

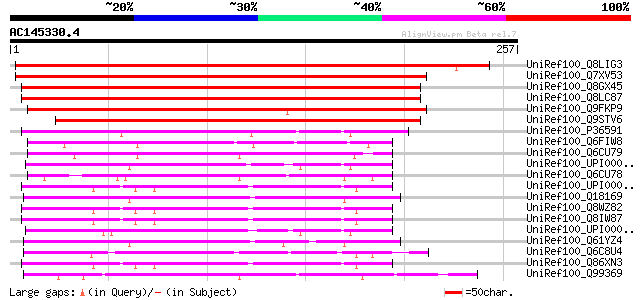
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145330.4 + phase: 0
(257 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LIG3 Hypothetical protein OJ1200_C08.123 [Oryza sativa] 363 3e-99
UniRef100_Q7XV53 OSJNBa0086B14.2 protein [Oryza sativa] 232 6e-60
UniRef100_Q8GX45 Hypothetical protein At4g24380/T22A6_210 [Arabi... 216 6e-55
UniRef100_Q8LC87 Hypothetical protein [Arabidopsis thaliana] 215 1e-54
UniRef100_Q9FKP9 Similarity to unknown protein [Arabidopsis thal... 207 3e-52
UniRef100_Q9STV6 Hypothetical protein T22A6.210 [Arabidopsis tha... 190 3e-47
UniRef100_P36591 Dihydrofolate reductase [Schizosaccharomyces po... 94 3e-18
UniRef100_Q6FIW8 Similar to sp|Q05015 Saccharomyces cerevisiae Y... 83 6e-15
UniRef100_Q6CU79 Similar to sp|Q05015 Saccharomyces cerevisiae Y... 80 4e-14
UniRef100_UPI0000014210 UPI0000014210 UniRef100 entry 78 2e-13
UniRef100_Q6CU78 Similar to sgd|S0005806 Saccharomyces cerevisia... 78 3e-13
UniRef100_UPI000036AA89 UPI000036AA89 UniRef100 entry 77 6e-13
UniRef100_Q18169 Hypothetical protein C25G4.2 [Caenorhabditis el... 76 7e-13
UniRef100_Q8WZ82 Candidate tumor suppressor OVCA2 [Homo sapiens] 76 1e-12
UniRef100_Q8IW87 Candidate tumor suppressor in ovarian cancer 2 ... 76 1e-12
UniRef100_UPI00002F9C06 UPI00002F9C06 UniRef100 entry 75 1e-12
UniRef100_Q61YZ4 Hypothetical protein CBG03338 [Caenorhabditis b... 75 2e-12
UniRef100_Q6C8U4 Yarrowia lipolytica chromosome D of strain CLIB... 75 2e-12
UniRef100_Q86XN3 Ovarian cancer gene-2 protein [Homo sapiens] 74 5e-12
UniRef100_Q99369 Hypothetical dihydrofolate reductase [Saccharom... 72 1e-11
>UniRef100_Q8LIG3 Hypothetical protein OJ1200_C08.123 [Oryza sativa]
Length = 247
Score = 363 bits (931), Expect = 3e-99
Identities = 174/242 (71%), Positives = 198/242 (80%), Gaps = 2/242 (0%)
Query: 4 ENGRKLKILCLHGFRTSGSFIKKQISKWDPSIFSQFHLEFPDGKFPAGGKSDIEGIFPPP 63
E +K K+LCLHGFRTSGSF+KKQISKW+PSIF QF + FPDG FPAGGKS+IEGIFPPP
Sbjct: 2 ELDKKFKVLCLHGFRTSGSFLKKQISKWNPSIFQQFDMVFPDGIFPAGGKSEIEGIFPPP 61
Query: 64 YFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQAQGKLL 123
YFEWFQF+KDFT YTNLDECISYL +Y++ NGPFDG LGFSQGATLSALLIGYQAQGK+L
Sbjct: 62 YFEWFQFNKDFTEYTNLDECISYLCDYMVKNGPFDGLLGFSQGATLSALLIGYQAQGKVL 121
Query: 124 KEHPPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDWLKIPSEELASAFDKPL 183
+HPPIKF+VSI+GSKFRDPSIC+VAYKDPIK KSVHFIGEKDWLK+PSEELA+AF+ P+
Sbjct: 122 NDHPPIKFMVSIAGSKFRDPSICNVAYKDPIKVKSVHFIGEKDWLKVPSEELAAAFEDPV 181
Query: 184 IIRHPQGHTVPRLDEVSTGQLQNFVAEILSQPKVGVSICEHE--SKVEVDGTNGDKGVNG 241
IIRHPQGHTVPRLDE S QL + + IL K + + K T + G N
