

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145109.3 - phase: 0
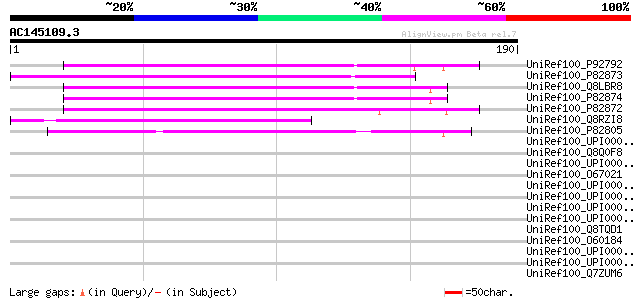
(190 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_P92792 Mitochondrial import receptor subunit TOM20 [So... 111 1e-23
UniRef100_P82873 Mitochondrial import receptor subunit TOM20-2 [... 107 1e-22
UniRef100_Q8LBR8 Putative TOM20 [Arabidopsis thaliana] 106 3e-22
UniRef100_P82874 Mitochondrial import receptor subunit TOM20-3 [... 106 3e-22
UniRef100_P82872 Mitochondrial import receptor subunit TOM20-1 [... 100 2e-20
UniRef100_Q8RZI8 Putative mitochondrial import receptor subunit ... 86 7e-16
UniRef100_P82805 Mitochondrial import receptor subunit TOM20-4 [... 80 2e-14
UniRef100_UPI00002F7C1F UPI00002F7C1F UniRef100 entry 44 0.003
UniRef100_Q8Q0F8 Conserved protein [Methanosarcina mazei] 40 0.032
UniRef100_UPI00001CEBE5 UPI00001CEBE5 UniRef100 entry 36 0.46
UniRef100_O67021 Hypothetical protein aq_854 [Aquifex aeolicus] 36 0.60
UniRef100_UPI000035F938 UPI000035F938 UniRef100 entry 35 0.78
UniRef100_UPI000035F937 UPI000035F937 UniRef100 entry 35 0.78
UniRef100_UPI000035F936 UPI000035F936 UniRef100 entry 35 0.78
UniRef100_UPI000035F935 UPI000035F935 UniRef100 entry 35 0.78
UniRef100_Q8TQD1 TPR-domain containing protein [Methanosarcina a... 35 0.78
UniRef100_O60184 Protein C23E6.09 in chromosome II [Schizosaccha... 35 0.78
UniRef100_UPI0000360266 UPI0000360266 UniRef100 entry 35 1.0
UniRef100_UPI00002DA2F7 UPI00002DA2F7 UniRef100 entry 35 1.0
UniRef100_Q7ZUM6 Small glutamine-rich tetratricopeptide repeat (... 35 1.3
>UniRef100_P92792 Mitochondrial import receptor subunit TOM20 [Solanum tuberosum]
Length = 204
Score = 111 bits (277), Expect = 1e-23
Identities = 69/165 (41%), Positives = 94/165 (56%), Gaps = 10/165 (6%)
Query: 21 AEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHW 80
AE Y +NP DA+NLTRWG ALL LSQ +S I + SKLEEA ++NP D W
Sbjct: 20 AETTYAQNPLDADNLTRWGGALLELSQFQPVAESKQMISDATSKLEEALTVNPEKHDALW 79
Query: 81 LLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLELPDTKDHEQHPK 140
LG A T LTPD +AK++F+ A F++AF DPS+ Y+ SLE+ K E H +
Sbjct: 80 CLGNAHTSHVFLTPDMDEAKVYFEKATQCFQQAFDADPSNDLYRKSLEV-TAKAPELHME 138
Query: 141 IVNHGLGQQS----KGSSSATKVIK-----HYKFLVFLFTLLVIG 176
I HG QQ+ +S++TK K K+ +F + +L +G
Sbjct: 139 IHRHGPMQQTMAAEPSTSTSTKSSKKTKSSDLKYDIFGWVILAVG 183
>UniRef100_P82873 Mitochondrial import receptor subunit TOM20-2 [Arabidopsis
thaliana]
Length = 210
Score = 107 bits (268), Expect = 1e-22
Identities = 58/152 (38%), Positives = 84/152 (55%), Gaps = 1/152 (0%)
Query: 1 MDYPLRKFKFHLPMKHECILAEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQV 60
M++ F+ + +H +E +Y +P D+ENL +WG ALL LSQ P++ +
Sbjct: 1 MEFSTADFERFIMFEHARKNSEAQYKNDPLDSENLLKWGGALLELSQFQPIPEAKLMLND 60
Query: 61 SISKLEEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSD 120
