

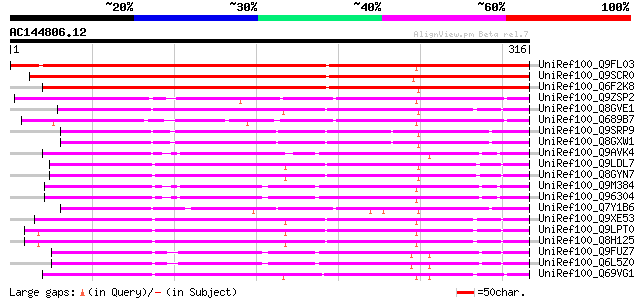
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144806.12 - phase: 1 /pseudo
(316 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FL03 SCARECROW gene regulator [Arabidopsis thaliana] 431 e-119
UniRef100_Q9SCR0 Scarecrow-like 7 [Arabidopsis thaliana] 397 e-109
UniRef100_Q6F2K8 Putative GRAS family transcription factor [Oryz... 359 5e-98
UniRef100_Q9ZSP2 Lateral suppressor protein [Lycopersicon escule... 190 4e-47
UniRef100_Q8GVE1 Chitin-inducible gibberellin-responsive protein... 181 3e-44
UniRef100_Q689B7 Lateral suppressor-like protein [Daucus carota] 180 4e-44
UniRef100_Q9SRP9 RGA1-like protein [Arabidopsis thaliana] 179 1e-43
UniRef100_Q8GXW1 Putative RGA1 [Arabidopsis thaliana] 179 1e-43
UniRef100_Q9AVK4 SCARECROW [Pisum sativum] 177 3e-43
UniRef100_Q9LDL7 SCARECROW gene regulator-like [Arabidopsis thal... 175 2e-42
UniRef100_Q8GYN7 Putative SCARECROW gene regulator [Arabidopsis ... 175 2e-42
UniRef100_Q9M384 SCARECROW1 [Arabidopsis thaliana] 174 2e-42
UniRef100_Q96304 SCARECROW [Arabidopsis thaliana] 173 6e-42
UniRef100_Q7Y1B6 GAI-like protein [Lycopersicon esculentum] 171 2e-41
UniRef100_Q9XE53 Scarecrow-like 5 [Arabidopsis thaliana] 170 4e-41
UniRef100_Q9LPT0 Putative transcription factor [Arabidopsis thal... 169 7e-41
UniRef100_Q8H125 Putative scarecrow protein [Arabidopsis thaliana] 169 7e-41
UniRef100_Q9FUZ7 SCARECROW [Zea mays] 169 9e-41
UniRef100_Q6L5Z0 SCARECROW [Oryza sativa] 169 9e-41
UniRef100_Q69VG1 Putative chitin-inducible gibberellin-responsiv... 169 1e-40
>UniRef100_Q9FL03 SCARECROW gene regulator [Arabidopsis thaliana]
Length = 584
Score = 431 bits (1107), Expect = e-119
Identities = 222/322 (68%), Positives = 259/322 (79%), Gaps = 9/322 (2%)
Query: 1 TNKITAQSSIASSNSSSTTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHI 60
+N+++ S SS+SSST E+L LSYK LNDACPYSKFAHLTANQAILEATE SN IHI
Sbjct: 265 SNRLSPNSPATSSSSSST--EDLILSYKTLNDACPYSKFAHLTANQAILEATEKSNKIHI 322
Query: 61 VDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKL 120
VDFGIVQGIQW ALLQA ATR+SGKP +R+SGIPA +LG SP S+ ATGNRL +FAK+
Sbjct: 323 VDFGIVQGIQWPALLQALATRTSGKPTQIRVSGIPAPSLGESPEPSLIATGNRLRDFAKV 382
Query: 121 LGLNFEFTPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLN 180
L LNF+F PILTPI LL+ SSF + PDE LAVNFMLQLY LLDE V+ ALRLAKSLN
Sbjct: 383 LDLNFDFIPILTPIHLLNGSSFRVDPDEVLAVNFMLQLYKLLDETPTIVDTALRLAKSLN 442
Query: 181 PKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRI 240
P++VTLGEYE SL RVGF R + A +++A FESLEPN+ DS ER +VE L GRRI
Sbjct: 443 PRVVTLGEYEVSL-NRVGFANRVKNALQFYSAVFESLEPNLGRDSEERVRVERELFGRRI 501
Query: 241 DGVIGV------RERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLV 294
G+IG RERME+KEQW+VLMEN GFESV LS+YA+SQAKILLWNY+YS+LYS+V
Sbjct: 502 SGLIGPEKTGIHRERMEEKEQWRVLMENAGFESVKLSNYAVSQAKILLWNYNYSNLYSIV 561
Query: 295 ESQPAFLSLAWKDVPLLTVSSW 316
ES+P F+SLAW D+PLLT+SSW
Sbjct: 562 ESKPGFISLAWNDLPLLTLSSW 583
>UniRef100_Q9SCR0 Scarecrow-like 7 [Arabidopsis thaliana]
Length = 542
Score = 397 bits (1021), Expect = e-109
Identities = 207/314 (65%), Positives = 245/314 (77%), Gaps = 11/314 (3%)
Query: 13 SNSSSTTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWA 72
S+SSS++ E+ LSYK LNDACPYSKFAHLTANQAILEAT SNNIHIVDFGI QGIQW+
Sbjct: 229 SSSSSSSLEDFILSYKTLNDACPYSKFAHLTANQAILEATNQSNNIHIVDFGIFQGIQWS 288
Query: 73 ALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILT 132
ALLQA ATRSSGKP +RISGIPA +LG SP S+ ATGNRL +FA +L LNFEF P+LT
