

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144759.6 + phase: 0
(579 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
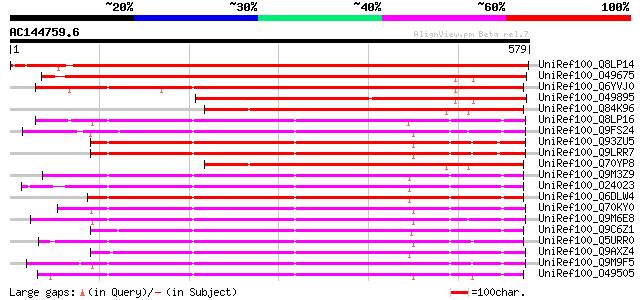
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LP14 Nine-cis-epoxycarotenoid dioxygenase4 [Pisum sa... 952 0.0
UniRef100_O49675 Neoxanthin cleavage enzyme-like protein [Arabid... 750 0.0
UniRef100_Q6YVJ0 Putative 9-cis-epoxycarotenoid dioxygenase [Ory... 631 e-179
UniRef100_O49895 Hypothetical protein [Malus domestica] 521 e-146
UniRef100_Q84K96 Zeaxanthin cleavage oxygenase [Crocus sativus] 388 e-106
UniRef100_Q8LP16 Nine-cis-epoxycarotenoid dioxygenase2 [Pisum sa... 387 e-106
UniRef100_Q9FS24 Neoxanthin cleavage enzyme [Vigna unguiculata] 387 e-106
UniRef100_Q93ZU5 Putative 9-cis-epoxycarotenoid dioxygenase [Ara... 385 e-105
UniRef100_Q9LRR7 9-cis-epoxycarotenoid dioxygenase [Arabidopsis ... 384 e-105
UniRef100_Q70YP8 Lycopene cleavage oxygenase [Bixa orellana] 380 e-104
UniRef100_Q9M3Z9 Putative 9-cis-epoxycarotenoid dioxygenase [Sol... 379 e-103
UniRef100_O24023 Nine-cis-epoxycarotenoid dioxygenase [Lycopersi... 378 e-103
UniRef100_Q6DLW4 9-cis-epoxy-carotenoid dioxygenase 1 [Solanum t... 378 e-103
UniRef100_Q70KY0 9-cis-epoxycarotenoid dioxygenase [Arachis hypo... 378 e-103
UniRef100_Q9M6E8 9-cis-epoxycarotenoid dioxygenase [Phaseolus vu... 377 e-103
UniRef100_Q9C6Z1 9-cis-epoxycarotenoid dioxygenase, putative [Ar... 376 e-103
UniRef100_Q5URR0 9-cis-epoxycarotenoid dioxygenase [Brassica rap... 370 e-101
UniRef100_Q9AXZ4 9-cis-epoxycarotenoid dioxygenase [Persea ameri... 368 e-100
UniRef100_Q9M9F5 F3F9.10 [Arabidopsis thaliana] 366 1e-99
UniRef100_O49505 Neoxanthin cleavage enzyme - like protein [Arab... 365 2e-99
>UniRef100_Q8LP14 Nine-cis-epoxycarotenoid dioxygenase4 [Pisum sativum]
Length = 574
Score = 952 bits (2461), Expect = 0.0
Identities = 464/583 (79%), Positives = 517/583 (88%), Gaps = 14/583 (2%)
Query: 1 MVPKPIIIIACKQPPTTYIPQFNTASVRTKEKPQTSQPKTLRTTPTKTLLALL-----TT 55
MVPK +III CKQPPT+ IP FN +S++TKEKPQT+Q +TL+TT TKT + TT
Sbjct: 1 MVPK-LIIITCKQPPTS-IPSFNISSIKTKEKPQTTQTQTLKTTTTKTKTSPPPPPPPTT 58
Query: 56 TTQKLKNTNKTLQTSSISALILNTLDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPP 115
TT K T S+ L+ NT DDIIN FIDPPIKP++DP+HVLSQNFAPVL ELPP
Sbjct: 59 TTNKKTET-------SLPGLLFNTFDDIINNFIDPPIKPALDPKHVLSQNFAPVLTELPP 111
Query: 116 TQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDGMLHAVKITNGKATLCSRYVQ 175
TQC++I+GTLPPSLNG YIRNGPNPQFLPRGPYHLFDGDGMLHA+KI+NG+ATLCSRYV+
Sbjct: 112 TQCQIIQGTLPPSLNGAYIRNGPNPQFLPRGPYHLFDGDGMLHAIKISNGEATLCSRYVK 171
Query: 176 TYKYKIENEAGYQILPNVFSGFNSLIGLAARGSVAVARVLTGQYNPINGIGLINTSLALF 235
TYKYK ENEAGY + PNVFSGFNSLI AARGS+ ARV TGQYNP NGIGL NTSLALF
Sbjct: 172 TYKYKTENEAGYPLFPNVFSGFNSLIASAARGSITAARVFTGQYNPSNGIGLANTSLALF 231
Query: 236 GNRLFALGESDLPYEIKVTPNGDIQTIGRYDFNGKLFMNMTAHPKIDSDTGECFAYRYGL 295
GNRLFALGESDLPYEI +TPNGDIQTIGRYDFNGKL M+MTAHPKID+DTGE FA+RYG
Sbjct: 232 GNRLFALGESDLPYEINLTPNGDIQTIGRYDFNGKLSMSMTAHPKIDADTGEAFAFRYGP 291
Query: 296 IRPFLTYFRFDSNGVKHNDVPVFSMKRPSFLHDFAITKKYALFADIQLEINPLDMIFGGS 355
I PFLTYFRFDSNGVKHNDVPVFSM PSFLHDFAITKKYA+FADIQL +NPLDMI GGS
Sbjct: 292 IPPFLTYFRFDSNGVKHNDVPVFSMTTPSFLHDFAITKKYAVFADIQLGMNPLDMISGGS 351
Query: 356 PVRSDPSKVPRIGILPRYADNESKMKWFDVPGLNIMHLINAWDEEDGKTVTIVAPNILCV 415
PV SDPSK+PRIGILPRYA++E KMKWF+VPG NI+H INAWDEEDGKTVT++APNIL V
Sbjct: 352 PVGSDPSKIPRIGILPRYANDEKKMKWFNVPGFNIVHAINAWDEEDGKTVTLIAPNILSV 411
Query: 416 EHMMERLELIHEMIEKVRINVETGIVTRQPLSARNLDLAVINNEFLGKKHRFIYAAIGNP 475
EH MERL+L+H MIEKV+INVETGIV+RQPLSARNLD AVIN++F+GK++RF+YAAIGNP