Sbjct: 182 IIRHPQGHTVPRLDEASVKQLSEWSSSILEDIKNADDVAKASIVEKPSEGNTVAESGENL 241
Query: 242 VE 243
VE
Sbjct: 242 VE 243
>UniRef100_Q7XV53 OSJNBa0086B14.2 protein [Oryza sativa]
Length = 245
Score = 232 bits (592), Expect = 6e-60
Identities = 109/209 (52%), Positives = 144/209 (68%), Gaps = 1/209 (0%)
Query: 4 ENGRKLKILCLHGFRTSGSFIKKQI-SKWDPSIFSQFHLEFPDGKFPAGGKSDIEGIFPP 62
E R+ + LCLHGFRTSG ++KQ+ KW + ++ L F D FPA GKSD+EGIF P
Sbjct: 17 EGARRPRFLCLHGFRTSGEIMRKQVVGKWPADVTARLDLVFADAPFPAEGKSDVEGIFDP 76
Query: 63 PYFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQAQGKL 122
PY+EWFQFDK+FT Y N DEC++Y+ E +I GPFDG +GFSQG+ LS L G Q QG
Sbjct: 77 PYYEWFQFDKNFTEYRNFDECLNYIEELMIKEGPFDGLMGFSQGSILSGALPGLQEQGLA 136
Query: 123 LKEHPPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDWLKIPSEELASAFDKP 182
L P IK+L+ I G+KF+ P++ + AY + IK SVHF+G+ D+LK E+L ++ P
Sbjct: 137 LTRVPKIKYLIIIGGAKFQSPTVAEKAYANNIKCPSVHFLGDTDFLKTHGEKLIESYVDP 196
Query: 183 LIIRHPQGHTVPRLDEVSTGQLQNFVAEI 211
IIRHP+GHTVPRLDE S + F+ +I
Sbjct: 197 FIIRHPKGHTVPRLDEKSLEIMLRFLDKI 225
>UniRef100_Q8GX45 Hypothetical protein At4g24380/T22A6_210 [Arabidopsis thaliana]
Length = 234
Score = 216 bits (549), Expect = 6e-55
Identities = 98/202 (48%), Positives = 137/202 (67%)
Query: 7 RKLKILCLHGFRTSGSFIKKQISKWDPSIFSQFHLEFPDGKFPAGGKSDIEGIFPPPYFE 66
RK + LCLHGFRTSG +K Q+ KW S+ + L F D FP GKSD+EGIF PPY+E
Sbjct: 10 RKPRFLCLHGFRTSGEIMKIQLHKWPKSVIDRLDLVFLDAPFPCQGKSDVEGIFDPPYYE 69
Query: 67 WFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQAQGKLLKEH 126
WFQF+K+FT YTN ++C+ YL + +I GPFDG +GFSQGA LS L G QA+G ++
Sbjct: 70 WFQFNKEFTEYTNFEKCLEYLEDRMIKLGPFDGLIGFSQGAILSGGLPGLQAKGIAFQKV 129
Query: 127 PPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDWLKIPSEELASAFDKPLIIR 186
P IKF++ I G+K + + + AY ++ S+HF+GE D+LK +L ++ P+++
Sbjct: 130 PKIKFVIIIGGAKLKSAKLAENAYSSSLETLSLHFLGETDFLKPYGTQLIESYKNPVVVH 189
Query: 187 HPQGHTVPRLDEVSTGQLQNFV 208
HP+GHTVPRLDE S ++ F+
Sbjct: 190 HPKGHTVPRLDEKSLEKVTAFI 211
>UniRef100_Q8LC87 Hypothetical protein [Arabidopsis thaliana]
Length = 234
Score = 215 bits (547), Expect = 1e-54
Identities = 98/202 (48%), Positives = 137/202 (67%)
Query: 7 RKLKILCLHGFRTSGSFIKKQISKWDPSIFSQFHLEFPDGKFPAGGKSDIEGIFPPPYFE 66
RK + LCLHGFRTSG +K Q+ KW S+ + L F D FP GKSD+EGIF PPY+E
Sbjct: 10 RKPRFLCLHGFRTSGEIMKIQLHKWPKSVIDRLDLVFLDAPFPCQGKSDVEGIFDPPYYE 69
Query: 67 WFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQAQGKLLKEH 126
WFQF+K+FT YTN ++C+ YL + +I GPFDG +GFSQGA LS L G QA+G ++
Sbjct: 70 WFQFNKEFTEYTNFEKCLEYLEDRMIKLGPFDGLIGFSQGAILSGGLPGLQAKGIEFQKV 129
Query: 127 PPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDWLKIPSEELASAFDKPLIIR 186