Sbjct: 289 ALLQALATRSSGKPTRIRISGIPAPSLGDSPGPSLIATGNRLRDFAAILDLNFEFYPVLT 348
Query: 133 PIELLDESSFCIQPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEAS 192
PI+LL+ SSF + PDE L VNFML+LY LLDE +V ALRLA+SLNP+IVTLGEYE S
Sbjct: 349 PIQLLNGSSFRVDPDEVLVVNFMLELYKLLDETATTVGTALRLARSLNPRIVTLGEYEVS 408
Query: 193 LTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVI-------- 244
L RV F R + + +++A FESLEPN+ DS ER +VE +L GRRI ++
Sbjct: 409 L-NRVEFANRVKNSLRFYSAVFESLEPNLDRDSKERLRVERVLFGRRIMDLVRSDDDNNK 467
Query: 245 -GVR-ERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLS 302
G R ME+KEQW+VLME GFE V S+YA+SQAK+LLWNY+YS+LYSLVES+P F+S
Sbjct: 468 PGTRFGLMEEKEQWRVLMEKAGFEPVKPSNYAVSQAKLLLWNYNYSTLYSLVESEPGFIS 527
Query: 303 LAWKDVPLLTVSSW 316
LAW +VPLLTVSSW
Sbjct: 528 LAWNNVPLLTVSSW 541
>UniRef100_Q6F2K8 Putative GRAS family transcription factor [Oryza sativa]
Length = 578
Score = 359 bits (922), Expect = 5e-98
Identities = 183/302 (60%), Positives = 222/302 (72%), Gaps = 7/302 (2%)
Query: 21 EELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFAT 80
+ELTL YK LNDACPYSKFAHLTANQAILEAT + IHIVDFGIVQGIQWAALLQA AT
Sbjct: 277 DELTLCYKTLNDACPYSKFAHLTANQAILEATGAATKIHIVDFGIVQGIQWAALLQALAT 336
Query: 81 RSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDES 140
R GKP +RI+G+P+ LG P +S++AT RL +FAKLLG++FEF P+L P+ L++S
Sbjct: 337 RPEGKPTRIRITGVPSPLLGPQPAASLAATNTRLRDFAKLLGVDFEFVPLLRPVHELNKS 396
Query: 141 SFCIQPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFV 200
F ++PDEA+AVNFMLQLY+LL ++ V + LRLAKSL+P +VTLGEYE SL R GFV
Sbjct: 397 DFLVEPDEAVAVNFMLQLYHLLGDSDELVRRVLRLAKSLSPAVVTLGEYEVSL-NRAGFV 455
Query: 201 ERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGVR------ERMEDKE 254
+RF A +Y+ + FESL+ M DSPER +VE + G RI +G ERM
Sbjct: 456 DRFANALSYYRSLFESLDVAMTRDSPERVRVERWMFGERIQRAVGPEEGADRTERMAGSS 515
Query: 255 QWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVS 314
+W+ LME CGFE V LS+YA SQA +LLWNY YSLVE PAFLSLAW+ PLLTVS
Sbjct: 516 EWQTLMEWCGFEPVPLSNYARSQADLLLWNYDSKYKYSLVELPPAFLSLAWEKRPLLTVS 575
Query: 315 SW 316
+W
Sbjct: 576 AW 577
>UniRef100_Q9ZSP2 Lateral suppressor protein [Lycopersicon esculentum]
Length = 428
Score = 190 bits (483), Expect = 4e-47
Identities = 123/326 (37%), Positives = 184/326 (55%), Gaps = 24/326 (7%)
Query: 4 ITAQSSIASSNSSSTTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNN-IHIVD 62
+T + + +SSS++ + SY +LN P+ +F LTANQAILEA G++ IHIVD
Sbjct: 113 MTPVETTPTDSSSSSSLALIQSSYLSLNQVTPFIRFTQLTANQAILEAINGNHQAIHIVD 172
Query: 63 FGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLG 122
F I G+QW L+QA A R ++RI+G + + ++ TG+RL++FA LG
Sbjct: 173 FDINHGVQWPPLMQALADRYPAP--TLRITGTG------NDLDTLRRTGDRLAKFAHSLG 224
Query: 123 LNFEFTPILTPIELLDE-------SSFCIQPDEALAVNFMLQLYNLLDENTNSVEKALRL 175
L F+F P+ D SS + PDE LA+N + L+ LL + + L
Sbjct: 225 LRFQFHPLYIANNNHDHDEDPSIISSIVLLPDETLAINCVFYLHRLLKDR-EKLRIFLHR 283
Query: 176 AKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLL 235
KS+NPKIVT+ E EA+ + F++RF A +Y+ A F+SLE + S ER VE +
Sbjct: 284 VKSMNPKIVTIAEKEANHNHPL-FLQRFIEALDYYTAVFDSLEATLPPGSRERMTVEQVW 342
Query: 236 LGRRIDGVIGV-----RERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSL 290
GR I ++ + +ER E W+V++ +CGF +V LS +A+SQAK+LL + S
Sbjct: 343 FGREIVDIVAMEGDKRKERHERFRSWEVMLRSCGFSNVALSPFALSQAKLLLRLHYPSEG 402
Query: 291 YSLVESQPAFLSLAWKDVPLLTVSSW 316
Y L S +F L W++ PL ++SSW
Sbjct: 403 YQLGVSSNSFF-LGWQNQPLFSISSW 427