Sbjct: 412 EHTMERLDLVHAMIEKVKINVETGIVSRQPLSARNLDFAVINSDFMGKRNRFVYAAIGNP 471
Query: 476 MPKFSGVVKIDVLKGEEVGCRLYGEDCYGGEPFFVAREDGEEEDDGYLVSYVHDEKKGES 535
MPK SGVVKIDVLKGEEVGCR++GE CYGGEPFFVARE G EEDDGYLVSY+HDEKKGES
Sbjct: 472 MPKISGVVKIDVLKGEEVGCRMFGEGCYGGEPFFVAREGGVEEDDGYLVSYMHDEKKGES 531
Query: 536 RFLVMDAKSPEFEIVAEVKLPRRVPYGFHGLFVKESDITRLSV 578
RFLVMDAKSPEF+IVAEVKLPRRVPYGFHGLFV+ESDI +LS+
Sbjct: 532 RFLVMDAKSPEFDIVAEVKLPRRVPYGFHGLFVRESDIRKLSL 574
>UniRef100_O49675 Neoxanthin cleavage enzyme-like protein [Arabidopsis thaliana]
Length = 595
Score = 750 bits (1937), Expect = 0.0
Identities = 358/551 (64%), Positives = 440/551 (78%), Gaps = 20/551 (3%)
Query: 36 SQPKTLRTTPTKTLLALLTTTTQKLKNTNKTLQTSSISALILNTLDDIINTFIDPPIKPS 95
++PKTL TL++ + K +++ + T++D+INTFIDPP +PS
Sbjct: 55 NKPKTLHNRTNHTLVS----------SPPKLRPEMTLATALFTTVEDVINTFIDPPSRPS 104
Query: 96 VDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDG 155
VDP+HVLS NFAPVLDELPPT C++I GTLP SLNG YIRNGPNPQFLPRGPYHLFDGDG
Sbjct: 105 VDPKHVLSDNFAPVLDELPPTDCEIIHGTLPLSLNGAYIRNGPNPQFLPRGPYHLFDGDG 164
Query: 156 MLHAVKITNGKATLCSRYVQTYKYKIENEAGYQILPNVFSGFNSLIGLAARGSVAVARVL 215
MLHA+KI NGKATLCSRYV+TYKY +E + G ++PNVFSGFN + ARG++ ARVL
Sbjct: 165 MLHAIKIHNGKATLCSRYVKTYKYNVEKQTGAPVMPNVFSGFNGVTASVARGALTAARVL 224
Query: 216 TGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQTIGRYDFNGKLFMNM 275
TGQYNP+NGIGL NTSLA F NRLFALGESDLPY +++T +GDI+TIGRYDF+GKL M+M
Sbjct: 225 TGQYNPVNGIGLANTSLAFFSNRLFALGESDLPYAVRLTESGDIETIGRYDFDGKLAMSM 284
Query: 276 TAHPKIDSDTGECFAYRYGLIRPFLTYFRFDSNGVKHNDVPVFSMKRPSFLHDFAITKKY 335
TAHPK D TGE FA+RYG + PFLTYFRFDS G K DVP+FSM PSFLHDFAITK++
Sbjct: 285 TAHPKTDPITGETFAFRYGPVPPFLTYFRFDSAGKKQRDVPIFSMTSPSFLHDFAITKRH 344
Query: 336 ALFADIQL--EINPLDMIF-GGSPVRSDPSKVPRIGILPRYADNESKMKWFDVPGLNIMH 392
A+FA+IQL +N LD++ GGSPV +D K PR+G++P+YA +ES+MKWF+VPG NI+H
Sbjct: 345 AIFAEIQLGMRMNMLDLVLEGGSPVGTDNGKTPRLGVIPKYAGDESEMKWFEVPGFNIIH 404
Query: 393 LINAWDEEDGKTVTIVAPNILCVEHMMERLELIHEMIEKVRINVETGIVTRQPLSARNLD 452
INAWDE+DG +V ++APNI+ +EH +ER++L+H ++EKV+I++ TGIV R P+SARNLD
Sbjct: 405 AINAWDEDDGNSVVLIAPNIMSIEHTLERMDLVHALVEKVKIDLVTGIVRRHPISARNLD 464
Query: 453 LAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGC----RLYGEDCYGGEPF 508
AVIN FLG+ R++YAAIG+PMPK SGVVK+DV KG+ C R+YG CYGGEPF
Sbjct: 465 FAVINPAFLGRCSRYVYAAIGDPMPKISGVVKLDVSKGDRDDCTVARRMYGSGCYGGEPF 524
Query: 509 FVAREDGE---EEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRRVPYGFHG 565
FVAR+ G EEDDGY+V+YVHDE GES+FLVMDAKSPE EIVA V+LPRRVPYGFHG
Sbjct: 525 FVARDPGNPEAEEDDGYVVTYVHDEVTGESKFLVMDAKSPELEIVAAVRLPRRVPYGFHG 584
Query: 566 LFVKESDITRL 576
LFVKESD+ +L
Sbjct: 585 LFVKESDLNKL 595
>UniRef100_Q6YVJ0 Putative 9-cis-epoxycarotenoid dioxygenase [Oryza sativa]
Length = 638
Score = 631 bits (1627), Expect = e-179
Identities = 313/570 (54%), Positives = 409/570 (70%), Gaps = 27/570 (4%)
Query: 29 TKEKPQTSQPKTLRTTPTKTLLALLTTTTQKLKNTNK----------TLQTSSISALILN 78
T+ QT Q TKT ++ TT + + +S+ N
Sbjct: 66 TRTPKQTEQEDEQLVAKTKTTRTVIATTNGRAAPSQSRPRRRPAPAAAASAASLPMTFCN 125
Query: 79 TLDDIINTFIDPP-IKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSL-NGVYIRN 136
L+++INTFIDPP ++P+VDPR+VL+ NF PV DELPPT C V++G +P L G YIRN
Sbjct: 126 ALEEVINTFIDPPALRPAVDPRNVLTSNFVPV-DELPPTPCPVVRGAIPRCLAGGAYIRN 184
Query: 137 GPNPQFLPRGPYHLFDGDGMLHAVKITNGKAT-----LCSRYVQTYKYKIENEAGYQILP 191
GPNPQ LPRGP+HLFDGDGMLH++ + + ++ LCSRYVQTYKY +E +AG +LP
Sbjct: 185 GPNPQHLPRGPHHLFDGDGMLHSLLLPSPASSGDDPVLCSRYVQTYKYLVERDAGAPVLP 244
Query: 192 NVFSGFNSLIGLAARGSVAVARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPYEI 251