P IKF++ I G+K + + + AY ++ S+HF+GE D+LK +L ++ P+++
Sbjct: 130 PKIKFVIIIGGAKLKSAKLAENAYSSSLETLSLHFLGETDFLKPYGTQLIESYKNPVVVH 189
Query: 187 HPQGHTVPRLDEVSTGQLQNFV 208
HP+GHTVPRLDE S ++ F+
Sbjct: 190 HPKGHTVPRLDEKSLEKVTAFI 211
>UniRef100_Q9FKP9 Similarity to unknown protein [Arabidopsis thaliana]
Length = 234
Score = 207 bits (526), Expect = 3e-52
Identities = 98/207 (47%), Positives = 134/207 (64%), Gaps = 5/207 (2%)
Query: 10 KILCLHGFRTSGSFIKKQISKWDPSIFSQFHLEFPDGKFPAGGKSDIEGIFPPPYFEWFQ 69
+ILCLHGFRTSG ++ I KW +I L+F D FPA GKSD+E F PPY+EW+Q
Sbjct: 13 RILCLHGFRTSGRILQAGIGKWPDTILRDLDLDFLDAPFPATGKSDVERFFDPPYYEWYQ 72
Query: 70 FDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQAQGKLLKEHPPI 129
+K F Y N +EC++Y+ +Y+I NGPFDG LGFSQGA L+A + G Q QG L + P +
Sbjct: 73 ANKGFKEYRNFEECLAYIEDYMIKNGPFDGLLGFSQGAFLTAAIPGMQEQGSALTKVPKV 132
Query: 130 KFLVSISGSK-----FRDPSICDVAYKDPIKAKSVHFIGEKDWLKIPSEELASAFDKPLI 184
KFLV ISG+K F +P A+ P++ S+HFIGE+D+LKI E L +F +P++
Sbjct: 133 KFLVIISGAKIPGLMFGEPKAAVNAFSSPVRCPSLHFIGERDFLKIEGEVLVESFVEPVV 192
Query: 185 IRHPQGHTVPRLDEVSTGQLQNFVAEI 211
I H GH +P+LD + + +F I
Sbjct: 193 IHHSGGHIIPKLDTKAEETMLSFFQSI 219
>UniRef100_Q9STV6 Hypothetical protein T22A6.210 [Arabidopsis thaliana]
Length = 208
Score = 190 bits (483), Expect = 3e-47
Identities = 86/185 (46%), Positives = 124/185 (66%)
Query: 24 IKKQISKWDPSIFSQFHLEFPDGKFPAGGKSDIEGIFPPPYFEWFQFDKDFTVYTNLDEC 83
+K Q+ KW S+ + L F D FP GKSD+EGIF PPY+EWFQF+K+FT YTN ++C
Sbjct: 1 MKIQLHKWPKSVIDRLDLVFLDAPFPCQGKSDVEGIFDPPYYEWFQFNKEFTEYTNFEKC 60
Query: 84 ISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQAQGKLLKEHPPIKFLVSISGSKFRDP 143
+ YL + +I GPFDG +GFSQGA LS L G QA+G ++ P IKF++ I G+K +
Sbjct: 61 LEYLEDRMIKLGPFDGLIGFSQGAILSGGLPGLQAKGIAFQKVPKIKFVIIIGGAKLKSA 120
Query: 144 SICDVAYKDPIKAKSVHFIGEKDWLKIPSEELASAFDKPLIIRHPQGHTVPRLDEVSTGQ 203
+ + AY ++ S+HF+GE D+LK +L ++ P+++ HP+GHTVPRLDE S +
Sbjct: 121 KLAENAYSSSLETLSLHFLGETDFLKPYGTQLIESYKNPVVVHHPKGHTVPRLDEKSLEK 180
Query: 204 LQNFV 208
+ F+
Sbjct: 181 VTAFI 185
>UniRef100_P36591 Dihydrofolate reductase [Schizosaccharomyces pombe]
Length = 461
Score = 94.0 bits (232), Expect = 3e-18
Identities = 63/214 (29%), Positives = 96/214 (44%), Gaps = 20/214 (9%)
Query: 7 RKLKILCLHGFRTSGSFIKKQISKWDPSIFSQFHLEFPDGKFPAGGKSD----------- 55
+ LK+LCLHG+ SG K++ + L FP G A ++D
Sbjct: 3 KPLKVLCLHGWIQSGPVFSKKMGSVQKYLSKYAELHFPTGPVVADEEADPNDEEEKKRLA 62
Query: 56 -IEGIFPPPYFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLI 114
+ G F WF+ + Y + DE + + +Y+ GPFDG +GFSQGA + A+L
Sbjct: 63 ALGGEQNGGKFGWFEVEDFKNTYGSWDESLECINQYMQEKGPFDGLIGFSQGAGIGAMLA 122
