

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145222.8 + phase: 0
(209 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
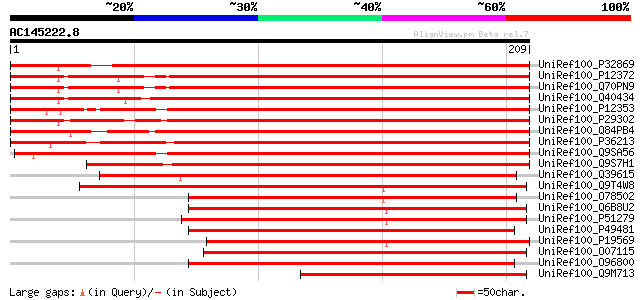
UniRef100_P32869 Photosystem I reaction center subunit II, chlor... 328 7e-89
UniRef100_P12372 Photosystem I reaction center subunit II, chlor... 327 9e-89
UniRef100_Q70PN9 Putative photosystem I reaction centre PSI-D su... 327 1e-88
UniRef100_Q40434 PSI-D1 precursor [Nicotiana sylvestris] 322 3e-87
UniRef100_P12353 Photosystem I reaction center subunit II, chlor... 317 1e-85
UniRef100_P29302 Photosystem I reaction center subunit II, chlor... 315 4e-85
UniRef100_Q84PB4 Chloroplast photosystem I reaction center subun... 297 1e-79
UniRef100_P36213 Photosystem I reaction center subunit II, chlor... 295 5e-79
UniRef100_Q9SA56 Photosystem I reaction center subunit II-2, chl... 293 2e-78
UniRef100_Q9S7H1 Photosystem I reaction center subunit II-1, chl... 288 5e-77
UniRef100_Q39615 Photosystem I reaction center subunit II, chlor... 224 1e-57
UniRef100_Q9T4W8 Photosystem I reaction centre subunit II precur... 212 6e-54
UniRef100_O78502 Photosystem I reaction center subunit II [Guill... 189 3e-47
UniRef100_Q6B8U2 Photosystem I reaction center subunit II [Graci... 189 4e-47
UniRef100_P51279 Photosystem I reaction center subunit II [Porph... 186 4e-46
UniRef100_P49481 Photosystem I reaction center subunit II [Odont... 182 6e-45
UniRef100_P19569 Photosystem I reaction center subunit II [Synec... 181 1e-44
UniRef100_O07115 Photosystem I reaction center subunit II [Masti... 181 1e-44
UniRef100_O96800 Photosystem I reaction center subunit II [Skele... 178 7e-44
UniRef100_Q9M713 Ferredoxin-binding subunit [Zea mays] 176 3e-43
>UniRef100_P32869 Photosystem I reaction center subunit II, chloroplast precursor
[Cucumis sativus]
Length = 207
Score = 328 bits (840), Expect = 7e-89
Identities = 166/215 (77%), Positives = 177/215 (82%), Gaps = 14/215 (6%)
Query: 1 MAMATQASLFTPPLSSPK------PWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAAS 54
MAMATQA+LFTP LS+PK PWKQ STLSF++ KP + AA
Sbjct: 1 MAMATQATLFTPSLSTPKSTGISIPWKQSSTLSFLTSKP--------HLKAASSSRSFKV 52
Query: 55 VTTKEEAPAAPVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMP 114
E + AP GF+PPELDP+TPSPIF GSTGGLLRKAQVEEFYVITWESPKEQIFEMP
Sbjct: 53 SAEAETSVEAPAGFSPPELDPSTPSPIFAGSTGGLLRKAQVEEFYVITWESPKEQIFEMP 112
