

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144765.9 + phase: 0
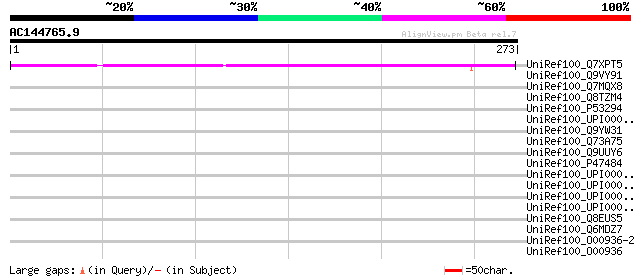
(273 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XPT5 OSJNBa0083N12.5 protein [Oryza sativa] 165 1e-39
UniRef100_Q9VY91 CG10990-PA [Drosophila melanogaster] 36 0.94
UniRef100_Q7MQX8 Hypothetical protein [Wolinella succinogenes] 35 1.6
UniRef100_Q8TZM4 Hypothetical protein PF1966 [Pyrococcus furiosus] 35 1.6
UniRef100_P53294 Hypothetical 46.8 kDa protein in CLC1-PDS2 inte... 35 1.6
UniRef100_UPI000032A071 UPI000032A071 UniRef100 entry 35 2.1
UniRef100_Q9YW31 ORF MSV061 putative LINE reverse transcriptase,... 35 2.1
UniRef100_Q73A75 Tagatose-6-phosphate kinase [Bacillus cereus] 34 4.7
UniRef100_Q9UUY6 Transmembrane protein [Erysiphe pisi] 33 6.1
UniRef100_P47484 Hypothetical protein MG242 [Mycoplasma genitalium] 33 6.1
UniRef100_UPI000049A12F UPI000049A12F UniRef100 entry 33 8.0
UniRef100_UPI0000498DE5 UPI0000498DE5 UniRef100 entry 33 8.0
UniRef100_UPI00003256BC UPI00003256BC UniRef100 entry 33 8.0
UniRef100_UPI0000305C4B UPI0000305C4B UniRef100 entry 33 8.0
UniRef100_Q8EUS5 Hypothetical protein MYPE8450 [Mycoplasma penet... 33 8.0
UniRef100_Q6MDZ7 Probable DNA-directed DNA polymerase III subuni... 33 8.0
UniRef100_O00936-2 Splice isoform Myosin C of O00936 [Toxoplasma... 33 8.0
UniRef100_O00936 Myosin B/C [Toxoplasma gondii] 33 8.0
>UniRef100_Q7XPT5 OSJNBa0083N12.5 protein [Oryza sativa]
Length = 2646
Score = 165 bits (418), Expect = 1e-39
Identities = 102/276 (36%), Positives = 156/276 (55%), Gaps = 8/276 (2%)
Query: 1 MAAQVYARGSFFSDCLNVCAKGGLLDTGSHYIQCWKQNERADPGWANSHDLYAIEQKFME 60
+AA Y R ++ CL++C+KG L G +Q +++ + + AI F+E
Sbjct: 1719 LAADAYFRAKCYAKCLSMCSKGKLFQKGLLLLQQLEEHLLENSSLVK---VAAIRNTFLE 1775
Query: 61 NCALNYFDKNDYRSMMKFVRAFHSIDLKRGFLQSLNLPDELLELEEESGNFMEAAVNIAK 120
+CAL+YF+ D + MM FV++F S+D R FL S NL DELL +E + GNF+EAA IAK
Sbjct: 1776 DCALHYFECGDIKHMMPFVKSFSSMDHIRVFLNSKNLVDELLSVEMDMGNFVEAA-GIAK 1834
Query: 121 TMGDILREADLLGKAGEFLDAYELVFFYVFAKSLWSGGSKAWPLKQFTQKAGLLGKALTF 180
G+IL EADLL KAG +A +L+ +F SLW+ S WP K+F +K LL KA
Sbjct: 1835 HTGNILLEADLLEKAGFLENATQLILLQLFVNSLWASHSTGWPPKRFAEKEQLLAKAKEM 1894
Query: 181 AKEVSSSFYELASTKVE-LSNKHDNIFEIVNQLKSSRIHSSIRGEILCLWELLDSHFRLN 239
++ VS SFY L ++ + LS++H ++ I L ++ E++ +LD H +
Sbjct: 1895 SRNVSESFYCLVCSEADALSDEHKSLASITYNLIEGNKCGNLLVELIASRLILDVHLQAE 1954
Query: 240 SSKYVWQD---SMFDVSVEGMIMKNQLSVETLFCCW 272
+S Y ++ S + M++ NQ+S+ETL W