Query: 115 GYQAQGK---LLKEHPPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDWLKIP 171
G+ +HPP KF+V + G + P D Y + S+H G D L +P
Sbjct: 123 QMLQPGQPPNPYVQHPPFKFVVFVGGFRAEKPEF-DHFYNPKLTTPSLHIAGTSDTL-VP 180
Query: 172 ---SEELASAFDKPLIIRHPQGHTVPRLDEVSTG 202
S++L + ++ HP H VP+ TG
Sbjct: 181 LARSKQLVERCENAHVLLHPGQHIVPQQAVYKTG 214
>UniRef100_Q6FIW8 Similar to sp|Q05015 Saccharomyces cerevisiae YMR222c [Candida
glabrata]
Length = 219
Score = 83.2 bits (204), Expect = 6e-15
Identities = 65/203 (32%), Positives = 94/203 (46%), Gaps = 21/203 (10%)
Query: 10 KILCLHGFRTSGSFIKK-------QISKWDPSIF-SQFHLEFPDGKFPAGGKSDIEGIFP 61
K+L LHG SG + ++ K D ++ FP P G +I+
Sbjct: 4 KVLMLHGLAQSGEYFASKTKGFAAELEKLDYELYYPTAPNSFPPADLPNGLLDEIDDKVS 63
Query: 62 PP--YFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQAQ 119
W Q + Y + I+YL YII NGPFDG +GFSQGA L+ L+
Sbjct: 64 SQSGVIAWLQNKETGDGYYIPETTINYLHNYIIENGPFDGIVGFSQGAGLAGYLV-TDFN 122
Query: 120 GKL---LKEHPPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDWLKIPSEELA 176
G L +E PP+KFL++ SG +FR YK+PI K++H GE D + P E++
Sbjct: 123 GLLNLTEEEQPPLKFLMAFSGFRFRGEE-HQKQYKNPISIKTLHVHGELDTVTEP-EKVQ 180
Query: 177 SAFD-----KPLIIRHPQGHTVP 194
+ +D + H GH +P
Sbjct: 181 ALYDSCDSSSRTFLTHKGGHFIP 203
>UniRef100_Q6CU79 Similar to sp|Q05015 Saccharomyces cerevisiae YMR222c
[Kluyveromyces lactis]
Length = 218
Score = 80.5 bits (197), Expect = 4e-14
Identities = 61/203 (30%), Positives = 90/203 (44%), Gaps = 23/203 (11%)
Query: 10 KILCLHGFRTSGSFIKKQISKW----DPSIFSQFHLEFPDGKFPAGGKSDIEGIFPPP-- 63
K+L LHG SG + + + + +P + F+ P PA SD+ I
Sbjct: 4 KVLMLHGLAQSGPYFESKTKGFRRVLEPLGYEFFYPTAPINLSPADLPSDVADIGAASGD 63
Query: 64 -YFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIG--YQAQG 120
Y W Q D Y D + +L EY++ NGPFDG GFSQGA ++ L+ G
Sbjct: 64 DYHAWLQPDPLHGEYRLPDVTLKFLKEYVVENGPFDGICGFSQGAGVTGYLMTDFNNLLG 123
Query: 121 KLLKEHPPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDWLKIPSE------- 173
++ P IKF ++ SG +FR + + PI +S+H G+ D + S+
Sbjct: 124 LTEEQQPAIKFFIAFSGFRFRPEIYQEQYIQHPITVRSLHVQGQLDTVTESSQVKGLYTS 183
Query: 174 --ELASAFDKPLIIRHPQGHTVP 194
E S F + H GH VP
Sbjct: 184 CKEGTSTF-----LEHSGGHFVP 201
>UniRef100_UPI0000014210 UPI0000014210 UniRef100 entry
Length = 212
Score = 78.2 bits (191), Expect = 2e-13
Identities = 58/202 (28%), Positives = 95/202 (46%), Gaps = 23/202 (11%)
Query: 9 LKILCLHGFRTSGSFIKKQISKWDPSIFSQFHLEFPDGKFPAGGKSDIEGI--------- 59
L+ILC+HG++ +GS +++ + + Q L + + A K+ G
Sbjct: 2 LRILCIHGYKQNGSTFREKTGAFRKLLKKQVELIYLNAPLKAPEKASTSGAGGDEASRAW 61
Query: 60 -FPPPYFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQA 118