Query: 115 TGGAAIMREGPNLLKLARKEQCLALGNRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYPE 174
TGGAAIMREGPNLLKLARKEQCLALG RLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYPE
Sbjct: 113 TGGAAIMREGPNLLKLARKEQCLALGTRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYPE 172
Query: 175 KVNPGRQGVGQNFRSIGKNVSPIEVKFTGKQPFDV 209
KVNPGR+GVGQNFRSIGKNVSPIEVKFTGKQ +D+
Sbjct: 173 KVNPGREGVGQNFRSIGKNVSPIEVKFTGKQVYDL 207
>UniRef100_P12372 Photosystem I reaction center subunit II, chloroplast precursor
[Lycopersicon esculentum]
Length = 208
Score = 327 bits (839), Expect = 9e-89
Identities = 171/214 (79%), Positives = 182/214 (84%), Gaps = 11/214 (5%)
Query: 1 MAMATQASLFTPPLSSPK----PWKQPSTLSFISLKPIKFTTKTTK-ISAADEKTEAASV 55
MAMATQASLFTPPLS PK PWKQ S +SF + K +K T T+ I A E+ AA+
Sbjct: 1 MAMATQASLFTPPLSVPKSTTAPWKQ-SLVSFSTPKQLKSTVSVTRPIRAMAEEAPAAT- 58
Query: 56 TTKEEAPAAPVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPT 115
EE PA P GFTPP+LDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPT
Sbjct: 59 ---EEKPA-PAGFTPPQLDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPT 114
Query: 116 GGAAIMREGPNLLKLARKEQCLALGNRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYPEK 175
GGAAIMR+GPNLLKLARKEQCLALG RLRSKYKI YQFYRVFPNGEVQYLHPKDGVYPEK
Sbjct: 115 GGAAIMRQGPNLLKLARKEQCLALGTRLRSKYKINYQFYRVFPNGEVQYLHPKDGVYPEK 174
Query: 176 VNPGRQGVGQNFRSIGKNVSPIEVKFTGKQPFDV 209
VNPGR+GVGQNFRSIGKN S IEVKFTGKQ +D+
Sbjct: 175 VNPGREGVGQNFRSIGKNKSAIEVKFTGKQVYDI 208
>UniRef100_Q70PN9 Putative photosystem I reaction centre PSI-D subunit precursor
[Solanum tuberosum]
Length = 207
Score = 327 bits (838), Expect = 1e-88
Identities = 172/213 (80%), Positives = 181/213 (84%), Gaps = 10/213 (4%)
Query: 1 MAMATQASLFTPPLSSPK---PWKQPSTLSFISLKPIKFTTKTTK-ISAADEKTEAASVT 56
MAMATQASLFTP LS PK PWKQ S LSF + K +K T T+ I A E+ AA+
Sbjct: 1 MAMATQASLFTPALSVPKSTAPWKQ-SLLSFSTPKQLKSTVSVTRPIRAMAEEAPAAT-- 57
Query: 57 TKEEAPAAPVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTG 116
EE PA P GFTPP+LDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTG
Sbjct: 58 --EEKPA-PAGFTPPQLDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTG 114
Query: 117 GAAIMREGPNLLKLARKEQCLALGNRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYPEKV 176
GAAIMREGPNLLKLARKEQCLALG RLRSKYKI YQFYRVFPNGEVQYLHPKDGVYPEKV