NVFSGF+ + G+A RG+V ARVLTGQ NP+ G+GL NTSLA F RL+ALGESDLPY +
Sbjct: 245 NVFSGFHGVAGMA-RGAVVAARVLTGQMNPLEGVGLANTSLAYFAGRLYALGESDLPYAV 303
Query: 252 KVTPN-GDIQTIGRYDFNGKLFMNMTAHPKIDSDTGECFAYRYGLIRPFLTYFRFDSNGV 310
+V P+ G++ T GR DF G+L M MTAHPK D TGE FA+RYG + PF+TYFRFD G
Sbjct: 304 RVHPDTGEVTTHGRCDFGGRLVMGMTAHPKKDPVTGELFAFRYGPVPPFVTYFRFDPAGN 363
Query: 311 KHNDVPVFSMKRPSFLHDFAITKKYALFADIQLEINPLDMIFGG-SPVRSDPSKVPRIGI 369
K DVP+FS+++PSFLHDFAIT++YA+F +IQ+ + P+DM+ GG SPV SDP KVPR+G+
Sbjct: 364 KGADVPIFSVQQPSFLHDFAITERYAIFPEIQIVMKPMDMVVGGGSPVGSDPGKVPRLGV 423
Query: 370 LPRYADNESKMKWFDVPGLNIMHLINAWDEEDGKTVTIVAPNILCVEHMMERLELIHEMI 429
+PRYA +ES+M+WF+VPG NIMH +NAW+E G+ + +VAPN+L +EH +E +EL+H +
Sbjct: 424 IPRYATDESEMRWFEVPGFNIMHSVNAWEEAGGEELVLVAPNVLSIEHALEHMELVHSCV 483
Query: 430 EKVRINVETGIVTRQPLSARNLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLK 489
EKVRIN+ TG+VTR PL+A N D VIN FLG+++R+ Y +G+P PK GV K+D +
Sbjct: 484 EKVRINLRTGVVTRTPLAAGNFDFPVINPAFLGRRNRYGYFGVGDPAPKIGGVAKLDFDR 543
Query: 490 GEEVGC----RLYGEDCYGGEPFFVAR--EDGEEEDDGYLVSYVHDEKKGESRFLVMDAK 543
E C R +G C+ GEPFFVA E EDDGYLV YVHDE GE+RF+VMDA+
Sbjct: 544 AGEGDCTVAQRDFGPGCFAGEPFFVADDVEGNGNEDDGYLVCYVHDEATGENRFVVMDAR 603
Query: 544 SPEFEIVAEVKLPRRVPYGFHGLFVKESDI 573
SP+ EIVAEV+LP RVPYGFHGLFV ++++
Sbjct: 604 SPDLEIVAEVQLPGRVPYGFHGLFVTQAEL 633
>UniRef100_O49895 Hypothetical protein [Malus domestica]
Length = 446
Score = 521 bits (1342), Expect = e-146
Identities = 250/377 (66%), Positives = 312/377 (82%), Gaps = 10/377 (2%)
Query: 208 SVAVARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQTIGRYDF 267
+++ ARVLTGQYNP NGIGL NTSLA FG+RL+ALGESDLPY +++T NGDI+T+GR+DF
Sbjct: 72 ALSAARVLTGQYNPANGIGLANTSLAFFGDRLYALGESDLPYSLRLTSNGDIETLGRHDF 131
Query: 268 NGKLFMNMTAHPKIDSDTGECFAYRYGLIRPFLTYFRFDSNGVKHNDVPVFSMKRPSFLH 327
+GKL MNMTAHPKID DTGE FA+RYG IRPFLTYFRFDSNGVK DVP+FSM P+FLH
Sbjct: 132 DGKLSMNMTAHPKIDPDTGEAFAFRYGFIRPFLTYFRFDSNGVKQPDVPIFSMVTPTFLH 191
Query: 328 DFAITKKYALFADIQLEINPLDMIF-GGSPVRSDPSKVPRIGILPRYADNESKMKWFDVP 386
DFAITKK+A+FADIQ+ +N +DMI +P DPSKVPRIG++P YA +ES+M+WF+VP
Sbjct: 192 DFAITKKHAIFADIQIGLNLIDMITKRATPFGLDPSKVPRIGVIPLYAKDESEMRWFEVP 251
Query: 387 GLNIMHLINAWDEEDGKTVTIVAPNILCVEHMMERLELIHEMIEKVRINVETGIVTRQPL 446
G N +H NAWDE+D + +VAPN+L EH++ER++L+H ++EKVRI+++TGIVTRQP+
Sbjct: 252 GFNGVHATNAWDEDD--AIVMVAPNVLSAEHVLERVDLVHCLVEKVRIDLKTGIVTRQPI 309
Query: 447 SARNLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGC----RLYGEDC 502
S RNLD AVIN +LG+K++++YAA G+PMPK SGVVK+DV E C R++G C
Sbjct: 310 STRNLDFAVINPAYLGRKNKYVYAAEGDPMPKISGVVKLDVSNVEHKECIVASRMFGPGC 369
Query: 503 YGGEPFFVAREDGE---EEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRRV 559
YGGEPFFVARE +ED+G+LVSYVHDEK GESRFLVMDAKSP+ +IVA V++PRRV
Sbjct: 370 YGGEPFFVAREPENPEADEDNGFLVSYVHDEKAGESRFLVMDAKSPQLDIVAAVRMPRRV 429
Query: 560 PYGFHGLFVKESDITRL 576
PYGFHGLFV+ESD+ L
Sbjct: 430 PYGFHGLFVRESDLNNL 446
>UniRef100_Q84K96 Zeaxanthin cleavage oxygenase [Crocus sativus]
Length = 369
Score = 388 bits (997), Expect = e-106
Identities = 193/365 (52%), Positives = 264/365 (71%), Gaps = 10/365 (2%)
Query: 218 QYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTP-NGDIQTIGRYDFNGKLFMNMT 276
Q +P GIGL NTSL RL AL E DLPY ++++P +GDI T+GR + N + T
Sbjct: 2 QVDPTKGIGLANTSLQFSNGRLHALCEYDLPYVVRLSPEDGDISTVGRIE-NNVSTKSTT 60
Query: 277 AHPKIDSDTGECFAYRYGLIRPFLTYFRFDSNGVKHN-DVPVFSMKRPSFLHDFAITKKY 335
AHPK D TGE F++ YG I+P++TY R+D +G K DVP+FS K PSF+HDFAIT+ Y
Sbjct: 61 AHPKTDPVTGETFSFSYGPIQPYVTYSRYDCDGKKSGPDVPIFSFKEPSFVHDFAITEHY 120
Query: 336 ALFADIQLEINPLDMIFGGSPVRSDPSKVPRIGILPRYADNESKMKWFDVPGLNIMHLIN 395
