

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145767.1 - phase: 0
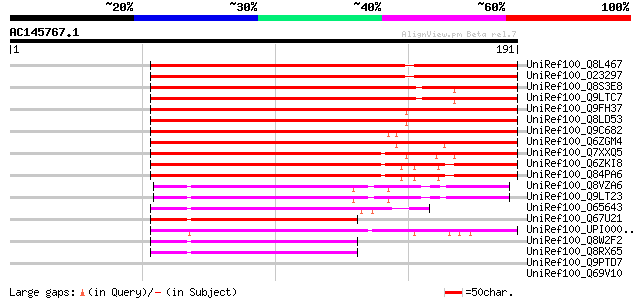
(191 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8L467 AT4g14410/dl3245w [Arabidopsis thaliana] 198 7e-50
UniRef100_O23297 Hypothetical protein dl3245w [Arabidopsis thali... 198 7e-50
UniRef100_Q8S3E8 Putative bHLH transcription factor [Arabidopsis... 160 1e-38
UniRef100_Q9LTC7 Emb|CAB10220.1 [Arabidopsis thaliana] 160 1e-38
UniRef100_Q9FH37 Arabidopsis thaliana genomic DNA, chromosome 5,... 144 2e-33
UniRef100_Q8LD53 BHLH transcription factor, putative [Arabidopsi... 142 5e-33
UniRef100_Q9C682 BHLH transcription factor, putative [Arabidopsi... 132 4e-30
UniRef100_Q6ZGM4 Putative bHLH protein [Oryza sativa] 132 5e-30
UniRef100_Q7XXQ5 Hypothetical protein [Oryza sativa] 129 5e-29
UniRef100_Q6ZKI8 Helix-loop-helix-like protein [Oryza sativa] 122 4e-27
UniRef100_Q84PA6 Helix-loop-helix-like protein [Oryza sativa] 122 4e-27
UniRef100_Q8VZA6 Hypothetical protein At3g19860; MPN9.10 [Arabid... 68 1e-10
UniRef100_Q9LT23 Emb|CAA18500.1 [Arabidopsis thaliana] 68 1e-10
UniRef100_O65643 Putative Myc-type transcription factor [Arabido... 62 8e-09
UniRef100_Q67U21 Basic helix-loop-helix-like protein [Oryza sativa] 60 2e-08
UniRef100_UPI00002C6501 UPI00002C6501 UniRef100 entry 59 5e-08
UniRef100_Q8W2F2 Putative transcription factor BHLH11 [Arabidops... 57 3e-07
UniRef100_Q8RX65 AT4g36060/T19K4_190 [Arabidopsis thaliana] 57 3e-07
UniRef100_Q9PTD7 Cingulin [Xenopus laevis] 45 0.001
UniRef100_Q69V10 Amelogenin-like protein [Oryza sativa] 44 0.002
>UniRef100_Q8L467 AT4g14410/dl3245w [Arabidopsis thaliana]
Length = 283
Score = 198 bits (503), Expect = 7e-50
Identities = 100/138 (72%), Positives = 117/138 (84%), Gaps = 3/138 (2%)
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
RF DLS+VLEPGR +TDKPAILDDAIR+L+QL+ EA +L+E+N+KLLEEIK LKAEKNE
Sbjct: 149 RFMDLSSVLEPGRTPKTDKPAILDDAIRILNQLRDEALKLEETNQKLLEEIKSLKAEKNE 208
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAGFMPPPPMAAYQASVNKMAVYPNYGYIPMWHYL 173
LREEKLVLKADKEK E+QLKSM +GF+P P A + NKMAVYP+YGY+PMWHY+
Sbjct: 209 LREEKLVLKADKEKTEQQLKSMTAPSSGFIPHIPAA---FNHNKMAVYPSYGYMPMWHYM 265
Query: 174 PQSARDTSQDHELRPPAA 191
PQS RDTS+D ELRPPAA
Sbjct: 266 PQSVRDTSRDQELRPPAA 283
>UniRef100_O23297 Hypothetical protein dl3245w [Arabidopsis thaliana]
Length = 277
Score = 198 bits (503), Expect = 7e-50
Identities = 100/138 (72%), Positives = 117/138 (84%), Gaps = 3/138 (2%)
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
RF DLS+VLEPGR +TDKPAILDDAIR+L+QL+ EA +L+E+N+KLLEEIK LKAEKNE