Sbjct: 115 GAAIMREGPNLLKLARKEQCLALGTRLRSKYKINYQFYRVFPNGEVQYLHPKDGVYPEKV 174
Query: 177 NPGRQGVGQNFRSIGKNVSPIEVKFTGKQPFDV 209
NPGR+GVGQNFRSIGKN S IEVKFTGKQ +D+
Sbjct: 175 NPGREGVGQNFRSIGKNKSAIEVKFTGKQVYDI 207
>UniRef100_Q40434 PSI-D1 precursor [Nicotiana sylvestris]
Length = 214
Score = 322 bits (826), Expect = 3e-87
Identities = 169/218 (77%), Positives = 181/218 (82%), Gaps = 13/218 (5%)
Query: 1 MAMATQASLFTPPLSSPK--------PWKQPSTLSFISLKPIKFTTKTTKISA-ADEKTE 51
MAMA+QASLFTP +S+ K PWKQ S SF + K K I A A EK +
Sbjct: 1 MAMASQASLFTPSISTSKTADPRVVAPWKQ-SASSFSAPKLSKSVVAYRPIKAMAVEKAQ 59
Query: 52 AASVTTKEEAPAAPVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIF 111
+A TKE PAAPVGFTPP+LDP+TPSPIFGGSTGGLLRKAQV+EFYVITWESPKEQIF
Sbjct: 60 SA---TKEAEPAAPVGFTPPQLDPSTPSPIFGGSTGGLLRKAQVDEFYVITWESPKEQIF 116
Query: 112 EMPTGGAAIMREGPNLLKLARKEQCLALGNRLRSKYKIKYQFYRVFPNGEVQYLHPKDGV 171
EMPTGGAAIMREGPNLLKLARKEQCLALG RLRSKYKI Y+FYRVFPNGEVQYLHPKDGV
Sbjct: 117 EMPTGGAAIMREGPNLLKLARKEQCLALGTRLRSKYKINYRFYRVFPNGEVQYLHPKDGV 176
Query: 172 YPEKVNPGRQGVGQNFRSIGKNVSPIEVKFTGKQPFDV 209
YPEKVNPGRQGVGQNFRSIGKN SPIEVKFTGKQ +D+
Sbjct: 177 YPEKVNPGRQGVGQNFRSIGKNKSPIEVKFTGKQVYDI 214
>UniRef100_P12353 Photosystem I reaction center subunit II, chloroplast precursor
[Spinacia oleracea]
Length = 212
Score = 317 bits (813), Expect = 1e-85
Identities = 169/218 (77%), Positives = 181/218 (82%), Gaps = 15/218 (6%)
Query: 1 MAMATQASLFTPP-LSSPKP--------WKQPSTLSFISLKPIKFTTKTTKISAADEKTE 51
MAMATQA+LF+P LSS KP +KQPS ++F S KP + + +AA E
Sbjct: 1 MAMATQATLFSPSSLSSAKPIDTRLTTSFKQPSAVTFAS-KPAS-RHHSIRAAAAAEGKA 58
Query: 52 AASVTTKEEAPAAPVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIF 111
AA+ TKE AP GFTPPELDPNTPSPIF GSTGGLLRKAQVEEFYVITWESPKEQIF
Sbjct: 59 AAATETKE----APKGFTPPELDPNTPSPIFAGSTGGLLRKAQVEEFYVITWESPKEQIF 114
Query: 112 EMPTGGAAIMREGPNLLKLARKEQCLALGNRLRSKYKIKYQFYRVFPNGEVQYLHPKDGV 171
EMPTGGAAIMREGPNLLKLARKEQCLALG RLRSKYKIKYQFYRVFP+GEVQYLHPKDGV
Sbjct: 115 EMPTGGAAIMREGPNLLKLARKEQCLALGTRLRSKYKIKYQFYRVFPSGEVQYLHPKDGV 174
Query: 172 YPEKVNPGRQGVGQNFRSIGKNVSPIEVKFTGKQPFDV 209
YPEKVNPGRQGVG N RSIGKNVSPIEVKFTGKQP+D+
Sbjct: 175 YPEKVNPGRQGVGLNMRSIGKNVSPIEVKFTGKQPYDL 212
>UniRef100_P29302 Photosystem I reaction center subunit II, chloroplast precursor
[Nicotiana sylvestris]
Length = 204
Score = 315 bits (808), Expect = 4e-85
