

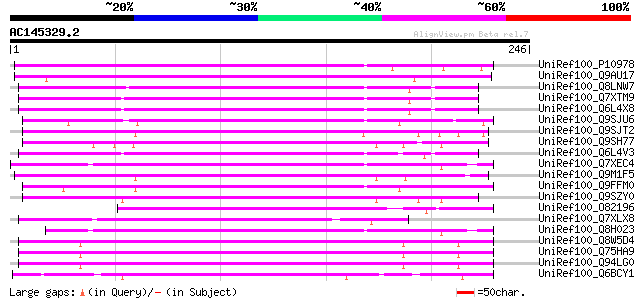
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145329.2 + phase: 0 /pseudo
(246 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_P10978 Retrovirus-related Pol polyprotein from transpo... 152 1e-35
UniRef100_Q9AU17 Polyprotein-like [Lycopersicon chilense] 137 3e-31
UniRef100_Q8LNW7 Putative polyprotein [Oryza sativa] 136 4e-31
UniRef100_Q7XTM9 OSJNBa0033G05.13 protein [Oryza sativa] 136 4e-31
UniRef100_Q6L4X8 Putative polyprotein [Oryza sativa] 134 2e-30
UniRef100_Q9SJU6 Putative retroelement pol polyprotein [Arabidop... 132 6e-30
UniRef100_Q9SJT2 Putative retroelement pol polyprotein [Arabidop... 132 1e-29
UniRef100_Q9SH77 Putative retroelement pol polyprotein [Arabidop... 132 1e-29
UniRef100_Q6L4V3 Putative polyprotein [Oryza sativa] 130 4e-29
UniRef100_Q7XEC4 Putative polyprotein [Oryza sativa] 129 7e-29
UniRef100_Q9M1F5 Copia-like polyprotein [Arabidopsis thaliana] 128 2e-28
UniRef100_Q9FFM0 Copia-like retrotransposable element [Arabidops... 125 7e-28
UniRef100_Q9SZY0 Putative retrotransposon [Arabidopsis thaliana] 123 5e-27
UniRef100_O82196 Copia-like retroelement pol polyprotein [Arabid... 121 1e-26
UniRef100_Q7XLX8 OSJNBa0042I15.12 protein [Oryza sativa] 119 9e-26
UniRef100_Q8H023 Putative retrovirus-related pol polyprotein [Or... 117 3e-25
UniRef100_Q8W5D4 Putative retrotransposon-related protein [Oryza... 116 5e-25
UniRef100_Q75HA9 Putative polyprotein [Oryza sativa] 115 1e-24
UniRef100_Q94LG0 Putative retroelement pol polyprotein [Oryza sa... 115 1e-24
UniRef100_Q6BCY1 Gag-Pol [Ipomoea batatas] 110 2e-23
>UniRef100_P10978 Retrovirus-related Pol polyprotein from transposon TNT 1-94
[Contains: Protease (EC 3.4.23.-); Reverse transcriptase
(EC 2.7.7.49); Endonuclease] [Nicotiana tabacum]
Length = 1328
Score = 152 bits (383), Expect = 1e-35
Identities = 89/236 (37%), Positives = 137/236 (57%), Gaps = 10/236 (4%)
Query: 3 GSKWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGLLPVTMSQVEKTDMVDKVRSA 62
G K+++ KF GDN F W+ +M +LIQQ K L + P TM + D+ ++ SA
Sbjct: 3 GVKYEVAKFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAEDWADLDERAASA 62
Query: 63 IVLCLGDKVLREVAKEPTAASIWAKLESLYMTKSLAHRQFLK*QLYLFRMVESKAIIEQL 122
I L L D V+ + E TA IW +LESLYM+K+L ++ +LK QLY M E + L
Sbjct: 63 IRLHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKKQLYALHMSEGTNFLSHL 122
Query: 123 MEFNKILDDLENIEVQLEDEDKAILLFCAIPKSFESFKDTMLYGKEGTVILEEVQAAL-- 180
FN ++ L N+ V++E+EDKAILL ++P S+++ T+L+GK T+ L++V +AL
Sbjct: 123 NVFNGLITQLANLGVKIEEEDKAILLLNSLPSSYDNLATTILHGKT-TIELKDVTSALLL 181
Query: 181 --RTKKLTKSKDLRVHEHGEGLSVLR--GNDGGRGNRRKSGNKSRS---ECFNCHK 229
+ +K +++ + G G S R N G G R KS N+S+S C+NC++
Sbjct: 182 NEKMRKKPENQGQALITEGRGRSYQRSSNNYGRSGARGKSKNRSKSRVRNCYNCNQ 237