A+F DIQ+ + P +++ G + D KVPR+G+LPRYA ++S+M+WFDVPG N++H++N
Sbjct: 121 AVFPDIQIVMKPAEIVRGRRMIGPDLEKVPRLGLLPRYATSDSEMRWFDVPGFNMVHVVN 180
Query: 396 AWDEEDGKTVTIVAPNILCVEHMMERLELIHEMIEKVRINVETGIVTRQPLSARNLDLAV 455
AW+EE G+ V IVAPN+ +E+ ++R +L+H +E RI +++G V+R LSA NLD V
Sbjct: 181 AWEEEGGEVVVIVAPNVSPIENAIDRFDLLHVSVEMARIELKSGSVSRTLLSAENLDFGV 240
Query: 456 INNEFLGKKHRFIYAAIGNPMPKFSGVVKID---VLKGE-EVGCRLYGEDCYGGEPFFV- 510
I+ + G+K R+ Y +G+PMPK GVVK+D +GE V R +G C+GGEPFFV
Sbjct: 241 IHRGYSGRKSRYAYLGVGDPMPKIRGVVKVDFELAGRGECVVARREFGVGCFGGEPFFVP 300
Query: 511 --AREDGEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRRVPYGFHGLFV 568
+++ G EEDDGY+VSY+HDE KGES F+VMDA+SPE EI+AEV LPRRVPYGFHGLFV
Sbjct: 301 ASSKKSGGEEDDGYVVSYLHDEGKGESSFVVMDARSPELEILAEVVLPRRVPYGFHGLFV 360
Query: 569 KESDI 573
E+++
Sbjct: 361 TEAEL 365
>UniRef100_Q8LP16 Nine-cis-epoxycarotenoid dioxygenase2 [Pisum sativum]
Length = 601
Score = 387 bits (995), Expect = e-106
Identities = 215/556 (38%), Positives = 316/556 (56%), Gaps = 18/556 (3%)
Query: 30 KEKPQTSQPKTLRTTPTKTLLALLTTTTQKLKNTNKTLQTSSISALILNTLDDIINTFID 89
K +PQ++ T PT+ L + T+ L + + + + TLD + T I
Sbjct: 52 KYQPQSTNTTTTTLIPTRETKPNLPSNTKPLHKQEQ--KWNLLQQAAATTLDFVETTLIK 109
Query: 90 P----PIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPR 145
P+ + DPR ++ NFAPV E P TQ I G LP ++GVY+RNG NP P
Sbjct: 110 QESKHPLPKTSDPRVQIAGNFAPV-PEHPVTQNLPITGKLPKGIDGVYLRNGANPLHEPV 168
Query: 146 GPYHLFDGDGMLHAVKITNGKATLCSRYVQTYKYKIENEAGYQILPNVFSGFNSLIGLAA 205
+H FDGDGM+HAVK TNG + R+ +T++ E G + P + G+A
Sbjct: 169 AGHHFFDGDGMVHAVKFTNGSVSYSCRFTETHRLAQEKALGRPVFPKAIGELHGHSGIA- 227
Query: 206 RGSVAVARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQTIGRY 265
R + AR L G + +G+G+ N L F NRL A+ E D+PY ++VTPNGD+ T+ RY
Sbjct: 228 RLMLYYARSLCGLVDGTHGMGVANAGLVYFNNRLLAMSEDDIPYHVRVTPNGDLTTVCRY 287
Query: 266 DFNGKLFMNMTAHPKIDSDTGECFAYRYGLI-RPFLTYFRFDSNGVKHNDVPVFSMKRPS 324
DFN +L M AHPK+D +A Y ++ +P+L YFRFDSNGVK DV + + P+
Sbjct: 288 DFNDQLKSTMIAHPKVDPVDKNLYALSYDVVQKPYLKYFRFDSNGVKSPDVEI-PLAEPT 346
Query: 325 FLHDFAITKKYALFADIQLEINPLDMIFGGSPVRSDPSKVPRIGILPRYADNESKMKWFD 384
+HDFAIT+ + + D Q+ +MI GGSPV D KV R G+L + A+N S+MKW D
Sbjct: 347 MMHDFAITENFVVVPDQQVVFKLGEMIRGGSPVVYDKEKVSRFGVLSKNAENASEMKWID 406
Query: 385 VPGLNIMHLINAWDEEDGKTVTIVAPNILCVEHMMERL-ELIHEMIEKVRINVETGIVTR 443
P HL NAW+E + V ++ + + + E + ++ ++R+N++TG TR
Sbjct: 407 APECFCFHLWNAWEEPENDEVVVIGSCMTPADSIFNECDESLKSVLSEIRLNLKTGKSTR 466
Query: 444 ----QPLSARNLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGCRLYG 499
Q NL+ ++N LG+K +F Y A+ P PK SG K+D+ G EV LYG
Sbjct: 467 RAIIQESEHMNLEAGMVNKNKLGRKTQFAYLALAEPWPKVSGFAKVDLFSG-EVKKYLYG 525
Query: 500 EDCYGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRRV 559
E+ +GGEP F+ ED E EDDGY++++VHDEK+ +S +++A + + E A + LP RV
Sbjct: 526 ENRFGGEPLFLPNEDSENEDDGYILTFVHDEKEWKSEIQIVNAVNLKLE--ACIPLPSRV 583
Query: 560 PYGFHGLFVKESDITR 575
PYGFHG F+ ++ +
Sbjct: 584 PYGFHGTFIHSKELEK 599
>UniRef100_Q9FS24 Neoxanthin cleavage enzyme [Vigna unguiculata]
Length = 612
Score = 387 bits (994), Expect = e-106
Identities = 215/572 (37%), Positives = 325/572 (56%), Gaps = 25/572 (4%)
Query: 15 PTTYIPQFNTASVRTKEKPQTSQPKTLRTTPTKTLLALLTTTTQKLKNTNKTLQTSSISA 74
P Y P + S T P + T+ TT L+ T Q L LQ ++ +A
Sbjct: 53 PKQYQPTSTSTSTATTTTPTPIKTTTITTTTPPRETNPLSDTNQPLPQKWNFLQKAAATA 112
Query: 75 LILNTLDDIINTFI-----DPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSL 129
L D++ T + P+ + DPR ++ NFAPV + V+ G +P +
Sbjct: 113 L------DLVETALVSHERKHPLPKTADPRVQIAGNFAPVPEHAADQGLPVV-GKIPKCI 165
