

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145329.3 + phase: 0 /partial
(121 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
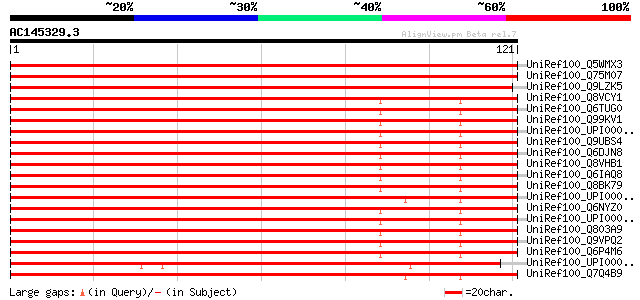
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q5WMX3 Hypothetical protein P0431G05.15 [Oryza sativa] 201 3e-51
UniRef100_Q75M07 Hypothetical protein P0676G05.7 [Oryza sativa] 201 3e-51
UniRef100_Q9LZK5 Hypothetical protein F26K9_30 [Arabidopsis thal... 192 2e-48
UniRef100_Q8VCY1 DnaJ (Hsp40) homolog, subfamily B, member 11 [M... 114 7e-25
UniRef100_Q6TUG0 LRRGT00084 [Rattus norvegicus] 114 7e-25
UniRef100_Q99KV1 DnaJ homolog subfamily B member 11 precursor [M... 114 7e-25
UniRef100_UPI00003622FB UPI00003622FB UniRef100 entry 113 1e-24
UniRef100_Q9UBS4 DnaJ homolog subfamily B member 11 precursor [H... 111 3e-24
UniRef100_Q6DJN8 MGC81924 protein [Xenopus laevis] 111 4e-24
UniRef100_Q8VHB1 Apobec-1 binding protein 2 [Mus musculus] 111 4e-24
UniRef100_Q6IAQ8 DNAJB11 protein [Homo sapiens] 111 4e-24
UniRef100_Q8BK79 Mus musculus 10 day old male pancreas cDNA, RIK... 110 6e-24
UniRef100_UPI00004322C0 UPI00004322C0 UniRef100 entry 109 1e-23
UniRef100_Q6NYZ0 Dnajb11 protein [Brachydanio rerio] 109 2e-23
UniRef100_UPI00003AED76 UPI00003AED76 UniRef100 entry 108 2e-23
UniRef100_Q803A9 DnaJ (Hsp40) homolog, subfamily B, member 11 [B... 108 2e-23
UniRef100_Q9VPQ2 CG4164-PA [Drosophila melanogaster] 108 2e-23
UniRef100_Q6P4M6 Hypothetical protein MGC75796 [Xenopus tropicalis] 106 1e-22
UniRef100_UPI00002D7590 UPI00002D7590 UniRef100 entry 103 7e-22
UniRef100_Q7Q4B9 ENSANGP00000018254 [Anopheles gambiae str. PEST] 102 2e-21
>UniRef100_Q5WMX3 Hypothetical protein P0431G05.15 [Oryza sativa]
Length = 127
Score = 201 bits (511), Expect = 3e-51
Identities = 95/121 (78%), Positives = 111/121 (91%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
EV F+E+GEP IDGE GDL+FRIRTAPHE F+REGNDLHTTVTI+L+QALVGFEKTIKHL
Sbjct: 6 EVSFFEEGEPKIDGEPGDLKFRIRTAPHERFRREGNDLHTTVTISLLQALVGFEKTIKHL 65
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMPLHTSTKKGDLYVTFEVLFPTTLSEEQKTKIKSI 120
D H+V+I +KGIT PK+VRKFKGEGMPL+ S KKGDLYVTFEVLFP TL+++QK+K+KSI
Sbjct: 66 DNHMVEIGTKGITKPKEVRKFKGEGMPLYQSNKKGDLYVTFEVLFPKTLTDDQKSKLKSI 125
Query: 121 L 121
L
Sbjct: 126 L 126
>UniRef100_Q75M07 Hypothetical protein P0676G05.7 [Oryza sativa]
Length = 347
Score = 201 bits (511), Expect = 3e-51
Identities = 95/121 (78%), Positives = 111/121 (91%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
EV F+E+GEP IDGE GDL+FRIRTAPHE F+REGNDLHTTVTI+L+QALVGFEKTIKHL