>UniRef100_Q9AU17 Polyprotein-like [Lycopersicon chilense]
Length = 1328
Score = 137 bits (344), Expect = 3e-31
Identities = 78/232 (33%), Positives = 126/232 (53%), Gaps = 6/232 (2%)
Query: 3 GSKWDIEKFTGDND-FGLWKVKMEAVLIQQKCEKALKGEGLLPVTMSQVEKTDMVDKVRS 61
G K+++ KF GD F +W+ +M+ +LIQQ KAL G+ P +M + ++ +K S
Sbjct: 3 GVKYEVAKFNGDKPVFSMWQRRMKDLLIQQGLHKALGGKSKKPESMKLEDWEELDEKAAS 62
Query: 62 AIVLCLGDKVLREVAKEPTAASIWAKLESLYMTKSLAHRQFLK*QLYLFRMVESKAIIEQ 121
AI L L D V+ + E +A IW KLE+LYM+K+L ++ +LK QLY M E +
Sbjct: 63 AIRLHLTDDVVNNIVDEESACGIWTKLENLYMSKTLTNKLYLKKQLYTLHMDEGTNFLSH 122
Query: 122 LMEFNKILDDLENIEVQLEDEDKAILLFCAIPKSFESFKDTMLYGKEGTVILEEVQAALR 181
L N ++ L N+ V++E+EDK I+L ++P S+++ T+L+GK+ + + A L
Sbjct: 123 LNVLNGLITQLANLGVKIEEEDKRIVLLNSLPSSYDTLSTTILHGKDSIQLKDVTSALLL 182
Query: 182 TKKLTKSKD-----LRVHEHGEGLSVLRGNDGGRGNRRKSGNKSRSECFNCH 228
+K+ K + G N G G R KS +S+S+ NC+
Sbjct: 183 NEKMRKKPENHGQVFITESRGRSYQRSSSNYGRSGARGKSKVRSKSKARNCY 234
>UniRef100_Q8LNW7 Putative polyprotein [Oryza sativa]
Length = 1280
Score = 136 bits (343), Expect = 4e-31
Identities = 84/220 (38%), Positives = 127/220 (57%), Gaps = 5/220 (2%)
Query: 5 KWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGLLPVTMSQVEKTDMVDKVRSAIV 64
K+D+ D F LW+VKM AVL QQ + AL G S EK K S I
Sbjct: 40 KYDLPLLDRDTRFSLWQVKMRAVLAQQDLDDALSGFDKRTQDWSNDEKKKD-RKAMSYIH 98
Query: 65 LCLGDKVLREVAKEPTAASIWAKLESLYMTKSLAHRQFLK*QLYLFRMVESKAIIEQLME 124
L L + +L+EV KE TAA +W KLE + MTK L + LK +L+L ++ + ++++ L
Sbjct: 99 LHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLST 158
Query: 125 FNKILDDLENIEVQLEDEDKAILLFCAIPKSFESFKDTMLYGKEGTVILEEVQAALRTKK 184
F +I+ DLE+IEV+ ++ED ++L C++P S+ +F+DT+LY + T+IL+EV AL K+
Sbjct: 159 FKEIVADLESIEVKYDEEDLGLILLCSLPSSYANFRDTILYSHD-TLILKEVYDALHAKE 217
Query: 185 LTKS--KDLRVHEHGEGLSVLRGNDGGRGNRRKSGNKSRS 222
K + EGL V+RG + + +S +KS S
Sbjct: 218 KMKKMVPSEGSNSQAEGL-VVRGRQQEKNTKNQSRDKSSS 256
>UniRef100_Q7XTM9 OSJNBa0033G05.13 protein [Oryza sativa]
Length = 1181
Score = 136 bits (343), Expect = 4e-31
Identities = 83/220 (37%), Positives = 129/220 (57%), Gaps = 5/220 (2%)
Query: 5 KWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGLLPVTMSQVEKTDMVDKVRSAIV 64
K+D+ D F LW+VKM AVL QQ+ + AL G S EK K S I
Sbjct: 5 KYDLPLLDRDTRFSLWQVKMRAVLAQQELDDALSGFDKRTQDWSNDEKK-RDRKAMSYIH 63
Query: 65 LCLGDKVLREVAKEPTAASIWAKLESLYMTKSLAHRQFLK*QLYLFRMVESKAIIEQLME 124
L L + +L+EV KE TAA +W KLE + MTK L + LK +L+L ++ + ++++ L
Sbjct: 64 LHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLSA 123
Query: 125 FNKILDDLENIEVQLEDEDKAILLFCAIPKSFESFKDTMLYGKEGTVILEEVQAALRTKK 184
F +I+ DLE++EV+ +++D A++L C++P S+ +F+DT+LY ++ T+ L+EV AL K+
Sbjct: 124 FKEIVADLESMEVKYDEKDLALILLCSLPSSYANFRDTILYSRD-TLTLKEVYDALHAKE 182
Query: 185 LTKS--KDLRVHEHGEGLSVLRGNDGGRGNRRKSGNKSRS 222