Identities = 166/212 (78%), Positives = 175/212 (82%), Gaps = 11/212 (5%)
Query: 1 MAMATQASLFTPPLSSPK---PWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTT 57
MAMATQASLFTP LS+PK PWKQ +L+ S K +K T + A A T
Sbjct: 1 MAMATQASLFTPALSAPKSSAPWKQ--SLASFSPKQLKSTVSAPRPIRA----MAEEAAT 54
Query: 58 KEEAPAAPVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGG 117
KE APVGFTPP+LDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGG
Sbjct: 55 KEAE--APVGFTPPQLDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGG 112
Query: 118 AAIMREGPNLLKLARKEQCLALGNRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYPEKVN 177
AAIMREG NLLKLARKEQCLALG RLRSKYKI Y+FYRVFPNGEVQYLHPKDGVYPEKVN
Sbjct: 113 AAIMREGANLLKLARKEQCLALGTRLRSKYKINYRFYRVFPNGEVQYLHPKDGVYPEKVN 172
Query: 178 PGRQGVGQNFRSIGKNVSPIEVKFTGKQPFDV 209
GRQGVGQNFRSIGKN SPIEVKFTGKQ +D+
Sbjct: 173 AGRQGVGQNFRSIGKNKSPIEVKFTGKQVYDL 204
>UniRef100_Q84PB4 Chloroplast photosystem I reaction center subunit II-like protein
[Oryza sativa]
Length = 203
Score = 297 bits (760), Expect = 1e-79
Identities = 158/211 (74%), Positives = 167/211 (78%), Gaps = 10/211 (4%)
Query: 1 MAMATQASLFTPPLSSPKPWKQP--STLSFISLKPIKFTTKTTKISAADEKTEAASVTTK 58
MAMATQAS L + +P STLS + + T + AA E AA+ T
Sbjct: 1 MAMATQASAAKCHLLAAWAPAKPRSSTLSMPTSRA------PTSLRAAAEDQPAAAAT-- 52
Query: 59 EEAPAAPVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGGA 118
EE AP GF PP+LDPNTPSPIFGGSTGGLLRKAQVEEFYVITW SPKEQ+FEMPTGGA
Sbjct: 53 EEKKPAPAGFVPPQLDPNTPSPIFGGSTGGLLRKAQVEEFYVITWTSPKEQVFEMPTGGA 112
Query: 119 AIMREGPNLLKLARKEQCLALGNRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYPEKVNP 178
AIMREGPNLLKLARKEQCLALG RLRSKYKI YQFYRVFPNGEVQYLHPKDGVYPEKVN
Sbjct: 113 AIMREGPNLLKLARKEQCLALGTRLRSKYKINYQFYRVFPNGEVQYLHPKDGVYPEKVNA 172
Query: 179 GRQGVGQNFRSIGKNVSPIEVKFTGKQPFDV 209
GRQGVGQNFRSIGKNVSPIEVKFTGK FD+
Sbjct: 173 GRQGVGQNFRSIGKNVSPIEVKFTGKNVFDI 203
>UniRef100_P36213 Photosystem I reaction center subunit II, chloroplast precursor
[Hordeum vulgare]
Length = 205
Score = 295 bits (755), Expect = 5e-79
Identities = 157/213 (73%), Positives = 166/213 (77%), Gaps = 12/213 (5%)
Query: 1 MAMATQASLFTPPLS----SPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVT 56
MAMATQAS T L SP +P+TL+ S + AA T A +
Sbjct: 1 MAMATQASAATRHLITAAWSPSAKPRPATLAMPSS-----ARGPAPLFAAAPDTPAPAAP 55
Query: 57 TKEEAPAAPVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTG 116
