

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145222.11 + phase: 0 /pseudo
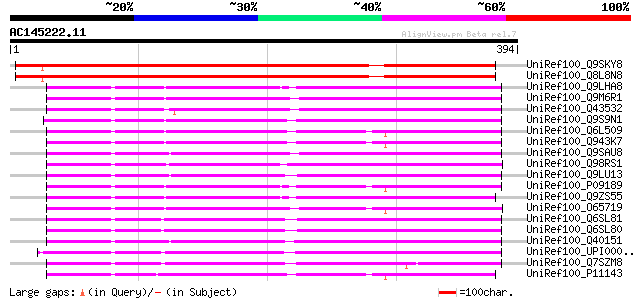
(394 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SKY8 70kD heat shock protein [Arabidopsis thaliana] 564 e-159
UniRef100_Q8L8N8 70kD heat shock protein [Arabidopsis thaliana] 562 e-159
UniRef100_Q9LHA8 70 kDa heat shock protein [Arabidopsis thaliana] 192 1e-47
UniRef100_Q9M6R1 High molecular weight heat shock protein [Malus... 191 2e-47
UniRef100_Q43532 HSP70 [Lilium longiflorum] 191 4e-47
UniRef100_Q9S9N1 T24D18.14 protein [Arabidopsis thaliana] 190 5e-47
UniRef100_Q6L509 Putative hsp70 [Oryza sativa] 190 7e-47
UniRef100_Q943K7 Putative HSP70 [Oryza sativa] 189 2e-46
UniRef100_Q9SAU8 HSP70 [Triticum aestivum] 187 3e-46
UniRef100_Q98RS1 Heat shock protein 70KD [Guillardia theta] 187 4e-46
UniRef100_Q9LU13 Hsp70 protein [Blastocystis hominis] 187 6e-46
UniRef100_P09189 Heat shock cognate 70 kDa protein [Petunia hybr... 186 8e-46
UniRef100_Q9ZS55 Heat shock protein 70 [Arabidopsis thaliana] 186 1e-45
UniRef100_O65719 Heat shock cognate 70 kDa protein 3 [Arabidopsi... 185 2e-45
UniRef100_Q6SL81 Heat shock protein 70 [Phytophthora nicotianae] 184 3e-45
UniRef100_Q6SL80 Heat shock protein 70 [Phytophthora nicotianae] 184 3e-45
UniRef100_Q40151 Hsc70 protein [Lycopersicon esculentum] 184 3e-45
UniRef100_UPI0000438C23 UPI0000438C23 UniRef100 entry 184 4e-45
UniRef100_Q7SZM8 Constitutive heat shock protein HSC70-1 [Cyprin... 184 4e-45
UniRef100_P11143 Heat shock 70 kDa protein [Zea mays] 184 4e-45
>UniRef100_Q9SKY8 70kD heat shock protein [Arabidopsis thaliana]
Length = 563
Score = 564 bits (1454), Expect = e-159
Identities = 283/376 (75%), Positives = 324/376 (85%), Gaps = 14/376 (3%)
Query: 5 EPAYTVASDSETTGEEKSSP---LPEIAIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLM 61
E AYTVASDSE TGEEKSS LPEIA+GIDIGTSQCS+AVWNGS+V +L+N RNQKL+
Sbjct: 3 EAAYTVASDSENTGEEKSSSSPSLPEIALGIDIGTSQCSIAVWNGSQVHILRNTRNQKLI 62
Query: 62 KSFVTFKDEAPSGGVTSQFSHEHEMLFGDTIFNMKRLIGRVDTDPVVHASKNLPFLVQTL 121
KSFVTFKDE P+GGV++Q +HE EML G IFNMKRL+GRVDTDPVVHASKNLPFLVQTL
Sbjct: 63 KSFVTFKDEVPAGGVSNQLAHEQEMLTGAAIFNMKRLVGRVDTDPVVHASKNLPFLVQTL 122
Query: 122 DIGVRPFIAALVNNVWRSTTPEEVLAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQL 181
DIGVRPFIAALVNN WRSTTPEEVLA+FLVELRLM E LKRP+RNVVLTVPVSFSRFQL
Sbjct: 123 DIGVRPFIAALVNNAWRSTTPEEVLAIFLVELRLMAEAQLKRPVRNVVLTVPVSFSRFQL 182
Query: 182 TRLERACAMAGLHVLRLMPEPTAVALLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDV 241
TR ERACAMAGLHVLRLMPEPTA+ALLY QQQQ + + MGSGSE++A+IFNMGAGYCDV
Sbjct: 183 TRFERACAMAGLHVLRLMPEPTAIALLYAQQQQMTTHDNMGSGSERLAVIFNMGAGYCDV 242
Query: 242 AVTATAGGVSQIKALAGSTIGGEDLLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQ 301
AVTATAGGVSQIKALAGS IGGED+LQN +RH+ P +E ++ LLRVA Q
Sbjct: 243 AVTATAGGVSQIKALAGSPIGGEDILQNTIRHIAPPNE-----------EASGLLRVAAQ 291
Query: 302 DAIHQLSTQSSVQVDVDLGNSLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVG 361
DAIH+L+ Q +VQ++VDLGN KI KV+DR EFEEVN++VFE+CE L++QCL DA+V G
Sbjct: 292 DAIHRLTDQENVQIEVDLGNGNKISKVLDRLEFEEVNQKVFEECERLVVQCLRDARVNGG 351
Query: 362 DINDVILVGGCSYIPQ 377
DI+D+I+VGGCSYIP+
Sbjct: 352 DIDDLIMVGGCSYIPK 367
>UniRef100_Q8L8N8 70kD heat shock protein [Arabidopsis thaliana]
Length = 563
Score = 562 bits (1449), Expect = e-159
Identities = 282/376 (75%), Positives = 324/376 (86%), Gaps = 14/376 (3%)