K + EGL V+RG+ + KS +KS S
Sbjct: 183 KMKKMVPSEGSNSQAEGL-VVRGSQQEKNTNNKSRDKSSS 221
>UniRef100_Q6L4X8 Putative polyprotein [Oryza sativa]
Length = 1211
Score = 134 bits (338), Expect = 2e-30
Identities = 84/220 (38%), Positives = 125/220 (56%), Gaps = 5/220 (2%)
Query: 5 KWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGLLPVTMSQVEKTDMVDKVRSAIV 64
K+D+ D F LW+VKM AVL QQ + AL G S EK K S I
Sbjct: 5 KYDLPLLDRDTRFSLWQVKMRAVLAQQDLDDALSGFDKRTQDWSNDEKK-RDRKTMSYIH 63
Query: 65 LCLGDKVLREVAKEPTAASIWAKLESLYMTKSLAHRQFLK*QLYLFRMVESKAIIEQLME 124
L L + +L+EV KE TAA +W KLE + MTK L + LK +L+L ++ + ++++ L
Sbjct: 64 LHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLSA 123
Query: 125 FNKILDDLENIEVQLEDEDKAILLFCAIPKSFESFKDTMLYGKEGTVILEEVQAALRTKK 184
F KI+ DLE++EV+ ++ED ++L C++P S+ +F+DT+LY + T+ L+EV AL K+
Sbjct: 124 FKKIVADLESMEVKYDEEDLCLILLCSLPSSYANFRDTILYSCD-TLTLKEVYDALHAKE 182
Query: 185 LTKS--KDLRVHEHGEGLSVLRGNDGGRGNRRKSGNKSRS 222
K + EGL V+RG + KS +KS S
Sbjct: 183 KIKKMVPSEGSNSQAEGL-VVRGRQQEKNTNSKSRDKSSS 221
>UniRef100_Q9SJU6 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 838
Score = 132 bits (333), Expect = 6e-30
Identities = 90/247 (36%), Positives = 142/247 (57%), Gaps = 28/247 (11%)
Query: 7 DIEKFTGDNDFGLWKVKMEA------VLIQQKCEKALKGEGLLPVTMSQVEKTDMVDKV- 59
++EK G+ D+ LWK K+ A +L + ++A++ E T S + KT+ DKV
Sbjct: 7 EVEKLDGEGDYVLWKEKLLAHIELLGLLEGLEEDEAIEEEESTAETDSLLTKTE--DKVL 64
Query: 60 -------RSAIVLCLGDKVLREVAKEPTAASIWAKLESLYMTKSLAHRQFLK*QLYLFRM 112
RS ++L LG+ VLR+V KE TAA + L+ L+M KSL +R +LK +LY ++M
Sbjct: 65 KEKRGKARSTVILSLGNHVLRKVIKEKTAAGMIRVLDKLFMAKSLPNRIYLKQRLYGYKM 124
Query: 113 VESKAIIEQLMEFNKILDDLENIEVQLEDEDKAILLFCAIPKSFESFKDTMLYGKEGTVI 172
+S I E + +F K++ DLEN++V + DED+AI+L ++PK F+ KDT+ YGK T+
Sbjct: 125 SDSMTIEENVNDFFKLISDLENVKVSVPDEDQAIVLLMSLPKQFDQLKDTLKYGKT-TLA 183
Query: 173 LEEVQAALRTK--------KLTKSKDLRVHEHGEGLSVLRGNDGGRGNRRKSGNKSRSE- 223
L+E+ A+R+K K+ K+ + G S R R N+ +S +KSR +
Sbjct: 184 LDEITGAIRSKVLELGASGKMLKNSSDALFVQDRGRSEKRDKSSER-NKSQSRSKSREKK 242
Query: 224 -CFNCHK 229
C+ C K
Sbjct: 243 VCWVCGK 249
>UniRef100_Q9SJT2 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1333
Score = 132 bits (331), Expect = 1e-29
Identities = 92/248 (37%), Positives = 142/248 (57%), Gaps = 27/248 (10%)
Query: 7 DIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGLLPVTMSQVEKTDMVDK-------- 58
++EKF G D+ +WK K+ A L ALK E L +++++ T+ +K
Sbjct: 7 EVEKFDGRGDYTMWKEKLMAHLDILGLSVALKEEDDLVEKVAEMQLTEEEEKEEVLRREL 66
Query: 59 -------VRSAIVLCLGDKVLREVAKEPTAASIWAKLESLYMTKSLAHRQFLK*QLYLFR 111
RSAIVL + D+VLR++ KE +AA++ L+ LYM+K+L +R + K +LY F+
Sbjct: 67 LEEKRRKARSAIVLSVTDRVLRKIKKEQSAAAMLGVLDKLYMSKALPNRIYQKQKLYSFK 126
Query: 112 MVESKAIIEQLMEFNKILDDLENIEVQLEDEDKAILLFCAIPKSFESFKDTMLYG-KEGT 170
M E+ +I + EF +I+ DLEN V + DED+AILL ++PK F+ +DT+ YG T