Sbjct: 143 RFMDLSSVLEPGRTPKTDKPAILDDAIRILNQLRDEALKLEETNQKLLEEIKSLKAEKNE 202
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAGFMPPPPMAAYQASVNKMAVYPNYGYIPMWHYL 173
LREEKLVLKADKEK E+QLKSM +GF+P P A + NKMAVYP+YGY+PMWHY+
Sbjct: 203 LREEKLVLKADKEKTEQQLKSMTAPSSGFIPHIPAA---FNHNKMAVYPSYGYMPMWHYM 259
Query: 174 PQSARDTSQDHELRPPAA 191
PQS RDTS+D ELRPPAA
Sbjct: 260 PQSVRDTSRDQELRPPAA 277
>UniRef100_Q8S3E8 Putative bHLH transcription factor [Arabidopsis thaliana]
Length = 291
Score = 160 bits (406), Expect = 1e-38
Identities = 89/142 (62%), Positives = 109/142 (76%), Gaps = 6/142 (4%)
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
+F DLS+VLEPGR +TDK AILDDAIRV++QL+ EA EL+E+N+KLLEEIK LKA+KNE
Sbjct: 152 KFMDLSSVLEPGRTPKTDKSAILDDAIRVVNQLRGEAHELQETNQKLLEEIKSLKADKNE 211
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAGFMPPPPMAAYQASVNKMAVYPNYGY----IPM 169
LREEKLVLKA+KEK+E+QLKSM V GFMP AA+ + +KMAV YGY +PM
Sbjct: 212 LREEKLVLKAEKEKMEQQLKSMVVPSPGFMPSQHPAAFHS--HKMAVAYPYGYYPPNMPM 269
Query: 170 WHYLPQSARDTSQDHELRPPAA 191
W LP + RDTS+D + PP A
Sbjct: 270 WSPLPPADRDTSRDLKNLPPVA 291
>UniRef100_Q9LTC7 Emb|CAB10220.1 [Arabidopsis thaliana]
Length = 320
Score = 160 bits (406), Expect = 1e-38
Identities = 89/142 (62%), Positives = 109/142 (76%), Gaps = 6/142 (4%)
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
+F DLS+VLEPGR +TDK AILDDAIRV++QL+ EA EL+E+N+KLLEEIK LKA+KNE
Sbjct: 181 KFMDLSSVLEPGRTPKTDKSAILDDAIRVVNQLRGEAHELQETNQKLLEEIKSLKADKNE 240
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAGFMPPPPMAAYQASVNKMAVYPNYGY----IPM 169
LREEKLVLKA+KEK+E+QLKSM V GFMP AA+ + +KMAV YGY +PM
Sbjct: 241 LREEKLVLKAEKEKMEQQLKSMVVPSPGFMPSQHPAAFHS--HKMAVAYPYGYYPPNMPM 298
Query: 170 WHYLPQSARDTSQDHELRPPAA 191
W LP + RDTS+D + PP A
Sbjct: 299 WSPLPPADRDTSRDLKNLPPVA 320
>UniRef100_Q9FH37 Arabidopsis thaliana genomic DNA, chromosome 5, TAC clone:K5F14
[Arabidopsis thaliana]
Length = 234
Score = 144 bits (362), Expect = 2e-33
Identities = 75/145 (51%), Positives = 99/145 (67%), Gaps = 7/145 (4%)
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
+F +L A+LEPG P +TDK AIL DA+R+++QL+ EAQ+LK+SN L ++IK LK EKNE
Sbjct: 90 KFMELGAILEPGNPPKTDKAAILVDAVRMVTQLRGEAQKLKDSNSSLQDKIKELKTEKNE 149
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAGFMPPPPM-------AAYQASVNKMAVYPNYGY 166
LR+EK LK +KEK+E+QLK+M F P PPM A QA NKM +Y
Sbjct: 150 LRDEKQRLKTEKEKLEQQLKAMNAPQPSFFPAPPMMPTAFASAQGQAPGNKMVPIISYPG 209
Query: 167 IPMWHYLPQSARDTSQDHELRPPAA 191
+ MW ++P ++ DTSQDH LRPP A
Sbjct: 210 VAMWQFMPPASVDTSQDHVLRPPVA 234
>UniRef100_Q8LD53 BHLH transcription factor, putative [Arabidopsis thaliana]
Length = 234