>UniRef100_Q8GVE1 Chitin-inducible gibberellin-responsive protein [Oryza sativa]
Length = 544
Score = 181 bits (458), Expect = 3e-44
Identities = 114/295 (38%), Positives = 159/295 (53%), Gaps = 13/295 (4%)
Query: 30 LNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSV 89
L +ACPY KF +++AN AI EA +G + IHI+DF I QG QW +LLQA A R G P +V
Sbjct: 254 LYEACPYFKFGYMSANGAIAEAVKGEDRIHIIDFHISQGAQWISLLQALAARPGGPP-TV 312
Query: 90 RISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDESSFCIQPDEA 149
RI+GI + + G RLS A L + FEF P+ ++ + + P EA
Sbjct: 313 RITGIDDSVSAYARGGGLELVGRRLSHIASLCKVPFEFHPLAISGSKVEAAHLGVIPGEA 372
Query: 150 LAVNFMLQLYNLLDEN---TNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETA 206
LAVNF L+L+++ DE+ N ++ LR+ KSL+PK++TL E E++ T F +RF
Sbjct: 373 LAVNFTLELHHIPDESVSTANHRDRLLRMVKSLSPKVLTLVEMESN-TNTAPFPQRFAET 431
Query: 207 FNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLME 261
+Y+ A FES++ + D ER +E L R I +I ER E +WK +
Sbjct: 432 LDYYTAIFESIDLTLPRDDRERINMEQHCLAREIVNLIACEGEERAERYEPFGKWKARLT 491
Query: 262 NCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSW 316
GF LS + + LL SYS Y L E A L L WK PL+ S+W
Sbjct: 492 MAGFRPSPLSSLVNATIRTLL--QSYSDNYKLAERDGA-LYLGWKSRPLVVSSAW 543
>UniRef100_Q689B7 Lateral suppressor-like protein [Daucus carota]
Length = 431
Score = 180 bits (457), Expect = 4e-44
Identities = 118/326 (36%), Positives = 184/326 (56%), Gaps = 32/326 (9%)
Query: 8 SSIASSNSSSTTWEELTL---SYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFG 64
SS S + S +++ ++ +Y +LN P+ +F HLTANQAILE+ EG + IHI+DF
Sbjct: 120 SSAGSGSGSQQVFDDESVVQSAYLSLNQITPFIRFTHLTANQAILESVEGHHAIHILDFN 179
Query: 65 IVQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLN 124
I+ G+QW L+QA A + P +RI+G ++ + TG+RL++FA LGL
Sbjct: 180 IMHGVQWPPLMQAMAEKF--PPPMLRITGT------GDNLTILRRTGDRLAKFAHTLGLR 231
Query: 125 FEFTPILTPIELLDESSFC---------IQPDEALAVNFMLQLYNLLDENTNSVEKALRL 175
F+F P+L + +ESS +QPD+ LAVN +L L+ L + + L
Sbjct: 232 FQFHPVL--LLENEESSITSFFASFAAYLQPDQTLAVNCVLYLHRL---SLERLSLCLHQ 286
Query: 176 AKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLL 235
K+LNP+++TL E EA+ + F++RF A +++ A F+SLE + +S +R +VE +
Sbjct: 287 IKALNPRVLTLSEREANHNLPI-FLQRFVEALDHYTALFDSLEATLPPNSRQRIEVEQIW 345
Query: 236 LGRRIDGVIGV-----RERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSL 290
GR I +I RER E W++++ GF ++ LS +A+SQAK+LL Y S
Sbjct: 346 FGREIADIIASEGETRRERHERFRAWELMLRGSGFHNLALSPFALSQAKLLLRLYYPSEG 405
Query: 291 YSLVESQPAFLSLAWKDVPLLTVSSW 316
Y L +F W++ L +VSSW
Sbjct: 406 YKLHILNDSFF-WGWQNQHLFSVSSW 430
>UniRef100_Q9SRP9 RGA1-like protein [Arabidopsis thaliana]
Length = 547
Score = 179 bits (453), Expect = 1e-43
Identities = 110/292 (37%), Positives = 168/292 (56%), Gaps = 14/292 (4%)
Query: 32 DACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRI 91
++CPY KFAH TANQAILEA + +H++D G+ QG+QW AL+QA A R G P S R+
Sbjct: 260 ESCPYLKFAHFTANQAILEAVTTARRVHVIDLGLNQGMQWPALMQALALRPGGPP-SFRL 318
Query: 92 SGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILT-PIELLDESSFCIQPD-EA 149
+GI T S+ G +L++FA+ +G+ FEF + + L+ F +P+ E
Sbjct: 319 TGIGPPQ--TENSDSLQQLGWKLAQFAQNMGVEFEFKGLAAESLSDLEPEMFETRPESET 376
Query: 150 LAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNY 209
L VN + +L+ LL + S+EK L K++ P IVT+ E EA+ + F++RF A +Y
Sbjct: 377 LVVNSVFELHRLL-ARSGSIEKLLNTVKAIKPSIVTVVEQEAN-HNGIVFLDRFNEALHY 434
Query: 210 FAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLMENCG 264