Sbjct: 127 MSENLSIEGNIDEFLRIIADLENTNVLVSDEDQAILLLMSLPKPFDQLRDTLKYGLGRVT 186
Query: 171 VILEEVQAALRTKKLTKSKDLR-VHEHGEGLSV-----LRGNDGGRG---NRRKSGNKSR 221
+ L+EV AA+ +K+L + + + EGL V RG RG N +KS +KSR
Sbjct: 187 LSLDEVVAAIYSKELELGSNKKSIKGQAEGLFVKEKTETRGRTEQRGNNNNNKKSRSKSR 246
Query: 222 SE--CFNC 227
S+ C+ C
Sbjct: 247 SKKGCWIC 254
>UniRef100_Q9SH77 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1356
Score = 132 bits (331), Expect = 1e-29
Identities = 89/247 (36%), Positives = 143/247 (57%), Gaps = 28/247 (11%)
Query: 7 DIEKFTGDNDFGLWKVKMEAVLIQQKCEKALK-----GEGLLPVTMS------QVEKTDM 55
++EKF G D+ +WK K+ A + ALK GE + S ++EK +
Sbjct: 7 EVEKFDGRGDYTMWKEKLLAHMDILGLNTALKESESTGEKKSVLDESDEDYEEKLEKFEA 66
Query: 56 VD----KVRSAIVLCLGDKVLREVAKEPTAASIWAKLESLYMTKSLAHRQFLK*QLYLFR 111
++ K RSAIVL + D+VLR++ KE TAA++ L+ LYM+K+L +R + K +LY F+
Sbjct: 67 LEEKKKKARSAIVLSVTDRVLRKIKKESTAAAMLLALDKLYMSKALPNRIYPKQKLYSFK 126
Query: 112 MVESKAIIEQLMEFNKILDDLENIEVQLEDEDKAILLFCAIPKSFESFKDTMLYGKEGTV 171
M E+ ++ + EF +I+ DLEN+ V + DED+AILL A+PK+F+ KDT+ Y ++
Sbjct: 127 MSENLSVEGNIDEFLQIITDLENMNVIISDEDQAILLLTALPKAFDQLKDTLKYSSGKSI 186
Query: 172 I-LEEVQAALRTKKL---TKSKDLRVHEHGEGLSVL-------RGNDGGRGNRRKSGNKS 220
+ L+EV AA+ +K+L + K ++V EGL V +G G+G +K +K
Sbjct: 187 LTLDEVAAAIYSKELELGSVKKSIKV--QAEGLYVKDKNENKGKGEQKGKGKGKKGKSKK 244
Query: 221 RSECFNC 227
+ C+ C
Sbjct: 245 KPGCWTC 251
>UniRef100_Q6L4V3 Putative polyprotein [Oryza sativa]
Length = 1243
Score = 130 bits (326), Expect = 4e-29
Identities = 81/222 (36%), Positives = 128/222 (57%), Gaps = 9/222 (4%)
Query: 5 KWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGLLPVTMSQVEKTDMVDKVRSAIV 64
K+D+ D F LW+VKM AVL QQ + AL G S EK K S I
Sbjct: 5 KYDLPLLDRDTRFSLWQVKMRAVLAQQDLDDALSGFDKRTQDWSNDEKK-RDRKAISYIH 63
Query: 65 LCLGDKVLREVAKEPTAASIWAKLESLYMTKSLAHRQFLK*QLYLFRMVESKAIIEQLME 124
L L + +L+EV KE TAA +W KLE + MTK L + LK +L+L ++ + +++++ L
Sbjct: 64 LHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDESVMDHLSA 123
Query: 125 FNKILDDLENIEVQLEDEDKAILLFCAIPKSFESFKDTMLYGKEGTVILEEVQAALRTKK 184
F +I+ DLE++EV+ +++D ++L C++P S+ +F+ T+LY ++ T+ L+EV A K+
Sbjct: 124 FKEIVADLESMEVKYDEDDLGLILLCSLPSSYANFRGTILYSRD-TLTLKEVYDAFHAKE 182
Query: 185 LTKSKDLRVHE----HGEGLSVLRGNDGGRGNRRKSGNKSRS 222
K K + E EGL V+RG + + +S +KS S
Sbjct: 183 --KMKKMVTSEGSNSQAEGL-VVRGRQQKKNTKNQSRDKSSS 221
>UniRef100_Q7XEC4 Putative polyprotein [Oryza sativa]
Length = 382
Score = 129 bits (324), Expect = 7e-29
Identities = 76/233 (32%), Positives = 134/233 (56%), Gaps = 11/233 (4%)
Query: 1 TMGSKWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGLLPVTMSQVEKTDMVDKVR 60
T SK+++ KF G +F LW+++++ +L QQ KAL + +P + + +M +
Sbjct: 4 TAVSKFEVVKFDGTGNFVLWQMRLKDLLAQQGISKAL--QETMPEKIDADKWNEMKAQAA 61