Sbjct: 1955 ASGYCFESEPGSHDGRHCKDMLVLNQISLETLVYDW 1990
>UniRef100_Q9VY91 CG10990-PA [Drosophila melanogaster]
Length = 509
Score = 36.2 bits (82), Expect = 0.94
Identities = 41/139 (29%), Positives = 69/139 (49%), Gaps = 17/139 (12%)
Query: 86 DLKRGFLQSL-NLPDELLELEEES---GNFMEAAVN--------IAKTMGDILREADLLG 133
D+++GF L NLPD +L+ E GNFM AV +AK+ G+ LR L
Sbjct: 268 DIEKGFNMLLANLPDLVLDTPEAPIMLGNFMARAVADDCIPPKFVAKS-GEELRHLGLGE 326
Query: 134 KAGEFLD-AYELVFFYVFAK--SLWSGGSKAWPLKQFTQKAGLLGKALTFAKEVSSSFYE 190
A + L A L++ +V+A ++W G P+K T + LL K +++V+ +
Sbjct: 327 HAEQALRRADSLIYKHVWAHLDNVWGMGGPLRPVKTITMQMELLLKEYLSSRDVAEAQRC 386
Query: 191 LASTKVELSNKHDNIFEIV 209
L + +V H+ ++E +
Sbjct: 387 LRALEVP-HYHHELVYEAI 404
>UniRef100_Q7MQX8 Hypothetical protein [Wolinella succinogenes]
Length = 480
Score = 35.4 bits (80), Expect = 1.6
Identities = 40/194 (20%), Positives = 80/194 (40%), Gaps = 49/194 (25%)
Query: 67 FDKNDYRSMMKFVRAFHSIDLKRGFLQSLNLPD------------ELLELEEESG----N 110
+DK R +++ I+ +GF++S L + +LL +E++ +
Sbjct: 165 YDKVFLRQQGNYLKLEEKIEATQGFIRSATLKERPLADRLKEKEKKLLAIEDKKSQAYFD 224
Query: 111 FMEAAVNIAKTMGDILR----EADLLGKAG------------EFLDAYELVFFYVFAKSL 154
F + + K D+L + DLL + EF+D Y+ + + K L
Sbjct: 225 FEKEVKELRKRYVDLLHFISLQRDLLAELSALSKMFVERYFQEFVDLYQPLSMELERKLL 284
Query: 155 WSGGSKAWPL--------------KQFTQKAGLLGKALTFAKEVSSSFYELASTKVELSN 200
W +KA+ L +QF +G+ G T++ + +Y + K +L N
Sbjct: 285 WLLNTKAYELDSNLWDRAKQSKLIRQFFMDSGITG---TYSSKTFLKYYLRSLDKDKLRN 341
Query: 201 KHDNIFEIVNQLKS 214
++ +F ++ L+S
Sbjct: 342 ENKELFSLLAYLES 355
>UniRef100_Q8TZM4 Hypothetical protein PF1966 [Pyrococcus furiosus]
Length = 334
Score = 35.4 bits (80), Expect = 1.6
Identities = 40/160 (25%), Positives = 66/160 (41%), Gaps = 28/160 (17%)
Query: 93 QSLNLPDELLELEEESGNFMEAAVNIAK--TMGDILREAD----LLGKAGEFLDAYELVF 146
++L + ELLE+E+E GN E A+ +A + D L EAD L+ +A + +
Sbjct: 108 EALKIYQELLEIEKELGNEREIALTLANIAVVKDELGEADEAIKLMSEAKDIFEKINDTK 167
Query: 147 FYVFAKSLWSGGSKAWPLKQFTQKAGLLGKALTFAKEVSSSFYELASTKVELSNKHDNIF 206
Y A L F + G KA+ +EV L N D+
Sbjct: 168 NYHIA---------LIDLAHFYYELGEYEKAMQIIEEV-------------LRNPLDDEI 205
Query: 207 EIVNQLKSSRIHSSIRGEILCLWELLDSHFRLNSSKYVWQ 246
E+ +L S IHS++ L ++ R +Y+++
Sbjct: 206 EVHGRLVESEIHSALENYKKAALSLRNALLRSGDDEYLFE 245
>UniRef100_P53294 Hypothetical 46.8 kDa protein in CLC1-PDS2 intergenic region
[Saccharomyces cerevisiae]
Length = 404
Score = 35.4 bits (80), Expect = 1.6