Query: 5 EPAYTVASDSETTGEEKSSP---LPEIAIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLM 61
E AYTVASDSE TGEEKSS LPEIA+GIDIGTSQCS+AVWNGS+V +L+N RNQKL+
Sbjct: 3 EAAYTVASDSENTGEEKSSSSPSLPEIALGIDIGTSQCSIAVWNGSQVHILRNTRNQKLI 62
Query: 62 KSFVTFKDEAPSGGVTSQFSHEHEMLFGDTIFNMKRLIGRVDTDPVVHASKNLPFLVQTL 121
KSFVTFKDE P+GGV++Q +HE EML G IFNMKRL+GRVDTDPVVHASKNLPFLVQTL
Sbjct: 63 KSFVTFKDEVPAGGVSNQLAHEQEMLTGAAIFNMKRLVGRVDTDPVVHASKNLPFLVQTL 122
Query: 122 DIGVRPFIAALVNNVWRSTTPEEVLAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQL 181
DIGVRPFIAALVNN WRSTTPEEVLA+FLVELRLM E LKRP+RNVVLTVPVSFSRFQL
Sbjct: 123 DIGVRPFIAALVNNAWRSTTPEEVLAIFLVELRLMAEAQLKRPVRNVVLTVPVSFSRFQL 182
Query: 182 TRLERACAMAGLHVLRLMPEPTAVALLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDV 241
TR ERACAMAGLHVLRLMPEPTA+ALLY QQQQ + + MGSGSE++A+IFNMGAGYCDV
Sbjct: 183 TRFERACAMAGLHVLRLMPEPTAIALLYAQQQQMTTHDNMGSGSERLAVIFNMGAGYCDV 242
Query: 242 AVTATAGGVSQIKALAGSTIGGEDLLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQ 301
AVTATAGGVSQIKALAGS IGGED+LQ+ +RH+ P +E ++ LLRVA Q
Sbjct: 243 AVTATAGGVSQIKALAGSPIGGEDILQSTIRHIAPPNE-----------EASGLLRVAAQ 291
Query: 302 DAIHQLSTQSSVQVDVDLGNSLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVG 361
DAIH+L+ Q +VQ++VDLGN KI KV+DR EFEEVN++VFE+CE L++QCL DA+V G
Sbjct: 292 DAIHRLTDQENVQIEVDLGNGNKISKVLDRLEFEEVNQKVFEECERLVVQCLRDARVNGG 351
Query: 362 DINDVILVGGCSYIPQ 377
DI+D+I+VGGCSYIP+
Sbjct: 352 DIDDLIMVGGCSYIPK 367
>UniRef100_Q9LHA8 70 kDa heat shock protein [Arabidopsis thaliana]
Length = 650
Score = 192 bits (488), Expect = 1e-47
Identities = 122/357 (34%), Positives = 199/357 (55%), Gaps = 12/357 (3%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKN-LPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +DP V A K+ PF V + G +P I + + EE+ +
Sbjct: 67 TNTVFDAKRLIGRRYSDPSVQADKSHWPFKVVS-GPGEKPMIVVNHKGEEKQFSAEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E L P++N V+TVP F+ Q + A ++GL+V+R++ EPTA A+
Sbjct: 126 MVLIKMREIAEAFLGSPVKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG +KAS S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYG-LDKKAS-----SVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 239
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
M+ H + + + K+ +++ LR A + A LS+ + +++D L +
Sbjct: 240 DNRMVNHFVQEFKRKNKKDITGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGIDF 299
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++L
Sbjct: 300 YTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSSVHDVVLVGGSTRIPKVQQLL 356
>UniRef100_Q9M6R1 High molecular weight heat shock protein [Malus domestica]
Length = 650
Score = 191 bits (486), Expect = 2e-47
Identities = 121/357 (33%), Positives = 195/357 (53%), Gaps = 12/357 (3%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +DP V K+ PF V G +P I + + EE+ +
Sbjct: 67 VNTVFDAKRLIGRRYSDPSVQGDMKHWPFKVIP-GPGDKPMIVVIYKGEEKQFAAEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M LV++R + E +L I+N V+TVP F+ Q + A +AGL+VLR++ EPTA A+
Sbjct: 126 MVLVKMREIAEAYLGSSIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYGLDKKATSV------GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 239
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
M+ H + + + K+ +++ LR A + A LS+ + +++D L +
Sbjct: 240 DNRMVNHFVQEFKRKHKKDISSSPRALRRLRTACERAKRTLSSTAQTTIEIDSLYEGVDF 299
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++L