Sbjct: 226 EVSFFEEGEPKIDGEPGDLKFRIRTAPHERFRREGNDLHTTVTISLLQALVGFEKTIKHL 285
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMPLHTSTKKGDLYVTFEVLFPTTLSEEQKTKIKSI 120
D H+V+I +KGIT PK+VRKFKGEGMPL+ S KKGDLYVTFEVLFP TL+++QK+K+KSI
Sbjct: 286 DNHMVEIGTKGITKPKEVRKFKGEGMPLYQSNKKGDLYVTFEVLFPKTLTDDQKSKLKSI 345
Query: 121 L 121
L
Sbjct: 346 L 346
>UniRef100_Q9LZK5 Hypothetical protein F26K9_30 [Arabidopsis thaliana]
Length = 346
Score = 192 bits (487), Expect = 2e-48
Identities = 89/120 (74%), Positives = 107/120 (89%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
EV FYEDGEPI+DG+ GDL+FRIRTAPH F+R+GNDLH V ITLV+ALVGFEK+ KHL
Sbjct: 225 EVSFYEDGEPILDGDPGDLKFRIRTAPHARFRRDGNDLHMNVNITLVEALVGFEKSFKHL 284
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMPLHTSTKKGDLYVTFEVLFPTTLSEEQKTKIKSI 120
D+H VDISSKGIT PK+V+KFKGEGMPLH STKKG+L+VTFEVLFP++L+++QK KIK +
Sbjct: 285 DDHEVDISSKGITKPKEVKKFKGEGMPLHYSTKKGNLFVTFEVLFPSSLTDDQKKKIKEV 344
>UniRef100_Q8VCY1 DnaJ (Hsp40) homolog, subfamily B, member 11 [Mus musculus]
Length = 358
Score = 114 bits (284), Expect = 7e-25
Identities = 63/123 (51%), Positives = 79/123 (64%), Gaps = 2/123 (1%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
E F +GEP +DGE GDLRFRI+ H +F+R G+DL+T VT++LV+ALVGFE I HL
Sbjct: 221 EYPFIGEGEPHVDGEPGDLRFRIKVVKHRIFERRGDDLYTNVTVSLVEALVGFEMDITHL 280
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMP-LHTSTKKGDLYVTFEVLFP-TTLSEEQKTKIK 118
D H V IS IT P KGEG+P + KG L +TF+V FP L+EE K IK
Sbjct: 281 DGHKVHISRDKITRPGAKLWKKGEGLPNFDNNNIKGSLIITFDVDFPKEQLTEEAKEGIK 340
Query: 119 SIL 121
+L
Sbjct: 341 QLL 343
>UniRef100_Q6TUG0 LRRGT00084 [Rattus norvegicus]
Length = 358
Score = 114 bits (284), Expect = 7e-25
Identities = 63/123 (51%), Positives = 79/123 (64%), Gaps = 2/123 (1%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
E F +GEP +DGE GDLRFRI+ H +F+R G+DL+T VT++LV+ALVGFE I HL
Sbjct: 221 EYPFIGEGEPHVDGEPGDLRFRIKVVKHRIFERRGDDLYTNVTVSLVEALVGFEMDITHL 280
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMP-LHTSTKKGDLYVTFEVLFP-TTLSEEQKTKIK 118
D H V IS IT P KGEG+P + KG L +TF+V FP L+EE K IK
Sbjct: 281 DGHKVHISRDKITRPGAKLWKKGEGLPNFDNNNIKGSLIITFDVDFPKEQLTEEAKEGIK 340
Query: 119 SIL 121
+L
Sbjct: 341 QLL 343
>UniRef100_Q99KV1 DnaJ homolog subfamily B member 11 precursor [Mus musculus]
Length = 358
Score = 114 bits (284), Expect = 7e-25
Identities = 63/123 (51%), Positives = 79/123 (64%), Gaps = 2/123 (1%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
E F +GEP +DGE GDLRFRI+ H +F+R G+DL+T VT++LV+ALVGFE I HL