+ISKLEEA ++NP W + A T A D +AK HFD A YF+RA +DP +
Sbjct: 61 AISKLEEALTINPGKHQALWCIANAYTAHAFYVHDPEEAKEHFDKATEYFQRAENEDPGN 120
Query: 121 PTYQISLELPDTKDHEQHPKIVNHGLGQQSKG 152
TY+ SL+ K E H + +N G+GQQ G
Sbjct: 121 DTYRKSLD-SSLKAPELHMQFMNQGMGQQILG 151
>UniRef100_Q8LBR8 Putative TOM20 [Arabidopsis thaliana]
Length = 202
Score = 106 bits (265), Expect = 3e-22
Identities = 61/149 (40%), Positives = 82/149 (54%), Gaps = 6/149 (4%)
Query: 21 AEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHW 80
AE Y NP DA+NLTRWG LL LSQ HS D+ IQ +I+K EEA ++P + W
Sbjct: 20 AENTYKSNPLDADNLTRWGGVLLELSQFHSISDAKQMIQEAITKFEEALLIDPKKDEAVW 79
Query: 81 LLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLELPDTKDHEQHPK 140
+G A T A LTPD +AK +FD A +F++A + P + Y SLE+ K + H +
Sbjct: 80 CIGNAYTSFAFLTPDETEAKHNFDLATQFFQQAVDEQPDNTHYLKSLEM-TAKAPQLHAE 138
Query: 141 IVNHGLGQQSKGSSSA-----TKVIKHYK 164
GLG Q G A +K +K+ K
Sbjct: 139 AYKQGLGSQPMGRVEAPAPPSSKAVKNKK 167
>UniRef100_P82874 Mitochondrial import receptor subunit TOM20-3 [Arabidopsis
thaliana]
Length = 202
Score = 106 bits (265), Expect = 3e-22
Identities = 61/149 (40%), Positives = 82/149 (54%), Gaps = 6/149 (4%)
Query: 21 AEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHW 80
AE Y NP DA+NLTRWG LL LSQ HS D+ IQ +I+K EEA ++P + W
Sbjct: 20 AENTYKSNPLDADNLTRWGGVLLELSQFHSISDAKQMIQEAITKFEEALLIDPKKDEAVW 79
Query: 81 LLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLELPDTKDHEQHPK 140
+G A T A LTPD +AK +FD A +F++A + P + Y SLE+ K + H +
Sbjct: 80 CIGNAYTSFAFLTPDETEAKHNFDLATQFFQQAVDEQPDNTHYLKSLEM-TAKAPQLHAE 138
Query: 141 IVNHGLGQQSKGSSSA-----TKVIKHYK 164
GLG Q G A +K +K+ K
Sbjct: 139 AYKQGLGSQPMGRVEAPAPPSSKAVKNKK 167
>UniRef100_P82872 Mitochondrial import receptor subunit TOM20-1 [Arabidopsis
thaliana]
Length = 188
Score = 100 bits (250), Expect = 2e-20
Identities = 65/164 (39%), Positives = 87/164 (52%), Gaps = 8/164 (4%)
Query: 21 AEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHW 80
AE+ Y NP DA+NL RWG ALL LSQ + DSL IQ +ISKLE+A ++P D W
Sbjct: 14 AEETYKLNPEDADNLMRWGEALLELSQFQNVIDSLKMIQDAISKLEDAILIDPMKHDAVW 73
Query: 81 LLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLELPDTKDHEQ--- 137
LG A T A LTPD A+L+F A ++F A Q P + Y SLE+ D
Sbjct: 74 CLGNAYTSYARLTPDDTQARLNFGLAYLFFGIAVAQQPDNQVYHKSLEMADKAPQLHTGF 133
Query: 138 HPKIVNHGLGQQSKGSSSATKVIKH-----YKFLVFLFTLLVIG 176
H + LG + + KV+K+ K++V + +L IG
Sbjct: 134 HKNRLLSLLGGVETLAIPSPKVVKNKKSSDEKYIVMGWVILAIG 177
>UniRef100_Q8RZI8 Putative mitochondrial import receptor subunit TOM20 [Oryza sativa]
Length = 237
Score = 85.5 bits (210), Expect = 7e-16
Identities = 49/113 (43%), Positives = 63/113 (55%), Gaps = 4/113 (3%)
Query: 1 MDYPLRKFKFHLPMKHECILAEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQV 60
M P R F F L C A+ Y +NPHDA+NLTRWG ALL LSQ + P+SL ++