Query: 61 SAIVLCLGDKVLREVAKEPTAASIWAKLESLYMTKSLAHRQFLK*QLYLFRMVESKAIIE 120
+ I L L D V+ +V E + IW KL SL+M+KSL + +LK QLY ++ E + +
Sbjct: 62 ATIRLSLSDSVMYQVMDEKSPKEIWDKLASLHMSKSLTSKLYLKQQLYGLQVQEESDLRK 121
Query: 121 QLMEFNKILDDLENIEVQLEDEDKAILLFCAIPKSFESFKDTMLYGKEGTVILEEVQAAL 180
+ FN+++ DL ++V+L+DEDKAI+L C++P S+E T+ +GK+ TV EE+ ++L
Sbjct: 122 HVDVFNQLVVDLSKLDVKLDDEDKAIILLCSLPLSYEHVVTTLTHGKD-TVKTEEIISSL 180
Query: 181 RTKKLTKSKDLRVHEHGEGLSVL----RGNDGGRGNRRKSGNKSRSECFNCHK 229
+ L +SK + +G S+L ++ G ++ G + C+ CH+
Sbjct: 181 LARDLRRSKKNEATKASQGKSLLVKDKHDHEAGVSKSKEKG----ARCYKCHE 229
>UniRef100_Q9M1F5 Copia-like polyprotein [Arabidopsis thaliana]
Length = 1363
Score = 128 bits (321), Expect = 2e-28
Identities = 86/251 (34%), Positives = 135/251 (53%), Gaps = 28/251 (11%)
Query: 3 GSKWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGLLPVTMSQVEKTDMVDK---- 58
G++ ++EKF G D+ +WK K+ A + L+ EK+D +K
Sbjct: 3 GARIEVEKFDGRGDYTMWKEKLLAHIDMLGLSAVLRESETPMGKERDSEKSDEDEKEERE 62
Query: 59 -----------VRSAIVLCLGDKVLREVAKEPTAASIWAKLESLYMTKSLAHRQFLK*QL 107
RS IVL + D+VLR++ KE +AA++ L+ LYM+K+L +R +LK +L
Sbjct: 63 KMEAFEEKKRKARSTIVLSVSDRVLRKIKKETSAAAMLEALDRLYMSKALPNRIYLKQKL 122
Query: 108 YLFRMVESKAIIEQLMEFNKILDDLENIEVQLEDEDKAILLFCAIPKSFESFKDTMLYGK 167
Y F+M E+ +I + EF I+ DLEN+ V + DED+AILL ++PK F+ KDT+ Y
Sbjct: 123 YSFKMSENLSIEGNIDEFLHIVADLENLNVLVSDEDQAILLLMSLPKPFDQLKDTLKYSS 182
Query: 168 EGTVI-LEEVQAALRTKKLT----------KSKDLRVHEHGEGLSVLRGNDGGRGNRRKS 216
TV+ L+EV AA+ +++L +++ L V + E D G+G R KS
Sbjct: 183 GKTVLSLDEVAAAIYSRELEFGSVKKSIKGQAEGLYVKDKAENRGRSEQKDKGKGKRSKS 242
Query: 217 GNKSRSECFNC 227
KS+ C+ C
Sbjct: 243 --KSKRGCWIC 251
>UniRef100_Q9FFM0 Copia-like retrotransposable element [Arabidopsis thaliana]
Length = 1342
Score = 125 bits (315), Expect = 7e-28
Identities = 83/251 (33%), Positives = 132/251 (52%), Gaps = 29/251 (11%)
Query: 7 DIEKFTGDNDFGLWKVKM-----------------EAVLIQQKCEKALKGEGLLPVTMSQ 49
++EKF GD D+ LWK K+ EAV+ E + G S+
Sbjct: 7 EVEKFDGDGDYILWKEKLLAHMEMLGLLEGLGEEEEAVVEDSTTEISDGGNQDPETATSK 66
Query: 50 VEKTDMVDK---VRSAIVLCLGDKVLREVAKEPTAASIWAKLESLYMTKSLAHRQFLK*Q 106
+E + +K RS I+L LG+ VLR+V K+ TAA + L+ L+M KSL +R +LK +
Sbjct: 67 LEDKILKEKRGKARSTIILSLGNNVLRKVIKQKTAAGMIKVLDQLFMAKSLPNRIYLKQR 126
Query: 107 LYLFRMVESKAIIEQLMEFNKILDDLENIEVQLEDEDKAILLFCAIPKSFESFKDTMLYG 166
LY ++M E+ + E + +F K++ DLEN++V + DED+AI+L ++P+ F+ K+T+ Y
Sbjct: 127 LYGYKMSENMTMEENVNDFFKLISDLENVKVVVPDEDQAIVLLMSLPRQFDQLKETLKYC 186
Query: 167 KEGTVILEEVQAALRTK--------KLTKSKDLRVHEHGEGLSVLRGNDGGRGNRRKSGN 218
K T+ LEE+ +A+R+K KL K+ + G S RG + R
Sbjct: 187 KT-TLHLEEITSAIRSKILELGASGKLLKNNSDGLFVQDRGRSETRGKGPNKNKSRSKSK 245
Query: 219 KSRSECFNCHK 229
+ C+ C K
Sbjct: 246 GAGKTCWICGK 256
>UniRef100_Q9SZY0 Putative retrotransposon [Arabidopsis thaliana]
Length = 1230