F + F + LDE ++ + + GPFDG LGFSQGA A+L Q
Sbjct: 62 WFSDVQAQTFNALQQCEESLGLDESVAAVRGAVKEQGPFDGILGFSQGAAFVAMLCSLQE 121
Query: 119 QGKLLKEHPPIKFLVSISGSKFRDPSIC---DVAYKDPIKAKSVHFIGEKDWLKIP---S 172
+G L+ +F V ++G + S C V Y P+ S+H G +D + IP S
Sbjct: 122 RG--LEPDFNFRFAVIVAGFR----SACQEHQVFYNLPLHIPSLHVFGLEDRV-IPDNMS 174
Query: 173 EELASAFDKPLIIRHPQGHTVP 194
+L F++P+++ HP GH +P
Sbjct: 175 RDLLPTFEEPVLLIHPGGHFIP 196
>UniRef100_Q6CU78 Similar to sgd|S0005806 Saccharomyces cerevisiae YOR280c
[Kluyveromyces lactis]
Length = 261
Score = 77.8 bits (190), Expect = 3e-13
Identities = 66/206 (32%), Positives = 97/206 (47%), Gaps = 28/206 (13%)
Query: 10 KILCLHG-------FRTSGSFIKKQISKWDPSIFSQFHLEFPDGKFPAGGK----SDIE- 57
K+L LHG FR + ++KQ+ K + L +P G + K S+++
Sbjct: 6 KVLMLHGYSQSDVIFRAKTAGMRKQLQK------KGYELIYPCGPYKLHSKDFNDSEVDL 59
Query: 58 --GIFPPPYFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIG 115
+ WF D + YT E + YL +YI NGPFDG GFSQGA L+ L
Sbjct: 60 RDSEKSTGMYGWFLKDPETHSYTLEPELLEYLAQYIKENGPFDGIGGFSQGAGLAGYLST 119
Query: 116 --YQAQGKLLKEHPPIKFLVSISGSKFRDPSICDVAYK-DPIKAKSVHFIGEKDWL--KI 170
+ +E PP KF + SG KF +P Y+ PI+ K++H +GE D L +
Sbjct: 120 DLWSILPLNKEEQPPFKFAMYFSGFKF-EPEQFQAPYETHPIQLKTLHIVGELDSLVTEE 178
Query: 171 PSEELASAFDKP--LIIRHPQGHTVP 194
S +L A D +++H GH +P
Sbjct: 179 RSMKLFEACDPSTRTMVKHSGGHYIP 204
>UniRef100_UPI000036AA89 UPI000036AA89 UniRef100 entry
Length = 227
Score = 76.6 bits (187), Expect = 6e-13
Identities = 63/209 (30%), Positives = 95/209 (45%), Gaps = 25/209 (11%)
Query: 7 RKLKILCLHGFRTSGSFIKKQISKWDPSIFSQFHL-------EFPDGKFPAGGKSDIEGI 59
R L++LCL GFR S +++ ++ + L PD P G +SD G
Sbjct: 5 RPLRVLCLAGFRQSERGFREKTGALRKALRGRAELVCLSGPHPVPDPPGPEGARSDF-GS 63
Query: 60 FPP---PYFEWFQFDK--------DFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGAT 108
PP P WF + + V L+E + + + + GPFDG LGFSQGA
Sbjct: 64 CPPEEQPRGWWFSEQEADVFSALEEPAVCRGLEESLGMVAQALNRLGPFDGLLGFSQGAA 123
Query: 109 LSALLIGYQAQGKLLKEHPPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDWL 168
L+AL+ G P +F++ +SG R + + P+ S+H G+ D +
Sbjct: 124 LAALVCALGQAGD--PRFPLPRFIILVSGFCPRGIGFKESILQRPLSLPSLHVFGDTDKV 181
Query: 169 KIPSEE---LASAFDKPLIIRHPQGHTVP 194
IPS+E LAS F + + H GH +P
Sbjct: 182 -IPSQESMQLASQFPGAITLTHSGGHFIP 209
>UniRef100_Q18169 Hypothetical protein C25G4.2 [Caenorhabditis elegans]
Length = 221
Score = 76.3 bits (186), Expect = 7e-13
Identities = 57/198 (28%), Positives = 91/198 (45%), Gaps = 9/198 (4%)
Query: 8 KLKILCLHGFRTSGSFIKKQISKWDPSIFSQFHLEFPDGKFPAGGKSDIEGI----FPPP 63
KL+ILCLHG+R +++ + S EF +G ++ F
Sbjct: 6 KLRILCLHGYRQCDQSFRQKTGSTRKLVKSLAEFEFVNGVHSVAVDEHVDSSRAWWFSNN 65