Query: 130 NGVYIRNGPNPQFLPRGPYHLFDGDGMLHAVKITNGKATLCSRYVQTYKYKIENEAGYQI 189
+GVY+RNG NP + P +H FDGDGM+HAVK TNG A+ R+ +T + E G +
Sbjct: 166 DGVYVRNGANPLYEPVAGHHFFDGDGMVHAVKFTNGAASYACRFTETQRLSQEKSLGRPV 225
Query: 190 LPNVFSGFNSLIGLAARGSVAVARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPY 249
P + G+A R + AR L G + G+G+ N L F N L A+ E DLPY
Sbjct: 226 FPKAIGELHGHSGIA-RLLLFYARGLFGLVDGSQGMGVANAGLVYFNNHLLAMSEDDLPY 284
Query: 250 EIKVTPNGDIQTIGRYDFNGKLFMNMTAHPKIDSDTGECFAYRYGLI-RPFLTYFRFDSN 308
+++TPNGD+ T+GRYDFNG+L M AHPK+D G+ A Y +I +P+L YFRF +
Sbjct: 285 HVRITPNGDLTTVGRYDFNGQLNSTMIAHPKLDPVDGDLHALSYDVIQKPYLKYFRFSPD 344
Query: 309 GVKHNDVPVFSMKRPSFLHDFAITKKYALFADIQLEINPLDMIFGGSPVRSDPSKVPRIG 368
GVK DV + +K P+ +HDFAIT+ + + D Q+ +MI GGSPV D +K R G
Sbjct: 345 GVKSPDVEI-PLKEPTMMHDFAITENFVVVPDQQVVFKLTEMITGGSPVVYDKNKTSRFG 403
Query: 369 ILPRYADNESKMKWFDVPGLNIMHLINAWDEEDGKTVTIVAPNILCVEHMM-ERLELIHE 427
IL + A + + M+W D P HL NAW+E + + V ++ + + + E E +
Sbjct: 404 ILHKNAKDANAMRWIDAPDCFCFHLWNAWEEPETEEVVVIGSCMTPADSIFNECEESLKS 463
Query: 428 MIEKVRINVETGIVTRQPLSAR----NLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVV 483
++ ++R+N+ TG TR+P+ + NL+ ++N LG+K +F Y A+ P PK SG
Sbjct: 464 VLSEIRLNLRTGKSTRRPIISDAEQVNLEAGMVNRNKLGRKTQFAYLALAEPWPKVSGFA 523
Query: 484 KIDVLKGEEVGCRLYGEDCYGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESRFLVMDAK 543
K+D+L G EV +YGE+ +GGEP F+ +G++EDDGY++++VHDEK+ +S +++A+
Sbjct: 524 KVDLLSG-EVKKYMYGEEKFGGEPLFL--PNGQKEDDGYILAFVHDEKEWKSELQIVNAQ 580
Query: 544 SPEFEIVAEVKLPRRVPYGFHGLFVKESDITR 575
+ + E A +KLP RVPYGFHG F+ D+ +
Sbjct: 581 NLKLE--ASIKLPSRVPYGFHGTFIHSKDLRK 610
>UniRef100_Q93ZU5 Putative 9-cis-epoxycarotenoid dioxygenase [Arabidopsis thaliana]
Length = 599
Score = 385 bits (989), Expect = e-105
Identities = 200/492 (40%), Positives = 299/492 (60%), Gaps = 14/492 (2%)
Query: 91 PIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHL 150
P+ + DP ++ NFAPV +E P + + G LP S+ GVY+RNG NP P +H
Sbjct: 112 PLPKTADPSVQIAGNFAPV-NEQPVRRNLPVVGKLPDSIKGVYVRNGANPLHEPVTGHHF 170
Query: 151 FDGDGMLHAVKITNGKATLCSRYVQTYKYKIENEAGYQILPNVFSGFNSLIGLAARGSVA 210
FDGDGM+HAVK +G A+ R+ QT ++ E + G + P + G+A R +
Sbjct: 171 FDGDGMVHAVKFEHGSASYACRFTQTNRFVQERQLGRPVFPKAIGELHGHTGIA-RLMLF 229
Query: 211 VARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQTIGRYDFNGK 270
AR G +P +G G+ N L F RL A+ E DLPY++++TPNGD++T+GR+DF+G+
Sbjct: 230 YARAAAGIVDPAHGTGVANAGLVYFNGRLLAMSEDDLPYQVQITPNGDLKTVGRFDFDGQ 289
Query: 271 LFMNMTAHPKIDSDTGECFAYRYGLI-RPFLTYFRFDSNGVKHNDVPVFSMKRPSFLHDF 329
L M AHPK+D ++GE FA Y ++ +P+L YFRF +G K DV + + +P+ +HDF
Sbjct: 290 LESTMIAHPKVDPESGELFALSYDVVSKPYLKYFRFSPDGTKSPDVEI-QLDQPTMMHDF 348
Query: 330 AITKKYALFADIQLEINPLDMIFGGSPVRSDPSKVPRIGILPRYADNESKMKWFDVPGLN 389
AIT+ + + D Q+ +MI GGSPV D +KV R GIL +YA++ S +KW D P
Sbjct: 349 AITENFVVVPDQQVVFKLPEMILGGSPVVYDKNKVARFGILDKYAEDSSNIKWIDAPDCF 408
Query: 390 IMHLINAWDEEDGKTVTIVAPNILCVEHMM-ERLELIHEMIEKVRINVETGIVTRQPLSA 448
HL NAW+E + V ++ + + + E E + ++ ++R+N++TG TR+P+ +
Sbjct: 409 CFHLWNAWEEPETDEVVVIGSCMTPPDSIFNESDENLKSVLSEIRLNLKTGESTRRPIIS 468
Query: 449 R-----NLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGCRLYGEDCY 503
NL+ ++N LG+K +F Y A+ P PK SG K+D+ G EV LYG++ Y
Sbjct: 469 NEDQQVNLEAGMVNRNMLGRKTKFAYLALAEPWPKVSGFAKVDLTTG-EVKKHLYGDNRY 527
Query: 504 GGEPFFVAREDGEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRRVPYGF 563
GGEP F+ E G EED+GY++ +VHDEK +S +++A S E+ A VKLP RVPYGF
Sbjct: 528 GGEPLFLPGEGG-EEDEGYILCFVHDEKTWKSELQIVNAVS--LEVEATVKLPSRVPYGF 584
Query: 564 HGLFVKESDITR 575
HG F+ D+ +
Sbjct: 585 HGTFIGADDLAK 596