Score = 123 bits (308), Expect = 5e-27
Identities = 80/232 (34%), Positives = 128/232 (54%), Gaps = 16/232 (6%)
Query: 7 DIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGLLPVTMSQVEK-------------T 53
++EKF G D+ LWK K+ A + AL+ + + E+
Sbjct: 7 EMEKFDGHGDYTLWKEKLMAHMDLLGLTVALRETQSVSDPLESEEEGKESEKGDKEALME 66
Query: 54 DMVDKVRSAIVLCLGDKVLREVAKEPTAASIWAKLESLYMTKSLAHRQFLK*QLYLFRMV 113
+ K RS IVL + D+VLR+ KE TA S+ L+ LYM+K+L +R +LK +LY ++M
Sbjct: 67 EKRQKARSTIVLSVSDQVLRKSKKEKTAPSMLEALDKLYMSKALPNRIYLKQKLYSYKMQ 126
Query: 114 ESKAIIEQLMEFNKILDDLENIEVQLEDEDKAILLFCAIPKSFESFKDTMLYGKEGTVI- 172
E+ ++ + EF +++ DLEN V + DED+AILL ++PK F+ KDT+ YG T +
Sbjct: 127 ENLSVEGNIDEFLRLIADLENTNVLVSDEDQAILLLMSLPKQFDQLKDTLKYGSGRTTLS 186
Query: 173 LEEVQAALRTKKLTKSKDLR-VHEHGEGLSVL-RGNDGGRGNRRKSGNKSRS 222
++EV AA+ +K+L + + + EGL V + G +++ GNK RS
Sbjct: 187 VDEVVAAIYSKELELGSNKKSIRGQAEGLYVKDKPETRGMSEQKEKGNKGRS 238
>UniRef100_O82196 Copia-like retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1137
Score = 121 bits (304), Expect = 1e-26
Identities = 67/185 (36%), Positives = 107/185 (57%), Gaps = 15/185 (8%)
Query: 52 KTDMVDKVRSAIVLCLGDKVLREVAKEPTAASIWAKLESLYMTKSLAHRQFLK*QLYLFR 111
+ D +K I + +GDKVLR + TAA WA L+ LY+ KSL +R +L+ ++Y +R
Sbjct: 37 RIDQDEKAMDMIFINVGDKVLRNIENSKTAAEAWATLDKLYLVKSLPNRVYLQLKVYNYR 96
Query: 112 MVESKAIIEQLMEFNKILDDLENIEVQLEDEDKAILLFCAIPKSFESFKDTMLYGKEGTV 171
M +SK + E + EF K++ DL N+++Q+ DE +AIL+ A+P S++ K+T+ YG+EG
Sbjct: 97 MQDSKTLEENVDEFQKMISDLNNLQIQVPDEVQAILILSALPDSYDMLKETLKYGREGIK 156
Query: 172 ILEEVQAALRTKKLTKSKDLRVHEH-------GEGLSVLRGNDGGRGNRRKSGNKSRSEC 224
+ + + AA KSK+L + + GEGL V RG RG+ + + C
Sbjct: 157 LDDVISAA-------KSKELELRDSSGGSRPVGEGLYV-RGKSQARGSDGPKSTEGKKVC 208
Query: 225 FNCHK 229
+ C K
Sbjct: 209 WICGK 213
>UniRef100_Q7XLX8 OSJNBa0042I15.12 protein [Oryza sativa]
Length = 432
Score = 119 bits (297), Expect = 9e-26
Identities = 66/191 (34%), Positives = 115/191 (59%), Gaps = 11/191 (5%)
Query: 5 KWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGLLPVTMSQVEKTDMVDKVRSAIV 64
K+D+ KF G +FGLW+ +++ +L QQ+ KALKG + P M + +M + + I
Sbjct: 43 KFDLVKFDGFGNFGLWQTRVKDLLAQQEVSKALKG--VKPAKMEDDDCEEMQLQAAATIR 100
Query: 65 LCLGDKVLREVAKEPTAASIWAKLESLYMTKSLAHRQFLK*QLYLFRMVESKAIIEQLME 124
CL D+++ V +E + IW KLE+ +M+K+L ++++LK +LY +M E + +
Sbjct: 101 RCLSDQIMYHVMEETSPKIIWEKLEAQFMSKTLTNKRYLKQKLYGLKMQEGADLTAHVNV 160
Query: 125 FNKILDDLENIEVQLEDEDKAILLFCAIPKSFESFKDTMLYGKEGT------VILEEVQA 178
FN+++ DL ++V+++DEDKAI+L C++P ES++ M KEG ++++E A
Sbjct: 161 FNQLVTDLVKMDVEVDDEDKAIVLLCSLP---ESYEHVMRKNKEGETSQAEGLLVKEDHA 217
Query: 179 ALRTKKLTKSK 189
+ KK K K
Sbjct: 218 GHKVKKNKKKK 228
>UniRef100_Q8H023 Putative retrovirus-related pol polyprotein [Oryza sativa]
Length = 556
Score = 117 bits (293), Expect = 3e-25