Sbjct: 300 YSTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLL 356
>UniRef100_Q43532 HSP70 [Lilium longiflorum]
Length = 649
Score = 191 bits (484), Expect = 4e-47
Identities = 126/359 (35%), Positives = 196/359 (54%), Gaps = 16/359 (4%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVR--PFIAALVNNVWRSTTPEEV 145
+T+F+ KRLIGR +DP V A L PF V + G R P I + EEV
Sbjct: 67 TNTVFDAKRLIGRHFSDPSVQADMKLWPFKVIS---GTRDKPMIVVQYKGEEKQFAVEEV 123
Query: 146 LAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAV 205
+M LV++R + +L R I N V+TVP F+ Q + A A+AGL+VLR++ EPTA
Sbjct: 124 SSMVLVKMREIAVAYLGRSINNAVVTVPAYFNDSQRQATKDAGAIAGLNVLRIINEPTAA 183
Query: 206 ALLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGE 264
A+ YG ++ S SEK LIF++G G DV++ G+ ++KA AG T +GGE
Sbjct: 184 AIAYGLDKKATSP------SEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGE 237
Query: 265 DLLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSL 323
D M+ H + + K+ +++ LR A + A LS+ + +++D L + +
Sbjct: 238 DFDNRMVNHFAQEFKRKHKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLCDGI 297
Query: 324 KICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++L
Sbjct: 298 DFYSTITRARFEELNIDLFRKCMDPVEKCLTDAKMDKSSVHDVVLVGGSTRIPKVQQLL 356
>UniRef100_Q9S9N1 T24D18.14 protein [Arabidopsis thaliana]
Length = 646
Score = 190 bits (483), Expect = 5e-47
Identities = 119/359 (33%), Positives = 197/359 (54%), Gaps = 12/359 (3%)
Query: 27 EIAIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEM 86
E AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ +
Sbjct: 6 EKAIGIDLGTTYSCVGVWMNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAK--NQVAL 63
Query: 87 LFGDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEV 145
+T+F+ KRLIGR +DP V + + PF V + G +P I N + +PEE+
Sbjct: 64 NPQNTVFDAKRLIGRKFSDPSVQSDILHWPFKVVS-GPGEKPMIVVSYKNEEKQFSPEEI 122
Query: 146 LAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAV 205
+M LV+++ + E L R ++N V+TVP F+ Q + A A++GL+VLR++ EPTA
Sbjct: 123 SSMVLVKMKEVAEAFLGRTVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEPTAA 182
Query: 206 ALLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGE 264
A+ YG ++ EK LIF++G G DV++ GV ++KA AG T +GGE
Sbjct: 183 AIAYGLDKKGT------KAGEKNVLIFDLGGGTFDVSLLTIEEGVFEVKATAGDTHLGGE 236
Query: 265 DLLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSL 323
D ++ H + + K+ +++ LR A + A LS+ + +++D L +
Sbjct: 237 DFDNRLVNHFVAEFRRKHKKDIAGNARALRRLRTACERAKRTLSSTAQTTIEIDSLHEGI 296
Query: 324 KICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + + L DAK++ ++DV+LVGG + IP+ ++L
Sbjct: 297 DFYATISRARFEEMNMDLFRKCMDPVEKVLKDAKLDKSSVHDVVLVGGSTRIPKIQQLL 355
>UniRef100_Q6L509 Putative hsp70 [Oryza sativa]
Length = 646
Score = 190 bits (482), Expect = 7e-47
Identities = 122/361 (33%), Positives = 197/361 (53%), Gaps = 20/361 (5%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 8 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 65
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +DP V + L PF V G +P I + EE+ +
Sbjct: 66 TNTVFDAKRLIGRRFSDPSVQSDMKLWPFKVVP-GPGDKPMIVVQYKGEEKQFAAEEISS 124
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E +L I+N V+TVP F+ Q + A +AGL+V+R++ EPTA A+
Sbjct: 125 MVLIKMREIAEAYLGSSIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAI 184
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 185 AYGLDKKAT------SSGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 238
