

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145512.9 - phase: 0
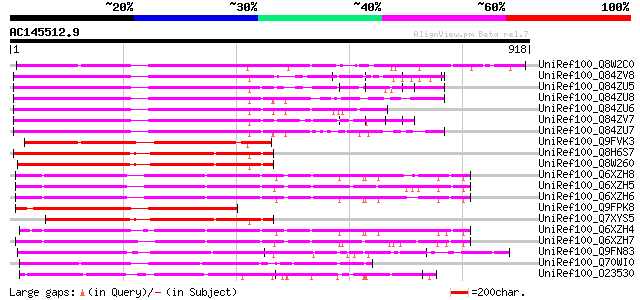
(918 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8W2C0 Functional candidate resistance protein KR1 [Gl... 625 e-177
UniRef100_Q84ZV8 R 3 protein [Glycine max] 474 e-132
UniRef100_Q84ZU5 R 8 protein [Glycine max] 472 e-131
UniRef100_Q84ZU8 R 10 protein [Glycine max] 467 e-130
UniRef100_Q84ZU6 R 1 protein [Glycine max] 443 e-122
UniRef100_Q84ZV7 R 12 protein [Glycine max] 432 e-119
UniRef100_Q84ZU7 R 5 protein [Glycine max] 421 e-116
UniRef100_Q9FVK3 Resistance protein MG63 [Glycine max] 411 e-113
UniRef100_Q8H6S7 Resistance protein KR3 [Glycine max] 357 1e-96
UniRef100_Q8W260 Functional resistance protein KR2 [Glycine max] 352 3e-95
UniRef100_Q6XZH8 Nematode resistance protein [Solanum tuberosum] 337 1e-90
UniRef100_Q6XZH5 Nematode resistance-like protein [Solanum tuber... 333 1e-89
UniRef100_Q6XZH6 Nematode resistance-like protein [Solanum tuber... 332 2e-89
UniRef100_Q9FPK8 Putative resistance protein [Glycine max] 332 4e-89
UniRef100_Q7XYS5 Disease resistance-like protein KR7 [Glycine max] 330 2e-88
UniRef100_Q6XZH4 Nematode resistance-like protein [Solanum tuber... 327 1e-87
UniRef100_Q6XZH7 Nematode resistance-like protein [Solanum tuber... 318 4e-85
UniRef100_Q9FN83 Disease resistance protein-like [Arabidopsis th... 313 2e-83
UniRef100_Q70WI0 Putative resistance gene analogue protein [Lens... 290 2e-76
UniRef100_O23530 Disease resistance RPP5 like protein [Arabidops... 270 2e-70
>UniRef100_Q8W2C0 Functional candidate resistance protein KR1 [Glycine max]
Length = 1124
Score = 625 bits (1611), Expect = e-177
Identities = 406/964 (42%), Positives = 548/964 (56%), Gaps = 139/964 (14%)
Query: 12 YVHEFIEKIVEEVLRNIKPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKVGILGIGG 71
Y +EFI++IVE V + I L D+ VG+E + Q V LL+VGSDD +H +GI G+GG
Sbjct: 169 YEYEFIQRIVELVSKKINRAPLHVADYPVGLESRIQEVKALLDVGSDDVVHMLGIHGLGG 228
Query: 72 IGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEITSVGE 131
+GKTTLA VYNSI FE CFL+ VRE S K+GL +LQ+ LLS + GE ++ V +
Sbjct: 229 VGKTTLAAAVYNSIADHFEALCFLQNVRETSKKHGLQHLQRNLLSEMAGED--KLIGVKQ 286
Query: 132 GISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISSGVER 191
GISI++ RL +KKVLL+LDDVDK+EQL+A+AG + F GSRVIITTR K LL GVER
Sbjct: 287 GISIIEHRLRQKKVLLILDDVDKREQLQALAGRPDLFGPGSRVIITTRDKQLLACHGVER 346
Query: 192 IYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLKNDEGYA 251
YEV LN+E A +L+ WK K + K D Y
Sbjct: 347 TYEVNELNEEYALELLNWKAFKLE-----------------------------KVDPFYK 377
Query: 252 NVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYDSLQE 311
+VL R YASG PLALEVIGS+ S K IEQ ALDRYKR+P+ IQ L++SYD+L+E
Sbjct: 378 DVLNRAATYASGLPLALEVIGSNLSGKNIEQWISALDRYKRIPNKEIQEILKVSYDALEE 437
Query: 312 EEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESGNVTLHDLV 371
+E+ +FLDIACCFK + L V+ ILHAHHGH MK I VLVEK LIKI+ G VTLHDL+
Sbjct: 438 DEQSIFLDIACCFKKYDLAEVQDILHAHHGHCMKHHIGVLVEKSLIKISLDGYVTLHDLI 497
Query: 372 EDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIEMIHFD-----GVIEVQWD 426
EDMGKEIVR+E+P++PGKRSRLW DI+QVLEEN GTS I +I + +E+QWD
Sbjct: 498 EDMGKEIVRKESPQEPGKRSRLWLPTDIVQVLEENKGTSHIGIICMNFYSSFEEVEIQWD 557
Query: 427 GEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLE---CRNHM*ELDLSYCINLESFSH 483
G+AFKKM+NLKTLI + FS+ PKH P SLRVLE +H D L F+
Sbjct: 558 GDAFKKMKNLKTLIIRSG-HFSKGPKHFPKSLRVLEWWRYPSHYFPYDFQ-MEKLAIFNL 615
Query: 484 VVDGFGDK------------LRTMSVRGCFKLKSIPPLK-LDSLETMDLSCCFRLESFPL 530
GF + L +++ C L IP + + L+ + C L +
Sbjct: 616 PDCGFTSRELAAMLKKKFVNLTSLNFDSCQHLTLIPDVSCVPHLQKLSFKDCDNLYAIHP 675
Query: 531 VVDGILGKIKTLNVESCHNLRSIPPLKLDSLEKLDISYCGSLESFPQVEGRFLGKLKTLN 590
V G L K++ L+ E C L++ PP+KL SLE+L + +C SLE+FP++ LGK++ +
Sbjct: 676 SV-GFLEKLRILDAEGCSRLKNFPPIKLTSLEQLKLGFCHSLENFPEI----LGKMENIT 730
Query: 591 VKSCRIMISIPTLMLSLLEELDLSYCLNLENFPLVVDGFLGKLKTLSAKSCRNLRSIPSL 650
ELDL ++ FPL L +L+T+ L P
Sbjct: 731 -------------------ELDLEQ-TPVKKFPLSFQN-LTRLETV-------LLCFPRN 762
Query: 651 KLDLLETLDLSNCVSLESLPLVVD----GFLGKL--------KTLLVTNCHNLKSIPPLK 698
+ + + LSN ++ P +++ G+ G L + + +T N++ +
Sbjct: 763 QANGCTGIFLSNICPMQESPELINVIGVGWEGCLFRKEDEGAENVSLTTSSNVQFLDLRN 822
Query: 699 C---DALETLDLSCCYSLQSFSLVADR-------------LWKLVLDDCKELQEIKVIPP 742
C D + L C ++ +L + L L L+ C+ L+EI+ IPP
Sbjct: 823 CNLSDDFFPIALPCFANVMELNLSGNNFTVIPECIKECRFLTTLYLNYCERLREIRGIPP 882
Query: 743 CLRMLSAVNCTSLTSSCTSKLLNQELHKAGNTWFCLPRVPKIPEWFDHKYEAGLSISFWF 802
L+ A C SLTSSC S LL+QELH+AG T+F LP KIPEWFD + + ISFWF
Sbjct: 883 NLKYFYAEECLSLTSSCRSMLLSQELHEAGRTFFYLPGA-KIPEWFDFQ-TSEFPISFWF 940
Query: 803 RNKFPAIALCVV--------SPLTWDGSRRHSIRVIINGNTFIYTDGLKMGTKSPLNMYH 854
RNKFPAIA+C + S W +VIINGN ++ + + +G+
Sbjct: 941 RNKFPAIAICHIIKRVAEFSSSRGWTFRPNIRTKVIINGNANLF-NSVVLGSDCTC---- 995
Query: 855 LHLFHMKMENFNDNMEKVLFENMWNHAEV-------DFGLPFQKSGIHVSKEKSNMKDIR 907
LF ++ E DN+++ L EN WNHAEV F F K+G+HV K++SNM+DIR
Sbjct: 996 --LFDLRGERVTDNLDEALLENEWNHAEVTCPGFTFTFAPTFIKTGLHVLKQESNMEDIR 1053
Query: 908 FTNP 911
F++P
Sbjct: 1054 FSDP 1057
>UniRef100_Q84ZV8 R 3 protein [Glycine max]
Length = 897
Score = 474 bits (1220), Expect = e-132
Identities = 320/816 (39%), Positives = 429/816 (52%), Gaps = 133/816 (16%)
Query: 8 DRDGYVHEFIEKIVEEVLRNIKPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKVGIL 67
D D Y ++FI+ IVE+V R I L D+ VG+ Q V LL+VGS D +H +GI
Sbjct: 157 DGDAYEYKFIQSIVEQVSREINRTPLHVADYPVGLGSQVIEVRKLLDVGSHDVVHIIGIH 216
Query: 68 GIGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEIT 127
G+GG+GKTTLAL VYN I F+ SCFL+ VRE S+K+GL +LQ I+LS + GEK+ +T
Sbjct: 217 GMGGLGKTTLALAVYNLIALHFDESCFLQNVREESNKHGLKHLQSIILSKLLGEKDINLT 276
Query: 128 SVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISS 187
S EG S+++ RL KKVLL+LDDVDK++QLKAI G +WF GSRVIITTR K++L
Sbjct: 277 SWQEGASMIQHRLQRKKVLLILDDVDKRQQLKAIVGRPDWFGPGSRVIITTRDKHILKYH 336
Query: 188 GVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLKND 247
VER YEVK LN A L+ W K + KND
Sbjct: 337 EVERTYEVKVLNQSAALQLLKWNAFKRE-----------------------------KND 367
Query: 248 EGYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYD 307
Y +VL RVV YASG PLALE+IGS+ KT+ + A++ YKR+P + I L++S+D
Sbjct: 368 PSYEDVLNRVVTYASGLPLALEIIGSNLFGKTVAEWESAMEHYKRIPSDEILEILKVSFD 427
Query: 308 SLQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESGNVTL 367
+L EE+K VFLDIACC KG KLT VE +L + + MK I+VLV+K L K+ G V +
Sbjct: 428 ALGEEQKNVFLDIACCLKGCKLTEVEHMLRGLYDNCMKHHIDVLVDKSLTKVRH-GIVEM 486
Query: 368 HDLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIEMIHFDGVIE----- 422
HDL++DMG+EI RQ +PE+PGKR RLW +DIIQVL+ NTGTSKIE+I+ D I
Sbjct: 487 HDLIQDMGREIERQRSPEEPGKRKRLWSPKDIIQVLKHNTGTSKIEIIYVDFSISDKEET 546
Query: 423 VQWDGEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLECRNHM*ELDLSYCINLESFS 482
V+W+ AF KMENLK LI N F S+ P + P LRVLE + S C+
Sbjct: 547 VEWNENAFMKMENLKILIIRNGKF-SKGPNYFPQGLRVLEWHRYP-----SNCLP----- 595
Query: 483 HVVDGFGDKLRTMSVRGCFKLKSIPPLKLDSLETMDLSCCFRLESFPLVVDGILGKIKTL 542
+ P+ L + D S + SF LG + L
Sbjct: 596 ---------------------SNFDPINLVICKLPDSS----MTSFEFHGSSKLGHLTVL 630
Query: 543 NVESCHNLRSIPPLK-LDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMISIP 601
+ C L IP + L +L +L +C SL + G FL KLK LN CR + S P
Sbjct: 631 KFDWCKFLTQIPDVSDLPNLRELSFQWCESLVAVDDSIG-FLNKLKKLNAYGCRKLTSFP 689
Query: 602 TLMLSLLEELDLSYCLNLENFPLVVDGFLGKLKTLSAKSCRNLRSIPSLKLDLLETLDLS 661
L L+ LE L+LS+C +LE FP + LG+++ + +L +P +L
Sbjct: 690 PLHLTSLETLELSHCSSLEYFPEI----LGEMENIERL---DLHGLPIKELPF----SFQ 738
Query: 662 NCVSLESLPLVVDGF---------LGKLKTLLVTNCHNLK-------------------- 692
N + L+ L + G + KL NC+ +
Sbjct: 739 NLIGLQQLSMFGCGIVQLRCSLAMMPKLSAFKFVNCNRWQWVESEEAEEKVGSIISSEAR 798
Query: 693 ----SIPPLKCDALETLDLS----------CCYSLQSFSLVADR------LWKLVLDDCK 732
S C+ + L+ S +F+++ + L L + CK
Sbjct: 799 FWTHSFSAKNCNLCDDFFLTGFKKFAHVGYLNLSRNNFTILPEFFKELQFLGSLNVSHCK 858
Query: 733 ELQEIKVIPPCLRMLSAVNCTSLTSSCTSKLLNQEL 768
LQEI+ IP LR+ +A NC SLTSS S LLNQ L
Sbjct: 859 HLQEIRGIPQNLRLFNARNCASLTSSSKSMLLNQVL 894
Score = 65.9 bits (159), Expect = 5e-09
Identities = 50/131 (38%), Positives = 67/131 (50%), Gaps = 14/131 (10%)