Identities = 74/216 (34%), Positives = 121/216 (55%), Gaps = 11/216 (5%)
Query: 18 GLWKVKMEAVLIQQKCEKALKGEGLLPVTMSQVEKTDMVDKVRSAIVLCLGDKVLREVAK 77
G +++++ +L QQ KAL E +P M + +M + + I L L D V+ +V
Sbjct: 152 GSSQMRLKDLLAQQGISKAL--EETMPEKMDVGKWVEMKAQATAIIRLSLSDFVMYQVMD 209
Query: 78 EPTAASIWAKLESLYMTKSLAHRQFLK*QLYLFRMVESKAIIEQLMEFNKILDDLENIEV 137
E T IW KL SLYM+KSL + +LK QLY +M E + + + FN+++ DL ++V
Sbjct: 210 EKTPKEIWDKLASLYMSKSLTSKLYLKQQLYGLQMQEESDLRKHVDVFNQLVVDLSKLDV 269
Query: 138 QLEDEDKAILLFCAIPKSFESFKDTMLYGKEGTVILEEVQAALRTKKLTKSKDLRVHEHG 197
+L+DEDKAI+L C++P SFE T+ +GK+ TV EE+ ++L + L +SK E
Sbjct: 270 KLDDEDKAIILLCSLPPSFEHVVTTLTHGKD-TVKTEEIISSLLARDLRRSKKNEAMEAS 328
Query: 198 EGLSVL----RGNDGGRGNRRKSGNKSRSECFNCHK 229
+ S+L ++ G ++ G + C+ CH+
Sbjct: 329 QAESLLVKAKHDHEAGVSKSKEKG----ARCYKCHE 360
>UniRef100_Q8W5D4 Putative retrotransposon-related protein [Oryza sativa]
Length = 1229
Score = 116 bits (291), Expect = 5e-25
Identities = 73/237 (30%), Positives = 125/237 (51%), Gaps = 12/237 (5%)
Query: 5 KWDIEKFTGDNDFGLWKVKMEAVLIQQK-CEKALKGEGLLPVTMSQVEKTDMVDKVRSAI 63
K+D+ F LW+VKM AVL Q ++AL+ G T E+ K S I
Sbjct: 2 KYDLPLQDYKTRFSLWQVKMRAVLAQTSDLDEALESFGKKKTTEWTAEEKRKDRKALSLI 61
Query: 64 VLCLGDKVLREVAKEPTAASIWAKLESLYMTKSLAHRQFLK*QLYLFRMVESKAIIEQLM 123
L L + +L++V +E TAA +W KLES+ M+K L + +K +L+ ++ ES +++ +
Sbjct: 62 QLHLSNDILQKVLQEKTAAELWFKLESICMSKDLTSKMHIKMKLFSHKLQESGSVLNHIS 121
Query: 124 EFNKILDDLENIEVQLEDEDKAILLFCAIPKSFESFKDTMLYGKEGTVILEEVQAALRTK 183
F +I+ DL ++EVQ +DED +LL C++P + +F+DT+L ++ + E +A +
Sbjct: 122 VFKEIIADLVSMEVQFDDEDLGLLLLCSLPSLYANFRDTILLSRDELTLAEVYEALQNRE 181
Query: 184 KL----------TKSKDLRVHEHGEGLSVLRGNDGGRG-NRRKSGNKSRSECFNCHK 229
K+ +K K L+V E + ND + +R +S ++ + C C K
Sbjct: 182 KMKGMVQSDASSSKGKALQVRGRSEQRTYNDSNDRDKSQSRGRSKSRGKKFCKYCKK 238
>UniRef100_Q75HA9 Putative polyprotein [Oryza sativa]
Length = 1322
Score = 115 bits (288), Expect = 1e-24
Identities = 72/237 (30%), Positives = 125/237 (52%), Gaps = 12/237 (5%)
Query: 5 KWDIEKFTGDNDFGLWKVKMEAVLIQQK-CEKALKGEGLLPVTMSQVEKTDMVDKVRSAI 63
K+D+ F LW+VKM AVL Q ++AL+ G T E+ K S I
Sbjct: 5 KYDLPLLDYKTRFSLWQVKMRAVLAQTSDLDEALESFGKKKTTEWTAEEKRKDRKALSLI 64
Query: 64 VLCLGDKVLREVAKEPTAASIWAKLESLYMTKSLAHRQFLK*QLYLFRMVESKAIIEQLM 123
L L + +L+EV ++ TAA +W KLES+ M+K L + +K +L+ ++ ES +++ +
Sbjct: 65 QLHLSNDILQEVLQKKTAAELWLKLESICMSKDLTSKMHIKMKLFSHKLHESGSVLNHIS 124
Query: 124 EFNKILDDLENIEVQLEDEDKAILLFCAIPKSFESFKDTMLYGKEGTVILEEVQAALRTK 183
F +I+ DL ++EVQ +DED +LL C++P S+ +F+ T+L ++ + E +A +
Sbjct: 125 VFKEIVADLVSMEVQFDDEDLGLLLLCSLPSSYANFRHTILLSRDELTLAEVYEALQNRE 184
Query: 184 KL----------TKSKDLRVHEHGEGLSVLRGNDGGRG-NRRKSGNKSRSECFNCHK 229
K+ +K + L+V E + ND + +R +S ++ + C C K
Sbjct: 185 KMKGMVQSYASSSKGEALQVRGRSEQRTYNDSNDHDKSQSRGRSKSRGKKFCKYCKK 241