Query: 64 YFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQAQGKLL 123
F + V +E ++ + ++I NGPFDG LGFSQGA++ LLI G++
Sbjct: 66 EAMSFSSRESTEVAVGFEESVAAVVKFIEENGPFDGLLGFSQGASMVHLLIAKAQLGEI- 124
Query: 124 KEHPPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDWL--KIPSEELASAFD- 180
+ P I+F + SG + + S+H G+ D + + SE++A FD
Sbjct: 125 -KLPGIRFAIFFSGFLSLSSKHDSLTLLRIKEFPSMHVFGDADEIVARPKSEKMADMFDV 183
Query: 181 KPLIIRHPQGHTVPRLDE 198
+PL I H GH VP + +
Sbjct: 184 EPLRIAHDGGHVVPSMSK 201
>UniRef100_Q8WZ82 Candidate tumor suppressor OVCA2 [Homo sapiens]
Length = 227
Score = 75.9 bits (185), Expect = 1e-12
Identities = 63/209 (30%), Positives = 95/209 (45%), Gaps = 25/209 (11%)
Query: 7 RKLKILCLHGFRTSGSFIKKQISKWDPSIFSQFHL-------EFPDGKFPAGGKSDIEGI 59
R L++LCL GFR S +++ ++ + L PD P G +SD G
Sbjct: 5 RPLRVLCLAGFRQSERGFREKTGALRKALRGRAELVCLSGPHPVPDPPGPEGARSDF-GS 63
Query: 60 FPP---PYFEWFQFDK--------DFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGAT 108
PP P WF + + V L+E + + + + GPFDG LGFSQGA
Sbjct: 64 CPPEEQPRGWWFSEQEADVFSALEEPAVCRGLEESLGMVAQALNRLGPFDGLLGFSQGAA 123
Query: 109 LSALLIGYQAQGKLLKEHPPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDWL 168
L+AL+ G P +F++ +SG R + + P+ S+H G+ D +
Sbjct: 124 LAALVCALGQAGD--PRFPLPRFILLVSGFCPRGIGFKESILQRPLSLPSLHVFGDTDKV 181
Query: 169 KIPSEE---LASAFDKPLIIRHPQGHTVP 194
IPS+E LAS F + + H GH +P
Sbjct: 182 -IPSQESVQLASQFPGAITLTHSGGHFIP 209
>UniRef100_Q8IW87 Candidate tumor suppressor in ovarian cancer 2 [Homo sapiens]
Length = 227
Score = 75.9 bits (185), Expect = 1e-12
Identities = 63/209 (30%), Positives = 95/209 (45%), Gaps = 25/209 (11%)
Query: 7 RKLKILCLHGFRTSGSFIKKQISKWDPSIFSQFHL-------EFPDGKFPAGGKSDIEGI 59
R L++LCL GFR S +++ ++ + L PD P G +SD G
Sbjct: 5 RPLRVLCLAGFRQSERGFREKTGALRKALRGRAELVCLSGPHPVPDPPGPEGARSDF-GS 63
Query: 60 FPP---PYFEWFQFDK--------DFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGAT 108
PP P WF + + V L+E + + + + GPFDG LGFSQGA
Sbjct: 64 CPPEEQPRGWWFSEQEADVFCALEEPAVCRGLEESLGMVAQALNRLGPFDGLLGFSQGAA 123
Query: 109 LSALLIGYQAQGKLLKEHPPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDWL 168
L+AL+ G P +F++ +SG R + + P+ S+H G+ D +
Sbjct: 124 LAALVCALGQAGD--PRFPLPRFILLVSGFCPRGIGFKESILQRPLSLPSLHVFGDTDKV 181
Query: 169 KIPSEE---LASAFDKPLIIRHPQGHTVP 194
IPS+E LAS F + + H GH +P
Sbjct: 182 -IPSQESVQLASQFPGAITLTHSGGHFIP 209
>UniRef100_UPI00002F9C06 UPI00002F9C06 UniRef100 entry
Length = 228
Score = 75.5 bits (184), Expect = 1e-12
Identities = 59/216 (27%), Positives = 97/216 (44%), Gaps = 37/216 (17%)
Query: 9 LKILCLHGFRTSGSFIKKQISKWDPSIFSQFHLEFPDG-------KFPA----------- 50
L++LC+HG+R +GS +++ + + Q L F D + PA