+++ F+SLE + +L S +R E + LGR+I V+ ER E QW++ M++ G
Sbjct: 435 YSSLFDSLEDSYSLPSQDRVMSE-VYLGRQILNVVAAEGSDRVERHETAAQWRIRMKSAG 493
Query: 265 FESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSW 316
F+ + L A QA +LL Y+ Y VE L + W+ PL+T S+W
Sbjct: 494 FDPIHLGSSAFKQASMLLSLYATGDGYR-VEENDGCLMIGWQTRPLITTSAW 544
>UniRef100_Q8GXW1 Putative RGA1 [Arabidopsis thaliana]
Length = 547
Score = 179 bits (453), Expect = 1e-43
Identities = 110/292 (37%), Positives = 168/292 (56%), Gaps = 14/292 (4%)
Query: 32 DACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRI 91
++CPY KFAH TANQAILEA + +H++D G+ QG+QW AL+QA A R G P S R+
Sbjct: 260 ESCPYLKFAHFTANQAILEAVTTARRVHVIDLGLNQGMQWPALMQALALRPGGPP-SFRL 318
Query: 92 SGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILT-PIELLDESSFCIQPD-EA 149
+GI T S+ G +L++FA+ +G+ FEF + + L+ F +P+ E
Sbjct: 319 TGIGPPQ--TENSDSLQQLGWKLAQFAQNMGVEFEFKGLAAESLSDLEPEMFETRPESET 376
Query: 150 LAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNY 209
L VN + +L+ LL + S+EK L K++ P IVT+ E EA+ + F++RF A +Y
Sbjct: 377 LVVNSVFELHRLL-ARSGSIEKLLNTVKAIKPSIVTVVEQEAN-HNGIVFLDRFNEALHY 434
Query: 210 FAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLMENCG 264
+++ F+SLE + +L S +R E + LGR+I V+ ER E QW++ M++ G
Sbjct: 435 YSSLFDSLEDSYSLPSQDRVMSE-VYLGRQILNVVAAEGSDRVERHETAAQWRIRMKSAG 493
Query: 265 FESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSW 316
F+ + L A QA +LL Y+ Y VE L + W+ PL+T S+W
Sbjct: 494 FDPIHLGSSAFKQASMLLSLYATGDGYR-VEENDGCLMIGWQTRPLITTSAW 544
>UniRef100_Q9AVK4 SCARECROW [Pisum sativum]
Length = 819
Score = 177 bits (449), Expect = 3e-43
Identities = 114/301 (37%), Positives = 167/301 (54%), Gaps = 20/301 (6%)
Query: 21 EELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFAT 80
+++ +++ N P+ KF+H TANQAI EA E +HI+D I+QG+QW L A+
Sbjct: 520 QKVASAFQVFNGISPFVKFSHFTANQAIQEAFEREERVHIIDLDIMQGLQWPGLFHILAS 579
Query: 81 RSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDES 140
R G P VR++G LGTS + ++ ATG RLS+FA LGL FEF P+ + +D
Sbjct: 580 RPGGPP-YVRLTG-----LGTS-METLEATGKRLSDFANKLGLPFEFFPVAEKVGNIDVE 632
Query: 141 SFCIQPDEALAVNFML-QLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGF 199
+ EA+AV+++ LY++ +TN+ L L + L PK+VT+ E L+ F
Sbjct: 633 KLNVSKSEAVAVHWLQHSLYDVTGSDTNT----LWLLQRLAPKVVTV--VEQDLSNAGSF 686
Query: 200 VERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGVRERMEDKE----Q 255
+ RF A +Y++A F+SL + +S ER VE LL R I V+ V E
Sbjct: 687 LGRFVEAIHYYSALFDSLGSSYGEESEERHVVEQQLLSREIRNVLAVGGPSRSGEIKFHN 746
Query: 256 WKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSS 315
W+ ++ CGF V L+ A +QA +LL + S Y+LVE L L WKD+ LLT S+
Sbjct: 747 WREKLQQCGFRGVSLAGNAATQASLLLGMFP-SEGYTLVEDN-GILKLGWKDLCLLTASA 804
Query: 316 W 316
W
Sbjct: 805 W 805
>UniRef100_Q9LDL7 SCARECROW gene regulator-like [Arabidopsis thaliana]
Length = 490
Score = 175 bits (443), Expect = 2e-42
Identities = 114/301 (37%), Positives = 157/301 (51%), Gaps = 14/301 (4%)
Query: 25 LSYK-ALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSS 83
LSY L + CPY KF +++AN AI EA + N +HI+DF I QG QW L+QAFA R
Sbjct: 194 LSYMHILYEVCPYFKFGYMSANGAIAEAMKEENRVHIIDFQIGQGSQWVTLIQAFAARPG 253
Query: 84 GKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDESSFC 143
G P +RI+GI M + +S GNRL++ AK + FEF + + + +
Sbjct: 254 GPPR-IRITGIDDMTSAYARGGGLSIVGNRLAKLAKQFNVPFEFNSVSVSVSEVKPKNLG 312
Query: 144 IQPDEALAVNFMLQLYNLLDENT---NSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFV 200