>UniRef100_Q9LRR7 9-cis-epoxycarotenoid dioxygenase [Arabidopsis thaliana]
Length = 599
Score = 384 bits (986), Expect = e-105
Identities = 200/492 (40%), Positives = 299/492 (60%), Gaps = 14/492 (2%)
Query: 91 PIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHL 150
P+ + DP ++ NFAPV +E P + + G LP S+ GVY+RNG NP P +H
Sbjct: 112 PLPKTADPSVQIAGNFAPV-NEQPVRRNLPVVGKLPDSIKGVYVRNGANPLHEPVTGHHF 170
Query: 151 FDGDGMLHAVKITNGKATLCSRYVQTYKYKIENEAGYQILPNVFSGFNSLIGLAARGSVA 210
FDGDGM+HAVK +G A+ R+ QT ++ E + G + P + G+A R +
Sbjct: 171 FDGDGMVHAVKFEHGSASYACRFTQTNRFVQERQLGRPVFPKAIGELHGHTGIA-RLMLF 229
Query: 211 VARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQTIGRYDFNGK 270
AR G +P +G G+ N L F RL A+ E DLPY++++TPNGD++T+GR+DF+G+
Sbjct: 230 YARAAAGIVDPAHGTGVANAGLVYFNGRLLAMSEDDLPYQVQITPNGDLKTVGRFDFDGQ 289
Query: 271 LFMNMTAHPKIDSDTGECFAYRYGLI-RPFLTYFRFDSNGVKHNDVPVFSMKRPSFLHDF 329
L M AHPK+D ++GE FA Y ++ +P+L YFRF +G K DV + + +P+ +HDF
Sbjct: 290 LESTMIAHPKVDPESGELFALSYDVVSKPYLKYFRFSPDGTKSPDVEI-QLDQPTMMHDF 348
Query: 330 AITKKYALFADIQLEINPLDMIFGGSPVRSDPSKVPRIGILPRYADNESKMKWFDVPGLN 389
AIT+ + + D Q+ +MI GGSPV D +KV R GIL +YA++ S +KW D P
Sbjct: 349 AITENFVVVPDQQVVFKLPEMIRGGSPVVYDKNKVARFGILDKYAEDSSNIKWIDAPDCF 408
Query: 390 IMHLINAWDEEDGKTVTIVAPNILCVEHMM-ERLELIHEMIEKVRINVETGIVTRQPLSA 448
HL NAW+E + V ++ + + + E E + ++ ++R+N++TG TR+P+ +
Sbjct: 409 CFHLWNAWEEPETDEVVVIGSCMTPPDSIFNESDENLKSVLSEIRLNLKTGESTRRPIIS 468
Query: 449 R-----NLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGCRLYGEDCY 503
NL+ ++N LG+K +F Y A+ P PK SG K+D+ G EV LYG++ Y
Sbjct: 469 NEDQQVNLEAGMVNRNMLGRKTKFAYLALAEPWPKVSGFAKVDLTTG-EVKKHLYGDNRY 527
Query: 504 GGEPFFVAREDGEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRRVPYGF 563
GGEP F+ E G EED+GY++ +VHDEK +S +++A S E+ A VKLP RVPYGF
Sbjct: 528 GGEPLFLPGEGG-EEDEGYILCFVHDEKTWKSELQIVNAVS--LEVEATVKLPSRVPYGF 584
Query: 564 HGLFVKESDITR 575
HG F+ D+ +
Sbjct: 585 HGTFIGADDLAK 596
>UniRef100_Q70YP8 Lycopene cleavage oxygenase [Bixa orellana]
Length = 369
Score = 380 bits (976), Expect = e-104
Identities = 190/365 (52%), Positives = 259/365 (70%), Gaps = 10/365 (2%)
Query: 218 QYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTP-NGDIQTIGRYDFNGKLFMNMT 276
Q P GIGL NTSL RL AL E DLPY ++++P +GDI T+GR + N + T
Sbjct: 2 QVEPTRGIGLANTSLQFSNGRLHALCEYDLPYVVRLSPEDGDISTVGRIE-NNVSTKSTT 60
Query: 277 AHPKIDSDTGECFAYRYGLIRPFLTYFRFDSNGVKHN-DVPVFSMKRPSFLHDFAITKKY 335
AHPK D TGE F++ YG I+P++TY R+D + K DVP+FS K PSF+HDFAIT+ Y
Sbjct: 61 AHPKTDPVTGETFSFSYGPIQPYVTYSRYDCDDKKSGPDVPIFSFKEPSFVHDFAITEHY 120
Query: 336 ALFADIQLEINPLDMIFGGSPVRSDPSKVPRIGILPRYADNESKMKWFDVPGLNIMHLIN 395
A+F DIQ+ + P +++ G + D KVPR+G+LPRYA ++S+M+WFDVPG N++H++N
Sbjct: 121 AVFPDIQIVMKPAEIVRGRRMIGPDLEKVPRLGLLPRYATSDSEMRWFDVPGFNMVHVVN 180
Query: 396 AWDEEDGKTVTIVAPNILCVEHMMERLELIHEMIEKVRINVETGIVTRQPLSARNLDLAV 455
AW+EE G+ V IVAPN+ +E+ ++R +L+H +E RI +++G V R LSA NLD V
Sbjct: 181 AWEEEGGEVVVIVAPNVSPIENAIDRFDLLHVSVEMARIELKSGSVPRTLLSAENLDFGV 240
Query: 456 INNEFLGKKHRFIYAAIGNPMPKFSGVVKID---VLKGE-EVGCRLYGEDCYGGEPFFV- 510
I+ + G+K R+ Y +G+PMPK GVVK+D +GE V R +G C+GGEPFFV
Sbjct: 241 IHRGYSGRKSRYAYLGVGDPMPKIRGVVKVDFELAGRGECVVARREFGVGCFGGEPFFVP 300
Query: 511 --AREDGEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRRVPYGFHGLFV 568
+++ G EEDDGY+VSY+HDE KGES F+VMDA+S + EI+AEV LPRRVPYGFHGLFV
Sbjct: 301 ASSKKSGGEEDDGYVVSYLHDEGKGESSFVVMDARSAQLEILAEVVLPRRVPYGFHGLFV 360
Query: 569 KESDI 573
+ D+
Sbjct: 361 TDKDL 365
>UniRef100_Q9M3Z9 Putative 9-cis-epoxycarotenoid dioxygenase [Solanum tuberosum]
Length = 604
Score = 379 bits (974), Expect = e-103