Sbjct: 4 LRVLCIHGYRQNGSTFREKTGAFRKLLKKQVELIFVDAPLSVQHIRSPAFIVCLEAPETT 63
Query: 51 ------GGKSDIEGIFPPPYFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFS 104
G ++ F F + +DE ++ + + GPFDG LGFS
Sbjct: 64 SASGAEGDEASRAWWFSDVQARSFNAQQQCEESLGMDESVAAVRAAVKEQGPFDGILGFS 123
Query: 105 QGATLSALLIGYQAQGKLLKEHPPIKFLVSISGSKFRDPSIC---DVAYKDPIKAKSVHF 161
QGA L A+L Q +G P +F ++ + FR S C Y+ P++ S+H
Sbjct: 124 QGAALVAMLCSLQERGL----EPDFRFRFAVLVAGFR--SACQEHQEFYRVPLQMPSLHV 177
Query: 162 IGEKDWLKIP---SEELASAFDKPLIIRHPQGHTVP 194
G +D + IP S +L F++P+++ HP GH +P
Sbjct: 178 FGLEDRV-IPDSMSRDLLPTFEEPVVLIHPGGHFIP 212
>UniRef100_Q61YZ4 Hypothetical protein CBG03338 [Caenorhabditis briggsae]
Length = 220
Score = 74.7 bits (182), Expect = 2e-12
Identities = 60/201 (29%), Positives = 94/201 (45%), Gaps = 15/201 (7%)
Query: 8 KLKILCLHGFRTSGSFIKKQISKWDPSIFSQFHLEFPDGKFPAGGKSDIEGI----FPPP 63
KL+ILCLHG+R +++ + + EF +G E F
Sbjct: 6 KLRILCLHGYRQCDQSFRQKTGSTRKLVKALADFEFVNGIHSVAVDEHSETSRAWWFSNA 65
Query: 64 YFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQAQGKLL 123
F + V DE ++ + ++I NGPFDG LGFSQGA++ LLI G++
Sbjct: 66 DQMSFSSREPTDVSVGFDESVNAVVKFIEDNGPFDGLLGFSQGASMVHLLIAKAQLGEI- 124
Query: 124 KEHPPIKFLVSISG---SKFRDPSICDVAYKDPIKAKSVHFIGEKDWL--KIPSEELASA 178
+ P I+F + SG + ++ + KD S+H G+ D + + SE+LA
Sbjct: 125 -KLPGIRFAIFFSGFLSLSSKHDTLTSLRIKD---FPSMHVFGDSDEIVARPKSEKLADQ 180
Query: 179 FD-KPLIIRHPQGHTVPRLDE 198
FD +P+ I H GH VP + +
Sbjct: 181 FDMEPIRIAHEGGHLVPSMSK 201
>UniRef100_Q6C8U4 Yarrowia lipolytica chromosome D of strain CLIB99 of Yarrowia
lipolytica [Yarrowia lipolytica]
Length = 238
Score = 74.7 bits (182), Expect = 2e-12
Identities = 64/221 (28%), Positives = 103/221 (45%), Gaps = 32/221 (14%)
Query: 8 KLKILCLHGFRTSGSFIKKQISKWDPSIFSQFH----------LEFPDGKFPAGGKSDIE 57
K K+L LHGF SGS K+ S ++ Q + L PD F S+++
Sbjct: 5 KGKLLFLHGFTQSGSLFAKKTSALRKALQKQGYQCFYIDAPVELSAPDLPFDT---SNLD 61
Query: 58 GIFPPPYFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATLSALLIGYQ 117
+ W+ +++ Y LD+ + + I +GPFDG +GFSQGA ++ +L
Sbjct: 62 SSADTDWKSWWVTNQNKPDYYKLDKAFDSVRDAIEKDGPFDGVMGFSQGAAMAGVLC--S 119
Query: 118 AQGKLLKEHPPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDWLKIPSEE--- 174
L ++ PP+K+ V G + P ++ PI ++H +G D + SEE
Sbjct: 120 QIHNLHEKQPPVKYGVLFCGFRIA-PEEYQKFFEPPIATNTLHVLGSLD--TVVSEERSL 176
Query: 175 -LASAFDKP--LIIRHPQGHTVPRLDEVSTGQLQNFVAEIL 212
L +A D+ +I+HP GH VP +NFV +I+
Sbjct: 177 GLWNACDEKTRTMIKHPGGHFVP--------NSKNFVTDII 209
>UniRef100_Q86XN3 Ovarian cancer gene-2 protein [Homo sapiens]
Length = 227
Score = 73.6 bits (179), Expect = 5e-12
Identities = 62/209 (29%), Positives = 94/209 (44%), Gaps = 25/209 (11%)