++P EALAVNF L+++ DE+ N ++ LR+ KSL+PK+VTL E E++ T F
Sbjct: 313 VRPGEALAVNFAFVLHHMPDESVSTENHRDRLLRMVKSLSPKVVTLVEQESN-TNTAAFF 371
Query: 201 ERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQ 255
RF NY+AA FES++ + D +R VE L R + +I ER E +
Sbjct: 372 PRFMETMNYYAAMFESIDVTLPRDHKQRINVEQHCLARDVVNIIACEGADRVERHELLGK 431
Query: 256 WKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSS 315
W+ GF LS S K LL N YS Y L E A L L W L+ +
Sbjct: 432 WRSRFGMAGFTPYPLSPLVNSTIKSLLRN--YSDKYRLEERDGA-LYLGWMHRDLVASCA 488
Query: 316 W 316
W
Sbjct: 489 W 489
>UniRef100_Q8GYN7 Putative SCARECROW gene regulator [Arabidopsis thaliana]
Length = 411
Score = 175 bits (443), Expect = 2e-42
Identities = 114/301 (37%), Positives = 157/301 (51%), Gaps = 14/301 (4%)
Query: 25 LSYK-ALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSS 83
LSY L + CPY KF +++AN AI EA + N +HI+DF I QG QW L+QAFA R
Sbjct: 115 LSYMHILYEVCPYFKFGYMSANGAIAEAMKEENRVHIIDFQIGQGSQWVTLIQAFAARPG 174
Query: 84 GKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDESSFC 143
G P +RI+GI M + +S GNRL++ AK + FEF + + + +
Sbjct: 175 GPPR-IRITGIDDMTSAYARGGGLSIVGNRLAKLAKQFNVPFEFNSVSVSVSEVKPKNLG 233
Query: 144 IQPDEALAVNFMLQLYNLLDENT---NSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFV 200
++P EALAVNF L+++ DE+ N ++ LR+ KSL+PK+VTL E E++ T F
Sbjct: 234 VRPGEALAVNFAFVLHHMPDESVSTENHRDRLLRMVKSLSPKVVTLVEQESN-TNTAAFF 292
Query: 201 ERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQ 255
RF NY+AA FES++ + D +R VE L R + +I ER E +
Sbjct: 293 PRFMETMNYYAAMFESIDVTLPRDHKQRINVEQHCLARDVVNIIACEGADRVERHELLGK 352
Query: 256 WKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSS 315
W+ GF LS S K LL N YS Y L E A L L W L+ +
Sbjct: 353 WRSRFGMAGFTPYPLSPLVNSTIKSLLRN--YSDKYRLEERDGA-LYLGWMHRDLVASCA 409
Query: 316 W 316
W
Sbjct: 410 W 410
>UniRef100_Q9M384 SCARECROW1 [Arabidopsis thaliana]
Length = 653
Score = 174 bits (442), Expect = 2e-42
Identities = 114/299 (38%), Positives = 168/299 (56%), Gaps = 18/299 (6%)
Query: 22 ELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATR 81
++ +++ N P KF+H TANQAI EA E +++HI+D I+QG+QW L A+R
Sbjct: 365 KMVSAFQVFNGISPLVKFSHFTANQAIQEAFEKEDSVHIIDLDIMQGLQWPGLFHILASR 424
Query: 82 SSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDESS 141
G P+ VR++G LGTS + ++ ATG RLS+FA LGL FEF P+ + LD
Sbjct: 425 PGGPPH-VRLTG-----LGTS-MEALQATGKRLSDFADKLGLPFEFCPLAEKVGNLDTER 477
Query: 142 FCIQPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVE 201
++ EA+AV++ L + L + T S L L + L PK+VT+ E L+ F+
Sbjct: 478 LNVRKREAVAVHW---LQHSLYDVTGSDAHTLWLLQRLAPKVVTV--VEQDLSHAGSFLG 532
Query: 202 RFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGV----RERMEDKEQWK 257
RF A +Y++A F+SL + +S ER VE LL + I V+ V R E W+
Sbjct: 533 RFVEAIHYYSALFDSLGASYGEESEERHVVEQQLLSKEIRNVLAVGGPSRSGEVKFESWR 592
Query: 258 VLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSW 316
M+ CGF+ + L+ A +QA +LL + S Y+LV+ L L WKD+ LLT S+W
Sbjct: 593 EKMQQCGFKGISLAGNAATQATLLLGMFP-SDGYTLVDDN-GTLKLGWKDLSLLTASAW 649
>UniRef100_Q96304 SCARECROW [Arabidopsis thaliana]
Length = 653
Score = 173 bits (438), Expect = 6e-42
Identities = 113/299 (37%), Positives = 167/299 (55%), Gaps = 18/299 (6%)
Query: 22 ELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATR 81
++ +++ N P KF+H TANQAI EA E +++HI+D I+QG+QW L A+R
Sbjct: 365 KMVSAFQVFNGISPLVKFSHFTANQAIQEAFEKEDSVHIIDLDIMQGLQWPGLFHILASR 424
Query: 82 SSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDESS 141