Query: 571 SLESFPQVEGRFLGKLKTLNVKSCRIMISIPTLM-LSLLEELDLSYCLNLENFPLVVD-- 627
S+ SF LG L L C+ + IP + L L EL +C +L + VD
Sbjct: 612 SMTSFEFHGSSKLGHLTVLKFDWCKFLTQIPDVSDLPNLRELSFQWCESL----VAVDDS 667
Query: 628 -GFLGKLKTLSAKSCRNLRSIPSLKLDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVT 686
GFL KLK L+A CR L S P L L LETL+LS+C SLE P + LG+++ +
Sbjct: 668 IGFLNKLKKLNAYGCRKLTSFPPLHLTSLETLELSHCSSLEYFPEI----LGEMENIERL 723
Query: 687 NCHNL--KSIP 695
+ H L K +P
Sbjct: 724 DLHGLPIKELP 734
Score = 54.7 bits (130), Expect = 1e-05
Identities = 47/142 (33%), Positives = 64/142 (44%), Gaps = 9/142 (6%)
Query: 630 LGKLKTLSAKSCRNLRSIPSLK-LDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNC 688
LG L L C+ L IP + L L L C SL ++ + GFL KLK L C
Sbjct: 624 LGHLTVLKFDWCKFLTQIPDVSDLPNLRELSFQWCESLVAVDDSI-GFLNKLKKLNAYGC 682
Query: 689 HNLKSIPPLKCDALETLDLSCCYSLQSFSLVADRLWKLVLDDCKELQEIKVIPPC----- 743
L S PPL +LETL+LS C SL+ F + + + D L IK +P
Sbjct: 683 RKLTSFPPLHLTSLETLELSHCSSLEYFPEILGEMENIERLDLHGL-PIKELPFSFQNLI 741
Query: 744 -LRMLSAVNCTSLTSSCTSKLL 764
L+ LS C + C+ ++
Sbjct: 742 GLQQLSMFGCGIVQLRCSLAMM 763
>UniRef100_Q84ZU5 R 8 protein [Glycine max]
Length = 892
Score = 472 bits (1215), Expect = e-131
Identities = 321/804 (39%), Positives = 439/804 (53%), Gaps = 114/804 (14%)
Query: 8 DRDGYVHEFIEKIVEEVLRNIKPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKVGIL 67
D D Y +EFI IVEE+ R +L D+ VG+E + V LL+VGS D +H +GI
Sbjct: 157 DGDSYEYEFIGSIVEEISRKFSRASLHVADYPVGLESEVTEVMKLLDVGSHDVVHIIGIH 216
Query: 68 GIGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEIT 127
G+GG+GKTTLAL V+N I F+ SCFL+ VRE S+K+GL +LQ ILLS + GEK+ +T
Sbjct: 217 GMGGLGKTTLALAVHNFIALHFDESCFLQNVREESNKHGLKHLQSILLSKLLGEKDITLT 276
Query: 128 SVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISS 187
S EG S+++ RL KKVLL+LDDVDK++QLKAI G +WF GSRVIITTR K+LL
Sbjct: 277 SWQEGASMIQHRLQRKKVLLILDDVDKRQQLKAIVGRPDWFGPGSRVIITTRDKHLLKYH 336
Query: 188 GVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLKND 247
VER YEVK LN A L+ W K + K D
Sbjct: 337 EVERTYEVKVLNQSAALQLLTWNAFKRE-----------------------------KID 367
Query: 248 EGYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYD 307
Y +VL RVV YASG PLALEVIGS+ KT+ + A++ YKR+P + IQ L++S+D
Sbjct: 368 PSYEDVLNRVVTYASGLPLALEVIGSNLFEKTVAEWESAMEHYKRIPSDEIQEILKVSFD 427
Query: 308 SLQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESGNVTL 367
+L EE+K VFLDIACCFKG++ T V+ IL +G+ K I VLVEK L+K++ V +
Sbjct: 428 ALGEEQKNVFLDIACCFKGYEWTEVDNILRDLYGNCTKHHIGVLVEKSLVKVSCCDTVEM 487
Query: 368 HDLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIEMIHFDGVIE----- 422
HD+++DMG+EI RQ +PE+PGK RL +DIIQVL++NTGTSKIE+I D I
Sbjct: 488 HDMIQDMGREIERQRSPEEPGKCKRLLLPKDIIQVLKDNTGTSKIEIICLDFSISDKEET 547
Query: 423 VQWDGEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLECRNHM*ELDLSYCI--NLES 480
V+W+ AF KM+NLK LI N FS+ P + P LRVLE + S C+ N +
Sbjct: 548 VEWNENAFMKMKNLKILIIRN-CKFSKGPNYFPEGLRVLEWHRYP-----SNCLPSNFDP 601
Query: 481 FSHVVDGFGDKLRTMSVRGCFKLKSIPPLKLDSLETMDLSCCFRLESFPLVVDGILGKIK 540
+ V+ C +P + S E S LG +
Sbjct: 602 INLVI--------------C----KLPDSSITSFEFHGSS-------------KKLGHLT 630
Query: 541 TLNVESCHNLRSIPPLK-LDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMIS 599
LN + C L IP + L +L++L ++C SL + G FL KLKTL+ CR + S
Sbjct: 631 VLNFDRCEFLTKIPDVSDLPNLKELSFNWCESLVAVDDSIG-FLNKLKTLSAYGCRKLTS 689
Query: 600 IPTLMLSLLEELDLSYCLNLENFPLVVDGFLGKLKTLSAKSCRNL--RSIPSLKLDLLET 657
P L L+ LE L+L C +LE FP + LG++K ++ + +L + +P +L+
Sbjct: 690 FPPLNLTSLETLNLGGCSSLEYFPEI----LGEMKNITVLALHDLPIKELPFSFQNLIGL 745
Query: 658 LDL--SNC------VSLESLPLVVD--------------------GFLGKLKTLLVTNCH 689
L L +C SL ++P + + +G + + T+C+
Sbjct: 746 LFLWLDSCGIVQLRCSLATMPKLCEFCITDSCNRWQWVESEEGEEKVVGSILSFEATDCN 805
Query: 690 ---NLKSIPPLKCDALETLDL--SCCYSLQSFSLVADRLWKLVLDDCKELQEIKVIPPCL 744
+ I + + L+L + L F L LV+ DCK LQEI+ +PP L
Sbjct: 806 LCDDFFFIGSKRFAHVGYLNLPGNNFTILPEFFKELQFLTTLVVHDCKHLQEIRGLPPNL 865
Query: 745 RMLSAVNCTSLTSSCTSKLLNQEL 768
+ A NC SLTSS S LLNQ L
Sbjct: 866 KHFDARNCASLTSSSKSMLLNQVL 889
Score = 73.9 bits (180), Expect = 2e-11
Identities = 51/119 (42%), Positives = 65/119 (53%), Gaps = 14/119 (11%)
Query: 583 LGKLKTLNVKSCRIMISIPTLM-LSLLEELDLSYCLNLENFPLVVD---GFLGKLKTLSA 638
LG L LN C + IP + L L+EL ++C +L + VD GFL KLKTLSA
Sbjct: 626 LGHLTVLNFDRCEFLTKIPDVSDLPNLKELSFNWCESL----VAVDDSIGFLNKLKTLSA 681
Query: 639 KSCRNLRSIPSLKLDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNL--KSIP 695
CR L S P L L LETL+L C SLE P + LG++K + V H+L K +P
Sbjct: 682 YGCRKLTSFPPLNLTSLETLNLGGCSSLEYFPEI----LGEMKNITVLALHDLPIKELP 736
Score = 56.6 bits (135), Expect = 3e-06
Identities = 37/88 (42%), Positives = 47/88 (53%), Gaps = 2/88 (2%)
Query: 630 LGKLKTLSAKSCRNLRSIPSLK-LDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNC 688
LG L L+ C L IP + L L+ L + C SL ++ + GFL KLKTL C
Sbjct: 626 LGHLTVLNFDRCEFLTKIPDVSDLPNLKELSFNWCESLVAVDDSI-GFLNKLKTLSAYGC 684
Query: 689 HNLKSIPPLKCDALETLDLSCCYSLQSF 716
L S PPL +LETL+L C SL+ F
Sbjct: 685 RKLTSFPPLNLTSLETLNLGGCSSLEYF 712
>UniRef100_Q84ZU8 R 10 protein [Glycine max]
Length = 901
Score = 467 bits (1202), Expect = e-130
Identities = 324/828 (39%), Positives = 422/828 (50%), Gaps = 158/828 (19%)
Query: 8 DRDGYVHEFIEKIVEEVLRNIKPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKVGIL 67
D D Y ++FI+ IVE+V R I L D+ VG+ Q V LL+VGS D +H +GI
Sbjct: 157 DGDAYEYKFIQSIVEQVSREINRAPLHVADYPVGLGSQVIEVRKLLDVGSHDVVHIIGIH 216
Query: 68 GIGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEIT 127
G+GG+GKTTLAL VYN I F+ SCFL+ VRE S+K+GL +LQ ILLS + GEK+ +T
Sbjct: 217 GMGGLGKTTLALAVYNLIALHFDESCFLQNVREESNKHGLKHLQSILLSKLLGEKDITLT 276
Query: 128 SVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISS 187
S EG S+++ RL KKVLL+LDDVDK+EQLKAI G +WF GSRVIITTR K+LL
Sbjct: 277 SWQEGASMIQHRLQRKKVLLILDDVDKREQLKAIVGRPDWFGPGSRVIITTRDKHLLKYH 336
Query: 188 GVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLKND 247
VER YEVK LN A L+ W K + K D
Sbjct: 337 EVERTYEVKVLNQSAALQLLKWNAFKRE-----------------------------KID 367
Query: 248 EGYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYD 307
Y +VL RVV YASG PLALEVIGS+ KT+ + A++ YKR+P + I L++S+D
Sbjct: 368 PSYEDVLNRVVTYASGLPLALEVIGSNLFGKTVAEWESAMEHYKRIPSDEILEILKVSFD 427
Query: 308 SLQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESG--NV 365
+L EE+K VFLDIACCF+G+K T V+ IL A +G+ K I VLVEK LIK+N G V
Sbjct: 428 ALGEEQKNVFLDIACCFRGYKWTEVDDILRALYGNCKKHHIGVLVEKSLIKLNCYGTDTV 487
Query: 366 TLHDLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIEMIHFDGVIE--- 422
+HDL++DM +EI R+ +P++PGK RLW +DIIQV ++NTGTSKIE+I D I
Sbjct: 488 EMHDLIQDMAREIERKRSPQEPGKCKRLWLPKDIIQVFKDNTGTSKIEIICLDSSISDKE 547
Query: 423 --VQWDGEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLE------------------ 462
V+W+ AF KMENLK LI ND FS+ P + P LRVLE
Sbjct: 548 ETVEWNENAFMKMENLKILIIRNDK-FSKGPNYFPEGLRVLEWHRYPSNCLPSNFHPNNL 606
Query: 463 --CR-----------------NHM*ELDLSYCINLESFSHVVD----------------- 486
C+ H+ L C L V D
Sbjct: 607 VICKLPDSCMTSFEFHGPSKFGHLTVLKFDNCKFLTQIPDVSDLPNLRELSFEECESLVA 666
Query: 487 -----GFGDKLRTMSVRGCFKLKSIPPLKLDSLETMDLSCCFRLESFPLVVDGILGKIKT 541
GF +KL+ +S GC KLKS PPL L SL+T++LS C LE FP ++
Sbjct: 667 VDDSIGFLNKLKKLSAYGCSKLKSFPPLNLTSLQTLELSQCSSLEYFPEIIG-------- 718
Query: 542 LNVESCHNLRSIPPLKLDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIM-ISI 600
+ +I L L L ++S+ SF + G L+ L ++SC I+ +
Sbjct: 719 -------EMENIKHLFLYGLPIKELSF-----SFQNLIG-----LRWLTLRSCGIVKLPC 761
Query: 601 PTLMLSLLEELDLSYCLNLENFPLVVDGFLGKLKTLSAKSCRNLRSIPSLKLDLLETLDL 660
M+ L E + YC + S + + + SIPS K D
Sbjct: 762 SLAMMPELFEFHMEYCNRWQ-------------WVESEEGEKKVGSIPSSKAHRFSAKDC 808
Query: 661 SNCVSLESLPLVVDGFLGKLKTLLVTNCHNLKSIPPLKCDALETLDLSCCYSLQSFSLVA 720
+ C D FL KT NL + L F
Sbjct: 809 NLC---------DDFFLTGFKTFARVGHLNLSG--------------NNFTILPEFFKEL 845
Query: 721 DRLWKLVLDDCKELQEIKVIPPCLRMLSAVNCTSLTSSCTSKLLNQEL 768
L L++ DC+ LQEI+ +PP L A NC SLTSS + LLNQ L
Sbjct: 846 QLLRSLMVSDCEHLQEIRGLPPNLEYFDARNCASLTSSSKNMLLNQVL 893
>UniRef100_Q84ZU6 R 1 protein [Glycine max]
Length = 902
Score = 443 bits (1140), Expect = e-122