Query: 7 RKLKILCLHGFRTSGSFIKKQISKWDPSIFSQFHL-------EFPDGKFPAGGKSDIEGI 59
R L++LCL GFR S +++ ++ + L PD P G +SD G
Sbjct: 5 RPLRVLCLAGFRQSERGFREKTGALRKALRGRAELVCLSGPHPVPDPPGPEGARSDF-GS 63
Query: 60 FPP---PYFEWFQFDK--------DFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGAT 108
PP P WF + + V L+E + + + + GPFDG LGFSQGA
Sbjct: 64 CPPEEQPRGWWFSEQEADVFSALEEPAVCRGLEESLGMVAQALNRLGPFDGLLGFSQGAA 123
Query: 109 LSALLIGYQAQGKLLKEHPPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDWL 168
L+AL+ G P +F++ +S R + + P+ S+H G+ D +
Sbjct: 124 LAALVCALGQAGD--PRFPLPRFILLVSSFCPRGIGFKESILQRPLSLPSLHVFGDTDKV 181
Query: 169 KIPSEE---LASAFDKPLIIRHPQGHTVP 194
IPS+E LAS F + + H GH +P
Sbjct: 182 -IPSQESVQLASQFPGAITLTHSGGHFIP 209
>UniRef100_Q99369 Hypothetical dihydrofolate reductase [Saccharomyces cerevisiae]
Length = 266
Score = 72.4 bits (176), Expect = 1e-11
Identities = 69/254 (27%), Positives = 108/254 (42%), Gaps = 32/254 (12%)
Query: 8 KLKILCLHGFRTSGSF-------IKKQISKWDPSIF-----------SQFHLEFPDGKFP 49
K K+L LHGF S ++K + K ++ + F E G+
Sbjct: 4 KKKVLMLHGFVQSDKIFSAKTGGLRKNLKKLGYDLYYPCAPHSIDKKALFQSESEKGR-D 62
Query: 50 AGGKSDIEGIFPPPYFEWFQFDKDFTVYTNLDECISYLTEYIIANGPFDGFLGFSQGATL 109
A + + Y +F+ + F + + +YL Y++ NGPFDG +GFSQGA L
Sbjct: 63 AAKEFNTSATSDEVYGWFFRNPESFNSFQIDQKVFNYLRNYVLENGPFDGVIGFSQGAGL 122
Query: 110 SALLIG--YQAQGKLLKEHPPIKFLVSISGSKFRDPSICDVAYKDPIKAKSVHFIGEKDW 167
L+ + ++ P +KF +S SG K D S Y I+ S+H GE D
Sbjct: 123 GGYLVTDFNRILNLTDEQQPALKFFISFSGFKLEDQSY-QKEYHRIIQVPSLHVRGELDE 181
Query: 168 LKIPSEELAS----AFDKPLIIRHPQGHTVPRLDEVSTGQLQNFVAEILSQPKVGVSICE 223
+ S +A +K ++ HP H VP + Q+ N++ I S+ E
Sbjct: 182 VVAESRIMALYESWPDNKRTLLVHPGAHFVPN-SKPFVSQVCNWIQGITSKEGQ-----E 235
Query: 224 HESKVEVDGTNGDK 237
H ++ EVD DK
Sbjct: 236 HNAQPEVDRKQFDK 249
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.140 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 485,425,040
Number of Sequences: 2790947
Number of extensions: 21397567
Number of successful extensions: 44350
Number of sequences better than 10.0: 89
Number of HSP's better than 10.0 without gapping: 55
Number of HSP's successfully gapped in prelim test: 34
Number of HSP's that attempted gapping in prelim test: 44202
Number of HSP's gapped (non-prelim): 102
length of query: 257
length of database: 848,049,833
effective HSP length: 125
effective length of query: 132
effective length of database: 499,181,458
effective search space: 65891952456
effective search space used: 65891952456
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 73 (32.7 bits)
Medicago: description of AC145330.4