G P+ VR++G LGTS + ++ ATG RLS+F LGL FEF P+ + LD
Sbjct: 425 PGGPPH-VRLTG-----LGTS-MEALQATGKRLSDFTDKLGLPFEFCPLAEKVGNLDTER 477
Query: 142 FCIQPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVE 201
++ EA+AV++ L + L + T S L L + L PK+VT+ E L+ F+
Sbjct: 478 LNVRKREAVAVHW---LQHSLYDVTGSDAHTLWLLQRLAPKVVTV--VEQDLSHAGSFLG 532
Query: 202 RFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGV----RERMEDKEQWK 257
RF A +Y++A F+SL + +S ER VE LL + I V+ V R E W+
Sbjct: 533 RFVEAIHYYSALFDSLGASYGEESEERHVVEQQLLSKEIRNVLAVGGPSRSGEVKFESWR 592
Query: 258 VLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSW 316
M+ CGF+ + L+ A +QA +LL + S Y+LV+ L L WKD+ LLT S+W
Sbjct: 593 EKMQQCGFKGISLAGNAATQATLLLGMFP-SDGYTLVDDN-GTLKLGWKDLSLLTASAW 649
>UniRef100_Q7Y1B6 GAI-like protein [Lycopersicon esculentum]
Length = 588
Score = 171 bits (434), Expect = 2e-41
Identities = 114/307 (37%), Positives = 164/307 (53%), Gaps = 29/307 (9%)
Query: 32 DACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRI 91
+ CPY KFAH TANQAILEA G N +H++DF + QG+QW AL+QA A R G P + R+
Sbjct: 274 ETCPYLKFAHFTANQAILEAFTGCNKVHVIDFSLKQGMQWPALMQALALRPGGPP-AFRL 332
Query: 92 SGI-PAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTP-ILTPIELLDESSFCIQPD-- 147
+GI P T + + G +L++ A+ +G+ FEF + + LD + I+P
Sbjct: 333 TGIGPPQPDNTDALQQV---GWKLAQLAETIGVEFEFRGFVANSLADLDATILDIRPSET 389
Query: 148 EALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAF 207
EA+A+N + +L+ LL ++EK L K +NPKIVTL E EA+ V F++RF A
Sbjct: 390 EAVAINSVFELHRLL-SRPGAIEKVLNSIKQINPKIVTLVEQEANHNAGV-FIDRFNEAL 447
Query: 208 NYFAAFFESLE---------PNMALDSP----ERFQVESLLLGRRIDGVIGVR-----ER 249
+Y++ F+SLE P L P + + + LGR+I V+ ER
Sbjct: 448 HYYSTMFDSLESSGSSSSASPTGILPQPPVNNQDLVMSEVYLGRQICNVVACEGSDRVER 507
Query: 250 MEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVP 309
E QW+V M + GF+ V L A QA +LL ++ Y VE L L W P
Sbjct: 508 HETLNQWRVRMNSSGFDPVHLGSNAFKQASMLLALFAGGDGYR-VEENDGCLMLGWHTRP 566
Query: 310 LLTVSSW 316
L+ S+W
Sbjct: 567 LIATSAW 573
>UniRef100_Q9XE53 Scarecrow-like 5 [Arabidopsis thaliana]
Length = 306
Score = 170 bits (431), Expect = 4e-41
Identities = 114/309 (36%), Positives = 161/309 (51%), Gaps = 13/309 (4%)
Query: 16 SSTTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALL 75
+S T EL L +ACPY KF + +AN AI EA + + +HI+DF I QG QW +L+
Sbjct: 2 TSPTGPELLTYMHILYEACPYFKFGYESANGAIAEAVKNESFVHIIDFQISQGGQWVSLI 61
Query: 76 QAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIE 135
+A R G PN VRI+GI + + G RL + A++ G+ FEF
Sbjct: 62 RALGARPGGPPN-VRITGIDDPRSSFARQGGLELVGQRLGKLAEMCGVPFEFHGAALCCT 120
Query: 136 LLDESSFCIQPDEALAVNFMLQLYNLLDENT---NSVEKALRLAKSLNPKIVTLGEYEAS 192
++ ++ EALAVNF L L+++ DE+ N ++ LRL K L+P +VTL E EA+
Sbjct: 121 EVEIEKLGVRNGEALAVNFPLVLHHMPDESVTVENHRDRLLRLVKHLSPNVVTLVEQEAN 180
Query: 193 LTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGV-----R 247
T F+ RF N++ A FES++ +A D ER VE L R + +I
Sbjct: 181 -TNTAPFLPRFVETMNHYLAVFESIDVKLARDHKERINVEQHCLAREVVNLIACEGVERE 239
Query: 248 ERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKD 307
ER E +W+ GF+ LS Y + K LL SYS Y+L E A L L WK+
Sbjct: 240 ERHEPLGKWRSRFHMAGFKPYPLSSYVNATIKGLL--ESYSEKYTLEERDGA-LYLGWKN 296
Query: 308 VPLLTVSSW 316
PL+T +W
Sbjct: 297 QPLITSCAW 305
>UniRef100_Q9LPT0 Putative transcription factor [Arabidopsis thaliana]
Length = 526
Score = 169 bits (429), Expect = 7e-41
Identities = 118/323 (36%), Positives = 165/323 (50%), Gaps = 21/323 (6%)