Sbjct: 52 MSDPERMFFFDLA----CQNAKVTYEQNPHDADNLTRWGGALLELSQMRNGPESLKCLED 107
Query: 61 SISKLEEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRA 113
+ SKLEEA ++P D W LG A T T D+ A F+ A F++A
Sbjct: 108 AESKLEEALKIDPMKADALWCLGNAQTSHGFFTSDTVKANEFFEKATQCFQKA 160
>UniRef100_P82805 Mitochondrial import receptor subunit TOM20-4 [Arabidopsis
thaliana]
Length = 187
Score = 80.5 bits (197), Expect = 2e-14
Identities = 56/161 (34%), Positives = 80/161 (48%), Gaps = 9/161 (5%)
Query: 15 KHECILAEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPN 74
+H +AE Y KNP DAENLTRW ALL LSQ + P + I +I KL EA ++P
Sbjct: 14 EHARKVAEATYVKNPLDAENLTRWAGALLELSQFQTEPKQM--ILEAILKLGEALVIDPK 71
Query: 75 NPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLELPDTKD 134
D WL+G A L+ D +A +F+ A +F+ A + P Y+ SL L
Sbjct: 72 KHDALWLIGNAHLSFGFLSSDQTEASDNFEKASQFFQLAVEEQPESELYRKSLTLA---- 127
Query: 135 HEQHPKIVNHGLGQQSKGSSSATKVIK--HYKFLVFLFTLL 173
+ P++ G S S+ K K +K+ VF + +L
Sbjct: 128 -SKAPELHTGGTAGPSSNSAKTMKQKKTSEFKYDVFGWVIL 167
>UniRef100_UPI00002F7C1F UPI00002F7C1F UniRef100 entry
Length = 313
Score = 43.5 bits (101), Expect = 0.003
Identities = 36/126 (28%), Positives = 60/126 (47%), Gaps = 18/126 (14%)
Query: 61 SISKLEEAFSLNPNNPDVHWLLGMALTMQALLTP--DSHDAKLHF--DSADVYFKRAFRQ 116
+I E++ +L+P+N D++ LG+A Q LT S++ + D+ADVY+
Sbjct: 176 AIQSYEKSIALDPDNADIYVNLGLAYKNQGNLTKAIQSYEKSIALDPDNADVYYNLGIVY 235
Query: 117 DPSDPT------YQISLEL-PDTKDHEQHPKIVNHGLGQQSKGSSSATKVIKHYKFLVFL 169
D T Y+ S+ L PD D V + LG K + TK I+ Y+ ++ L
Sbjct: 236 DNQGNTTKAIQSYEKSIALDPDNAD-------VYYNLGLAYKNQGNLTKAIQSYEKVIEL 288
Query: 170 FTLLVI 175
F ++
Sbjct: 289 FEKAIV 294
>UniRef100_Q8Q0F8 Conserved protein [Methanosarcina mazei]
Length = 1711
Score = 40.0 bits (92), Expect = 0.032
Identities = 24/75 (32%), Positives = 41/75 (54%), Gaps = 14/75 (18%)
Query: 17 ECILAEQRYNK----NPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLN 72
EC AE+ + + NP D ++L G++LL+L + + ++ LE+ SL
Sbjct: 453 ECEKAEEAFAEVLKINPEDIDSLYNRGISLLKLGRK----------ETALEYLEKVVSLR 502
Query: 73 PNNPDVHWLLGMALT 87
P+ PD+ + LG+ALT
Sbjct: 503 PDYPDLSYSLGVALT 517
>UniRef100_UPI00001CEBE5 UPI00001CEBE5 UniRef100 entry
Length = 505
Score = 36.2 bits (82), Expect = 0.46
Identities = 28/98 (28%), Positives = 44/98 (44%), Gaps = 19/98 (19%)
Query: 22 EQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHWL 81
E+ K+P + E+ + W +A RL + D I LE+A SL+P+N V L
Sbjct: 203 EKALEKDPKNPESTSGWAIANYRLDDWPASNDY-------IDSLEQAISLSPDNTYVKVL 255
Query: 82 LGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPS 119
L M L ++H + A + A ++DPS
Sbjct: 256 LAMKL------------EEVHENRAKELVEEALKKDPS 281
>UniRef100_O67021 Hypothetical protein aq_854 [Aquifex aeolicus]
Length = 545
Score = 35.8 bits (81), Expect = 0.60
Identities = 21/71 (29%), Positives = 34/71 (47%), Gaps = 8/71 (11%)
Query: 53 DSLHTIQVSISKLEEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKR 112