Sbjct: 221 EYPFIGEGEPHVDGEPGDLRFRIKVVKHRIFERRGDDLYTNVTVSLVEALVGFEMDITHL 280
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMP-LHTSTKKGDLYVTFEVLFP-TTLSEEQKTKIK 118
D H V IS IT P KGEG+P + KG L +TF+V FP L+EE K IK
Sbjct: 281 DGHKVHISRDKITRPGAKLWKKGEGLPNFDNNNIKGSLIITFDVDFPKEQLTEEAKEGIK 340
Query: 119 SIL 121
+L
Sbjct: 341 QLL 343
>UniRef100_UPI00003622FB UPI00003622FB UniRef100 entry
Length = 360
Score = 113 bits (282), Expect = 1e-24
Identities = 64/123 (52%), Positives = 79/123 (64%), Gaps = 2/123 (1%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
E F +GEP IDGE GDLRFRI+ H LF+R G+DL+T VTI+LV+ALVGFE I HL
Sbjct: 223 EYPFIGEGEPHIDGEPGDLRFRIKVLKHPLFERRGDDLYTNVTISLVEALVGFEMDIVHL 282
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMP-LHTSTKKGDLYVTFEVLFP-TTLSEEQKTKIK 118
D H V I IT P KGEG+P + +G L +TF+V FP T L E+QK I+
Sbjct: 283 DGHKVHIVRDKITKPGARMWKKGEGLPNFDNNNIRGSLIITFDVEFPGTQLDEQQKDGIR 342
Query: 119 SIL 121
+L
Sbjct: 343 GLL 345
>UniRef100_Q9UBS4 DnaJ homolog subfamily B member 11 precursor [Homo sapiens]
Length = 358
Score = 111 bits (278), Expect = 3e-24
Identities = 62/123 (50%), Positives = 79/123 (63%), Gaps = 2/123 (1%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
E F +GEP +DGE GDLRFRI+ H +F+R G+DL+T VTI+LV++LVGFE I HL
Sbjct: 221 EYPFIGEGEPHVDGEPGDLRFRIKVVKHPIFERRGDDLYTNVTISLVESLVGFEMDITHL 280
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMP-LHTSTKKGDLYVTFEVLFP-TTLSEEQKTKIK 118
D H V IS IT P KGEG+P + KG L +TF+V FP L+EE + IK
Sbjct: 281 DGHKVHISRDKITRPGAKLWKKGEGLPNFDNNNIKGSLIITFDVDFPKEQLTEEAREGIK 340
Query: 119 SIL 121
+L
Sbjct: 341 QLL 343
>UniRef100_Q6DJN8 MGC81924 protein [Xenopus laevis]
Length = 360
Score = 111 bits (277), Expect = 4e-24
Identities = 61/123 (49%), Positives = 79/123 (63%), Gaps = 2/123 (1%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
E F +GEP IDGE GDLRFRI+ H +F+R G+DL+T V+I+LV+AL+GFE I HL
Sbjct: 223 EYPFIGEGEPHIDGEPGDLRFRIKVLKHPIFERRGDDLYTNVSISLVEALIGFEMDITHL 282
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMP-LHTSTKKGDLYVTFEVLFP-TTLSEEQKTKIK 118
D H V I IT P KGEG+P + KG L +TF+V FP L+EEQ+ +K
Sbjct: 283 DGHKVHIMRDKITKPGAKLWKKGEGLPNFDNNNIKGSLIITFDVEFPKEQLTEEQRQGVK 342
Query: 119 SIL 121
+L
Sbjct: 343 QLL 345
>UniRef100_Q8VHB1 Apobec-1 binding protein 2 [Mus musculus]
Length = 358
Score = 111 bits (277), Expect = 4e-24
Identities = 62/123 (50%), Positives = 78/123 (63%), Gaps = 2/123 (1%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
E F +GEP +DGE GDL FRI+ H +F+R G+DL+T VT++LV+ALVGFE I HL