Query: 10 IASSNSS--------STTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIV 61
+ASS SS T EL L +ACPY KF + +AN AI EA + + +HI+
Sbjct: 208 LASSGSSIYKALRCKDPTGPELLTYMHILYEACPYFKFGYESANGAIAEAVKNESFVHII 267
Query: 62 DFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLL 121
DF I QG QW +L++A R G PN VRI+GI + + G RL + A++
Sbjct: 268 DFQISQGGQWVSLIRALGARPGGPPN-VRITGIDDPRSSFARQGGLELVGQRLGKLAEMC 326
Query: 122 GLNFEFTPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDENT---NSVEKALRLAKS 178
G+ FEF ++ ++ EALAVNF L L+++ DE+ N ++ LRL K
Sbjct: 327 GVPFEFHGAALCCTEVEIEKLGVRNGEALAVNFPLVLHHMPDESVTVENHRDRLLRLVKH 386
Query: 179 LNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGR 238
L+P +VTL E EA+ T F+ RF N++ A FES++ +A D ER VE L R
Sbjct: 387 LSPNVVTLVEQEAN-TNTAPFLPRFVETMNHYLAVFESIDVKLARDHKERINVEQHCLAR 445
Query: 239 RIDGVIGV-----RERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSL 293
+ +I ER E +W+ GF+ LS Y + K LL SYS Y+L
Sbjct: 446 EVVNLIACEGVEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIKGLL--ESYSEKYTL 503
Query: 294 VESQPAFLSLAWKDVPLLTVSSW 316
E A L L WK+ PL+T +W
Sbjct: 504 EERDGA-LYLGWKNQPLITSCAW 525
>UniRef100_Q8H125 Putative scarecrow protein [Arabidopsis thaliana]
Length = 597
Score = 169 bits (429), Expect = 7e-41
Identities = 118/323 (36%), Positives = 165/323 (50%), Gaps = 21/323 (6%)
Query: 10 IASSNSS--------STTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIV 61
+ASS SS T EL L +ACPY KF + +AN AI EA + + +HI+
Sbjct: 279 LASSGSSIYKALRCKDPTGPELLTYMHILYEACPYFKFGYESANGAIAEAVKNESFVHII 338
Query: 62 DFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLL 121
DF I QG QW +L++A R G PN VRI+GI + + G RL + A++
Sbjct: 339 DFQISQGGQWVSLIRALGARPGGPPN-VRITGIDDPRSSFARQGGLELVGQRLGKLAEMC 397
Query: 122 GLNFEFTPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDENT---NSVEKALRLAKS 178
G+ FEF ++ ++ EALAVNF L L+++ DE+ N ++ LRL K
Sbjct: 398 GVPFEFHGAALCCTEVEIEKLGVRNGEALAVNFPLVLHHMPDESVTVENHRDRLLRLVKH 457
Query: 179 LNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGR 238
L+P +VTL E EA+ T F+ RF N++ A FES++ +A D ER VE L R
Sbjct: 458 LSPNVVTLVEQEAN-TNTAPFLPRFVETMNHYLAVFESIDVKLARDHKERINVEQHCLAR 516
Query: 239 RIDGVIGV-----RERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSL 293
+ +I ER E +W+ GF+ LS Y + K LL SYS Y+L
Sbjct: 517 EVVNLIACEGVEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIKGLL--ESYSEKYTL 574
Query: 294 VESQPAFLSLAWKDVPLLTVSSW 316
E A L L WK+ PL+T +W
Sbjct: 575 EERDGA-LYLGWKNQPLITSCAW 596
>UniRef100_Q9FUZ7 SCARECROW [Zea mays]
Length = 668
Score = 169 bits (428), Expect = 9e-41
Identities = 112/295 (37%), Positives = 161/295 (53%), Gaps = 18/295 (6%)
Query: 26 SYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGK 85
+++ N P+ KF+H TANQAI EA E +HI+D I+QG+QW L A+R G
Sbjct: 379 AFQVFNGISPFVKFSHFTANQAIQEAFEREERVHIIDLDIMQGLQWPGLFHILASRPGGP 438
Query: 86 PNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDESSFCIQ 145
P VR++G+ A + ++ ATG RLS+FA LGL FEF + +D +
Sbjct: 439 PR-VRLTGLGA------SMEALEATGKRLSDFADTLGLPFEFCAVAEKAGNVDPEKLGVT 491
Query: 146 PDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFET 205
EA+AV++ L++ L + T S L L + L PK+VT+ E L+ F+ RF
Sbjct: 492 RREAVAVHW---LHHSLYDVTGSDSNTLWLIQRLAPKVVTM--VEQDLSHSGSFLARFVE 546
Query: 206 AFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGV--IGVRERMEDKE--QWKVLME 261
A +Y++A F+SL+ + DSPER VE LL R I V +G R D + W+ +
Sbjct: 547 AIHYYSALFDSLDASYGEDSPERHVVEQQLLSREIRNVLAVGGPARTGDVKFGSWREKLA 606