E APA GF PP+LDP+TPSPIFGGSTGGLLRKAQVEEFYVITW SPKEQ+FEMPTG
Sbjct: 56 PAEPAPA---GFVPPQLDPSTPSPIFGGSTGGLLRKAQVEEFYVITWTSPKEQVFEMPTG 112
Query: 117 GAAIMREGPNLLKLARKEQCLALGNRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYPEKV 176
GAAIMREGPNLLKLARKEQCLALGNRLRSKYKI YQFYRVFPNGEVQYLHPKDGVYPEKV
Sbjct: 113 GAAIMREGPNLLKLARKEQCLALGNRLRSKYKIAYQFYRVFPNGEVQYLHPKDGVYPEKV 172
Query: 177 NPGRQGVGQNFRSIGKNVSPIEVKFTGKQPFDV 209
N GRQGVGQNFRSIGKNVSPIEVKFTGK FD+
Sbjct: 173 NAGRQGVGQNFRSIGKNVSPIEVKFTGKNSFDI 205
>UniRef100_Q9SA56 Photosystem I reaction center subunit II-2, chloroplast precursor
[Arabidopsis thaliana]
Length = 204
Score = 293 bits (750), Expect = 2e-78
Identities = 150/208 (72%), Positives = 167/208 (80%), Gaps = 5/208 (2%)
Query: 3 MATQAS-LFTPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEA 61
MATQA+ +F+P +++ + L S +P + T I A ++ +A+ KE
Sbjct: 1 MATQAAGIFSPAITTTTSAVKKLHLFSSSHRPKSLSFTKTAIRAEKTESSSAAPAVKE-- 58
Query: 62 PAAPVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGGAAIM 121
APVGFTPP+LDPNTPSPIF GSTGGLLRKAQVEEFYVITW SPKEQIFEMPTGGAAIM
Sbjct: 59 --APVGFTPPQLDPNTPSPIFAGSTGGLLRKAQVEEFYVITWNSPKEQIFEMPTGGAAIM 116
Query: 122 REGPNLLKLARKEQCLALGNRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYPEKVNPGRQ 181
REGPNLLKLARKEQCLALG RLRSKYKI YQFYRVFPNGEVQYLHPKDGVYPEK NPGR+
Sbjct: 117 REGPNLLKLARKEQCLALGTRLRSKYKITYQFYRVFPNGEVQYLHPKDGVYPEKANPGRE 176
Query: 182 GVGQNFRSIGKNVSPIEVKFTGKQPFDV 209
GVG N RSIGKNVSPIEVKFTGKQ +D+
Sbjct: 177 GVGLNMRSIGKNVSPIEVKFTGKQSYDL 204
>UniRef100_Q9S7H1 Photosystem I reaction center subunit II-1, chloroplast precursor
[Arabidopsis thaliana]
Length = 208
Score = 288 bits (738), Expect = 5e-77
Identities = 141/178 (79%), Positives = 149/178 (83%), Gaps = 3/178 (1%)
Query: 32 KPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTPPELDPNTPSPIFGGSTGGLLR 91
K + FT + D AA+ +EAP VGFTPP+LDPNTPSPIF GSTGGLLR
Sbjct: 34 KSLSFTKTAIRAEKTDSSAAAAAAPATKEAP---VGFTPPQLDPNTPSPIFAGSTGGLLR 90
Query: 92 KAQVEEFYVITWESPKEQIFEMPTGGAAIMREGPNLLKLARKEQCLALGNRLRSKYKIKY 151
KAQVEEFYVITW SPKEQIFEMPTGGAAIMREGPNLLKLARKEQCLALG RLRSKYKI Y
Sbjct: 91 KAQVEEFYVITWNSPKEQIFEMPTGGAAIMREGPNLLKLARKEQCLALGTRLRSKYKITY 150
Query: 152 QFYRVFPNGEVQYLHPKDGVYPEKVNPGRQGVGQNFRSIGKNVSPIEVKFTGKQPFDV 209
QFYRVFPNGEVQYLHPKDGVYPEK NPGR+GVG N RSIGKNVSPIEVKFTGKQ +D+
Sbjct: 151 QFYRVFPNGEVQYLHPKDGVYPEKANPGREGVGLNMRSIGKNVSPIEVKFTGKQSYDL 208