Identities = 26/74 (35%), Positives = 34/74 (45%), Gaps = 11/74 (14%)
Query: 43 PGWANSHDLYAIE-----QKFMENCALNYFDKNDYRSMMKFVRAFHSIDLKRGFLQSLNL 97
P W N DL A E QKF++ KN+ R+M F H++DLK L L L
Sbjct: 322 PRWENQQDLDAEELKVRFQKFVDET------KNNCRTMETFCPECHTVDLKDPVLSDLEL 375
Query: 98 PDELLELEEESGNF 111
+ EE +G F
Sbjct: 376 WLHAWKYEEINGKF 389
>UniRef100_UPI000032A071 UPI000032A071 UniRef100 entry
Length = 235
Score = 35.0 bits (79), Expect = 2.1
Identities = 24/65 (36%), Positives = 35/65 (52%), Gaps = 5/65 (7%)
Query: 195 KVELSNKHDNIFEIVNQLKSSRIHSSIRGEI-LCLWELLDSHF--RLNSSKYVWQ--DSM 249
K +LS D+IF ++N S+ + SSI ++ + LD HF N SK VW+ D
Sbjct: 47 KSDLSKSIDSIFNVINNSMSTYLESSIISDVNKNIKTQLDEHFIKVFNKSKEVWEESDKY 106
Query: 250 FDVSV 254
FD +V
Sbjct: 107 FDPTV 111
>UniRef100_Q9YW31 ORF MSV061 putative LINE reverse transcriptase, similar to
Caenorhabditis elegans GB:U00063 [Melanoplus sanguinipes
entomopoxvirus]
Length = 531
Score = 35.0 bits (79), Expect = 2.1
Identities = 19/73 (26%), Positives = 40/73 (54%), Gaps = 5/73 (6%)
Query: 188 FYELASTKVELSNKHDNIFEIVNQLKSSRIHSSIRGEILCLWELLDSHFRLNSSKYVWQD 247
FYEL+S + + +H+ F + + + I +IL +++L+D ++N SKY W++
Sbjct: 459 FYELSSEEENIITRHEMHF--LKNILDFNLEDDIIRQILDVYKLVD---KINESKYKWKE 513
Query: 248 SMFDVSVEGMIMK 260
+ + + + MK
Sbjct: 514 RLSSMDYDRLPMK 526
>UniRef100_Q73A75 Tagatose-6-phosphate kinase [Bacillus cereus]
Length = 307
Score = 33.9 bits (76), Expect = 4.7
Identities = 30/134 (22%), Positives = 59/134 (43%), Gaps = 7/134 (5%)
Query: 89 RGFLQSLNLPDELLELEEESGNFMEAAVNIAKTMGDILREADLLG--KAGEFLDAYELVF 146
RG L++L++ DE + ++EE+ N + + + ++L + + F+ YE +
Sbjct: 68 RGKLENLSIVDEFVSIKEETRNCIALITKKSASATEVLEAGPTISEDEIALFIKKYEKIV 127
Query: 147 ----FYVFAKSLWSGGSKAWPLKQFTQKAGLLGKALTFAKEVSSSFYELASTKVELSNKH 202
F V + SL G SK++ KQ QKA GK F+ + +
Sbjct: 128 KDFEFIVASGSLPKGISKSF-YKQLAQKAAENGKKFILDTSGEPLFHGIQGKPFLIKPNR 186
Query: 203 DNIFEIVNQLKSSR 216
I +++ + + S+
Sbjct: 187 QEICQLLQKKRVSQ 200
>UniRef100_Q9UUY6 Transmembrane protein [Erysiphe pisi]
Length = 1475
Score = 33.5 bits (75), Expect = 6.1
Identities = 22/71 (30%), Positives = 33/71 (45%), Gaps = 2/71 (2%)
Query: 39 ERADPGWANSHDLYAIEQKFMENCALNYFDKNDYRSMMKFVRAFHSIDLKRGFLQSLNLP 98
E A+P W + LY I F +N +FD ND+R + A S+D + + N
Sbjct: 492 ENANPKW--NETLYLIVTTFTDNLTFQFFDYNDFRKDKEIGTATLSLDTIEEYPELENEQ 549
Query: 99 DELLELEEESG 109
E+L + SG
Sbjct: 550 LEVLMNGKSSG 560
>UniRef100_P47484 Hypothetical protein MG242 [Mycoplasma genitalium]
Length = 630
Score = 33.5 bits (75), Expect = 6.1