Query: 262 NCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSW 316
GF + L+ A +QA +LL + S Y+LVE A L L WKD+ LLT S+W
Sbjct: 607 QSGFRAASLAGSAAAQASLLLGMFP-SDGYTLVEENGA-LKLGWKDLCLLTASAW 659
>UniRef100_Q6L5Z0 SCARECROW [Oryza sativa]
Length = 660
Score = 169 bits (428), Expect = 9e-41
Identities = 113/295 (38%), Positives = 161/295 (54%), Gaps = 18/295 (6%)
Query: 26 SYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGK 85
+++ N P+ KF+H TANQAI EA E +HI+D I+QG+QW L A+R G
Sbjct: 372 AFQVFNGISPFVKFSHFTANQAIQEAFEREERVHIIDLDIMQGLQWPGLFHILASRPGGP 431
Query: 86 PNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDESSFCIQ 145
P VR++G+ A + ++ ATG RLS+FA LGL FEF P+ LD +
Sbjct: 432 PR-VRLTGLGA------SMEALEATGKRLSDFADTLGLPFEFCPVADKAGNLDPEKLGVT 484
Query: 146 PDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFET 205
EA+AV++ L + L + T S L L + L PK+VT+ E L+ F+ RF
Sbjct: 485 RREAVAVHW---LRHSLYDVTGSDSNTLWLIQRLAPKVVTM--VEQDLSHSGSFLARFVE 539
Query: 206 AFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGV--IGVRERMEDKE--QWKVLME 261
A +Y++A F+SL+ + + DSPER VE LL R I V +G R D + W+ +
Sbjct: 540 AIHYYSALFDSLDASYSEDSPERHVVEQQLLSREIRNVLAVGGPARTGDVKFGSWREKLA 599
Query: 262 NCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSW 316
GF L+ A +QA +LL + S Y+L+E A L L WKD+ LLT S+W
Sbjct: 600 QSGFRVSSLAGSAAAQAVLLLGMFP-SDGYTLIEENGA-LKLGWKDLCLLTASAW 652
>UniRef100_Q69VG1 Putative chitin-inducible gibberellin-responsive protein [Oryza
sativa]
Length = 571
Score = 169 bits (427), Expect = 1e-40
Identities = 112/304 (36%), Positives = 157/304 (50%), Gaps = 13/304 (4%)
Query: 21 EELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFAT 80
+EL + L + CPY KF ++ AN AI EA NNIHI+DF I QG QW L+QA A
Sbjct: 272 KELLSYMRILYNICPYFKFGYMAANGAIAEALRTENNIHIIDFQIAQGTQWITLIQALAA 331
Query: 81 RSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDES 140
R G P VRI+GI + + G L ++ + EFTP+ + +
Sbjct: 332 RPGGPPR-VRITGIDDPVSEYARGEGLDIVGKMLKSMSEEFKIPLEFTPLSVYATQVTKE 390
Query: 141 SFCIQPDEALAVNFMLQLYNLLDEN---TNSVEKALRLAKSLNPKIVTLGEYEASLTTRV 197
I+P EAL+VNF LQL++ DE+ N + LR+ K L+PK+ TL E E S T
Sbjct: 391 MLEIRPGEALSVNFTLQLHHTPDESVDVNNPRDGLLRMVKGLSPKVTTLVEQE-SHTNTT 449
Query: 198 GFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGV--RERMEDKE- 254
F+ RF Y++A FES++ N+ D+ ER VE L + I +I ++R+E E
Sbjct: 450 PFLMRFGETMEYYSAMFESIDANLPRDNKERISVEQHCLAKDIVNIIACEGKDRVERHEL 509
Query: 255 --QWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLT 312
+WK + GF LS Y S + LL YS Y+L E A L L W+ L++
Sbjct: 510 LGKWKSRLTMAGFRPYPLSSYVNSVIRKLL--ACYSDKYTLDEKDGAML-LGWRSRKLIS 566
Query: 313 VSSW 316
S+W
Sbjct: 567 ASAW 570
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.131 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 483,161,956
Number of Sequences: 2790947
Number of extensions: 18827816
Number of successful extensions: 53114
Number of sequences better than 10.0: 184
Number of HSP's better than 10.0 without gapping: 154
Number of HSP's successfully gapped in prelim test: 30
Number of HSP's that attempted gapping in prelim test: 52420
Number of HSP's gapped (non-prelim): 193
length of query: 316
length of database: 848,049,833
effective HSP length: 127
effective length of query: 189
effective length of database: 493,599,564
effective search space: 93290317596
effective search space used: 93290317596
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 75 (33.5 bits)
Medicago: description of AC144806.12