Sbjct: 221 EYPFIGEGEPHVDGEPGDLPFRIKVVKHRIFERRGDDLYTNVTVSLVEALVGFEMDITHL 280
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMP-LHTSTKKGDLYVTFEVLFP-TTLSEEQKTKIK 118
D H V IS IT P KGEG+P + KG L +TF+V FP L+EE K IK
Sbjct: 281 DGHKVHISRDKITRPGAKLWKKGEGLPNFDNNNIKGSLIITFDVDFPKEQLTEEAKEGIK 340
Query: 119 SIL 121
+L
Sbjct: 341 QLL 343
>UniRef100_Q6IAQ8 DNAJB11 protein [Homo sapiens]
Length = 358
Score = 111 bits (277), Expect = 4e-24
Identities = 62/123 (50%), Positives = 79/123 (63%), Gaps = 2/123 (1%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
E F +GEP +DGE GDLRFRI+ H +F+R G+DL+T VTI+LV++LVGFE I HL
Sbjct: 221 EYPFIGEGEPHVDGEPGDLRFRIKVVRHPIFERRGDDLYTNVTISLVESLVGFEMDITHL 280
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMP-LHTSTKKGDLYVTFEVLFP-TTLSEEQKTKIK 118
D H V IS IT P KGEG+P + KG L +TF+V FP L+EE + IK
Sbjct: 281 DGHKVHISRDKITRPGAKLWKKGEGLPNFDNNNIKGSLIITFDVDFPKEQLTEEAREGIK 340
Query: 119 SIL 121
+L
Sbjct: 341 QLL 343
>UniRef100_Q8BK79 Mus musculus 10 day old male pancreas cDNA, RIKEN full-length
enriched library, clone:1810031F23 product:DNAJ HOMOLOG
SUBFAMILY B MEMBER 11, full insert sequence [Mus
musculus]
Length = 358
Score = 110 bits (276), Expect = 6e-24
Identities = 62/123 (50%), Positives = 78/123 (63%), Gaps = 2/123 (1%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
E F +GEP +DGE GDLRFRI+ H +F+R +DL+T VT++LV+ALVGFE I HL
Sbjct: 221 EYPFIGEGEPHVDGEPGDLRFRIKVVKHRIFERREDDLYTNVTVSLVEALVGFEMDITHL 280
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMP-LHTSTKKGDLYVTFEVLFP-TTLSEEQKTKIK 118
D H V IS IT P KGEG+P + KG L +TF+V FP L+EE K IK
Sbjct: 281 DGHKVHISRDKITRPGAKLWKKGEGLPNFDNNNIKGSLIITFDVDFPKEQLTEEAKEGIK 340
Query: 119 SIL 121
+L
Sbjct: 341 QLL 343
>UniRef100_UPI00004322C0 UPI00004322C0 UniRef100 entry
Length = 248
Score = 109 bits (273), Expect = 1e-23
Identities = 57/123 (46%), Positives = 81/123 (65%), Gaps = 2/123 (1%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
E F +GEP +DGE GDL +IRT PH +F+R G+DL+T +TI++ AL+GF+ I+HL
Sbjct: 112 ETKFTAEGEPHLDGEPGDLILKIRTQPHPVFERIGDDLYTNITISMQDALIGFKMDIEHL 171
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMPLHTSTK-KGDLYVTFEVLFP-TTLSEEQKTKIK 118
D H V I +T P + KGEGMP + + G LY+TF+V FP T ++ QK +IK
Sbjct: 172 DGHKVTIQRDKVTKPGARIRKKGEGMPNYENNNLHGTLYITFDVAFPETEFTDIQKEEIK 231
Query: 119 SIL 121
++L
Sbjct: 232 NLL 234
>UniRef100_Q6NYZ0 Dnajb11 protein [Brachydanio rerio]
Length = 360
Score = 109 bits (272), Expect = 2e-23
Identities = 62/123 (50%), Positives = 78/123 (63%), Gaps = 2/123 (1%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