Score = 142 bits (358), Expect = 5e-33
Identities = 74/145 (51%), Positives = 99/145 (68%), Gaps = 7/145 (4%)
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
+F +L A+LEPG P +TDK AIL DA+R+++QL+ EAQ+LK+SN L ++IK LK EKNE
Sbjct: 90 KFMELGAILEPGNPPKTDKAAILVDAVRMVTQLRGEAQKLKDSNSSLQDKIKELKTEKNE 149
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAGFMPPPPM-------AAYQASVNKMAVYPNYGY 166
LR+EK LK +KEK+E+QLK++ F P PPM A QA NKM +Y
Sbjct: 150 LRDEKQRLKTEKEKLEQQLKAINAPQPSFFPAPPMMPTAFASAQGQAPGNKMVPIISYPG 209
Query: 167 IPMWHYLPQSARDTSQDHELRPPAA 191
+ MW ++P ++ DTSQDH LRPP A
Sbjct: 210 VAMWQFMPPASVDTSQDHVLRPPVA 234
>UniRef100_Q9C682 BHLH transcription factor, putative [Arabidopsis thaliana]
Length = 226
Score = 132 bits (333), Expect = 4e-30
Identities = 70/142 (49%), Positives = 99/142 (69%), Gaps = 4/142 (2%)
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
+F +LS+VLEPGR +TDK AI++DAIR+++Q + EAQ+LK+ N L E+IK LK EKNE
Sbjct: 85 KFTELSSVLEPGRTPKTDKVAIINDAIRMVNQARDEAQKLKDLNSSLQEKIKELKDEKNE 144
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAG---FMP-PPPMAAYQASVNKMAVYPNYGYIPM 169
LR+EK LK +KE+I++QLK++ P F+P P ++ QA +K+ + Y M
Sbjct: 145 LRDEKQKLKVEKERIDQQLKAIKTQPQPQPCFLPNPQTLSQAQAPGSKLVPFTTYPGFAM 204
Query: 170 WHYLPQSARDTSQDHELRPPAA 191
W ++P +A DTSQDH LRPP A
Sbjct: 205 WQFMPPAAVDTSQDHVLRPPVA 226
>UniRef100_Q6ZGM4 Putative bHLH protein [Oryza sativa]
Length = 236
Score = 132 bits (332), Expect = 5e-30
Identities = 73/152 (48%), Positives = 102/152 (67%), Gaps = 14/152 (9%)
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
RF +LS+V+ P + + DK IL DA R+L++L+ EA++LKESNEKL E IK LK EKNE
Sbjct: 85 RFLELSSVINPDKQAKLDKANILSDAARLLAELRGEAEKLKESNEKLRETIKDLKVEKNE 144
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAGFMP-------------PPPMAAYQASVNKMAV 160
LR+EK+ LKA+KE++E+Q+K++ V+P GF+P PP + YQA NK A
Sbjct: 145 LRDEKVTLKAEKERLEQQVKALSVAPTGFVPHLPHPAAFHPAAFPPFIPPYQALGNKNAP 204
Query: 161 YP-NYGYIPMWHYLPQSARDTSQDHELRPPAA 191
P + + MW +LP +A DT+QD +L PP A
Sbjct: 205 TPAAFQGMAMWQWLPPTAVDTTQDPKLWPPNA 236
>UniRef100_Q7XXQ5 Hypothetical protein [Oryza sativa]
Length = 256
Score = 129 bits (323), Expect = 5e-29
Identities = 73/147 (49%), Positives = 102/147 (68%), Gaps = 10/147 (6%)
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
RF +L AVLEPG+ + DK +IL+DAIRV+++L++EAQ+LKESNE L E+IK LKAEKNE
Sbjct: 111 RFLELGAVLEPGKTPKMDKSSILNDAIRVMAELRSEAQKLKESNESLQEKIKELKAEKNE 170
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAGFMPPPPM---AAYQASVNKMA----VYPNYGY 166
LR+EK LKA+KE +E+Q+K + P+ F+P PP+ +A+ A + A + P GY
Sbjct: 171 LRDEKQKLKAEKESLEQQIKFLNARPS-FVPHPPVIPASAFTAPQGQAAGQKLMMPVIGY 229