Identities = 210/546 (38%), Positives = 314/546 (57%), Gaps = 15/546 (2%)
Query: 37 QPKTLRTTPTKTLLALLTTTTQKLKNTNKTLQTSSISALILNTLDDIINTF-IDPPIKPS 95
Q +T T T+ + +K +A+ L+ ++ + ++ P+ +
Sbjct: 61 QSSNYQTPKTSTISHPKQENNNSSSSISKWNLVQKAAAMALDAVEGALTKHELEHPLPKT 120
Query: 96 VDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDG 155
DPR +S NFAPV E P Q + G +P + GVY+RNG NP F P H FDGDG
Sbjct: 121 ADPRVQISGNFAPV-PENPVCQSLPVTGKIPKCVQGVYVRNGANPLFEPTAGRHFFDGDG 179
Query: 156 MLHAVKITNGKATLCSRYVQTYKYKIENEAGYQILPNVFSGFNSLIGLAARGSVAVARVL 215
M+HAV+ NG A+ R+ +T ++ E G + P + G+A R + AR L
Sbjct: 180 MVHAVQFKNGSASYACRFTETERFVQEKALGRPVFPKAIGELHGHSGIA-RLMLFYARGL 238
Query: 216 TGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQTIGRYDFNGKLFMNM 275
G + G G+ N L F NRL A+ E DLPY +KVTP GD++T GR+DF+G+L M
Sbjct: 239 FGLIDHSRGTGVANAGLVYFNNRLLAMSEDDLPYHVKVTPTGDLKTEGRFDFDGQLKSTM 298
Query: 276 TAHPKIDSDTGECFAYRYGLI-RPFLTYFRFDSNGVKHNDVPVFSMKRPSFLHDFAITKK 334
AHPK+D +GE FA Y +I +P+L YFRF NG K NDV + ++ P+ +HDFAIT+K
Sbjct: 299 IAHPKLDPVSGELFALSYDVIQKPYLKYFRFSKNGEKSNDVEI-PVEDPTMMHDFAITEK 357
Query: 335 YALFADIQLEINPLDMIFGGSPVRSDPSKVPRIGILPRYADNESKMKWFDVPGLNIMHLI 394
+ + D Q+ +MI GGSPV D +KV R GIL +YA + S +KW +VP HL
Sbjct: 358 FVIIPDQQVVFKMSEMIRGGSPVVYDKNKVSRFGILDKYAKDGSDLKWVEVPDCFCFHLW 417
Query: 395 NAWDEEDGKTVTIVAPNILCVEHMMERL-ELIHEMIEKVRINVETGIVTRQ-----PLSA 448
NAW+E + + ++ + + + E + ++ ++R+N++TG TR+ P
Sbjct: 418 NAWEEPETDEIVVIGSCMTPPDSIFNECDEGLKSVLSEIRLNLKTGKSTRKAIIENPDEQ 477
Query: 449 RNLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGCRLYGEDCYGGEPF 508
NL+ ++N LG+K ++ Y AI P PK SG K+D+ G EV +YG++ YGGEP
Sbjct: 478 VNLEAGMVNRNKLGRKTQYAYLAIAEPWPKVSGFAKVDLFTG-EVEKFIYGDNKYGGEPL 536
Query: 509 FVARE-DGEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRRVPYGFHGLF 567
F+ R+ + +EEDDGY++++VHDEK+ S +++A + + E A VKLP RVPYGFHG F
Sbjct: 537 FLPRDPNSKEEDDGYILAFVHDEKEWTSELQIVNAMTLKLE--ATVKLPSRVPYGFHGTF 594
Query: 568 VKESDI 573
+ +D+
Sbjct: 595 INANDL 600
>UniRef100_O24023 Nine-cis-epoxycarotenoid dioxygenase [Lycopersicon esculentum]
Length = 605
Score = 378 bits (971), Expect = e-103
Identities = 213/569 (37%), Positives = 324/569 (56%), Gaps = 29/569 (5%)
Query: 14 PPTTYIPQFNTASVRTKEKPQTSQPKTLRTTPTKTLLALLTTTTQKLKNTNKTLQTSSIS 73
PP + P+ +++ +T + S PK + + +T+K +
Sbjct: 53 PPILHFPK-QSSNYQTPKNNTISHPKQENNNSSSS-------------STSKWNLVQKAA 98
Query: 74 ALILNTLDDIINTF-IDPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGV 132
A+ L+ ++ + ++ P+ + DPR +S NFAPV E P Q + G +P + GV
Sbjct: 99 AMALDAVESALTKHELEHPLPKTADPRVQISGNFAPV-PENPVCQSLPVTGKIPKCVQGV 157
Query: 133 YIRNGPNPQFLPRGPYHLFDGDGMLHAVKITNGKATLCSRYVQTYKYKIENEAGYQILPN 192
Y+RNG NP F P +H FDGDGM+HAV+ NG A+ R+ +T + E G + P
Sbjct: 158 YVRNGANPLFEPTAGHHFFDGDGMVHAVQFKNGSASYACRFTETERLVQEKALGRPVFPK 217
Query: 193 VFSGFNSLIGLAARGSVAVARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIK 252
+ G+A R + AR L G + G G+ N L F NRL A+ E DLPY +K
Sbjct: 218 AIGELHGHSGIA-RLMLFYARGLFGLVDHSKGTGVANAGLVYFNNRLLAMSEDDLPYHVK 276
Query: 253 VTPNGDIQTIGRYDFNGKLFMNMTAHPKIDSDTGECFAYRYGLI-RPFLTYFRFDSNGVK 311
VTP GD++T GR+DF+G+L M AHPK+D +GE FA Y +I +P+L YFRF NG K
Sbjct: 277 VTPTGDLKTEGRFDFDGQLKSTMIAHPKLDPVSGELFALSYDVIQKPYLKYFRFSKNGEK 336
Query: 312 HNDVPVFSMKRPSFLHDFAITKKYALFADIQLEINPLDMIFGGSPVRSDPSKVPRIGILP 371
NDV + ++ P+ +HDFAIT+ + + D Q+ +MI GGSPV D +KV R GIL
Sbjct: 337 SNDVEI-PVEDPTMMHDFAITENFVVIPDQQVVFKMSEMIRGGSPVVYDKNKVSRFGILD 395
Query: 372 RYADNESKMKWFDVPGLNIMHLINAWDEEDGKTVTIVAPNILCVEHMMERL-ELIHEMIE 430
+YA + S +KW +VP HL NAW+E + + ++ + + + E + ++
Sbjct: 396 KYAKDGSDLKWVEVPDCFCFHLWNAWEEAETDEIVVIGSCMTPPDSIFNECDEGLKSVLS 455