E F +GEP IDGE GDLRFRI+ H +F+R G+DL+T VTI+LV+ALVGFE I HL
Sbjct: 223 EYPFIGEGEPHIDGEPGDLRFRIKVLKHPVFERRGDDLYTNVTISLVEALVGFEMDITHL 282
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMP-LHTSTKKGDLYVTFEVLFP-TTLSEEQKTKIK 118
D H V I IT P KGEG+P + +G L +TF+V FP L ++QK IK
Sbjct: 283 DGHKVHIVRDKITKPGSRIWKKGEGLPSFDNNNIRGSLIITFDVDFPKEQLDDQQKDGIK 342
Query: 119 SIL 121
+L
Sbjct: 343 QLL 345
>UniRef100_UPI00003AED76 UPI00003AED76 UniRef100 entry
Length = 358
Score = 108 bits (271), Expect = 2e-23
Identities = 59/123 (47%), Positives = 78/123 (62%), Gaps = 2/123 (1%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
E F +GEP +DGE GDLRFRI+ H +F+R G+DL+T VTI+LV+AL GFE I HL
Sbjct: 221 EYPFIGEGEPHVDGEPGDLRFRIKVLKHPVFERRGDDLYTNVTISLVEALTGFEMDITHL 280
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMP-LHTSTKKGDLYVTFEVLFP-TTLSEEQKTKIK 118
D H V ++ IT P KGEG+P + KG L +TF+V FP L+ EQ+ +K
Sbjct: 281 DGHKVHVARDKITKPGAKLWKKGEGLPNFDNNNIKGSLIITFDVEFPKEQLTSEQREGLK 340
Query: 119 SIL 121
+L
Sbjct: 341 QLL 343
>UniRef100_Q803A9 DnaJ (Hsp40) homolog, subfamily B, member 11 [Brachydanio rerio]
Length = 360
Score = 108 bits (271), Expect = 2e-23
Identities = 62/123 (50%), Positives = 78/123 (63%), Gaps = 2/123 (1%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
E F +GEP IDGE GDLRFRI+ H +F+R G+DL+T VTI+LV+ALVGFE I HL
Sbjct: 223 EYPFIGEGEPHIDGEPGDLRFRIKVLKHPVFERRGDDLYTNVTISLVEALVGFEVDITHL 282
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMP-LHTSTKKGDLYVTFEVLFP-TTLSEEQKTKIK 118
D H V I IT P KGEG+P + +G L +TF+V FP L ++QK IK
Sbjct: 283 DGHKVHIVRDKITKPGSRIWKKGEGLPSFDNNNIRGSLIITFDVDFPREQLDDQQKDGIK 342
Query: 119 SIL 121
+L
Sbjct: 343 QLL 345
>UniRef100_Q9VPQ2 CG4164-PA [Drosophila melanogaster]
Length = 354
Score = 108 bits (271), Expect = 2e-23
Identities = 60/123 (48%), Positives = 78/123 (62%), Gaps = 2/123 (1%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
E F +GEP IDGE GDL R++ PH F R+ +DL+T VTI+L ALVGF IKHL
Sbjct: 220 ETRFVAEGEPHIDGEPGDLIVRVQQMPHPRFLRKNDDLYTNVTISLQDALVGFSMEIKHL 279
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMP-LHTSTKKGDLYVTFEVLFP-TTLSEEQKTKIK 118
D HLV ++ + +T P + KGEGMP + G+LY+TF+V FP L+EE K +K
Sbjct: 280 DGHLVPVTREKVTWPGARIRKKGEGMPNFENNNLTGNLYITFDVEFPKKDLTEEDKEALK 339
Query: 119 SIL 121
IL
Sbjct: 340 KIL 342
>UniRef100_Q6P4M6 Hypothetical protein MGC75796 [Xenopus tropicalis]
Length = 360
Score = 106 bits (264), Expect = 1e-22
Identities = 59/123 (47%), Positives = 77/123 (61%), Gaps = 2/123 (1%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