Query: 267 LQNMMRHLLPDSENIFKRHGVEEI----KSMALLRVATQDAIHQLSTQSSVQVDVD-LGN 321
M+ H + + FKR ++I +++ LR A + A LS+ + +++D L
Sbjct: 239 DNRMVNHFVQE----FKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLYE 294
Query: 322 SLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRI 381
+ + RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++
Sbjct: 295 GIDFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSSVHDVVLVGGSTRIPKVQQL 354
Query: 382 L 382
L
Sbjct: 355 L 355
>UniRef100_Q943K7 Putative HSP70 [Oryza sativa]
Length = 648
Score = 189 bits (479), Expect = 2e-46
Identities = 122/361 (33%), Positives = 198/361 (54%), Gaps = 20/361 (5%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 8 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNP 65
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +DP V + L PF V G +P I + + EE+ +
Sbjct: 66 INTVFDAKRLIGRRFSDPSVQSDMKLWPFKVIP-GPGDKPMIVVQYKGEEKQFSAEEISS 124
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E +L I+N V+TVP F+ Q + A +AGL+V+R++ EPTA A+
Sbjct: 125 MVLIKMREIAEAYLGSNIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAI 184
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 185 AYGLDKKAT------SSGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 238
Query: 267 LQNMMRHLLPDSENIFKRHGVEEI----KSMALLRVATQDAIHQLSTQSSVQVDVD-LGN 321
M+ H + + FKR ++I +++ LR A + A LS+ + +++D L
Sbjct: 239 DNRMVNHFVLE----FKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLYE 294
Query: 322 SLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRI 381
+ + RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++
Sbjct: 295 GIDFYSTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSSVHDVVLVGGSTRIPKVQQL 354
Query: 382 L 382
L
Sbjct: 355 L 355
>UniRef100_Q9SAU8 HSP70 [Triticum aestivum]
Length = 648
Score = 187 bits (476), Expect = 3e-46
Identities = 119/357 (33%), Positives = 193/357 (53%), Gaps = 12/357 (3%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 8 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 65
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +DP V + L PF V +P I + EE+ +
Sbjct: 66 TNTVFDAKRLIGRRFSDPSVQSDMKLWPFKVIPGPAD-KPMIVVNYKGEEKQFAAEEISS 124
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E L ++N V+TVP F+ Q + A A+AGL+VLR++ EPTA A+
Sbjct: 125 MVLIKMREIAEAFLGNSVKNAVVTVPAYFNDSQRQATKDAGAIAGLNVLRIINEPTAAAI 184
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 185 AYGLDKKATST------GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 238
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
M+ H + + + K+ +++ LR A + A LS+ + +++D L +
Sbjct: 239 DNRMVNHFVQEFKRKHKKDITGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLYEGVDF 298
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++L
Sbjct: 299 YTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLL 355
>UniRef100_Q98RS1 Heat shock protein 70KD [Guillardia theta]
Length = 650
Score = 187 bits (475), Expect = 4e-46
Identities = 118/358 (32%), Positives = 200/358 (54%), Gaps = 11/358 (3%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V +W VE++ N + + S+V F + G +++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGIWQHDRVEIIANDQGNRTTPSYVAFTETERLIGDSAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR DP V K+ PF V D G +P I + PEE+ A
Sbjct: 67 HNTVFDAKRLIGRRFQDPAVQDDIKHFPFKVICKD-GDKPAIEVKFKGETKVFAPEEISA 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L++++ + E+ L + ++N V+TVP F+ Q + A A+ GL+VLR++ EPTA A+