Identities = 306/763 (40%), Positives = 398/763 (52%), Gaps = 139/763 (18%)
Query: 8 DRDGYVHEFIEKIVEEVLRNIKPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKVGIL 67
D D Y ++FI IVEEV R I L D+ VG+ Q V LL+VGSDD +H +GI
Sbjct: 157 DGDAYEYKFIGNIVEEVSRKINCAPLHVADYPVGLGSQVIEVMKLLDVGSDDLVHIIGIH 216
Query: 68 GIGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEIT 127
G+GG+GKTTLAL VYN I F+ SCFL+ VRE S+K+GL + Q ILLS + GEK+ +T
Sbjct: 217 GMGGLGKTTLALAVYNFIALHFDESCFLQNVREESNKHGLKHFQSILLSKLLGEKDITLT 276
Query: 128 SVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISS 187
S EG S+++ RL KKVLL+LDDVDK+EQL+AI G S+WF GSRVIITTR K+LL
Sbjct: 277 SWQEGASMIQHRLRRKKVLLILDDVDKREQLEAIVGRSDWFGPGSRVIITTRDKHLLKYH 336
Query: 188 GVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLKND 247
VER YEVK LN A L+ W K + K D
Sbjct: 337 EVERTYEVKVLNHNAALQLLTWNAFKRE-----------------------------KID 367
Query: 248 EGYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYD 307
Y +VL RVV YASG PLALEVIGS KT+ + A++ YKR+P + I L++S+D
Sbjct: 368 PIYDDVLNRVVTYASGLPLALEVIGSDLFGKTVAEWESAVEHYKRIPSDEILKILKVSFD 427
Query: 308 SLQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKIN--ESGNV 365
+L EE+K VFLDIACCFKG+K T V+ IL A +G+ K I VLVEK LIK+N +SG V
Sbjct: 428 ALGEEQKNVFLDIACCFKGYKWTEVDDILRAFYGNCKKHHIGVLVEKSLIKLNCYDSGTV 487
Query: 366 TLHDLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIEMIHFDGVIE--- 422
+HDL++DMG+EI RQ +PE+P K RLW +DI QVL+ NTGTSKIE+I D I
Sbjct: 488 EMHDLIQDMGREIERQRSPEEPWKCKRLWSPKDIFQVLKHNTGTSKIEIICLDFSISDKE 547
Query: 423 --VQWDGEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLECRN--------------- 465
V+W+ AF KMENLK LI N FS+ P + P L VLE
Sbjct: 548 ETVEWNENAFMKMENLKILIIRNGK-FSKGPNYFPEGLTVLEWHRYPSNCLPYNFHPNNL 606
Query: 466 ----------------------HM*ELDLSYCINLESFSHVVD----------------- 486
H+ L+ C L V D
Sbjct: 607 LICKLPDSSITSFELHGPSKFWHLTVLNFDQCEFLTQIPDVSDLPNLKELSFDWCESLIA 666
Query: 487 -----GFGDKLRTMSVRGCFKLKSIPPLKLDSLETMDLSCCFRLESFPLVVDGILGKIKT 541
GF +KL+ +S GC KL+S PPL L SLET+ LS C LE FP ++ G + IK
Sbjct: 667 VDDSIGFLNKLKKLSAYGCRKLRSFPPLNLTSLETLQLSGCSSLEYFPEIL-GEMENIKA 725
Query: 542 LNVESCHNLRSIP--PLKLDSLEKLDISYCG------SLESFPQVE-------------- 579
L+++ ++ +P L L +L ++ CG SL P++
Sbjct: 726 LDLDGL-PIKELPFSFQNLIGLCRLTLNSCGIIQLPCSLAMMPELSVFRIENCNRWHWVE 784
Query: 580 --------GRFLGKLK----TLNVKSCRIMISIPTLMLSLLEELDLSYCLNLENFPLVVD 627
G + + +N C + + +E LDLS NF ++ +
Sbjct: 785 SEEGEEKVGSMISSKELWFIAMNCNLCDDFFLTGSKRFTRVEYLDLSG----NNFTILPE 840
Query: 628 GF--LGKLKTLSAKSCRNLRSIPSLKLDLLETLDLSNCVSLES 668
F L L+ L C +L+ I L + LE D NC SL S
Sbjct: 841 FFKELQFLRALMVSDCEHLQEIRGLPPN-LEYFDARNCASLTS 882
>UniRef100_Q84ZV7 R 12 protein [Glycine max]
Length = 893
Score = 432 bits (1110), Expect = e-119
Identities = 282/674 (41%), Positives = 381/674 (55%), Gaps = 68/674 (10%)
Query: 8 DRDGYVHEFIEKIVEEVLRNIKPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKVGIL 67
D Y + I +IV++V R +L D+ VG+E Q V LL+VGSDD +H +GI
Sbjct: 152 DGGSYEYMLIGRIVKQVSRMFGLASLHVADYPVGLESQVTEVMKLLDVGSDDVVHIIGIH 211
Query: 68 GIGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEIT 127
G+GG+GKTTLA+ VYN I F+ SCFL+ VRE S+K+GL +LQ +LLS + GEK+ +T
Sbjct: 212 GMGGLGKTTLAMAVYNFIAPHFDESCFLQNVREESNKHGLKHLQSVLLSKLLGEKDITLT 271
Query: 128 SVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISS 187
S EG S+++ RL KK+LL+LDDVDK+EQLKAI G +WF GSRVIITTR K+LL
Sbjct: 272 SWQEGASMIQHRLRLKKILLILDDVDKREQLKAIVGKPDWFGPGSRVIITTRDKHLLKYH 331
Query: 188 GVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLKND 247
VER YEV LN +DAF L+ W K + K D
Sbjct: 332 EVERTYEVNVLNHDDAFQLLTWNAFKRE-----------------------------KID 362
Query: 248 EGYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYD 307
Y +VL RVV YASG PLALEVIGS+ KT+ + AL+ YKR+P N I L++S+D
Sbjct: 363 PSYKDVLNRVVTYASGLPLALEVIGSNLYGKTVAEWESALETYKRIPSNEILKILEVSFD 422
Query: 308 SLQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEK-YLIKINESGNVT 366
+L+EE+K VFLDIACCFKG+K T V I A + + I VLVEK L+K++ NV
Sbjct: 423 ALEEEQKNVFLDIACCFKGYKWTEVYDIFRALYSNCKMHHIGVLVEKSLLLKVSWRDNVE 482
Query: 367 LHDLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIEMIHFDGVIE---- 422
+HDL++DMG++I RQ +PE+PGK RLW +DIIQVL+ NTGTSK+E+I D I
Sbjct: 483 MHDLIQDMGRDIERQRSPEEPGKCKRLWSPKDIIQVLKHNTGTSKLEIICLDSSISDKEE 542
Query: 423 -VQWDGEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLECRNHM*ELDLSYCI--NLE 479
V+W+ AF KMENLK LI N FS+ P + P LRVLE + S C+ N +
Sbjct: 543 TVEWNENAFMKMENLKILIIRNGK-FSKGPNYFPEGLRVLEWHRYP-----SNCLPSNFD 596
Query: 480 SFSHVVDGFGD-KLRTMSVRGCFKLKSIPPLKLDSLETMDLSCCFRLESFPLVVDGILGK 538
+ V+ D + ++ G KL + LK D C L P V D L
Sbjct: 597 PINLVICKLPDSSITSLEFHGSSKLGHLTVLKFDK--------CKFLTQIPDVSD--LPN 646
Query: 539 IKTLNVESCHNLRSIPPL--KLDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRI 596
++ L+ C +L +I L+ LE L+ + C L SFP + L L+TL + C
Sbjct: 647 LRELSFVGCESLVAIDDSIGFLNKLEILNAAGCRKLTSFPPLN---LTSLETLELSHCSS 703
Query: 597 MISIPTLMLSLLEELDLSYCLNLENFPLVVDGF----LGKLKTLSAKSCRNLRSIPSL-K 651
+ P +L E++ L+LE P+ F L L+ ++ + CR +R SL
Sbjct: 704 LEYFP----EILGEMENITALHLERLPIKELPFSFQNLIGLREITLRRCRIVRLRCSLAM 759
Query: 652 LDLLETLDLSNCVS 665
+ L + NC S
Sbjct: 760 MPNLFRFQIRNCNS 773
Score = 61.2 bits (147), Expect = 1e-07
Identities = 44/119 (36%), Positives = 61/119 (50%), Gaps = 14/119 (11%)
Query: 583 LGKLKTLNVKSCRIMISIPTLM-LSLLEELDLSYCLNLENFPLVVD---GFLGKLKTLSA 638
LG L L C+ + IP + L L EL C +L + +D GFL KL+ L+A
Sbjct: 621 LGHLTVLKFDKCKFLTQIPDVSDLPNLRELSFVGCESL----VAIDDSIGFLNKLEILNA 676
Query: 639 KSCRNLRSIPSLKLDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNL--KSIP 695
CR L S P L L LETL+LS+C SLE P + LG+++ + + L K +P
Sbjct: 677 AGCRKLTSFPPLNLTSLETLELSHCSSLEYFPEI----LGEMENITALHLERLPIKELP 731
Score = 56.6 bits (135), Expect = 3e-06
Identities = 39/89 (43%), Positives = 47/89 (51%), Gaps = 4/89 (4%)
Query: 630 LGKLKTLSAKSCRNLRSIPSLKLDLLETLDLSNCVSLESLPLVVD--GFLGKLKTLLVTN 687
LG L L C+ L IP + DL +LS V ESL + D GFL KL+ L
Sbjct: 621 LGHLTVLKFDKCKFLTQIPDVS-DLPNLRELS-FVGCESLVAIDDSIGFLNKLEILNAAG 678
Query: 688 CHNLKSIPPLKCDALETLDLSCCYSLQSF 716
C L S PPL +LETL+LS C SL+ F
Sbjct: 679 CRKLTSFPPLNLTSLETLELSHCSSLEYF 707
>UniRef100_Q84ZU7 R 5 protein [Glycine max]
Length = 907
Score = 421 bits (1081), Expect = e-116
Identities = 312/838 (37%), Positives = 421/838 (50%), Gaps = 180/838 (21%)
Query: 8 DRDGYVHEFIEKIVEEVLRNIKPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKVGIL 67
DRD Y ++FI +IV V I P +L D VG+E + Q V LL+VG+ D + +GI
Sbjct: 155 DRDEYEYKFIGRIVASVSEKINPASLHVADLPVGLESKVQEVRKLLDVGNHDGVCMIGIH 214
Query: 68 GIGGIGKTTLALEVYNSIV--SQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTE 125
G+GGIGK+TLA VYN ++ F+ CFLE VRE+S+ +GL +LQ ILLS I GE + +
Sbjct: 215 GMGGIGKSTLARAVYNDLIITENFDGLCFLENVRESSNNHGLQHLQSILLSEILGE-DIK 273
Query: 126 ITSVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLI 185
+ S +GIS ++ L KKVLL+LDDVDK +QL+ IAG +WF GS +IITTR K LL
Sbjct: 274 VRSKQQGISKIQSMLKGKKVLLILDDVDKPQQLQTIAGRRDWFGPGSIIIITTRDKQLLA 333
Query: 186 SSGVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLK 245
GV++ YEV+ LN A L+ W K + K
Sbjct: 334 PHGVKKRYEVEVLNQNAALQLLTWNAFKRE-----------------------------K 364
Query: 246 NDEGYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLS 305
D Y +VL RVV YASG PLALEVIGS+ K + + A++ YKR+P++ I L++S
Sbjct: 365 IDPSYEDVLNRVVTYASGLPLALEVIGSNMFGKRVAEWKSAVEHYKRIPNDEILEILKVS 424
Query: 306 YDSLQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESGNV 365
+D+L EE+K VFLDIACCFKG KLT VE +L + + MK I+VLV+K LIK+ G V
Sbjct: 425 FDALGEEQKNVFLDIACCFKGCKLTEVEHMLRGLYNNCMKHHIDVLVDKSLIKVRH-GTV 483
Query: 366 TLHDLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIEMIHFDGVIE--- 422
+HDL++ +G+EI RQ +PE+PGK RLW +DIIQVL+ NTGTSKIE+I D I
Sbjct: 484 NMHDLIQVVGREIERQISPEEPGKCKRLWLPKDIIQVLKHNTGTSKIEIICLDFSISDKE 543
Query: 423 --VQWDGEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLE------------------ 462
V+W+ AF KMENLK LI N FS+ P + P LRVLE
Sbjct: 544 QTVEWNQNAFMKMENLKILIIRNGK-FSKGPNYFPEGLRVLEWHRYPSKCLPSNFHPNNL 602
Query: 463 --CR-----------------NHM*ELDLSYCINLESFSHVVD----------------- 486
C+ H+ L C L V D