Query: 167 --IPMWHYLPQSARDTSQDHELRPPAA 191
PMW ++P S DT+ D + PP A
Sbjct: 230 PGFPMWQFMPPSDVDTTDDTKSCPPVA 256
>UniRef100_Q6ZKI8 Helix-loop-helix-like protein [Oryza sativa]
Length = 253
Score = 122 bits (307), Expect = 4e-27
Identities = 71/157 (45%), Positives = 101/157 (64%), Gaps = 23/157 (14%)
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
RF +L LEPG+PV++DK AIL DA R++ QL+ EA++LK++NE L ++IK LKAEK+E
Sbjct: 101 RFLELGTTLEPGKPVKSDKAAILSDATRMVIQLRAEAKQLKDTNESLEDKIKELKAEKDE 160
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAGFMPPP----------PMAAY-----QASVNKM 158
LR+EK LK +KE +E+Q+K + +PA +MP P P+A + QA+ K+
Sbjct: 161 LRDEKQKLKVEKETLEQQVKILTATPA-YMPHPTLMPAPYPQAPLAPFHHAQGQAAGQKL 219
Query: 159 AV----YPNYGYIPMWHYLPQSARDTSQDHELRPPAA 191
+ YP Y PMW ++P S DTS+D E PP A
Sbjct: 220 MMPFVGYPGY---PMWQFMPPSEVDTSKDSEACPPVA 253
>UniRef100_Q84PA6 Helix-loop-helix-like protein [Oryza sativa]
Length = 216
Score = 122 bits (307), Expect = 4e-27
Identities = 71/157 (45%), Positives = 101/157 (64%), Gaps = 23/157 (14%)
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
RF +L LEPG+PV++DK AIL DA R++ QL+ EA++LK++NE L ++IK LKAEK+E
Sbjct: 64 RFLELGTTLEPGKPVKSDKAAILSDATRMVIQLRAEAKQLKDTNESLEDKIKELKAEKDE 123
Query: 114 LREEKLVLKADKEKIEKQLKSMPVSPAGFMPPP----------PMAAY-----QASVNKM 158
LR+EK LK +KE +E+Q+K + +PA +MP P P+A + QA+ K+
Sbjct: 124 LRDEKQKLKVEKETLEQQVKILTATPA-YMPHPTLMPAPYPQAPLAPFHHAQGQAAGQKL 182
Query: 159 AV----YPNYGYIPMWHYLPQSARDTSQDHELRPPAA 191
+ YP Y PMW ++P S DTS+D E PP A
Sbjct: 183 MMPFVGYPGY---PMWQFMPPSEVDTSKDSEACPPVA 216
>UniRef100_Q8VZA6 Hypothetical protein At3g19860; MPN9.10 [Arabidopsis thaliana]
Length = 284
Score = 67.8 bits (164), Expect = 1e-10
Identities = 52/145 (35%), Positives = 76/145 (51%), Gaps = 19/145 (13%)
Query: 55 FCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNEL 114
F +L VL+P RP + DK IL D +++L +L +E +LK L +E + L EKN+L
Sbjct: 25 FVELGNVLDPERP-KNDKATILTDTVQLLKELTSEVNKLKSEYTALTDESRELTQEKNDL 83
Query: 115 REEKLVLKADKEKI----EKQLKSMPVSPAG-------FMPPPPMAAYQASVNKMAVYPN 163
REEK LK+D E + +++L+SM SP G M PPP Y + A+ P
Sbjct: 84 REEKTSLKSDIENLNLQYQQRLRSM--SPWGAAMDHTVMMAPPPSFPYPMPI---AMPP- 137
Query: 164 YGYIPMWHYLPQSARDTSQDHELRP 188
G IPM +P +Q+ + P
Sbjct: 138 -GSIPMHPSMPSYTYFGNQNPSMIP 161
>UniRef100_Q9LT23 Emb|CAA18500.1 [Arabidopsis thaliana]
Length = 337
Score = 67.8 bits (164), Expect = 1e-10
Identities = 52/145 (35%), Positives = 76/145 (51%), Gaps = 19/145 (13%)
Query: 55 FCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNEL 114
F +L VL+P RP + DK IL D +++L +L +E +LK L +E + L EKN+L