>UniRef100_Q39615 Photosystem I reaction center subunit II, chloroplast precursor
[Chlamydomonas reinhardtii]
Length = 196
Score = 224 bits (570), Expect = 1e-57
Identities = 110/177 (62%), Positives = 134/177 (75%), Gaps = 9/177 (5%)
Query: 37 TTKTTKISAADEKTEAASVTTKEEAPAAPVG---------FTPPELDPNTPSPIFGGSTG 87
T +++++ A +V + EA AAP +T P L+P+TPSPIFGGSTG
Sbjct: 13 TRASSRVAVAARPAARRAVVVRAEAEAAPAAAKKAAEKPAWTVPTLNPDTPSPIFGGSTG 72
Query: 88 GLLRKAQVEEFYVITWESPKEQIFEMPTGGAAIMREGPNLLKLARKEQCLALGNRLRSKY 147
GLLRKAQ EEFYVITWE+ KEQIFEMPTGGAAIMR+GPNLLK +KEQCLAL +LR+K+
Sbjct: 73 GLLRKAQTEEFYVITWEAKKEQIFEMPTGGAAIMRQGPNLLKFGKKEQCLALTTQLRNKF 132
Query: 148 KIKYQFYRVFPNGEVQYLHPKDGVYPEKVNPGRQGVGQNFRSIGKNVSPIEVKFTGK 204
K+ FYRVFP+G+VQYLHP DGVYPEKVN GR G QN R IG+NV+PI+VKF+G+
Sbjct: 133 KLTPCFYRVFPDGKVQYLHPADGVYPEKVNAGRVGANQNMRRIGQNVNPIKVKFSGR 189
>UniRef100_Q9T4W8 Photosystem I reaction centre subunit II precursor [Cyanophora
paradoxa]
Length = 220
Score = 212 bits (539), Expect = 6e-54
Identities = 104/181 (57%), Positives = 130/181 (71%), Gaps = 1/181 (0%)
Query: 29 ISLKPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTPPELDPNTPSPIFGGSTGG 88
++ K F + E+ EAA E+ AAP F+ P L+ N P+PIFGGSTGG
Sbjct: 35 VAAKKTTFEAAPARFIVRAEEEEAAPAEKVEKKAAAPKPFSVPTLNLNAPTPIFGGSTGG 94
Query: 89 LLRKAQVEEFYVITWESPKEQIFEMPTGGAAIMREGPNLLKLARKEQCLALGNRLRSKYK 148
LLRKA+VEEFY ITW E +FE+PTGGAAIMR G NLL+LARKEQC+ALG +L+ K+K
Sbjct: 95 LLRKAEVEEFYSITWTGKSETVFELPTGGAAIMRAGENLLRLARKEQCIALGAQLKDKFK 154
Query: 149 I-KYQFYRVFPNGEVQYLHPKDGVYPEKVNPGRQGVGQNFRSIGKNVSPIEVKFTGKQPF 207
I Y+ YRV+P+GEVQ+LHPKDGV+PEKVNPGR VG N R IG+N P ++KF G++ F
Sbjct: 155 ITDYKIYRVYPSGEVQFLHPKDGVFPEKVNPGRVAVGSNKRRIGQNPDPAKLKFKGQETF 214
Query: 208 D 208
D
Sbjct: 215 D 215
>UniRef100_O78502 Photosystem I reaction center subunit II [Guillardia theta]
Length = 141
Score = 189 bits (481), Expect = 3e-47
Identities = 91/133 (68%), Positives = 108/133 (80%), Gaps = 1/133 (0%)
Query: 73 LDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGGAAIMREGPNLLKLAR 132
L+ PSP F GSTGG LR A+ EE Y ITW SPKEQ+FEMPTGGAAIMR+G NLL LAR
Sbjct: 5 LNLQIPSPTFEGSTGGWLRAAETEEKYAITWTSPKEQVFEMPTGGAAIMRKGENLLYLAR 64
Query: 133 KEQCLALGNRLRSKYKI-KYQFYRVFPNGEVQYLHPKDGVYPEKVNPGRQGVGQNFRSIG 191
KEQCLALG ++++ +KI Y+ YR+FP+GEVQYLHPKDGV+PEKVNPGR GVG SIG