Sbjct: 603 LICKLPDSSMASFEFHGSSKFGHLTVLKFDNCKFLTQIPDVSDLPNLRELSFKGCESLVA 662
Query: 487 -----GFGDKLRTMSVRGCFKLKSIPPLKLDSLETMDLSCCFRLESFPLVVDGILGKIKT 541
GF +KL+ ++ GC KL S PPL L SLET+ LS C LE FP ++ G + IK
Sbjct: 663 VDDSIGFLNKLKKLNAYGCRKLTSFPPLNLTSLETLQLSGCSSLEYFPEIL-GEMENIKQ 721
Query: 542 LNVESCHNLRSIPPLKLDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMISIP 601
L LR +P +++L S+ + L L+ L + SC +++ +P
Sbjct: 722 L------VLRDLP------IKELPFSF------------QNLIGLQVLYLWSC-LIVELP 756
Query: 602 --TLMLSLLEELDLSYC-----LNLENFPLVVDGFLGK----LKTLSAKSCRNLRSIPSL 650
+M+ L +L + YC + E V L + ++ C + S
Sbjct: 757 CRLVMMPELFQLHIEYCNRWQWVESEEGEEKVGSILSSKARWFRAMNCNLCDDFFLTGSK 816
Query: 651 KLDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNLKSIPPLKCDALETLDLSCC 710
+ +E LDLS G T+L LK L TLD+S C
Sbjct: 817 RFTHVEYLDLS----------------GNNFTILPEFFKELK--------FLRTLDVSDC 852
Query: 711 YSLQSFSLVADRLWKLVLDDCKELQEIKVIPPCLRMLSAVNCTSLTSSCTSKLLNQEL 768
L Q+I+ +PP L+ A+NC SLTSS S LLNQ L
Sbjct: 853 EHL---------------------QKIRGLPPNLKDFRAINCASLTSSSKSMLLNQVL 889
>UniRef100_Q9FVK3 Resistance protein MG63 [Glycine max]
Length = 459
Score = 411 bits (1056), Expect = e-113
Identities = 232/442 (52%), Positives = 292/442 (65%), Gaps = 45/442 (10%)
Query: 26 RNIKPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKVGILGIGGIGKTTLALEVYNSI 85
+ I L D+ VG+E + Q V +LL+VGSDD +H VGI G+GGIGKTTLA +YNSI
Sbjct: 2 KRINRAPLHVADYPVGLESRIQEVKMLLDVGSDDVVHMVGIHGLGGIGKTTLAAAIYNSI 61
Query: 86 VSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEITSVGEGISILKKRLPEKKV 145
FE CFLE VRE S +GL YLQ+ LLS GE E+ V +GISI++ RL +KKV
Sbjct: 62 ADHFEALCFLENVRETSKTHGLQYLQRNLLSETVGED--ELIGVKQGISIIQHRLQQKKV 119
Query: 146 LLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISSGVERIYEVKGLNDEDAFD 205
LL+LDDVDK+EQL+A+ G + F GSRVIITTR K LL GV+R YEV LN+E A
Sbjct: 120 LLILDDVDKREQLQALVGRPDLFCPGSRVIITTRDKQLLACHGVKRTYEVNELNEEYALQ 179
Query: 206 LVGWKTLKND-CSPRYKDVLLEQKYGRESDANKLRRLKDLKNDEGYANVLKRVVAYASGH 264
L+ WK K + +P YKDVL R V Y++G
Sbjct: 180 LLSWKAFKLEKVNPCYKDVL------------------------------NRTVTYSAGL 209
Query: 265 PLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYDSLQEEEKIVFLDIACCF 324
PLALEVIGS+ S + IEQ LDRYKR+P+ IQ L++SYD+L+E+E+ VFLDI+CC
Sbjct: 210 PLALEVIGSNLSGRNIEQWRSTLDRYKRIPNKEIQEILKVSYDALEEDEQSVFLDISCCL 269
Query: 325 KGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESGNVTLHDLVEDMGKEIVRQEAP 384
K + L V+ IL AH+GH M+ I VL+EK LIKI++ G +TLHDL+EDMGKEIVR+E+P
Sbjct: 270 KEYDLKEVQDILRAHYGHCMEHHIRVLLEKSLIKISD-GYITLHDLIEDMGKEIVRKESP 328
Query: 385 EDPGKRSRLWFSEDIIQVLEENTGTSKIEMIHFD----GVIEVQWDGEAFKKMENLKTLI 440
+PGKRSRLW DIIQ GTS+IE+I D +E++WD AFKKMENLKTLI
Sbjct: 329 REPGKRSRLWLHTDIIQ------GTSQIEIICTDFSLFEEVEIEWDANAFKKMENLKTLI 382
Query: 441 FSNDVFFSENPKHLPNSLRVLE 462
N F++ PKHLP++LRVLE
Sbjct: 383 IKNG-HFTKGPKHLPDTLRVLE 403
>UniRef100_Q8H6S7 Resistance protein KR3 [Glycine max]
Length = 636
Score = 357 bits (915), Expect = 1e-96
Identities = 210/470 (44%), Positives = 295/470 (62%), Gaps = 42/470 (8%)
Query: 8 DRDGYVHEFIEKIVEEVLRNIKPV--ALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKVG 65
D GY +FI KIV++V I ++ D VG+ + + + LL GS D+I +G
Sbjct: 178 DGAGYEFKFIRKIVDDVFDKINKAEASIYVADHPVGLHLEVEKIRKLLEAGSSDAISMIG 237
Query: 66 ILGIGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTE 125
I G+GG+GK+TLA VYN F+ SCFL+ VRE S+++GL LQ ILLS I +K
Sbjct: 238 IHGMGGVGKSTLARAVYNLHTDHFDDSCFLQNVREESNRHGLKRLQSILLSQIL-KKEIN 296
Query: 126 ITSVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRK--GSR--VIITTRYK 181
+ S +G S++K +L KKVLL+LDDVD+ +QL+AI G S W G+R +IITTR K
Sbjct: 297 LASEQQGTSMIKNKLKGKKVLLVLDDVDEHKQLQAIVGKSVWSESEFGTRLVLIITTRDK 356
Query: 182 NLLISSGVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRL 241
LL S GV+R +EVK L+ +DA L+ K K Y +V
Sbjct: 357 QLLTSYGVKRTHEVKELSKKDAIQLLKRKAFKT-----YDEV------------------ 393
Query: 242 KDLKNDEGYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTT 301
D+ Y VL VV + SG PLALEVIGS+ K+I++ A+ +Y+R+P+ I
Sbjct: 394 -----DQSYNQVLNDVVTWTSGLPLALEVIGSNLFGKSIKEWESAIKQYQRIPNKEILKI 448
Query: 302 LQLSYDSLQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINE 361
L++S+D+L+EEEK VFLDI CC KG+K +E ILH+ + + MK I VLV+K LI+I++
Sbjct: 449 LKVSFDALEEEEKSVFLDITCCLKGYKCREIEDILHSLYDNCMKYHIGVLVDKSLIQISD 508
Query: 362 SGNVTLHDLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIEMIHFDGVI 421
VTLHDL+E+MGKEI RQ++P++ GKR RLW +DIIQVL++N+GTS++++I D I
Sbjct: 509 D-RVTLHDLIENMGKEIDRQKSPKETGKRRRLWLLKDIIQVLKDNSGTSEVKIICLDFPI 567
Query: 422 E-----VQWDGEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLECRNH 466
++W+G AFK+M+NLK LI N + S+ P +LP SLR+LE H
Sbjct: 568 SDKQETIEWNGNAFKEMKNLKALIIRNGI-LSQGPNYLPESLRILEWHRH 616
>UniRef100_Q8W260 Functional resistance protein KR2 [Glycine max]
Length = 457
Score = 352 bits (903), Expect = 3e-95
Identities = 207/463 (44%), Positives = 292/463 (62%), Gaps = 42/463 (9%)
Query: 15 EFIEKIVEEVLRNIKPV--ALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKVGILGIGGI 72
+FI KIV++V I ++ D VG+ + + + LL GS D+I +GI G+GG+
Sbjct: 6 KFIRKIVDDVFDKINKAEASIYVADHPVGLHLEVEKIRKLLEAGSSDAISMIGIHGMGGV 65
Query: 73 GKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEITSVGEG 132
GK+TLA VYN F+ SCFL+ VRE S+++GL LQ ILLS I +K + S +G
Sbjct: 66 GKSTLARAVYNLHTDHFDDSCFLQNVREESNRHGLKRLQSILLSQIL-KKEINLASEQQG 124
Query: 133 ISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRK--GSR--VIITTRYKNLLISSG 188
S++K +L KKVLL+LDDVD+ +QL+AI G S W G+R +IITTR K LL S G
Sbjct: 125 TSMIKNKLKGKKVLLVLDDVDEHKQLQAIVGKSVWSESEFGTRLVLIITTRDKQLLTSYG 184
Query: 189 VERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLKNDE 248
V+R +EVK L+ +DA L+ K K Y +V D+
Sbjct: 185 VKRTHEVKELSKKDAIQLLKRKAFKT-----YDEV-----------------------DQ 216
Query: 249 GYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYDS 308
Y VL VV + SG PLALEVIGS+ K+I++ A+ +Y+R+P+ I L++S+D+
Sbjct: 217 SYNQVLNDVVTWTSGLPLALEVIGSNLFGKSIKEWESAIKQYQRIPNKEILKILKVSFDA 276
Query: 309 LQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESGNVTLH 368
L+EEEK VFLDI CC KG+K +E ILH+ + + MK I VLV+K LI+I++ VTLH
Sbjct: 277 LEEEEKSVFLDITCCLKGYKCREIEDILHSLYDNCMKYHIGVLVDKSLIQISDD-RVTLH 335
Query: 369 DLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIEMIHFDGVIE-----V 423
DL+E+MGKEI RQ++P++ GKR RLW +DIIQVL++N+GTS++++I D I +
Sbjct: 336 DLIENMGKEIDRQKSPKETGKRRRLWLLKDIIQVLKDNSGTSEVKIICLDFPISDKQETI 395
Query: 424 QWDGEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLECRNH 466
+W+G AFK+M+NLK LI N + S+ P +LP SLR+LE H
Sbjct: 396 EWNGNAFKEMKNLKALIIRNGI-LSQGPNYLPESLRILEWHRH 437
>UniRef100_Q6XZH8 Nematode resistance protein [Solanum tuberosum]
Length = 1136
Score = 337 bits (864), Expect = 1e-90
Identities = 289/923 (31%), Positives = 430/923 (46%), Gaps = 176/923 (19%)
Query: 10 DGYVHEFIEKIVEEVLRNIKPVALPAGDF-LVGVEHQKQHVTLLLNVGSDDSIHKVGILG 68
+G+ +EKI E+++ + + LVG+E V +L +GS +H +GILG
Sbjct: 165 NGHEARVMEKIAEDIMARLGSQRHASNARNLVGMESHMHKVYKMLGIGSG-GVHFLGILG 223
Query: 69 IGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEITS 128
+ G+GKTTLA +Y++I SQF+ +CFL +VR+ S K GL LQ+ILLS I K I
Sbjct: 224 MSGVGKTTLARVIYDNIRSQFQGACFLHEVRDRSAKQGLERLQEILLSEILVVKKLRIND 283
Query: 129 VGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISSG 188
EG ++ K+RL KKVLL+LDDVD +QL A+AG WF GSR+IITT+ K+LL+
Sbjct: 284 SFEGANMQKQRLQYKKVLLVLDDVDHIDQLNALAGEREWFGDGSRIIITTKDKHLLVKYE 343
Query: 189 VERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLKNDE 248
E+IY +K LN+ ++ L K K + +
Sbjct: 344 TEKIYRMKTLNNYESLQLF-----------------------------KQHAFKKNRPTK 374
Query: 249 GYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYDS 308
+ ++ +V+ + G PLAL+V+GS + +++ ++R K++P N I L+ S+
Sbjct: 375 EFEDLSAQVIKHTDGLPLALKVLGSFLYGRGLDEWISEVERLKQIPENEILKKLEQSFTG 434
Query: 309 LQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESGNVTLH 368
L E+ +FLDIAC F G K V IL + H + I VL+EK LI I + G +T+H
Sbjct: 435 LHNTEQKIFLDIACFFSGKKKDSVTRILESFHFCPVIG-IKVLMEKCLITILQ-GRITIH 492
Query: 369 DLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIE--MIHFDGVIEVQWD 426
L++DMG IVR+EA +DP SR+W EDI VLE N GT K E +H EV +