Sbjct: 78 FVELGNVLDPERP-KNDKATILTDTVQLLKELTSEVNKLKSEYTALTDESRELTQEKNDL 136
Query: 115 REEKLVLKADKEKI----EKQLKSMPVSPAG-------FMPPPPMAAYQASVNKMAVYPN 163
REEK LK+D E + +++L+SM SP G M PPP Y + A+ P
Sbjct: 137 REEKTSLKSDIENLNLQYQQRLRSM--SPWGAAMDHTVMMAPPPSFPYPMPI---AMPP- 190
Query: 164 YGYIPMWHYLPQSARDTSQDHELRP 188
G IPM +P +Q+ + P
Sbjct: 191 -GSIPMHPSMPSYTYFGNQNPSMIP 214
>UniRef100_O65643 Putative Myc-type transcription factor [Arabidopsis thaliana]
Length = 272
Score = 62.0 bits (149), Expect = 8e-09
Identities = 41/110 (37%), Positives = 62/110 (56%), Gaps = 12/110 (10%)
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
+F +L L+P RP ++DK ++L D I++L + + LK E L +E + L EK+E
Sbjct: 63 QFLELGNALDPNRP-KSDKASVLTDTIQMLKDVMNQVDRLKAEYETLSQESRELIQEKSE 121
Query: 114 LREEKLVLKADKEKIEKQ----LKSM-PVSPAGFMPPPPMAAYQASVNKM 158
LREEK LK+D E + Q +K+M P + F+P Y ASVN +
Sbjct: 122 LREEKATLKSDIEILNAQYQHRIKTMVPWGQSSFIP------YSASVNPL 165
>UniRef100_Q67U21 Basic helix-loop-helix-like protein [Oryza sativa]
Length = 343
Score = 60.5 bits (145), Expect = 2e-08
Identities = 33/78 (42%), Positives = 48/78 (61%), Gaps = 1/78 (1%)
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
+F +L + L+P RP R DK IL DAI++L L ++ +LK L EE + L EKNE
Sbjct: 58 QFQELGSTLDPDRP-RNDKATILSDAIQMLKDLTSQVNKLKAEYTSLSEEARELTQEKNE 116
Query: 114 LREEKLVLKADKEKIEKQ 131
LR+EK+ LK + + + Q
Sbjct: 117 LRDEKVSLKFEVDNLNTQ 134
>UniRef100_UPI00002C6501 UPI00002C6501 UniRef100 entry
Length = 177
Score = 59.3 bits (142), Expect = 5e-08
Identities = 49/164 (29%), Positives = 78/164 (46%), Gaps = 27/164 (16%)
Query: 54 RFCDLSAVLEPGR--PVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEK 111
+F +LSA+L+P P +T+K I+ +A RV+ QL+ E +L + E + E LK E
Sbjct: 15 KFMELSALLDPTAVGPFKTEKATIVTEAARVIKQLRAELAKLSATLESMQESNLALKKET 74
Query: 112 NELREEKLVLKADKEKIEKQLKSMPVSPAGFMPPPPMAAY------------------QA 153
+ + +K L+ DK K++ QL +S F PPP A + Q
Sbjct: 75 SCVAADKAALQQDKAKLQHQLYYF-MSSMPFASPPPGAVFASITGAYHLPRGTSALLAQQ 133
Query: 154 SVNKMAVYPNY---GYIP-MWHY--LPQSARDTSQDHELRPPAA 191
+ N + PN+ G P MW++ L + +D +LR P A
Sbjct: 134 NDNAEKLAPNHLAAGSTPAMWNFPRLIVQSTTAEEDAKLRAPVA 177
>UniRef100_Q8W2F2 Putative transcription factor BHLH11 [Arabidopsis thaliana]
Length = 286
Score = 57.0 bits (136), Expect = 3e-07
Identities = 31/78 (39%), Positives = 47/78 (59%), Gaps = 1/78 (1%)
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
+F +L L+P RP ++DK ++L D I++L + + LK E L +E + L EK+E
Sbjct: 63 QFLELGNALDPNRP-KSDKASVLTDTIQMLKDVMNQVDRLKAEYETLSQESRELIQEKSE 121
Query: 114 LREEKLVLKADKEKIEKQ 131
LREEK LK+D E + Q
Sbjct: 122 LREEKATLKSDIEILNAQ 139