E F +GEP IDGE GDLRFRI+ H +F+R G+DL+T V+I+LV+AL GFE I HL
Sbjct: 223 EYPFIGEGEPHIDGEPGDLRFRIKVLKHPIFERRGDDLYTNVSISLVEALTGFEMDIAHL 282
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMP-LHTSTKKGDLYVTFEVLFP-TTLSEEQKTKIK 118
D H V I IT P KGEG+P + KG L +TF+V FP L+ EQ+ +K
Sbjct: 283 DGHKVHILRDKITKPGAKLWKKGEGLPNFDNNNIKGSLIITFDVEFPKEQLTMEQRQGVK 342
Query: 119 SIL 121
++
Sbjct: 343 QLM 345
>UniRef100_UPI00002D7590 UPI00002D7590 UniRef100 entry
Length = 305
Score = 103 bits (258), Expect = 7e-22
Identities = 55/121 (45%), Positives = 77/121 (63%), Gaps = 4/121 (3%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHEL--FKREG-NDLHTTVTITLVQALVGFEKTI 57
E+LF+E+G+ +IDG+ GDL F ++T E KR G +DLH T ITL++AL GF K
Sbjct: 168 EILFFEEGDAMIDGDPGDLVFVVKTRDDEKNRIKRVGASDLHMTYEITLIEALNGFSKIF 227
Query: 58 KHLDEHLVDISSKGITNPKQVRKFKGEGMPLHTSTKK-GDLYVTFEVLFPTTLSEEQKTK 116
KH D H V I G+T P +GEG+P H KK GDL++TF+V FP +L++ Q+
Sbjct: 228 KHYDGHEVKIERDGVTVPFDKMTLRGEGLPKHNQFKKFGDLFITFQVQFPDSLNKAQRDA 287
Query: 117 I 117
+
Sbjct: 288 V 288
>UniRef100_Q7Q4B9 ENSANGP00000018254 [Anopheles gambiae str. PEST]
Length = 318
Score = 102 bits (254), Expect = 2e-21
Identities = 55/123 (44%), Positives = 79/123 (63%), Gaps = 2/123 (1%)
Query: 1 EVLFYEDGEPIIDGESGDLRFRIRTAPHELFKREGNDLHTTVTITLVQALVGFEKTIKHL 60
E F +GEP +DGE GDL +I+T PH F+R G+DL+T +TI+L ALVGF I HL
Sbjct: 196 ETRFSGEGEPHMDGEPGDLILKIKTVPHTRFERRGDDLYTNITISLQDALVGFTLDIVHL 255
Query: 61 DEHLVDISSKGITNPKQVRKFKGEGMPLHTSTK-KGDLYVTFEVLFP-TTLSEEQKTKIK 118
D H V ++ + +T P + GEGMP + + G LY+TF+V FP + L++ +K IK
Sbjct: 256 DGHKVTVTREKVTWPGARIRKNGEGMPNYENNNLHGTLYITFDVEFPKSQLTDTEKEDIK 315
Query: 119 SIL 121
++L
Sbjct: 316 NLL 318
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.138 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 200,758,375
Number of Sequences: 2790947
Number of extensions: 7772833
Number of successful extensions: 19493
Number of sequences better than 10.0: 1159
Number of HSP's better than 10.0 without gapping: 987
Number of HSP's successfully gapped in prelim test: 172
Number of HSP's that attempted gapping in prelim test: 17642
Number of HSP's gapped (non-prelim): 1415
length of query: 121
length of database: 848,049,833
effective HSP length: 97
effective length of query: 24
effective length of database: 577,327,974
effective search space: 13855871376
effective search space used: 13855871376
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 67 (30.4 bits)
Medicago: description of AC145329.3