Sbjct: 493 QLIQDMGWHIVRREATDDPRMCSRMWKREDICPVLERNLGTDKNEGMSLHLTNEEEVNFG 552
Query: 427 GEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLECRNHM*E---------------LD 471
G+AF +M L+ L F N + + P+ LP+ LR L+ + + L
Sbjct: 553 GKAFMQMTRLRFLKFRN-AYVCQGPEFLPDELRWLDWHGYPSKSLPNSFKGDQLVGLKLK 611
Query: 472 LSYCINLESFSHVVDGFGDKLRTMSVRGCFKLKSIPPLKLD-SLETMDLSCCFRLESFPL 530
S I L S + KL+ M++ KL P + +LE + L C L
Sbjct: 612 KSRIIQLWKTSKDLG----KLKYMNLSHSQKLIRTPDFSVTPNLERLVLEECTSLVEINF 667
Query: 531 VVDGILGKIKTLNVESCHNLRSIPP-LKLDSLEKLDISYCGSLESFPQVEGRF------- 582
++ LGK+ LN+++C NL+++P ++L+ LE L ++ C L +FP++E +
Sbjct: 668 SIEN-LGKLVLLNLKNCRNLKTLPKRIRLEKLEILVLTGCSKLRTFPEIEEKMNCLAELY 726
Query: 583 ---------------LGKLKTLNVKSCRIMISIPTLMLSL--LEELDLSYCLNLENFP-- 623
L + +N+ C+ + S+P+ + L L+ LD+S C L+N P
Sbjct: 727 LGATSLSELPASVENLSGVGVINLSYCKHLESLPSSIFRLKCLKTLDVSGCSKLKNLPDD 786
Query: 624 --LVVD------------------GFLGKLKTLSAKSCRNLRSIPSLK------------ 651
L+V L LK LS C L S S
Sbjct: 787 LGLLVGLEELHCTHTAIQTIPSSMSLLKNLKHLSLSGCNALSSQVSSSSHGQKSMGVNFQ 846
Query: 652 ----LDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNLKSIPPLKCDALETLDL 707
L L LDLS+C + L GFL L+ +L+ N +N +IP
Sbjct: 847 NLSGLCSLIMLDLSDCNISDGGILNNLGFLSSLE-ILILNGNNFSNIPAA---------- 895
Query: 708 SCCYSLQSFSLVADRLWKLVLDDCKELQEIKVIPPCLRMLSAVNCTSLTS---------- 757
S+ F+ RL +L L C L+ + +PP ++ + A CTSL S
Sbjct: 896 ----SISRFT----RLKRLKLHGCGRLESLPELPPSIKGIFANECTSLMSIDQLTKYPML 947
Query: 758 ------------------SCTSKLLNQELHKA-GNTWFCLPRVP--KIPEWFDHKYEAGL 796
S LL Q L N FCL VP +IPEWF +K
Sbjct: 948 SDATFRNCRQLVKNKQHTSMVDSLLKQMLEALYMNVRFCL-YVPGMEIPEWFTYKSWGTQ 1006
Query: 797 SISF-----WFRNKFPAIALCVV 814
S+S WF F +CV+
Sbjct: 1007 SMSVALPTNWFTPTFRGFTVCVI 1029
>UniRef100_Q6XZH5 Nematode resistance-like protein [Solanum tuberosum]
Length = 1136
Score = 333 bits (854), Expect = 1e-89
Identities = 284/909 (31%), Positives = 431/909 (47%), Gaps = 148/909 (16%)
Query: 10 DGYVHEFIEKIVEEVLRNIKPVALPAGDF-LVGVEHQKQHVTLLLNVGSDDSIHKVGILG 68
+G+ +EKI E+++ + + LVG+E V +L +GS +H +GILG
Sbjct: 165 NGHEARVMEKIAEDIMARLGSQRHASNARNLVGMESHMHQVYKMLGIGSG-GVHFLGILG 223
Query: 69 IGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEITS 128
+ G+GKTTLA +Y++I SQF+ +CFL +VR+ S K GL LQ+ILLS I K I
Sbjct: 224 MSGVGKTTLARVIYDNIRSQFQGACFLHEVRDRSAKQGLERLQEILLSEILVVKKLRIND 283
Query: 129 VGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISSG 188
EG ++ K+RL KKVLL+LDDVD +QL A+AG WF GSR+IITT+ K+LL+
Sbjct: 284 SFEGANMQKQRLQYKKVLLVLDDVDHIDQLNALAGEREWFGDGSRIIITTKDKHLLVKYE 343
Query: 189 VERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLKNDE 248
E+IY +K LN+ ++ L K K + +
Sbjct: 344 TEKIYRMKTLNNYESLQLF-----------------------------KQHAFKKNRPTK 374
Query: 249 GYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYDS 308
+ ++ +V+ + G PLAL+V+GS + +++ ++R K++P N I L+ S+
Sbjct: 375 EFEDLSAQVIKHTDGLPLALKVLGSFLYGRGLDEWISEVERLKQIPENEILKKLEQSFTG 434
Query: 309 LQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESGNVTLH 368
L E+ +FLDIAC F G K V IL + H + I VL+EK LI + G +T+H
Sbjct: 435 LHNTEQKIFLDIACFFSGKKKDSVTRILESFHFCPVIG-IKVLMEKCLITTLQ-GRITIH 492
Query: 369 DLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIE--MIHFDGVIEVQWD 426
L++DMG IVR+EA +DP SRLW EDI VLE N GT KIE +H EV +
Sbjct: 493 QLIQDMGWHIVRREATDDPRMCSRLWKREDICPVLERNLGTDKIEGMSLHLTNEEEVNFG 552
Query: 427 GEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLECRNH---------------M*ELD 471
G+AF +M L+ L F N + + P+ LP+ LR L+ + +L
Sbjct: 553 GKAFMQMTRLRFLKFQN-AYVCQGPEFLPDELRWLDWHGYPSKSLPNSFKGDQLVSLKLK 611
Query: 472 LSYCINLESFSHVVDGFGDKLRTMSVRGCFKLKSIPPLKLD-SLETMDLSCCFRLESFPL 530
S I L S + KL+ M++ KL +P + +LE + L C L
Sbjct: 612 KSRIIQLWKTSKDLG----KLKYMNLSHSQKLIRMPDFSVTPNLERLVLEECTSLVEINF 667
Query: 531 VVDGILGKIKTLNVESCHNLRSIPP-LKLDSLEKLDISYCGSLESFPQVEGRF--LGKLK 587
++ LGK+ LN+++C NL+++P ++L+ LE L ++ C L +FP++E + L +L
Sbjct: 668 SIEN-LGKLVLLNLKNCRNLKTLPKRIRLEKLEILVLTGCSKLRTFPEIEEKMNCLAELY 726
Query: 588 TLNVKSCRIMISIPTLMLSLLEELDLSYCLNLENFPLVVDGFLGKLKTLSAKSCRNLRSI 647
+ S+ L S + ++LSYC +LE+ P + L LKTL C L+++
Sbjct: 727 LDATSLSELPASVENL--SGVGVINLSYCKHLESLPSSIFR-LKCLKTLDVSGCSKLKNL 783
Query: 648 PSLKLDLLETLDLSNCV--SLESLPLVVDGFLGKLKTLLVTNCHNLKSIPPLKCD----- 700
P L LL L+ +C +++++P + L LK L ++ C+ L S
Sbjct: 784 PD-DLGLLVGLEQLHCTHTAIQTIPSSMS-LLKNLKRLSLSGCNALSSQVSSSSHGQKSM 841
Query: 701 -----------ALETLDLSCCY--------------SLQSFSLVAD-------------- 721
+L LDLS C SL+ L +
Sbjct: 842 GVNFQNLSGLCSLIMLDLSDCNISDGGILSNLGFLPSLERLILDGNNFSNIPAASISRLT 901
Query: 722 RLWKLVLDDCKELQEIKVIPPCLRMLSAVNCTSLTS------------------------ 757
RL L L C L+ + +PP ++ + A CTSL S
Sbjct: 902 RLKTLKLLGCGRLESLPELPPSIKGIYANECTSLMSIDQLTKYPMLSDASFRNCRQLVKN 961
Query: 758 ----SCTSKLLNQ---ELHKAGNTWFCLPRVPKIPEWFDHKYEAGLSISF-----WFRNK 805
S LL Q L+ F +P + +IPEWF +K S+S W
Sbjct: 962 KQHTSMVDSLLKQMLEALYMNVRFGFYVPGM-EIPEWFTYKSWGTQSMSVALPTNWLTPT 1020
Query: 806 FPAIALCVV 814
F +CVV
Sbjct: 1021 FRGFTVCVV 1029
>UniRef100_Q6XZH6 Nematode resistance-like protein [Solanum tuberosum]
Length = 1136
Score = 332 bits (852), Expect = 2e-89
Identities = 289/923 (31%), Positives = 426/923 (45%), Gaps = 176/923 (19%)
Query: 10 DGYVHEFIEKIVEEVLRNIKPVALPAGDF-LVGVEHQKQHVTLLLNVGSDDSIHKVGILG 68
+G+ +EKI E+++ + + LVG+E V +L +GS +H +GILG
Sbjct: 165 NGHEARVMEKIAEDIMARLGSQRHASNARNLVGMESHMLKVYKMLGIGSG-GVHFLGILG 223
Query: 69 IGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEITS 128
+ G+GKTTLA +Y++I SQF+ +CFL +VR+ S K GL LQ+ILLS I K I +
Sbjct: 224 MSGVGKTTLARVIYDNIRSQFQGACFLHEVRDRSAKQGLERLQEILLSEILVVKKLRINN 283
Query: 129 VGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISSG 188
EG ++ K+RL KKVLL+LDDVD +QL A+AG WF GSR+IITT+ K+LL+
Sbjct: 284 SFEGANMQKQRLQYKKVLLVLDDVDHIDQLNALAGEREWFGDGSRIIITTKDKHLLVKYE 343
Query: 189 VERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLKNDE 248
E+IY +K LN+ ++ L K K + +
Sbjct: 344 TEKIYRMKTLNNYESLQLF-----------------------------KQHAFKKNRPTK 374
Query: 249 GYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYDS 308
+ ++ +V+ + G PLAL+V+GS + +++ ++R K++P N I L+ S+
Sbjct: 375 EFEDLSAQVIKHTDGLPLALKVLGSFLYGRGLDEWISEVERLKQIPENEILKKLEQSFTG 434
Query: 309 LQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESGNVTLH 368
L E+ +FLDIAC F G K V IL + H + I VL+EK LI I + G +T+H
Sbjct: 435 LHNTEQKIFLDIACFFSGKKKDSVTRILESFHFCPVIG-IKVLMEKCLITILQ-GRITIH 492
Query: 369 DLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIE--MIHFDGVIEVQWD 426
L++DMG IVR+EA +DP SRLW EDI VLE N GT K E +H EV +
Sbjct: 493 QLIQDMGWHIVRREATDDPRMCSRLWKREDICPVLERNLGTDKNEGMSLHLTNEEEVNFG 552
Query: 427 GEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLECRNHM*E---------------LD 471
G+AF +M L+ L F N + + P+ LP+ LR L+ + + L
Sbjct: 553 GKAFMQMTRLRFLKFRN-AYVCQGPEFLPDELRWLDWHGYPSKSLPNSFKGDQLVGLKLK 611
Query: 472 LSYCINLESFSHVVDGFGDKLRTMSVRGCFKLKSIPPLKLD-SLETMDLSCCFRLESFPL 530
S I L S + KL+ M++ KL P + +LE + L C L
Sbjct: 612 KSRIIQLWKTSKDLG----KLKYMNLSHSQKLIRTPDFSVTPNLERLVLEECTSLVEINF 667
Query: 531 VVDGILGKIKTLNVESCHNLRSIPP-LKLDSLEKLDISYCGSLESFPQVEGRF------- 582
++ LGK+ LN+++C NL+++P ++L+ LE L ++ C L +FP++E +
Sbjct: 668 SIEN-LGKLVLLNLKNCRNLKTLPKRIRLEKLEILVLTGCSKLRTFPEIEEKMNCLAELY 726
Query: 583 ---------------LGKLKTLNVKSCRIMISIPTLMLSL--LEELDLSYCLNLENFP-- 623
L + +N+ C+ + S+P+ + L L+ LD+S C L+N P
Sbjct: 727 LGATSLSGLPASVENLSGVGVINLSYCKHLESLPSSIFRLKCLKTLDVSGCSKLKNLPDD 786
Query: 624 --LVVD------------------GFLGKLKTLSAKSCRNLRSIPSLK------------ 651
L+V L LK LS + C L S S
Sbjct: 787 LGLLVGLEKLHCTHTAIHTIPSSMSLLKNLKRLSLRGCNALSSQVSSSSHGQKSMGVNFQ 846
Query: 652 ----LDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNLKSIPPLKCDALETLDL 707
L L LDLS+C + L GFL LK LL+ +N +IP L
Sbjct: 847 NLSGLCSLIRLDLSDCDISDGGILRNLGFLSSLKVLLLDG-NNFSNIPAASISRLT---- 901
Query: 708 SCCYSLQSFSLVADRLWKLVLDDCKELQEIKVIPPCLRMLSAVNCTSLTS---------- 757
RL L L C L+ + +PP + + A +CTSL S
Sbjct: 902 --------------RLKSLALRGCGRLESLPELPPSITGIYAHDCTSLMSIDQLTKYPML 947
Query: 758 ------------------SCTSKLLNQELHKA-GNTWFCLPRVP--KIPEWFDHKYEAGL 796
S LL Q L N F L VP +IPEWF +K
Sbjct: 948 SDVSFRNCHQLVKNKQHTSMVDSLLKQMLEALYMNVRFGL-YVPGMEIPEWFTYKSWGTQ 1006
Query: 797 SISF-----WFRNKFPAIALCVV 814