>UniRef100_Q8RX65 AT4g36060/T19K4_190 [Arabidopsis thaliana]
Length = 268
Score = 57.0 bits (136), Expect = 3e-07
Identities = 31/78 (39%), Positives = 47/78 (59%), Gaps = 1/78 (1%)
Query: 54 RFCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNE 113
+F +L L+P RP ++DK ++L D I++L + + LK E L +E + L EK+E
Sbjct: 45 QFLELGNALDPNRP-KSDKASVLTDTIQMLKDVMNQVDRLKAEYETLSQESRELIQEKSE 103
Query: 114 LREEKLVLKADKEKIEKQ 131
LREEK LK+D E + Q
Sbjct: 104 LREEKATLKSDIEILNAQ 121
>UniRef100_Q9PTD7 Cingulin [Xenopus laevis]
Length = 1360
Score = 45.1 bits (105), Expect = 0.001
Identities = 24/68 (35%), Positives = 42/68 (61%)
Query: 68 VRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNELREEKLVLKADKEK 127
V+ D + LD+A R L +L E +EL+E ++ +++ LK KNEL E+K +L +K
Sbjct: 956 VKRDLESKLDEAQRSLKRLSLEYEELQECYQEEMKQKDHLKKTKNELEEQKRLLDKSMDK 1015
Query: 128 IEKQLKSM 135
+ ++L +M
Sbjct: 1016 LTRELDNM 1023
Score = 32.7 bits (73), Expect = 5.1
Identities = 16/60 (26%), Positives = 36/60 (59%)
Query: 76 LDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNELREEKLVLKADKEKIEKQLKSM 135
+ +A + + EA+ + ++ ++ EE++ LK EL+ EK ++ DK+ I ++L+S+
Sbjct: 1049 IGEAQKQAKEKTAEAERHQFNSSRMQEEVQKLKLALQELQVEKETVELDKQMISQRLQSL 1108
>UniRef100_Q69V10 Amelogenin-like protein [Oryza sativa]
Length = 265
Score = 44.3 bits (103), Expect = 0.002
Identities = 29/118 (24%), Positives = 58/118 (48%), Gaps = 9/118 (7%)
Query: 55 FCDLSAVLEPGRPVRTDKPAILDDAIRVLSQLKTEAQELKESNEKLLEEIKCLKAEKNEL 114
F +L +LEP R K +L + R+L L ++ + L++ N L E + E+NEL
Sbjct: 55 FNELGNLLEPDRQ-NNGKACVLGETTRILKDLLSQVESLRKENSSLKNESHYVALERNEL 113
Query: 115 REEKLVLKADKEKIEKQLKSMPVSPAGFMPPPPMAAYQASVNKMAV-YPNYGYIPMWH 171
++ +L+ + +++ +L++ M P+ ++ + + V YP G P+ H
Sbjct: 114 HDDNSMLRTEILELQNELRTR-------MEGNPVWSHVNTRPALRVPYPTTGVFPVQH 164
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.136 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 317,314,296
Number of Sequences: 2790947
Number of extensions: 12484060
Number of successful extensions: 104926
Number of sequences better than 10.0: 1032
Number of HSP's better than 10.0 without gapping: 490
Number of HSP's successfully gapped in prelim test: 547
Number of HSP's that attempted gapping in prelim test: 100774
Number of HSP's gapped (non-prelim): 4215
length of query: 191
length of database: 848,049,833
effective HSP length: 120
effective length of query: 71
effective length of database: 513,136,193
effective search space: 36432669703
effective search space used: 36432669703
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 71 (32.0 bits)
Medicago: description of AC145767.1