Sbjct: 126 MVLMKMKEIAESFLGKDVKNAVITVPAYFNDSQRQATKDAGAITGLNVLRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++T GS SE+ LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYG-----LDKKTAGSKSERNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 240
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
++ + + + + FK+ +S+ LR A + A LS+ + V++D L + +
Sbjct: 241 DSRLVNYFVSEFKRKFKKDVTTNARSLRRLRTACERAKRTLSSTTQTTVEIDSLVDGIDF 300
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRILL 383
+ RA+FEE+ ++F + + L D+K+ +I+DV+LVGG + IP+ ++L+
Sbjct: 301 YSSITRAKFEELCMDLFRGTLDPVEKVLRDSKIAKSEIDDVVLVGGSTRIPKVQQLLI 358
>UniRef100_Q9LU13 Hsp70 protein [Blastocystis hominis]
Length = 656
Score = 187 bits (474), Expect = 6e-46
Identities = 114/357 (31%), Positives = 199/357 (54%), Gaps = 15/357 (4%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
++GID+GT+ V VW +VE++ N + Q+ S+V F D G ++ ++ M
Sbjct: 9 SVGIDLGTTYSCVGVWQNDKVEIIANDQGQRTTPSYVAFTDTERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR DP V + K+ PF ++ G +P I ++ +PEE+ +
Sbjct: 67 TNTVFDAKRLIGRKFDDPSVQSDMKHWPFTLKAGPDG-KPLIEVEYKGEKKTFSPEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L +++ + E +L + ++N V+TVP F+ Q + A +AGL+V+R++ EPTA A+
Sbjct: 126 MVLSKMKQIAEAYLGKEVKNAVITVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG Q+ G EK LIF++G G DV++ + G+ ++KA AG T +GGED
Sbjct: 186 AYGLDQK---------GGEKNILIFDLGGGTFDVSLLSIEDGIFEVKATAGDTHLGGEDF 236
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
M+ + + + F++ +++ LR A + A LS+ + +++D L +
Sbjct: 237 DNRMVEYFCTEFQRKFRKDLTTSQRALRRLRTACERAKRTLSSSTQATIEIDSLFEGIDF 296
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N + F K + + + L DAK+ ++++V+LVGG S IP+ ++L
Sbjct: 297 NSQITRARFEELNMDYFRKTMAPVEKVLRDAKLSKSEVDEVVLVGGSSRIPKVQKLL 353
>UniRef100_P09189 Heat shock cognate 70 kDa protein [Petunia hybrida]
Length = 651
Score = 186 bits (473), Expect = 8e-46
Identities = 124/361 (34%), Positives = 198/361 (54%), Gaps = 20/361 (5%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +DP V + L PF V G +P I + EE+ +
Sbjct: 67 INTVFDAKRLIGRRFSDPSVQSDIKLWPFKVIP-GPGDKPMIVVTYKGEEKQFAAEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L +++ + E +L I+N V+TVP F+ Q + A +AGL+V+R++ EPTA A+
Sbjct: 126 MVLTKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG +KAS S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYG-LDKKAS-----SAGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 239
Query: 267 LQNMMRHLLPDSENIFKRHGVEEI----KSMALLRVATQDAIHQLSTQSSVQVDVD-LGN 321
M+ H + + FKR ++I +++ LR A + A LS+ + +++D L
Sbjct: 240 DNRMVNHFVQE----FKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLYE 295
Query: 322 SLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRI 381
+ + RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++
Sbjct: 296 GIDFYSTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSSVHDVVLVGGSTRIPKVQQL 355
Query: 382 L 382
L
Sbjct: 356 L 356
>UniRef100_Q9ZS55 Heat shock protein 70 [Arabidopsis thaliana]
Length = 650
Score = 186 bits (472), Expect = 1e-45
Identities = 118/352 (33%), Positives = 195/352 (54%), Gaps = 12/352 (3%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKN-LPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +DP V A K+ PF V + G +P I + + EE+ +
Sbjct: 67 TNTVFDAKRLIGRRYSDPSVQADKSHWPFKVVS-GPGEKPMIVVNHKGEEKQFSAEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