Sbjct: 65 KEQCLALGTQVKTSFKITDYKIYRIFPSGEVQYLHPKDGVFPEKVNPGRIGVGNVSHSIG 124
Query: 192 KNVSPIEVKFTGK 204
KN++P ++KFT K
Sbjct: 125 KNLNPAQIKFTNK 137
>UniRef100_Q6B8U2 Photosystem I reaction center subunit II [Gracilaria tenuistipitata
var. liui]
Length = 141
Score = 189 bits (480), Expect = 4e-47
Identities = 91/137 (66%), Positives = 107/137 (77%), Gaps = 1/137 (0%)
Query: 73 LDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGGAAIMREGPNLLKLAR 132
+D PSP FGGSTGG LR A++EE Y ITW S +EQIFEMP GGAAIMREG NLL LAR
Sbjct: 5 IDLKMPSPTFGGSTGGWLRSAEIEEKYAITWTSKQEQIFEMPIGGAAIMREGNNLLYLAR 64
Query: 133 KEQCLALGNRLRSKYKIK-YQFYRVFPNGEVQYLHPKDGVYPEKVNPGRQGVGQNFRSIG 191
KEQCLAL +L+ ++KIK ++ YR+FPN EVQYLHPKDGV+PEKVNPGR G+G SIG
Sbjct: 65 KEQCLALSTQLKMQFKIKDFKIYRIFPNSEVQYLHPKDGVFPEKVNPGRVGIGNIDHSIG 124
Query: 192 KNVSPIEVKFTGKQPFD 208
KN P+ KFTGK ++
Sbjct: 125 KNEDPVNKKFTGKATYE 141
>UniRef100_P51279 Photosystem I reaction center subunit II [Porphyra purpurea]
Length = 141
Score = 186 bits (471), Expect = 4e-46
Identities = 91/140 (65%), Positives = 104/140 (74%), Gaps = 1/140 (0%)
Query: 70 PPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGGAAIMREGPNLLK 129
P ++ N PSP FGGSTGG LR A+VEE Y ITW E FEMPTGG A MR+G NLL
Sbjct: 2 PDTINLNMPSPTFGGSTGGWLRAAEVEEKYAITWTGKNESKFEMPTGGTATMRDGENLLY 61
Query: 130 LARKEQCLALGNRLRSKYKIK-YQFYRVFPNGEVQYLHPKDGVYPEKVNPGRQGVGQNFR 188
LA+KEQCLALG +L+ K+KI Y+ YRVFPNGEVQYLHPKDGV+PEKVN GR +
Sbjct: 62 LAKKEQCLALGTQLKGKFKISDYKIYRVFPNGEVQYLHPKDGVFPEKVNAGRASINSVDH 121
Query: 189 SIGKNVSPIEVKFTGKQPFD 208
SIGKNV+PI VKFT K +D
Sbjct: 122 SIGKNVNPINVKFTNKATYD 141
>UniRef100_P49481 Photosystem I reaction center subunit II [Odontella sinensis]
Length = 139
Score = 182 bits (461), Expect = 6e-45
Identities = 90/131 (68%), Positives = 101/131 (76%)
Query: 73 LDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGGAAIMREGPNLLKLAR 132
L+ TP P FGGSTGG LR A+VEE Y ITW S KEQIFEMPTGGAAIMR G NLL LAR
Sbjct: 3 LNLQTPFPTFGGSTGGWLRAAEVEEKYAITWTSKKEQIFEMPTGGAAIMRNGENLLYLAR 62
Query: 133 KEQCLALGNRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYPEKVNPGRQGVGQNFRSIGK 192
KEQCLALG +LR+ Y+ YR+FP+GEVQYLHPKDGV+PEKVNPGR V SIGK
Sbjct: 63 KEQCLALGTQLRTFKINDYKIYRIFPSGEVQYLHPKDGVFPEKVNPGRTSVNSRGFSIGK 122
Query: 193 NVSPIEVKFTG 203
N +P +KF+G
Sbjct: 123 NPNPASIKFSG 133
>UniRef100_P19569 Photosystem I reaction center subunit II [Synechocystis sp.]