S+S WF F +CV+
Sbjct: 1007 SMSVVLPTNWFTPTFRGFTVCVL 1029
>UniRef100_Q9FPK8 Putative resistance protein [Glycine max]
Length = 522
Score = 332 bits (850), Expect = 4e-89
Identities = 181/394 (45%), Positives = 251/394 (62%), Gaps = 37/394 (9%)
Query: 10 DGYVHEFIEKIVEEVLRNIKPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKVGILGI 69
DGY +E IEKIVE V + I VG++++ + LL+ S +H +GI G+
Sbjct: 166 DGYEYELIEKIVEGVSKKINRP--------VGLQYRMLELNGLLDAASLSGVHLIGIYGV 217
Query: 70 GGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEITSV 129
GGIGKTTLA +Y+S+ QF+ CFL++VREN+ K+GL++LQ+ +L+ GEK+ + SV
Sbjct: 218 GGIGKTTLARALYDSVAVQFDALCFLDEVRENAMKHGLVHLQQTILAETVGEKDIRLPSV 277
Query: 130 GEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISSGV 189
+GI++LK+RL EK+VLL+LDD+++ EQLKA+ GS WF GSRVIITTR + LL S GV
Sbjct: 278 KQGITLLKQRLQEKRVLLVLDDINESEQLKALVGSPGWFGPGSRVIITTRDRQLLESHGV 337
Query: 190 ERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLKNDEG 249
E+IYEV+ L D +A +L+ WK K D K
Sbjct: 338 EKIYEVENLADGEALELLCWKAFKTD-----------------------------KVYPD 368
Query: 250 YANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYDSL 309
+ N + R + YASG PLALEVIGS+ + I + LD Y+++ IQ L++S+D+L
Sbjct: 369 FINKIYRALTYASGLPLALEVIGSNLFGREIVEWQYTLDLYEKIHDKDIQKILKISFDAL 428
Query: 310 QEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESGNVTLHD 369
E EK +FLDIAC FKG KL VE I+ +G ++K I+VL+EK LIKI+E G V +HD
Sbjct: 429 DEHEKDLFLDIACFFKGCKLAQVESIVSGRYGDSLKAIIDVLLEKTLIKIDEHGRVKMHD 488
Query: 370 LVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVL 403
L++ MG+EIVRQE+P+ PG SRLW ED+ VL
Sbjct: 489 LIQQMGREIVRQESPKHPGNCSRLWSPEDVADVL 522
>UniRef100_Q7XYS5 Disease resistance-like protein KR7 [Glycine max]
Length = 402
Score = 330 bits (845), Expect = 2e-88
Identities = 191/412 (46%), Positives = 266/412 (64%), Gaps = 40/412 (9%)
Query: 64 VGILGIGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKN 123
+GI G+GG+GK+TLA VYN F+ SCFL+ VRE S+++GL LQ ILLS I +K
Sbjct: 2 IGIHGMGGVGKSTLARAVYNLHTDHFDDSCFLQNVREESNRHGLKRLQSILLSQIL-KKE 60
Query: 124 TEITSVGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRK--GSR--VIITTR 179
+ S +G S++K +L KKVLL+LDDVD+ +QL+AI G S W G+R +IITTR
Sbjct: 61 INLASEQQGTSMIKNKLKGKKVLLVLDDVDEHKQLQAIVGKSVWSESEFGTRLVLIITTR 120
Query: 180 YKNLLISSGVERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLR 239
K LL S GV+R +EVK L+ +DA L+ K K Y +V
Sbjct: 121 DKQLLTSYGVKRTHEVKELSKKDAIQLLKRKAFKT-----YDEV---------------- 159
Query: 240 RLKDLKNDEGYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQ 299
D+ Y VL VV + SG PLALEVIGS+ K+I++ A+ +Y+R+P+ I
Sbjct: 160 -------DQSYNQVLNDVVTWTSGLPLALEVIGSNLFGKSIKEWESAIKQYQRIPNKEIL 212
Query: 300 TTLQLSYDSLQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKI 359
L++S+D+L+EEEK VFLDI CC KG+K +E ILH+ + + MK I VLV+K LI+I
Sbjct: 213 KILKVSFDALEEEEKSVFLDITCCLKGYKCREIEDILHSLYDNCMKYHIGVLVDKSLIQI 272
Query: 360 NESGNVTLHDLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIEMIHFDG 419
++ VTLHDL+E+MGKEI RQ++P++ GKR RLW +DIIQVL++N+GTS++++I D
Sbjct: 273 SDD-RVTLHDLIENMGKEIDRQKSPKETGKRRRLWLLKDIIQVLKDNSGTSEVKIICLDF 331
Query: 420 VIE-----VQWDGEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLECRNH 466
I ++W+G AFK+M+NLK LI N + S+ P +LP SLR+LE H
Sbjct: 332 PISDKQETIEWNGNAFKEMKNLKALIIRNGI-LSQGPNYLPESLRILEWHRH 382
>UniRef100_Q6XZH4 Nematode resistance-like protein [Solanum tuberosum]
Length = 980
Score = 327 bits (838), Expect = 1e-87
Identities = 286/899 (31%), Positives = 427/899 (46%), Gaps = 142/899 (15%)
Query: 17 IEKIVEEVLRNI--KPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKVGILGIGGIGK 74
IE+I E+++ + + A G+ LVG+E V +L VGS + +GILG+ G+GK
Sbjct: 6 IERIAEDIMARLGSQRHASNVGN-LVGMELHMHQVYKMLGVGSG-GVRFLGILGMSGVGK 63
Query: 75 TTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEITSVGEGIS 134
TTLA +Y++I SQF+ +CFL +VR+ S K GL LQ+ILLS I K I + EG +
Sbjct: 64 TTLARVIYDNIRSQFQGTCFLHEVRDRSAKQGLERLQEILLSEILVVKKLRINDLFEGAN 123
Query: 135 ILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISSGVERIYE 194
+ K+RL KKVLL+LDDVD +QL +AG WF GSR+IITT+ K+LL+ E+IY
Sbjct: 124 MQKQRLRYKKVLLVLDDVDHIDQLDTLAGEREWFGDGSRIIITTKDKHLLVKYETEKIYR 183
Query: 195 VKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLKNDEGYANVL 254
+ L+ ++ L K + + + +DL
Sbjct: 184 MGTLDKYESLQLFKQHAFKKN--------------------HPTKEFEDLS--------- 214
Query: 255 KRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYDSLQEEEK 314
+V+ + G P+AL+V+GS + +++ ++R K++P N I L+ S+ L E+
Sbjct: 215 AQVIEHTGGLPVALKVLGSFLYGRGLDEWLSEVERLKQIPQNEILKKLEPSFIGLNNIEQ 274
Query: 315 IVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESGNVTLHDLVEDM 374
+FLDIAC F G K V IL + H + I VL+EK LI I + G + +H L++DM
Sbjct: 275 KIFLDIACFFSGKKKDSVTRILESFHFSPVIG-IKVLMEKCLITILQ-GRIAIHQLIQDM 332
Query: 375 GKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIE--MIHFDGVIEVQWDGEAFKK 432
G IVR+EA +P SRLW EDI VLE N T KIE +H EV + G+AF +
Sbjct: 333 GWHIVRREASYNPRICSRLWKREDICPVLERNLATDKIEGISLHLTNEEEVNFGGKAFMQ 392
Query: 433 MENLKTLIFSNDVFFSENPKHLPNSLRVLECRNHM*E---------------LDLSYCIN 477
M +L+ L F N + + P+ LP+ LR L+ + + L S I
Sbjct: 393 MTSLRFLKFRN-AYVCQGPEFLPDELRWLDWHGYPSKSLPNSFKGDQLVSLTLKKSRIIQ 451
Query: 478 LESFSHVVDGFGDKLRTMSVRGCFKLKSIPPLK-LDSLETMDLSCCFRLESFPLVVDGIL 536
L S + KL+ M++ KL P + +LE + L C L + G L
Sbjct: 452 LWKTSKDL----GKLKYMNLSHSQKLIRTPDFSVMPNLERLVLEECKSLVEINFSI-GDL 506
Query: 537 GKIKTLNVESCHNLRSIPP-LKLDSLEKLDISYCGSLESFPQVEGRF------------- 582
GK+ LN+++C NL+++P ++L+ LE L +S C L +FP++E +
Sbjct: 507 GKLVLLNLKNCRNLKTLPKRIRLEKLEILVLSGCSKLRTFPEIEEKMNCLAELYLGATAL 566
Query: 583 ---------LGKLKTLNVKSCRIMISIPT--LMLSLLEELDLSYCLNLENFP----LVVD 627
L + +N+ C+ + S+P+ L L+ LD+S C L+N P L+V
Sbjct: 567 SELSASVENLSGVGVINLCYCKHLESLPSSIFRLKCLKTLDVSGCSKLKNLPDDLGLLVG 626
Query: 628 ------------------GFLGKLKTLSAKSCRNLRSIPSLK----------------LD 653
L LK LS + C L S S L
Sbjct: 627 LEEFHCTHTAIQTIPSSISLLKNLKHLSLRGCNALSSQVSSSSHGQKSVGVNFQNLSGLC 686
Query: 654 LLETLDLSNCVSLESLPLVVDGFLGKLKTLLV--TNCHNLKSIPPLKCDALETLDLSCCY 711
L LDLS+C + L GFL L L++ N N+ + + LE L L+ C
Sbjct: 687 SLIMLDLSDCNISDGGILSNLGFLPSLAGLILDGNNFSNIPAASISRLTRLEILALAGCR 746
Query: 712 SLQSFSLVADRLWKLVLDDCKELQEIKVIP--PCLRMLSAVNCTSLT-----SSCTSKLL 764
L+S + + ++ D+C L I + L +S C L +S LL
Sbjct: 747 RLESLPELPPSIKEIYADECTSLMSIDQLTKYSMLHEVSFTKCHQLVTNKQHASMVDSLL 806
Query: 765 NQELHKA----GNTWFCLPRVPKIPEWFDHKYEAGLSISF-----WFRNKFPAIALCVV 814
Q +HK G+ +P V +IPEWF +K SIS W+ F IA+CVV
Sbjct: 807 KQ-MHKGLYLNGSFSMYIPGV-EIPEWFTYKNSGTESISVALPKNWYTPTFRGIAICVV 863
>UniRef100_Q6XZH7 Nematode resistance-like protein [Solanum tuberosum]
Length = 1121
Score = 318 bits (816), Expect = 4e-85
Identities = 287/911 (31%), Positives = 436/911 (47%), Gaps = 151/911 (16%)
Query: 10 DGYVHEFIEKIVEEVLRNIKPVALPAGDF-LVGVEHQKQHVTLLLNVGSDDSIHKVGILG 68
+G+ IEKI E+++ + + +VG+E V +L +GS + +GILG
Sbjct: 165 NGHEARVIEKITEDIMVRLGSQRHASNARNVVGMESHMHQVYKMLGIGSG-GVRFLGILG 223
Query: 69 IGGIGKTTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEITS 128
+ G+GKTTLA +Y++I SQFE +CFL +VR+ S K GL +LQ+ILLS I K I
Sbjct: 224 MSGVGKTTLARVIYDNIQSQFEGACFLHEVRDRSAKQGLEHLQEILLSEILVVKKLRIND 283
Query: 129 VGEGISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISSG 188
EG ++ K+RL KKVLL+LDDVD +QL A+AG WF GSR+IITT+ K+LL+
Sbjct: 284 SFEGANMQKQRLQYKKVLLVLDDVDHIDQLNALAGEREWFGDGSRIIITTKDKHLLVKYE 343
Query: 189 VERIYEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLKNDE 248
E+IY + L+ ++ L K + S + + L Q
Sbjct: 344 TEKIYRMGTLDKYESLQLFKQHAFKKNHSTKEFEDLSAQ--------------------- 382
Query: 249 GYANVLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYDS 308
V+ + G PLAL+V+GS + +++ ++R K++P N I L+ S+
Sbjct: 383 --------VIEHTGGLPLALKVLGSFLYGRGLDEWISEVERLKQIPQNEILKKLEPSFTG 434
Query: 309 LQEEEKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESGNVTLH 368
L E+ +FLDIAC F G K V IL + H + I VL+EK LI I + G +T+H
Sbjct: 435 LNNIEQKIFLDIACFFSGKKKDSVTRILESFHFSPVIG-IKVLMEKCLITILK-GRITIH 492
Query: 369 DLVEDMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIE--MIHFDGVIEVQWD 426
L+++MG IVR+EA +P SRLW EDI VLE+N T KIE +H EV +