+ L+++R + E L P++N V+ VP F+ Q + A ++GL+V+R++ EPTA A+
Sbjct: 126 IVLIKMREIAEAFLGSPVKNAVVIVPAYFNDSQRQGTKDAGVISGLNVMRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG +KAS S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYG-LDKKAS-----SVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 239
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
M+ H + + + K+ +++ LR A + A LS+ + +++D L +
Sbjct: 240 DNRMVNHFVQEFKRKNKKDITGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGIDF 299
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQ 377
+ RA FEE+N ++F KC + +CL DAK++ ++DV++VGG + IP+
Sbjct: 300 YTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSSVHDVVVVGGSTRIPK 351
>UniRef100_O65719 Heat shock cognate 70 kDa protein 3 [Arabidopsis thaliana]
Length = 649
Score = 185 bits (470), Expect = 2e-45
Identities = 121/362 (33%), Positives = 200/362 (54%), Gaps = 20/362 (5%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR TD V + L PF +++ +P I + + EE+ +
Sbjct: 67 INTVFDAKRLIGRRFTDSSVQSDIKLWPFTLKS-GPAEKPMIVVNYKGEDKEFSAEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E +L I+N V+TVP F+ Q + A +AGL+V+R++ EPTA A+
Sbjct: 126 MILIKMREIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYGLDKKATSV------GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 239
Query: 267 LQNMMRHLLPDSENIFKRHGVEEI----KSMALLRVATQDAIHQLSTQSSVQVDVD-LGN 321
M+ H + + FKR ++I +++ LR A + A LS+ + +++D L +
Sbjct: 240 DNRMVNHFVQE----FKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFD 295
Query: 322 SLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRI 381
+ + RA FEE+N ++F KC + +CL DAK++ I+DV+LVGG + IP+ ++
Sbjct: 296 GIDFYAPITRARFEELNIDLFRKCMEPVEKCLRDAKMDKNSIDDVVLVGGSTRIPKVQQL 355
Query: 382 LL 383
L+
Sbjct: 356 LV 357
>UniRef100_Q6SL81 Heat shock protein 70 [Phytophthora nicotianae]
Length = 655
Score = 184 bits (468), Expect = 3e-45
Identities = 112/357 (31%), Positives = 197/357 (54%), Gaps = 15/357 (4%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
++GID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 SVGIDLGTTYSCVGVWQNDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNA 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +DP+V A K+ PF + T G +P I ++ PEE+ +
Sbjct: 67 ANTVFDAKRLIGRKFSDPIVQADIKHWPFKI-TSGPGDKPQITVQFKGESKTFQPEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E + + ++N V+TVP F+ Q + A A+AGL+VLR++ EPTA A+
Sbjct: 126 MVLIKMREVAEAFIGKEVKNAVITVPAYFNDSQRQATKDAGAIAGLNVLRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ G E+ LIF++G G DV++ + G+ ++KA AG T +GGED
Sbjct: 186 AYGLDKK---------GGERNVLIFDLGGGTFDVSLLSIEEGIFEVKATAGDTHLGGEDF 236
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
++ H + + ++ +++ LR A + A LS+ + +++D L + +
Sbjct: 237 DNRLVDHFTQEFKRKHRKDLTHNQRALRRLRTACERAKRTLSSSAQAYIEIDSLYDGVDF 296
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FE++ + F K + + L DAK+ G +++V+LVGG + IP+ ++L
Sbjct: 297 NSTITRARFEDLCADYFRKTMEPVEKVLRDAKLSKGQVHEVVLVGGSTRIPKVQQLL 353
>UniRef100_Q6SL80 Heat shock protein 70 [Phytophthora nicotianae]
Length = 655
Score = 184 bits (468), Expect = 3e-45
Identities = 112/357 (31%), Positives = 197/357 (54%), Gaps = 15/357 (4%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
++GID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 SVGIDLGTTYSCVGVWQNDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNA 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +DP+V A K+ PF + T G +P I ++ PEE+ +