>UniRef100_Q94LG0 Putative retroelement pol polyprotein [Oryza sativa]
Length = 1326
Score = 115 bits (288), Expect = 1e-24
Identities = 72/237 (30%), Positives = 125/237 (52%), Gaps = 12/237 (5%)
Query: 5 KWDIEKFTGDNDFGLWKVKMEAVLIQQK-CEKALKGEGLLPVTMSQVEKTDMVDKVRSAI 63
K+D+ F LW+VKM A+L Q ++AL+ G T E+ K I
Sbjct: 5 KYDLPLLDYKTRFSLWQVKMRAILAQTSDLDEALESFGKKKSTEWTAEEKRKDRKALLLI 64
Query: 64 VLCLGDKVLREVAKEPTAASIWAKLESLYMTKSLAHRQFLK*QLYLFRMVESKAIIEQLM 123
L L + +L+EV +E TAA +W KLES+ M+K L + +K +L+ ++ ES +++ +
Sbjct: 65 QLHLSNDILQEVLQEKTAAELWLKLESICMSKDLTSKMHIKMKLFSHKLQESGSVLNHIS 124
Query: 124 EFNKILDDLENIEVQLEDEDKAILLFCAIPKSFESFKDTMLYGKEGTVILEEVQAALRTK 183
F +I+ DL +IEVQ +DED +LL C++P S+ +F+DT+L ++ + E +A +
Sbjct: 125 VFKEIVVDLVSIEVQFDDEDLGLLLLCSLPSSYANFRDTILLSRDELTLAEVYEALQNRE 184
Query: 184 KL----------TKSKDLRVHEHGEGLSVLRGNDGGRG-NRRKSGNKSRSECFNCHK 229
K+ +K + L+V E + +D + +R +S ++ + C C K
Sbjct: 185 KMKGMVQSDASSSKGEALQVRGRSEQRTYNDSSDRDKSQSRGRSKSRGKKFCKYCKK 241
>UniRef100_Q6BCY1 Gag-Pol [Ipomoea batatas]
Length = 1298
Score = 110 bits (276), Expect = 2e-23
Identities = 74/242 (30%), Positives = 123/242 (50%), Gaps = 23/242 (9%)
Query: 2 MGSKWDIEKFTGDNDFGLWKVKMEAVLIQQKCEKALKGEGLLPVTMSQVEK-TDMVDKVR 60
M +K++IEKF G N F LWK+K++A+L + C A+ PV + +K ++M +
Sbjct: 1 MAAKFEIEKFNGKN-FSLWKLKVKAILRKDNCLAAISER---PVDFTDDKKWSEMNEDAM 56
Query: 61 SAIVLCLGDKVLREVAKEPTAASIWAKLESLYMTKSLAHRQFLK*QLYLFRMVESKAIIE 120
+ + L + D VL + ++ TA IW L LY KSL ++ FLK +LY RM ES ++ E
Sbjct: 57 ADLYLSIADGVLSSIEEKKTANEIWDHLNRLYEAKSLHNKIFLKRKLYTLRMSESTSVTE 116
Query: 121 QLMEFNKILDDLENIEVQLEDEDKAILLFCAIPKSFES---------FKDTMLYGKEGTV 171
L N + L ++ ++E +++A LL ++P S++ D +++
Sbjct: 117 HLNTLNTLFSQLTSLSCKIEPQERAELLLQSLPDSYDQLIINLTNNILTDYLVFDDVAAA 176
Query: 172 ILEEVQAALRTKKLTKSKDLRVHEHGEGLSVLRGNDGGRGNR----RKSGNKSRSECFNC 227
+LEE + R K + +L + E L+V+RG RG R +K C+NC
Sbjct: 177 VLEE--ESRRKNKEDRQVNL---QQAEALTVMRGRSTERGQSSGRGRSKSSKKNLTCYNC 231
Query: 228 HK 229
K
Sbjct: 232 GK 233
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.138 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 374,567,803
Number of Sequences: 2790947
Number of extensions: 14207785
Number of successful extensions: 44625
Number of sequences better than 10.0: 287
Number of HSP's better than 10.0 without gapping: 161
Number of HSP's successfully gapped in prelim test: 126
Number of HSP's that attempted gapping in prelim test: 44342
Number of HSP's gapped (non-prelim): 316
length of query: 246
length of database: 848,049,833
effective HSP length: 124
effective length of query: 122
effective length of database: 501,972,405
effective search space: 61240633410
effective search space used: 61240633410
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 73 (32.7 bits)
Medicago: description of AC145329.2