Identities = 23/81 (28%), Positives = 40/81 (48%), Gaps = 5/81 (6%)
Query: 36 KQNERADPGWANSHDLYA----IEQKFMENCALNYFDKNDYRSMMKFVRAFHSIDLKRGF 91
K R DP DLY IEQ +++ ++K + + + + + I+L+
Sbjct: 291 KNTLREDPDQNPETDLYELIQFIEQNYLKKDKKTSWNKKKVQDLEQLLEEINKINLETKN 350
Query: 92 LQSLNLPDELLELEEESGNFM 112
+SL PDE+ ELE ++ NF+
Sbjct: 351 -ESLAYPDEITELEIDNDNFV 370
>UniRef100_UPI000049A12F UPI000049A12F UniRef100 entry
Length = 1406
Score = 33.1 bits (74), Expect = 8.0
Identities = 27/95 (28%), Positives = 43/95 (44%), Gaps = 15/95 (15%)
Query: 182 KEVSSSFYELASTKVELSNKHDNIFEIVNQLKSSRIHSSIRGEILCLWELLDSHFRLNSS 241
K V +++ L +T +SN+ + V + +S H GE+ L D+HF
Sbjct: 146 KSVGDNYFTLLNTNGNISNE-----DFVRLISTS--HKYSLGELPVLVRDKDTHFAFVLL 198
Query: 242 KYVW--------QDSMFDVSVEGMIMKNQLSVETL 268
KY + DS+ V G+ MKN+L +E L
Sbjct: 199 KYFFLCLPKPMINDSVLKVFEMGLSMKNKLGIERL 233
>UniRef100_UPI0000498DE5 UPI0000498DE5 UniRef100 entry
Length = 1061
Score = 33.1 bits (74), Expect = 8.0
Identities = 19/45 (42%), Positives = 26/45 (57%), Gaps = 1/45 (2%)
Query: 182 KEVSSSFYELASTKVELSNKHDNIF-EIVNQLKSSRIHSSIRGEI 225
KE+ S F E K L K ++ E+VNQ+KSS + I+GEI
Sbjct: 692 KEILSHFEEFIKNKHSLDEKEFKLYSEMVNQMKSSYCNLIIKGEI 736
>UniRef100_UPI00003256BC UPI00003256BC UniRef100 entry
Length = 415
Score = 33.1 bits (74), Expect = 8.0
Identities = 15/48 (31%), Positives = 30/48 (62%), Gaps = 4/48 (8%)
Query: 84 SIDLKRGFLQSLNLPDELLELEEESGNFMEAAVNIAKTMGDILREADL 131
+++L +LQ LPD+ ++L +E+G + +AK MG +R++D+
Sbjct: 334 AVELSNKYLQDSKLPDKAIDLIDEAG----SEKKLAKNMGKTIRKSDI 377
>UniRef100_UPI0000305C4B UPI0000305C4B UniRef100 entry
Length = 667
Score = 33.1 bits (74), Expect = 8.0
Identities = 15/48 (31%), Positives = 30/48 (62%), Gaps = 4/48 (8%)
Query: 84 SIDLKRGFLQSLNLPDELLELEEESGNFMEAAVNIAKTMGDILREADL 131
+++L +LQ LPD+ ++L +E+G + +AK MG +R++D+
Sbjct: 299 AVELSNKYLQDSKLPDKAIDLIDEAG----SEKKLAKNMGKTIRKSDI 342
>UniRef100_Q8EUS5 Hypothetical protein MYPE8450 [Mycoplasma penetrans]
Length = 1084
Score = 33.1 bits (74), Expect = 8.0
Identities = 20/72 (27%), Positives = 38/72 (52%), Gaps = 1/72 (1%)
Query: 143 ELVFFYVFAKSLWSGGSKAWPLKQFTQKAGLLGKALTFAKEVSSSFYELASTKVELSNKH 202
+L +Y+ A S + + + KQ+ K + A +F+ E S + Y +S+ ++L+N
Sbjct: 266 QLFAYYIDASSYSNTNNTNYKYKQYLSKVSISSNAFSFSGEKSLNDYVSSSSTLDLNNFK 325
Query: 203 DNIFEIV-NQLK 213
+ F +V N LK
Sbjct: 326 NIAFPVVINNLK 337
>UniRef100_Q6MDZ7 Probable DNA-directed DNA polymerase III subunits gamma/tau dnaX
[Parachlamydia sp.]