Sbjct: 67 ANTVFDAKRLIGRKFSDPIVQADIKHWPFKI-TSGPGDKPQITVQFKGESKTFQPEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E + + ++N V+TVP F+ Q + A A+AGL+VLR++ EPTA A+
Sbjct: 126 MVLIKMREVAEAFIGKEVKNAVITVPAYFNDSQRQATKDAGAIAGLNVLRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ G E+ LIF++G G DV++ + G+ ++KA AG T +GGED
Sbjct: 186 AYGLDKK---------GGERNVLIFDLGGGTFDVSLLSIEEGIFEVKATAGDTHLGGEDF 236
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
++ H + + ++ +++ LR A + A LS+ + +++D L + +
Sbjct: 237 DNRLVDHFTQEFKRKHRKDLTHNQRALRRLRTACERAKRTLSSSAQAYIEIDSLYDGVDF 296
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FE++ + F K + + L DAK+ G +++V+LVGG + IP+ ++L
Sbjct: 297 NSTITRARFEDLCADYFRKTMEPVEKVLRDAKLSKGQVHEVVLVGGSTRIPKVQQLL 353
>UniRef100_Q40151 Hsc70 protein [Lycopersicon esculentum]
Length = 651
Score = 184 bits (468), Expect = 3e-45
Identities = 115/357 (32%), Positives = 194/357 (54%), Gaps = 12/357 (3%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +DP V + L PF V +P I + + EE+ +
Sbjct: 67 TNTVFDAKRLIGRRFSDPSVQSDMKLWPFKVIPGPAD-KPMIVVNYKGEEKQFSAEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L++++ + E L I+N V+TVP F+ Q + A ++GL+V+R++ EPTA A+
Sbjct: 126 MVLIKMKEIAEAFLGTTIKNAVVTVPAYFNDSQRQATKDAGTISGLNVMRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ + S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYGLDKK------ISSTGEKTVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 239
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
M+ H + + + K+ +++ LR A + A LS+ + +++D L +
Sbjct: 240 DNRMVNHFVQEFKRKHKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLYEGIDF 299
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + +CL DAK++ ++D++LVGG + IP+ ++L
Sbjct: 300 YTTITRARFEELNMDLFRKCMEPVEKCLRDAKIDKSGVHDIVLVGGSTRIPKVQQLL 356
>UniRef100_UPI0000438C23 UPI0000438C23 UniRef100 entry
Length = 361
Score = 184 bits (467), Expect = 4e-45
Identities = 121/364 (33%), Positives = 201/364 (54%), Gaps = 16/364 (4%)
Query: 22 SSPLPEIAIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFS 81
SSP EIAIGID+GT+ V V+ +VE++ N + + S+V F D G ++
Sbjct: 2 SSP-KEIAIGIDLGTTYSCVGVFQHGKVEIIANDQGNRTTPSYVAFTDTERLIGDAAK-- 58
Query: 82 HEHEMLFGDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRST 140
++ M +T+F+ KRLIGR DPVV + K+ F V + G +P +A ++
Sbjct: 59 NQVAMNPNNTVFDAKRLIGRRFDDPVVQSDMKHWSFKV--VSDGGKPKVAVEHKGENKTF 116
Query: 141 TPEEVLAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMP 200
PEE+ +M LV+++ + E +L + + N V+TVP F+ Q + A +AGL+VLR++
Sbjct: 117 NPEEISSMVLVKMKEIAEAYLGQKVTNAVITVPAYFNDSQRQATKDAGVIAGLNVLRIIN 176
Query: 201 EPTAVALLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST 260
EPTA A+ YG + G SE+ LIF++G G DV++ G+ ++KA AG T
Sbjct: 177 EPTAAAIAYGLDK--------GKSSERNVLIFDLGGGTFDVSILTIEDGIFEVKATAGDT 228
Query: 261 -IGGEDLLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD- 318
+GGED M+ H + + + K+ + +++ LR A + A LS+ S +++D
Sbjct: 229 HLGGEDFDNRMVNHFVEEFKRKHKKDISQNKRALRRLRTACERAKRTLSSSSQASIEIDS 288
Query: 319 LGNSLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQG 378
L + + RA FEE+ ++F + + L DAK++ I+D++LVGG + IP+
Sbjct: 289 LYEGIDFYTSITRARFEELCSDLFRGTLDPVEKALRDAKMDKAQIHDIVLVGGSTRIPKI 348
Query: 379 WRIL 382
++L
Sbjct: 349 QKLL 352
>UniRef100_Q7SZM8 Constitutive heat shock protein HSC70-1 [Cyprinus carpio]
Length = 642
Score = 184 bits (467), Expect = 4e-45