D+L I+ + L +A L+P NPD + LG +L + K + A+ K+
Sbjct: 400 DNLGDIKNAEKALRKAIELDPENPDYYNYLGYSLLLWY--------GKERVEEAEELIKK 451
Query: 113 AFRQDPSDPTY 123
A +DP +P Y
Sbjct: 452 ALEKDPENPAY 462
>UniRef100_UPI000035F938 UPI000035F938 UniRef100 entry
Length = 298
Score = 35.4 bits (80), Expect = 0.78
Identities = 30/113 (26%), Positives = 50/113 (43%), Gaps = 23/113 (20%)
Query: 28 NPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHWLLGMALT 87
NPH+A V + AHS L ++ E+A S++PN + +G+AL
Sbjct: 118 NPHNA-------VYFCNRAAAHS---KLGNYAGAVQDCEQAISIDPNYSKAYGRMGLAL- 166
Query: 88 MQALLTPDSHDAKLHFDSADV-YFKRAFRQDPSDPTYQISLELPDTKDHEQHP 139
A L+ S V Y+++A DP + TY+ +L++ + K P
Sbjct: 167 -----------ASLNKHSEAVGYYQKALELDPHNDTYKTNLKIAEEKMETSSP 208
>UniRef100_UPI000035F937 UPI000035F937 UniRef100 entry
Length = 308
Score = 35.4 bits (80), Expect = 0.78
Identities = 30/113 (26%), Positives = 50/113 (43%), Gaps = 23/113 (20%)
Query: 28 NPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHWLLGMALT 87
NPH+A V + AHS L ++ E+A S++PN + +G+AL
Sbjct: 116 NPHNA-------VYFCNRAAAHS---KLGNYAGAVQDCEQAISIDPNYSKAYGRMGLAL- 164
Query: 88 MQALLTPDSHDAKLHFDSADV-YFKRAFRQDPSDPTYQISLELPDTKDHEQHP 139
A L+ S V Y+++A DP + TY+ +L++ + K P
Sbjct: 165 -----------ASLNKHSEAVGYYQKALELDPHNDTYKTNLKIAEEKMETSSP 206
>UniRef100_UPI000035F936 UPI000035F936 UniRef100 entry
Length = 292
Score = 35.4 bits (80), Expect = 0.78
Identities = 30/113 (26%), Positives = 50/113 (43%), Gaps = 23/113 (20%)
Query: 28 NPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHWLLGMALT 87
NPH+A V + AHS L ++ E+A S++PN + +G+AL
Sbjct: 99 NPHNA-------VYFCNRAAAHS---KLGNYAGAVQDCEQAISIDPNYSKAYGRMGLAL- 147
Query: 88 MQALLTPDSHDAKLHFDSADV-YFKRAFRQDPSDPTYQISLELPDTKDHEQHP 139
A L+ S V Y+++A DP + TY+ +L++ + K P
Sbjct: 148 -----------ASLNKHSEAVGYYQKALELDPHNDTYKTNLKIAEEKMETSSP 189
>UniRef100_UPI000035F935 UPI000035F935 UniRef100 entry
Length = 341
Score = 35.4 bits (80), Expect = 0.78
Identities = 30/113 (26%), Positives = 50/113 (43%), Gaps = 23/113 (20%)
Query: 28 NPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHWLLGMALT 87
NPH+A V + AHS L ++ E+A S++PN + +G+AL
Sbjct: 122 NPHNA-------VYFCNRAAAHS---KLGNYAGAVQDCEQAISIDPNYSKAYGRMGLAL- 170
Query: 88 MQALLTPDSHDAKLHFDSADV-YFKRAFRQDPSDPTYQISLELPDTKDHEQHP 139
A L+ S V Y+++A DP + TY+ +L++ + K P
Sbjct: 171 -----------ASLNKHSEAVGYYQKALELDPHNDTYKTNLKIAEEKMETSSP 212
>UniRef100_Q8TQD1 TPR-domain containing protein [Methanosarcina acetivorans]
Length = 1885
Score = 35.4 bits (80), Expect = 0.78
Identities = 20/59 (33%), Positives = 33/59 (55%), Gaps = 10/59 (16%)
Query: 28 NPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHWLLGMAL 86
NP D + L G++LLRL + + ++ LE+ SL+P+ PD+ + LG+AL
Sbjct: 579 NPEDLDALYNRGISLLRLGRN----------ETALEYLEKVVSLSPDYPDLAYSLGVAL 627