Sbjct: 493 QLIQEMGWHIVRREASYNPRICSRLWKREDICPVLEQNLCTDKIEGMSLHLTNEEEVNFG 552
Query: 427 GEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLECRNH---------------M*ELD 471
G+A +M +L+ L F N + + P+ LP+ LR L+ + +L
Sbjct: 553 GKALMQMTSLRFLKFRN-AYVYQGPEFLPDELRWLDWHGYPSKNLPNSFKGDQLVSLKLK 611
Query: 472 LSYCINLESFSHVVDGFGDKLRTMSVRGCFKLKSIPPLKLD-SLETMDLSCCFRLESFPL 530
S I L S + KL+ M++ KL +P + +LE + L C L
Sbjct: 612 KSRIIQLWKTSKDLG----KLKYMNLSHSQKLIRMPDFSVTPNLERLVLEECTSLVEINF 667
Query: 531 VVDGILGKIKTLNVESCHNLRSIPP-LKLDSLEKLDISYCGSLESFPQVEGR-------F 582
+ G LGK+ LN+++C NL++IP ++L+ LE L +S C L +FP++E + +
Sbjct: 668 SI-GDLGKLVLLNLKNCRNLKTIPKRIRLEKLEVLVLSGCSKLRTFPEIEEKMNRLAELY 726
Query: 583 LGKLK---------------TLNVKSCRIMISIPTLMLSL--LEELDLSYCLNLENFPLV 625
LG +N+ C+ + S+P+ + L L+ LD+S C L+N P
Sbjct: 727 LGATSLSELPASVENFSGVGVINLSYCKHLESLPSSIFRLKCLKTLDVSGCSKLKNLP-- 784
Query: 626 VDGFLGKLKTLSAKSCRN--LRSIPSLK--LDLLETLDLSNCVSLESL---------PLV 672
LG L + C + +++IPS L L+ L LS C +L S +
Sbjct: 785 --DDLGLLVGIEKLHCTHTAIQTIPSSMSLLKNLKHLSLSGCNALSSQVSSSSHGQKSMG 842
Query: 673 VDGF-----LGKLKTLLVTNCH--------NLKSIPPLKCDALETLDLSCCYSLQSFSLV 719
++ F L L L +++C+ NL +P LK L+ + S + L
Sbjct: 843 INFFQNLSGLCSLIKLDLSDCNISDGGILSNLGLLPSLKVLILDGNNFSNIPAASISRLT 902
Query: 720 ADRLWKLVLDDCKELQEIKVIPPCLRMLSAVNCTSL---------------TSSCTSKLL 764
RL L L C L+ + +PP ++ + A TSL + + +L+
Sbjct: 903 --RLKCLALHGCTSLEILPKLPPSIKGIYANESTSLMGFDQLTEFPMLSEVSLAKCHQLV 960
Query: 765 NQELHKA--------------GNTWFCLPRVP--KIPEWFDHKYEAGLSISF-----WFR 803
+LH + N FCL VP +IPEWF +K SIS WF
Sbjct: 961 KNKLHTSMADLLLKEMLEALYMNFRFCL-YVPGMEIPEWFTYKNWGTESISVALPTNWFT 1019
Query: 804 NKFPAIALCVV 814
F +CVV
Sbjct: 1020 PTFRGFTVCVV 1030
>UniRef100_Q9FN83 Disease resistance protein-like [Arabidopsis thaliana]
Length = 1295
Score = 313 bits (802), Expect = 2e-83
Identities = 257/903 (28%), Positives = 425/903 (46%), Gaps = 112/903 (12%)
Query: 15 EFIEKIVEEVLRNIKPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKVGILGIGGIGK 74
+ I+KIV+++ + + L+G+ + ++++ D + +GI G+GG+GK
Sbjct: 162 KLIKKIVKDISDKLVSTSWDDSKGLIGMSSHMDFLQSMISI-VDKDVRMLGIWGMGGVGK 220
Query: 75 TTLALEVYNSIVSQFECSCFLEKVRENSDKNGLIYLQKILLSHIFGEKNTEITSVGEGIS 134
TT+A +YN + QF+ CF+E V+E ++ G+ LQ L +F E++ E S +
Sbjct: 221 TTIAKYLYNQLSGQFQVHCFMENVKEVCNRYGVRRLQVEFLCRMFQERDKEAWSSVSCCN 280
Query: 135 ILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISSGVERIYE 194
I+K+R K V ++LDDVD+ EQL + + WF GSR+I+TTR ++LL+S G+ +Y+
Sbjct: 281 IIKERFRHKMVFIVLDDVDRSEQLNELVKETGWFGPGSRIIVTTRDRHLLLSHGINLVYK 340
Query: 195 VKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLKNDEGYANVL 254
VK L ++A L C+ +++ ++ G+ +
Sbjct: 341 VKCLPKKEALQLF--------CNYAFREEIILP--------------------HGFEELS 372
Query: 255 KRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYDSLQEEEK 314
+ V YASG PLAL V+GS ++ + L R K PH+ I L++SYD L E+EK
Sbjct: 373 VQAVNYASGLPLALRVLGSFLYRRSQIEWESTLARLKTYPHSDIMEVLRVSYDGLDEQEK 432
Query: 315 IVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESGNVTLHDLVEDM 374
+FL I+C + ++ V +L G+ + I +L EK LI + +G V +HDL+E M
Sbjct: 433 AIFLYISCFYNMKQVDYVRKLLDL-CGYAAEIGITILTEKSLI-VESNGCVKIHDLLEQM 490
Query: 375 GKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIE--MIHFDGVIEVQWDGEAFKK 432
G+E+VRQ+A +P +R LW EDI +L EN+GT +E ++ + EV AF+
Sbjct: 491 GRELVRQQAVNNPAQRLLLWDPEDICHLLSENSGTQLVEGISLNLSEISEVFASDRAFEG 550
Query: 433 MENLKTLIFSNDVFFSENPKHLPNSLRVLECR----------------NHM*ELDLSYCI 476
+ NLK L F + F E HLPN L L + E + C+
Sbjct: 551 LSNLKLLNFYDLSFDGETRVHLPNGLSYLPRKLRYLRWDGYPLKTMPSRFFPEFLVELCM 610
Query: 477 NLESFSHVVDGFGD--KLRTMSVRGCFKLKSIPPL-KLDSLETMDLSCCFRLESFPLVVD 533
+ + + DG L+ M + C L +P L K +LE ++LS C L +
Sbjct: 611 SNSNLEKLWDGIQPLRNLKKMDLSRCKYLVEVPDLSKATNLEELNLSYCQSLVEVTPSIK 670
Query: 534 GILGKIKTLNVESCHNLRSIP-PLKLDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVK 592
+ G + + +C L+ IP + L SLE + +S C SL+ FP++ + L +
Sbjct: 671 NLKG-LSCFYLTNCIQLKDIPIGIILKSLETVGMSGCSSLKHFPEIS----WNTRRLYLS 725
Query: 593 SCRIMISIPTLM--LSLLEELDLSYCLNLENFPLVVDGFLGKLKTLSAKSCRNLRSIPSL 650
S +I +P+ + LS L +LD+S C L P + G L LK+L+ CR L ++P
Sbjct: 726 STKIE-ELPSSISRLSCLVKLDMSDCQRLRTLPSYL-GHLVSLKSLNLDGCRRLENLPDT 783
Query: 651 --KLDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVTNCHNLKSIPPLKCD--ALETLD 706
L LETL++S C+++ P V ++ L ++ +++ IP C+ L +LD
Sbjct: 784 LQNLTSLETLEVSGCLNVNEFPRVST----SIEVLRISET-SIEEIPARICNLSQLRSLD 838
Query: 707 LSCCYSLQSFSLVADR---LWKLVLDDCKELQEIKVIPPCLRMLSAVNCTSLTSSCTSKL 763
+S L S + L KL L C L+ L + ++C
Sbjct: 839 ISENKRLASLPVSISELRSLEKLKLSGCSVLESFP-----LEICQTMSCL---------- 883
Query: 764 LNQELHKAGNTWFCLPR--VPKIPEWFDHKYEAGLSISFWFRNKFPAIALCVVSPLTWDG 821
WF L R + ++PE +I + + V+ W
Sbjct: 884 ----------RWFDLDRTSIKELPE----------NIGNLVALEVLQASRTVIRRAPWSI 923
Query: 822 SRRHSIRVIINGNTFIYTDGLKMGTKSPLNMYHLHLFHMKMENFNDNMEKVLFENMWNHA 881
+R ++V+ GN+F +GL PL+ + L + + N N N+WN
Sbjct: 924 ARLTRLQVLAIGNSFFTPEGLLHSLCPPLSRFD-DLRALSLSNMNMTEIPNSIGNLWNLL 982
Query: 882 EVD 884
E+D
Sbjct: 983 ELD 985
Score = 63.9 bits (154), Expect = 2e-08
Identities = 84/339 (24%), Positives = 148/339 (42%), Gaps = 60/339 (17%)
Query: 452 KHLPNSLRVLECRNHM*ELDLSYCINLESFSHVVDGFGDKLRTMSVRGCFKLKSIPPL-- 509
+ LP+S+ L C + +LD+S C L + + G L+++++ GC +L+++P
Sbjct: 730 EELPSSISRLSC---LVKLDMSDCQRLRTLPSYL-GHLVSLKSLNLDGCRRLENLPDTLQ 785
Query: 510 KLDSLETMDLSCCFRLESFPLVVDGI-------------------LGKIKTLNVESCHNL 550
L SLET+++S C + FP V I L ++++L++ L
Sbjct: 786 NLTSLETLEVSGCLNVNEFPRVSTSIEVLRISETSIEEIPARICNLSQLRSLDISENKRL 845
Query: 551 RSIPPL--KLDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRI---------MIS 599
S+P +L SLEKL +S C LESFP + + L+ ++ I +++
Sbjct: 846 ASLPVSISELRSLEKLKLSGCSVLESFPLEICQTMSCLRWFDLDRTSIKELPENIGNLVA 905
Query: 600 IPTLMLSL---------LEELDLSYCLNLENFPLVVDGFLGKL----------KTLSAKS 640
+ L S + L L + N +G L L + LS +
Sbjct: 906 LEVLQASRTVIRRAPWSIARLTRLQVLAIGNSFFTPEGLLHSLCPPLSRFDDLRALSLSN 965
Query: 641 CRNLRSIPSLKLDLLETLDLS-NCVSLESLPLVVDGFLGKLKTLLVTNCHNLKSIPPLKC 699
N+ IP+ +L L+L + + E +P + L +L L + NC L+++P
Sbjct: 966 M-NMTEIPNSIGNLWNLLELDLSGNNFEFIPASIKR-LTRLNRLNLNNCQRLQALPDELP 1023
Query: 700 DALETLDLSCCYSLQSFSLVADR--LWKLVLDDCKELQE 736
L + + C SL S S ++ L KLV +C +L +
Sbjct: 1024 RGLLYIYIHSCTSLVSISGCFNQYCLRKLVASNCYKLDQ 1062
Score = 41.2 bits (95), Expect = 0.14
Identities = 74/314 (23%), Positives = 136/314 (42%), Gaps = 53/314 (16%)
Query: 412 IEMIHFDGVIEVQWDGEAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLECRNHM*ELD 471
++ ++ DG ++ + + + +L+TL S + +E P+ + S+ VL E
Sbjct: 766 LKSLNLDGCRRLENLPDTLQNLTSLETLEVSGCLNVNEFPR-VSTSIEVLRISETSIEEI 824
Query: 472 LSYCINLESFSHVVDGFGDKLRTMSVRGCFKLKSIPPL--KLDSLETMDLSCCFRLESFP 529
+ NL +LR++ + +L S+P +L SLE + LS C LESFP
Sbjct: 825 PARICNLS-----------QLRSLDISENKRLASLPVSISELRSLEKLKLSGCSVLESFP 873
Query: 530 LVVDGILGKIKTLNV---------ESCHNLRSI------------PPLKLDSLEKLDISY 568
L + + ++ ++ E+ NL ++ P + L +L +
Sbjct: 874 LEICQTMSCLRWFDLDRTSIKELPENIGNLVALEVLQASRTVIRRAPWSIARLTRLQVLA 933
Query: 569 CGSLESFPQVEGRF---------LGKLKTLNVKSCRIMISIPTLMLSL--LEELDLSYCL 617
G+ SF EG L+ L++ + M IP + +L L ELDLS
Sbjct: 934 IGN--SFFTPEGLLHSLCPPLSRFDDLRALSLSNMN-MTEIPNSIGNLWNLLELDLS-GN 989
Query: 618 NLENFPLVVDGFLGKLKTLSAKSCRNLRSIPSLKLDLLETLDLSNCVSLESLPLVVDGFL 677
N E P + L +L L+ +C+ L+++P L + + +C SL S+ + +
Sbjct: 990 NFEFIPASIKR-LTRLNRLNLNNCQRLQALPDELPRGLLYIYIHSCTSLVSISGCFNQYC 1048
Query: 678 GKLKTLLVTNCHNL 691
L+ L+ +NC+ L
Sbjct: 1049 --LRKLVASNCYKL 1060
>UniRef100_Q70WI0 Putative resistance gene analogue protein [Lens culinaris]
Length = 810
Score = 290 bits (741), Expect = 2e-76
Identities = 218/637 (34%), Positives = 329/637 (51%), Gaps = 63/637 (9%)
Query: 17 IEKIVEEVLRNIKPVALPAGDFLVGVEHQKQHVTLLLNVGSDDSIHKVGILGIGGIGKTT 76
+E I E + + + P D LVG++ + + + LL + D + +GI G+GGIGKTT
Sbjct: 224 VETIAEHIHKKLIPKLPVCKDNLVGIDSRIEEIYSLLGMRLSD-VRFIGIWGMGGIGKTT 282
Query: 77 LALEVYNSIVSQFECSCFLEKVREN-SDKNGLIYLQKILLSHIFGEKNTEITSVGEGISI 135
+A VY++I +F+ SCFL +RE S NGL+ +Q LLSH+ N + ++ +G I
Sbjct: 283 IARSVYDAIKDEFQVSCFLADIRETISRTNGLVRIQTELLSHLTIRSN-DFYNIHDGKKI 341
Query: 136 LKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISSGVERIYEV 195