Identities = 120/358 (33%), Positives = 199/358 (55%), Gaps = 17/358 (4%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
A+GID+GT+ V V+ +VE++ N + + S+V F D G ++ ++ M
Sbjct: 1 AVGIDLGTTYSCVGVFQHGKVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 58
Query: 89 GDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR DPVV + K+ PF V ++ G +P + +S PEEV +
Sbjct: 59 NNTVFDAKRLIGRRFDDPVVQSDMKHWPFKV--INDGSKPKMEVEYKGEIKSFYPEEVSS 116
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L +++ ++E +L +P+ N V+TVP F+ Q + A +AGL VLR+ EPTA A+
Sbjct: 117 MVLTKMKEISEAYLGKPVSNAVITVPAYFNDSQRQATKDAGTIAGLDVLRITNEPTAAAI 176
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ G+E+ LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 177 AYGLDKK--------VGAERNVLIFDLGGGTFDVSILTIEDGIFEVKATAGDTHLGGEDF 228
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQL--STQSSVQVDVDLGNSLK 324
M+ H + + + FK+ +++ LR A + A L STQ+S+++D L
Sbjct: 229 DNRMVNHFIAEFKRKFKKDITGNKRAVRRLRTACERAKRTLSSSTQASIEID-SLYEGSD 287
Query: 325 ICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F + + L DAK++ I+D++LVGG + IP+ ++L
Sbjct: 288 FYTSITRARFEELNADLFRGTLEPVEKSLRDAKMDKAQIHDIVLVGGSTRIPKIQKLL 345
>UniRef100_P11143 Heat shock 70 kDa protein [Zea mays]
Length = 645
Score = 184 bits (467), Expect = 4e-45
Identities = 117/355 (32%), Positives = 194/355 (53%), Gaps = 19/355 (5%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V +W VE++ N + + S+V F D G ++ ++ M
Sbjct: 8 AIGIDLGTTYSCVGLWQHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAAK--NQVAMNP 65
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLAM 148
+T+F+ KRLIGR + P V +S L + + L +G +P I + EE+ +M
Sbjct: 66 TNTVFDAKRLIGRRFSSPAVQSSMKL-WPSRHLGLGDKPMIVFNYKGEEKQFAAEEISSM 124
Query: 149 FLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVALL 208
L++++ + E +L I+N V+TVP F+ Q + A +AGL+V+R++ EPTA A+
Sbjct: 125 VLIKMKEIAEAYLGSTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAIA 184
Query: 209 YGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDLL 267
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 185 YGLDKKAT------SSGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFD 238
Query: 268 QNMMRHLLPDSENIFKRHGVEEI----KSMALLRVATQDAIHQLSTQSSVQVDVD-LGNS 322
M+ H + + FKR ++I +++ LR A + A LS+ + +++D L
Sbjct: 239 NRMVNHFVQE----FKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEG 294
Query: 323 LKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQ 377
+ RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+
Sbjct: 295 IDFTPRSSRARFEELNMDLFRKCMEPVEKCLRDAKMDKSSVHDVVLVGGSTRIPK 349
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.134 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 615,365,855
Number of Sequences: 2790947
Number of extensions: 24493199
Number of successful extensions: 71773
Number of sequences better than 10.0: 2475
Number of HSP's better than 10.0 without gapping: 2157
Number of HSP's successfully gapped in prelim test: 319
Number of HSP's that attempted gapping in prelim test: 63017
Number of HSP's gapped (non-prelim): 2795
length of query: 394
length of database: 848,049,833
effective HSP length: 129
effective length of query: 265
effective length of database: 488,017,670
effective search space: 129324682550
effective search space used: 129324682550
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 76 (33.9 bits)
Medicago: description of AC145222.11