>UniRef100_O60184 Protein C23E6.09 in chromosome II [Schizosaccharomyces pombe]
Length = 1102
Score = 35.4 bits (80), Expect = 0.78
Identities = 37/134 (27%), Positives = 55/134 (40%), Gaps = 26/134 (19%)
Query: 18 CILAEQRYNKNPH--------DAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAF 69
C +A+Q+YNK D N T W + Q + + D+L A
Sbjct: 593 CYVAQQKYNKAYEAYQQAVYRDGRNPTFWCSIGVLYYQINQYQDALDAYS-------RAI 645
Query: 70 SLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLEL 129
LNP +V + LG D+ DA ++RA DP++P + L+L
Sbjct: 646 RLNPYISEVWYDLGTLYESCHNQISDALDA----------YQRAAELDPTNPHIKARLQL 695
Query: 130 PDTKDHEQHPKIVN 143
++EQH KIVN
Sbjct: 696 LRGPNNEQH-KIVN 708
>UniRef100_UPI0000360266 UPI0000360266 UniRef100 entry
Length = 306
Score = 35.0 bits (79), Expect = 1.0
Identities = 23/82 (28%), Positives = 39/82 (47%), Gaps = 12/82 (14%)
Query: 66 EEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQI 125
E A ++P + +G+ALT A + A YFK+A DP + TY+
Sbjct: 145 ERAIGIDPTYSKAYGRMGLALT-----------AMSKYPEAISYFKKALVLDPENDTYKS 193
Query: 126 SLELPDTKDHEQHPKIVNHGLG 147
+L++ + K H++ + GLG
Sbjct: 194 NLKIAEQK-HKEATSPIAAGLG 214
>UniRef100_UPI00002DA2F7 UPI00002DA2F7 UniRef100 entry
Length = 337
Score = 35.0 bits (79), Expect = 1.0
Identities = 23/82 (28%), Positives = 39/82 (47%), Gaps = 12/82 (14%)
Query: 66 EEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQI 125
E A ++P + +G+ALT A + A YFK+A DP + TY+
Sbjct: 138 ERAIGIDPTYSKAYGRMGLALT-----------AMSKYPEAISYFKKALVLDPENDTYKS 186
Query: 126 SLELPDTKDHEQHPKIVNHGLG 147
+L++ + K H++ + GLG
Sbjct: 187 NLKIAEQK-HKEATSPIAAGLG 207
>UniRef100_Q7ZUM6 Small glutamine-rich tetratricopeptide repeat (TPR)-containing
[Brachydanio rerio]
Length = 320
Score = 34.7 bits (78), Expect = 1.3
Identities = 23/88 (26%), Positives = 41/88 (46%), Gaps = 14/88 (15%)
Query: 61 SISKLEEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLH-FDSADVYFKRAFRQDPS 119
++ E A ++ N + +G+AL A L+ + A Y+K+A DP
Sbjct: 144 AVQDCERAIGIDANYSKAYGRMGLAL------------ASLNKYSEAVSYYKKALELDPD 191
Query: 120 DPTYQISLELPDTKDHEQHPKIVNHGLG 147
+ TY+++L++ + K E P GLG
Sbjct: 192 NDTYKVNLQVAEQKVKETQPSTAG-GLG 218
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.136 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 325,699,416
Number of Sequences: 2790947
Number of extensions: 12240841
Number of successful extensions: 27935
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 36
Number of HSP's that attempted gapping in prelim test: 27902
Number of HSP's gapped (non-prelim): 59
length of query: 190
length of database: 848,049,833
effective HSP length: 120
effective length of query: 70
effective length of database: 513,136,193
effective search space: 35919533510
effective search space used: 35919533510
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 71 (32.0 bits)
Medicago: description of AC145109.3