Length = 140
Score = 181 bits (459), Expect = 1e-44
Identities = 88/133 (66%), Positives = 103/133 (77%), Gaps = 3/133 (2%)
Query: 80 PIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGGAAIMREGPNLLKLARKEQCLAL 139
P FGGSTGGLL KA EE Y ITW S EQ+FEMPTGGAAIM EG NLL LARKEQCLAL
Sbjct: 8 PKFGGSTGGLLSKANREEKYAITWTSASEQVFEMPTGGAAIMNEGENLLYLARKEQCLAL 67
Query: 140 GNRLRSKYKIK---YQFYRVFPNGEVQYLHPKDGVYPEKVNPGRQGVGQNFRSIGKNVSP 196
G +LR+K+K K Y+ YRV+P+GEVQYLHP DGV+PEKVN GR+ G R IG+N P
Sbjct: 68 GTQLRTKFKPKIQDYKIYRVYPSGEVQYLHPADGVFPEKVNEGREAQGTKTRRIGQNPEP 127
Query: 197 IEVKFTGKQPFDV 209
+ +KF+GK P++V
Sbjct: 128 VTIKFSGKAPYEV 140
>UniRef100_O07115 Photosystem I reaction center subunit II [Mastigocladus laminosus]
Length = 142
Score = 181 bits (459), Expect = 1e-44
Identities = 85/130 (65%), Positives = 103/130 (78%)
Query: 79 SPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGGAAIMREGPNLLKLARKEQCLA 138
+PIFGGSTGGLL+KA+VEE Y ITW SPKEQ+FEMPTGGAA MR+G NLL LARKEQC+A
Sbjct: 9 TPIFGGSTGGLLKKAEVEEKYAITWTSPKEQVFEMPTGGAAKMRQGQNLLYLARKEQCIA 68
Query: 139 LGNRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYPEKVNPGRQGVGQNFRSIGKNVSPIE 198
LG++LR Y+ YR++PNGE Y+HP DGV+PEKVNPGRQ V N R IG+N P +
Sbjct: 69 LGSQLRRLKITDYKIYRIYPNGETAYIHPADGVFPEKVNPGRQKVRYNDRRIGQNPDPAK 128
Query: 199 VKFTGKQPFD 208
+KF+G +D
Sbjct: 129 LKFSGVATYD 138
>UniRef100_O96800 Photosystem I reaction center subunit II [Skeletonema costatum]
Length = 139
Score = 178 bits (452), Expect = 7e-44
Identities = 88/131 (67%), Positives = 100/131 (76%)
Query: 73 LDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGGAAIMREGPNLLKLAR 132
L+ TP P FGGSTGG LR A+VEE Y ITW S KEQIFEMPTGG+AIMR G NLL LAR
Sbjct: 3 LNLKTPFPTFGGSTGGWLRAAEVEEKYAITWTSAKEQIFEMPTGGSAIMRNGENLLYLAR 62
Query: 133 KEQCLALGNRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYPEKVNPGRQGVGQNFRSIGK 192
KEQCLAL +LR+ Y+ YR+FP+GEVQYLHPKDGV+PEKVNPGR V SIGK
Sbjct: 63 KEQCLALSTQLRTFKINDYKIYRIFPSGEVQYLHPKDGVFPEKVNPGRTSVNSRGFSIGK 122
Query: 193 NVSPIEVKFTG 203
N +P +KF+G
Sbjct: 123 NPNPASIKFSG 133
>UniRef100_Q9M713 Ferredoxin-binding subunit [Zea mays]
Length = 108
Score = 176 bits (446), Expect = 3e-43
Identities = 83/91 (91%), Positives = 87/91 (95%)
Query: 118 AAIMREGPNLLKLARKEQCLALGNRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYPEKVN 177
+AIMREGPNLLKLARKEQCLALG RLRSKYKI YQFYRVFPNGEVQYLHPKDGVYPEKVN
Sbjct: 18 SAIMREGPNLLKLARKEQCLALGTRLRSKYKITYQFYRVFPNGEVQYLHPKDGVYPEKVN 77
Query: 178 PGRQGVGQNFRSIGKNVSPIEVKFTGKQPFD 208
PGR+GVGQNFRSIGKNV+PI+VKFTGK FD
Sbjct: 78 PGREGVGQNFRSIGKNVNPIDVKFTGKSTFD 108
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 387,011,911
Number of Sequences: 2790947
Number of extensions: 17510032
Number of successful extensions: 38162
Number of sequences better than 10.0: 149
Number of HSP's better than 10.0 without gapping: 73
Number of HSP's successfully gapped in prelim test: 78
Number of HSP's that attempted gapping in prelim test: 37904
Number of HSP's gapped (non-prelim): 282
length of query: 209
length of database: 848,049,833
effective HSP length: 122
effective length of query: 87
effective length of database: 507,554,299
effective search space: 44157224013
effective search space used: 44157224013
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 72 (32.3 bits)
Medicago: description of AC145222.8


