SCSMGNGNREETAALQSVQAYLTKRKEESHKGKTDLVVFCNGPQASKNVD 173
S S+ N + QS A+L++ ES T V + +++ D
Sbjct: 1180 SEAISASESTSISLSESNSTSDSESQSASAFLSESLSESTSESTSESVSSSTSESTSLSD 1239
Query: 174 STKSEGEVLSELISSVEESGAKYAVLYVSD---------ISRSIQYPSYRDLQRFLAEST 224
ST G + L +S S + +S+ +S S+ + L + ST
Sbjct: 1240 STSESGSTSTSLSNSTSGSASISTSTSISESTSTFKSESVSTSLSMSTSTSLSNSTSLST 1299
Query: 225 TGNGSTNSTACDGV 238
+ + ST+ + D +
Sbjct: 1300 SLSDSTSDSKSDSL 1313
>UniRef100_Q812R4 Hypothetical Membrane Associated Protein [Bacillus cereus]
Length = 215
Score = 35.0 bits (79), Expect = 2.2
Identities = 21/61 (34%), Positives = 33/61 (53%), Gaps = 1/61 (1%)
Query: 17 LKDLAKSVLSEAGWSNFLCKGKKFQDPLDLAL-LFVGGELQSSDLSLNKHADSALSYLLK 75
++D+ ++L +G + K K QD L L L + GE+QSS L + +H D L LK
Sbjct: 129 IRDVEYAILEPSGKLSVFQKQKNKQDTLQFTLPLIIDGEIQSSHLQMIEHTDEWLIEKLK 188
Query: 76 D 76
+
Sbjct: 189 N 189
>UniRef100_Q8IR22 CG32580-PA [Drosophila melanogaster]
Length = 16223
Score = 35.0 bits (79), Expect = 2.2
Identities = 29/110 (26%), Positives = 49/110 (44%), Gaps = 5/110 (4%)
Query: 88 PYVSASEDVNLEDSLVSGFAEACGDDLGIGNVAFLGSCSMGNGNRE--ETAALQSVQAYL 145
P++S ++ E S G C L IG + + C++ E ++ S+ YL
Sbjct: 10452 PFLSTISNIKFEQSSFHGCDCTC--QLEIGKDSCICECNLKESTSERLKSVTASSLYIYL 10509
Query: 146 TKRKEESHKGKTDLVVFCNGPQASKNVDSTKSEGEVLSELISSVEESGAK 195
+ +H TDL N + + +KS ++ +E ISS +ES AK
Sbjct: 10510 PQSSTTAHSLLTDLSAE-NQTNQEETSELSKSLPQLTTEEISSFQESSAK 10558
>UniRef100_Q9Y5G8-2 Splice isoform 2 of Q9Y5G8 [Homo sapiens]
Length = 813
Score = 35.0 bits (79), Expect = 2.2
Identities = 27/96 (28%), Positives = 42/96 (43%), Gaps = 13/96 (13%)
Query: 143 AYLTKRKEESHKGKTDLVVFCNGPQASKNVDSTKS------EGEVLSELISSVEESGAKY 196
+Y T E + +G + V + P + N T S +G LS +S ++G Y
Sbjct: 456 SYSTSVTENNPRGVSIFSVTAHDPDSGDNARVTYSLAEDTFQGAPLSSYVSINSDTGVLY 515
Query: 197 AVLYVSDISRSIQYPSYRDLQRFLAESTTGNGSTNS 232
A+ RS Y RDLQ ++ S +GN +S
Sbjct: 516 AL-------RSFDYEQLRDLQLWVTASDSGNPPLSS 544
>UniRef100_Q9Y5G8 Protocadherin gamma A5 precursor [Homo sapiens]
Length = 931
Score = 35.0 bits (79), Expect = 2.2
Identities = 27/96 (28%), Positives = 42/96 (43%), Gaps = 13/96 (13%)
Query: 143 AYLTKRKEESHKGKTDLVVFCNGPQASKNVDSTKS------EGEVLSELISSVEESGAKY 196
+Y T E + +G + V + P + N T S +G LS +S ++G Y
Sbjct: 456 SYSTSVTENNPRGVSIFSVTAHDPDSGDNARVTYSLAEDTFQGAPLSSYVSINSDTGVLY 515
Query: 197 AVLYVSDISRSIQYPSYRDLQRFLAESTTGNGSTNS 232
A+ RS Y RDLQ ++ S +GN +S
Sbjct: 516 AL-------RSFDYEQLRDLQLWVTASDSGNPPLSS 544
>UniRef100_Q634D0 Hypothetical protein [Bacillus cereus]
Length = 215
Score = 34.7 bits (78), Expect = 2.8
Identities = 22/72 (30%), Positives = 36/72 (49%), Gaps = 1/72 (1%)
Query: 17 LKDLAKSVLSEAGWSNFLCKGKKFQDPLDLAL-LFVGGELQSSDLSLNKHADSALSYLLK 75
++D+ ++L +G + K K QD L L L + GE+QSS L + +H D L LK
Sbjct: 129 IRDVEYAILEPSGKLSVFQKQKGGQDALQFTLPLIIDGEIQSSHLQMIEHTDEWLIGKLK 188
Query: 76 DSFVRSNTSMAF 87
+ T + +
Sbjct: 189 NLGYNDTTQILY 200
>UniRef100_UPI00003CB97D UPI00003CB97D UniRef100 entry
Length = 215
Score = 34.3 bits (77), Expect = 3.7
Identities = 21/61 (34%), Positives = 33/61 (53%), Gaps = 1/61 (1%)
Query: 17 LKDLAKSVLSEAGWSNFLCKGKKFQDPLDLAL-LFVGGELQSSDLSLNKHADSALSYLLK 75
++D+ ++L +G + K K QD L L L + GE+QSS L + +H D L LK
Sbjct: 129 IRDVEYAILEPSGKLSVFQKQKGGQDALQFTLPLIIDGEIQSSHLQMIEHTDEWLVEKLK 188
Query: 76 D 76
+
Sbjct: 189 N 189
>UniRef100_Q5ZLV4 Hypothetical protein [Gallus gallus]
Length = 796
Score = 33.9 bits (76), Expect = 4.8
Identities = 15/61 (24%), Positives = 31/61 (50%)
Query: 138 LQSVQAYLTKRKEESHKGKTDLVVFCNGPQASKNVDSTKSEGEVLSELISSVEESGAKYA 197
+ V+ + KRKE +GKT+ V C Q + +D E + ++++ + ++E +
Sbjct: 697 MMGVEVFKAKRKEGESEGKTEEEVQCRPAQTEEGMDVEDKERDAVTKMEAEIDEESPRSP 756
Query: 198 V 198
V
Sbjct: 757 V 757
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.131 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 418,278,796
Number of Sequences: 2790947
Number of extensions: 15903368
Number of successful extensions: 37124
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 29
Number of HSP's that attempted gapping in prelim test: 37102
Number of HSP's gapped (non-prelim): 43
length of query: 279
length of database: 848,049,833
effective HSP length: 126
effective length of query: 153
effective length of database: 496,390,511
effective search space: 75947748183
effective search space used: 75947748183
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 74 (33.1 bits)
Medicago: description of AC146331.8