Length = 522
Score = 33.1 bits (74), Expect = 8.0
Identities = 42/161 (26%), Positives = 70/161 (43%), Gaps = 20/161 (12%)
Query: 90 GFLQSLNLPDELLELEEESGNFMEAAVNIAKTMGDILREA----DLLGKAGEFLDAYELV 145
G + +L D+++ E N A + +G + REA DL GK G F+ A+E+
Sbjct: 215 GLRDAESLFDQIVAFSEGKIN----AATVQSILGLMPREAYFELDLAGKEGNFVKAFEIA 270
Query: 146 FFYVFAKSLWSGGSKAWPLKQF-----TQKAGLLGKALTFAKEVSSSFYELAS---TKVE 197
+ +FA+ + F Q AG+ LT + + FYE A+ T+ +
Sbjct: 271 -YRIFAEGKDLNHFIEGLIDHFRHILLIQLAGITSPLLTISPK-EKQFYERAAHLYTQEQ 328
Query: 198 LSNKHDNIFEIVNQLKSSRIHSSIRGEILCLWELLDSHFRL 238
+ D + E Q++ + HS L ++ SHFRL
Sbjct: 329 CLDLIDYLIESAQQIRFT--HSGKIALEAILLHVIRSHFRL 367
>UniRef100_O00936-2 Splice isoform Myosin C of O00936 [Toxoplasma gondii]
Length = 1074
Score = 33.1 bits (74), Expect = 8.0
Identities = 30/103 (29%), Positives = 44/103 (42%), Gaps = 4/103 (3%)
Query: 22 GGLLDTGSHYIQCWKQNERADP-GWANSHDLYAI-EQKFMENCALNYFDKNDYRSMMKFV 79
G + T +H+I+C K NE P GW S L + +E L R+ +F
Sbjct: 664 GIVAQTEAHFIRCLKPNEEKKPLGWNGSKVLNQLFSLSILEALQLRQVGYAYRRNFSEFC 723
Query: 80 RAFHSIDLKRGFLQSLNLPDELLELEEESGNFMEAAVNIAKTM 122
F +DL G + S E+ +L E E++ I KTM
Sbjct: 724 SHFRWLDL--GLVNSDRDRKEVAQLLLEQSGIPESSWVIGKTM 764
>UniRef100_O00936 Myosin B/C [Toxoplasma gondii]
Length = 1171
Score = 33.1 bits (74), Expect = 8.0
Identities = 30/103 (29%), Positives = 44/103 (42%), Gaps = 4/103 (3%)
Query: 22 GGLLDTGSHYIQCWKQNERADP-GWANSHDLYAI-EQKFMENCALNYFDKNDYRSMMKFV 79
G + T +H+I+C K NE P GW S L + +E L R+ +F
Sbjct: 664 GIVAQTEAHFIRCLKPNEEKKPLGWNGSKVLNQLFSLSILEALQLRQVGYAYRRNFSEFC 723
Query: 80 RAFHSIDLKRGFLQSLNLPDELLELEEESGNFMEAAVNIAKTM 122
F +DL G + S E+ +L E E++ I KTM
Sbjct: 724 SHFRWLDL--GLVNSDRDRKEVAQLLLEQSGIPESSWVIGKTM 764
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.135 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 428,918,984
Number of Sequences: 2790947
Number of extensions: 17059887
Number of successful extensions: 44539
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 1
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 44535
Number of HSP's gapped (non-prelim): 18
length of query: 273
length of database: 848,049,833
effective HSP length: 125
effective length of query: 148
effective length of database: 499,181,458
effective search space: 73878855784
effective search space used: 73878855784
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 74 (33.1 bits)
Medicago: description of AC144765.9