L KKVLL+LDDV + QL+++AG WF G RVIIT+R K+LL++ GV Y+
Sbjct: 342 LANSFRNKKVLLVLDDVSELSQLESLAGKQEWFGSGIRVIITSRDKHLLMTHGVNETYKA 401
Query: 196 KGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLKNDEGYANVLK 255
KGL +A L K K + + E Y ++ K
Sbjct: 402 KGLVKNEALKLFCLKAFKQN-----------------------------QPKEEYLSLCK 432
Query: 256 RVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYDSLQEEEKI 315
VV YA G PLALEV+GSHF +T+E + AL++ + VPH+ I TL++SYDSLQ E+
Sbjct: 433 EVVEYARGLPLALEVLGSHFHGRTVEVWHSALEQMRNVPHSKIHDTLKISYDSLQPMERN 492
Query: 316 VFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESG-NVTLHDLVEDM 374
+FLDIAC FKG + V IL G+ K I++L+E+ L+ + + +HDL+E+M
Sbjct: 493 MFLDIACFFKGMDIDGVMEILE-DCGYYPKIGIDILIERSLVSFDRGDRKLWMHDLLEEM 551
Query: 375 GKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTSKIEMIHFDGV--IEVQWDGEAFKK 432
G+ IV QE+P DPGKRSRLW +DI QVL +N GT KI+ I + V E W+ EAF +
Sbjct: 552 GRNIVCQESPNDPGKRSRLWSQKDIDQVLTKNKGTDKIQGIALNLVQPYEAGWNIEAFSR 611
Query: 433 MENLKTLIFSNDVFFSENPKHLPNSLRVLECRNHM*ELDLSYCINLESFSHVVDGFGDKL 492
+ L+ L ++ +H ++ L Y + S + F L
Sbjct: 612 LSQLRLLKLC-EIKLPRGSRHELSAS----------PLGTQY---VNKTSRGLGCFPSSL 657
Query: 493 RTMSVRGCFKLKSIPPLK-LDSLETMDL--SCCFRLESFPLVVDGI--LGKIKTLNVESC 547
+ + RGC LK+ P D + + L S + ++ D I L + L +
Sbjct: 658 KVLDWRGC-PLKTPPQTNHFDEIVNLKLFHSKIEKTLAWNTGKDSINSLFQFMLLKLFKY 716
Query: 548 H-NLRSIPPLKLDSLEKLDISYCGSLESFPQVEGRFLGKLKTLNVKSCRIMISIPTLMLS 606
H N SI + L++L+ +++S+ L P G + L++L ++ C + I +LS
Sbjct: 717 HPNNSSILIMFLENLKSINLSFSKCLTRSPDFVG--VPNLESLVLEGCTSLTEIHPSLLS 774
Query: 607 --LLEELDLSYCLNLENFPLVVDGFLGKLKTLSAKSC 641
L L+L C L+ P ++ LK LS C
Sbjct: 775 HKTLILLNLKDCKRLKALPCKIE--TSSLKCLSLSGC 809
>UniRef100_O23530 Disease resistance RPP5 like protein [Arabidopsis thaliana]
Length = 1301
Score = 270 bits (689), Expect = 2e-70
Identities = 236/765 (30%), Positives = 379/765 (48%), Gaps = 104/765 (13%)
Query: 17 IEKIVEEVLRNIKPVALPAGDF--LVGVEHQKQHVTLLLNVGSDDSIHKVGILGIGGIGK 74
IE++ E+VLR P+ DF LVG+E+ + + +L + S ++ VGI G GIGK
Sbjct: 162 IEELAEDVLRK---TMTPSDDFGDLVGIENHIEAIKSVLCLESKEARIMVGIWGQSGIGK 218
Query: 75 TTLALEVYNSIVSQFECSCFLE-KVRENSDKNGL-IYLQKILLSHIFGEKNTEITSVGEG 132
+T+ +Y+ + QF F+ K SD +G+ + +K LLS I G+K+ +I G
Sbjct: 219 STIGRALYSKLSIQFHHRAFITYKSTSGSDVSGMKLRWEKELLSEILGQKDIKIEHFG-- 276
Query: 133 ISILKKRLPEKKVLLLLDDVDKKEQLKAIAGSSNWFRKGSRVIITTRYKNLLISSGVERI 192
++++RL ++KVL+LLDDVD E LK + G + WF GSR+I+ T+ + LL + ++ I
Sbjct: 277 --VVEQRLKQQKVLILLDDVDSLEFLKTLVGKAEWFGSGSRIIVITQDRQLLKAHEIDLI 334
Query: 193 YEVKGLNDEDAFDLVGWKTLKNDCSPRYKDVLLEQKYGRESDANKLRRLKDLKNDEGYAN 252
YEV+ ++ A + L +G++S + + L
Sbjct: 335 YEVEFPSEHLALTM-----------------LCRSAFGKDSPPDDFKEL----------- 366
Query: 253 VLKRVVAYASGHPLALEVIGSHFSNKTIEQCNDALDRYKRVPHNMIQTTLQLSYDSLQEE 312
V A PL L V+GS +T E + + R + + I TL++SYD L ++
Sbjct: 367 -AFEVAKLAGNLPLGLSVLGSSLKGRTKEWWMEMMPRLRNGLNGDIMKTLRVSYDRLHQK 425
Query: 313 EKIVFLDIACCFKGWKLTMVEGILHAHHGHTMKDQINVLVEKYLIKINESGNVTLHDLVE 372
++ +FL IAC F G++++ V+ +L + G TM L EK LI+I G + +H+L+E
Sbjct: 426 DQDMFLYIACLFNGFEVSYVKDLLKDNVGFTM------LTEKSLIRITPDGYIEMHNLLE 479
Query: 373 DMGKEIVRQEAPEDPGKRSRLWFSEDIIQVLEENTGTS-----KIEMIHFDGVIEVQWDG 427
+G+EI R ++ +PGKR L EDI +V+ E TGT ++ + + D
Sbjct: 480 KLGREIDRAKSKGNPGKRRFLTNFEDIHEVVTEKTGTETLLGIRLPFEEYFSTRPLLIDK 539
Query: 428 EAFKKMENLKTLIFSNDVFFSENPKHLPNSLRVLE-----CRNHM*ELDLSYCINLESFS 482
E+FK M NL+ L ++ +LP LR+L+ ++ Y +NL
Sbjct: 540 ESFKGMRNLQYLEIGYYGDLPQSLVYLPLKLRLLDWDDCPLKSLPSTFKAEYLVNLIMKY 599
Query: 483 HVVDGFGD------KLRTMSVRGCFKLKSIPPLKLD-SLETMDLSCCFRLESFPLVVDGI 535
++ + L+ M++R LK IP L L +LE +DL C L + P +
Sbjct: 600 SKLEKLWEGTLPLGSLKEMNLRYSNNLKEIPDLSLAINLEELDLVGCKSLVTLPSSIQNA 659
Query: 536 LGKIKTLNVESCHNLRSIP-PLKLDSLEKLDISYCGSLESFPQV----------EGRFLG 584
K+ L++ C L S P L L+SLE L+++ C +L +FP + EGR
Sbjct: 660 T-KLIYLDMSDCKKLESFPTDLNLESLEYLNLTGCPNLRNFPAIKMGCSDVDFPEGR--- 715
Query: 585 KLKTLNVKSCRIMISIPTLMLSLLEELD-LSYCLNLENFP-----LVVDGF--------- 629
+ V+ C ++P + L+ LD L+ C+ E P L V G+
Sbjct: 716 --NEIVVEDCFWNKNLP----AGLDYLDCLTRCMPCEFRPEQLAFLNVRGYKHEKLWEGI 769
Query: 630 --LGKLKTLSAKSCRNLRSIPSL-KLDLLETLDLSNCVSLESLPLVVDGFLGKLKTLLVT 686
LG L+ + NL IP L K LE+L L+NC SL +LP + G L +L L +
Sbjct: 770 QSLGSLEGMDLSESENLTEIPDLSKATKLESLILNNCKSLVTLPSTI-GNLHRLVRLEMK 828
Query: 687 NCHNLKSIP-PLKCDALETLDLSCCYSLQSFSLVADRLWKLVLDD 730
C L+ +P + +LETLDLS C SL+SF L++ + L L++
Sbjct: 829 ECTGLEVLPTDVNLSSLETLDLSGCSSLRSFPLISTNIVWLYLEN 873
Score = 105 bits (261), Expect = 8e-21
Identities = 107/333 (32%), Positives = 154/333 (46%), Gaps = 54/333 (16%)
Query: 470 LDLSYCINLESFSHVVDG-----FGDKLRTMSVRGCFKLKSIPPLKLDSLE--TMDLSCC 522
L+L+ C NL +F + G F + + V CF K++P LD L+ T + C
Sbjct: 688 LNLTGCPNLRNFPAIKMGCSDVDFPEGRNEIVVEDCFWNKNLPA-GLDYLDCLTRCMPCE 746
Query: 523 FRLESFPLVV----------DGI--LGKIKTLNVESCHNLRSIPPL-KLDSLEKLDISYC 569
FR E + +GI LG ++ +++ NL IP L K LE L ++ C
Sbjct: 747 FRPEQLAFLNVRGYKHEKLWEGIQSLGSLEGMDLSESENLTEIPDLSKATKLESLILNNC 806
Query: 570 GSLESFPQVEGRFLGKLKTLNVKSCRIMISIPT-LMLSLLEELDLSYCLNLENFPLVVD- 627
SL + P G L +L L +K C + +PT + LS LE LDLS C +L +FPL+
Sbjct: 807 KSLVTLPSTIGN-LHRLVRLEMKECTGLEVLPTDVNLSSLETLDLSGCSSLRSFPLISTN 865
Query: 628 ------------------GFLGKLKTLSAKSCRNLRSIPS-LKLDLLETLDLSNCVSLES 668
G L +L L K C L +P+ + L LETLDLS C SL S
Sbjct: 866 IVWLYLENTAIEEIPSTIGNLHRLVRLEMKKCTGLEVLPTDVNLSSLETLDLSGCSSLRS 925
Query: 669 LPLVVDGFLGKLKTLLVTNCHNLKSIPPL-KCDALETLDLSCCYSLQSFSLVADRLWKLV 727
PL+ + +K L + N ++ IP L K L+ L L+ C SL + L KLV
Sbjct: 926 FPLISES----IKWLYLENTA-IEEIPDLSKATNLKNLKLNNCKSLVTLPTTIGNLQKLV 980
Query: 728 LDDCKELQEIKVIP-----PCLRMLSAVNCTSL 755
+ KE ++V+P L +L C+SL
Sbjct: 981 SFEMKECTGLEVLPIDVNLSSLMILDLSGCSSL 1013
Score = 87.4 bits (215), Expect = 2e-15
Identities = 94/305 (30%), Positives = 139/305 (44%), Gaps = 55/305 (18%)
Query: 470 LDLSYCINLESF-----------------SHVVDGFGD--KLRTMSVRGCFKLKSIPP-L 509
LDLS C +L SF + G+ +L + ++ C L+ +P +
Sbjct: 848 LDLSGCSSLRSFPLISTNIVWLYLENTAIEEIPSTIGNLHRLVRLEMKKCTGLEVLPTDV 907
Query: 510 KLDSLETMDLSCCFRLESFPLVVDGILGKIKTLNVESCHNLRSIPPL-KLDSLEKLDISY 568
L SLET+DLS C L SFPL+ + IK L +E+ + IP L K +L+ L ++
Sbjct: 908 NLSSLETLDLSGCSSLRSFPLISE----SIKWLYLENT-AIEEIPDLSKATNLKNLKLNN 962
Query: 569 CGSLESFPQVEGRFLGKLKTLNVKSCRIMISIP-TLMLSLLEELDLSYCLNLENFPLV-- 625
C SL + P G L KL + +K C + +P + LS L LDLS C +L FPL+
Sbjct: 963 CKSLVTLPTTIGN-LQKLVSFEMKECTGLEVLPIDVNLSSLMILDLSGCSSLRTFPLIST 1021
Query: 626 -----------------VDGFLGKLKTLSAKSCRNLRSIPS-LKLDLLETLDLSNCVSLE 667
G L +L L K C L +P+ + L L LDLS C SL
Sbjct: 1022 NIVWLYLENTAIEEIPSTIGNLHRLVKLEMKECTGLEVLPTDVNLSSLMILDLSGCSSLR 1081
Query: 668 SLPLVVDGFLGKLKTLLVTNCHNLKSIPPLKCD--ALETLDLSCCYSLQSFSLVADRLWK 725
+ PL+ +++ L + N ++ +P D L L + CC L++ S RL +
Sbjct: 1082 TFPLIST----RIECLYLQNT-AIEEVPCCIEDFTRLTVLMMYCCQRLKTISPNIFRLTR 1136
Query: 726 LVLDD 730
L L D
Sbjct: 1137 LELAD 1141
Score = 35.4 bits (80), Expect = 7.9
Identities = 37/111 (33%), Positives = 52/111 (46%), Gaps = 12/111 (10%)
Query: 632 KLKTLSAKSCRNLRSIPS-LKLDLLETLDLSNCVSLESLPLVVDGFL--GKLKTLLVTNC 688
KL+ L C L+S+PS K + L L + L + +G L G LK + +
Sbjct: 569 KLRLLDWDDCP-LKSLPSTFKAEYLVNL----IMKYSKLEKLWEGTLPLGSLKEMNLRYS 623
Query: 689 HNLKSIPPLKCDA-LETLDLSCCYSLQSFSLVADRLWKLV---LDDCKELQ 735
+NLK IP L LE LDL C SL + KL+ + DCK+L+
Sbjct: 624 NNLKEIPDLSLAINLEELDLVGCKSLVTLPSSIQNATKLIYLDMSDCKKLE 674
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.140 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,539,049,056
Number of Sequences: 2790947
Number of extensions: 66888889
Number of successful extensions: 195876
Number of sequences better than 10.0: 2187
Number of HSP's better than 10.0 without gapping: 943
Number of HSP's successfully gapped in prelim test: 1258
Number of HSP's that attempted gapping in prelim test: 186325
Number of HSP's gapped (non-prelim): 4868
length of query: 918
length of database: 848,049,833
effective HSP length: 137
effective length of query: 781
effective length of database: 465,690,094
effective search space: 363703963414
effective search space used: 363703963414
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 80 (35.4 bits)
Medicago: description of AC145512.9