Query: 431 KVRINVETGIVTRQ-----PLSARNLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKI 485
++R+N++TG TR+ P NL+ ++N LG+K + Y AI P PK SG K+
Sbjct: 456 EIRLNLKTGKSTRKSIIENPDEQVNLEAGMVNRNKLGRKTEYAYLAIAEPWPKVSGFAKV 515
Query: 486 DVLKGEEVGCRLYGEDCYGGEPFFVARE-DGEEEDDGYLVSYVHDEKKGESRFLVMDAKS 544
++ G EV +YG++ YGGEP F+ R+ + +EEDDGY++++VHDEK+ +S +++A S
Sbjct: 516 NLFTG-EVEKFIYGDNKYGGEPLFLPRDPNSKEEDDGYILAFVHDEKEWKSELQIVNAMS 574
Query: 545 PEFEIVAEVKLPRRVPYGFHGLFVKESDI 573
+ E A VKLP RVPYGFHG F+ +D+
Sbjct: 575 LKLE--ATVKLPSRVPYGFHGTFINANDL 601
>UniRef100_Q6DLW4 9-cis-epoxy-carotenoid dioxygenase 1 [Solanum tuberosum]
Length = 603
Score = 378 bits (971), Expect = e-103
Identities = 203/494 (41%), Positives = 297/494 (60%), Gaps = 14/494 (2%)
Query: 88 IDPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGP 147
++ P+ + DPR +S NFAPV E P Q + G +P + GVY+RNG NP F P
Sbjct: 112 LEHPLPKTADPRVQISGNFAPV-PENPVCQSLPVTGKIPKCVQGVYVRNGANPLFEPTAG 170
Query: 148 YHLFDGDGMLHAVKITNGKATLCSRYVQTYKYKIENEAGYQILPNVFSGFNSLIGLAARG 207
+H FDGDGM+HAV+ NG A+ R+ +T + E G + P + G+A R
Sbjct: 171 HHFFDGDGMVHAVQFKNGSASYACRFTETERLVQEKALGRPVFPKAIGELHGHSGIA-RL 229
Query: 208 SVAVARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQTIGRYDF 267
+ AR L G + G G+ N L F NRL A+ E DLPY +KVTP GD++T GR+DF
Sbjct: 230 MLFYARGLFGLIDHSRGTGVANAGLVYFNNRLLAMSEDDLPYHVKVTPTGDLKTEGRFDF 289
Query: 268 NGKLFMNMTAHPKIDSDTGECFAYRYGLI-RPFLTYFRFDSNGVKHNDVPVFSMKRPSFL 326
+G+L M AHPK+D +GE FA Y +I +P+L YFRF NG K NDV + ++ P+ +
Sbjct: 290 DGQLKSTMIAHPKLDPVSGELFALSYDVIQKPYLKYFRFSKNGEKSNDVEI-PVEDPTMM 348
Query: 327 HDFAITKKYALFADIQLEINPLDMIFGGSPVRSDPSKVPRIGILPRYADNESKMKWFDVP 386
HDFAIT+K+ + D Q+ +MI GGSPV D +KV R GIL +YA + S +KW +VP
Sbjct: 349 HDFAITEKFVIIPDQQVVFKMSEMIRGGSPVVYDKNKVSRFGILDKYAKDGSDLKWVEVP 408
Query: 387 GLNIMHLINAWDEEDGKTVTIVAPNILCVEHMMERL-ELIHEMIEKVRINVETGIVTRQ- 444
HL NAW+E + + ++ + + + E + ++ ++R+N++TG TR+
Sbjct: 409 DCFCFHLWNAWEEPETDEIVVIGSCMTPPDSIFNECDEGLKSVLSEIRLNLKTGKSTRKA 468
Query: 445 ----PLSARNLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGCRLYGE 500
P NL+ ++N LG+K ++ Y AI P PK SG K+D+ G EV +YG+
Sbjct: 469 IIENPDEQVNLEAGMVNRNKLGRKTQYAYLAIAEPWPKVSGFAKVDLFTG-EVEKFIYGD 527
Query: 501 DCYGGEPFFVARE-DGEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRRV 559
+ YGGEP F+ R+ + +EEDDGY++++VHDEK+ S +++A + + E A VKLP RV
Sbjct: 528 NKYGGEPLFLPRDPNSKEEDDGYILAFVHDEKEWNSELQIVNAMTLKLE--ATVKLPSRV 585
Query: 560 PYGFHGLFVKESDI 573
PYGFHG F+ +D+
Sbjct: 586 PYGFHGTFINANDL 599
>UniRef100_Q70KY0 9-cis-epoxycarotenoid dioxygenase [Arachis hypogaea]
Length = 601
Score = 378 bits (970), Expect = e-103
Identities = 212/543 (39%), Positives = 311/543 (57%), Gaps = 27/543 (4%)
Query: 54 TTTTQKLKNT--NKTLQTSSISALILNTLDDIINTFID------------PPIKPSVDPR 99
TTTT K K T NKT T++ N + ID P+ + DPR
Sbjct: 63 TTTTTKPKPTKENKTASTTAAPVKDWNLIQKAAAMAIDFAESALASHERRHPLPKTADPR 122
Query: 100 HVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDGMLHA 159
++ NFAPV E P T I G LP ++GVY+RNG NP + P +HLFDGDGM+HA
Sbjct: 123 VQIAGNFAPV-PEHPSTHSLPIAGKLPECIDGVYVRNGANPMYEPLAGHHLFDGDGMVHA 181
Query: 160 VKITNGKATLCSRYVQTYKYKIENEAGYQILPNVFSGFNSLIGLAARGSVAVARVLTGQY 219
VK NG A+ R+ +T + E G + P + G+A R + AR L G
Sbjct: 182 VKFHNGSASYSCRFTETNRLVQEKSLGKPVFPKAIGELHGHSGIA-RLLLFYARGLFGLV 240
Query: 220 NPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQTIGRYDFNGKLFMNMTAHP 279
+ NG+G+ N L F NRL A+ E DLPY ++VTPNGD++T+GRYDFNG+L M AHP
Sbjct: 241 DGKNGMGVANAGLVYFNNRLLAMSEDDLPYHVRVTPNGDLKTVGRYDFNGQLKSTMIAHP 300
Query: 280 KIDSDTGECFAYRYGLI-RPFLTYFRFDSNGVKHNDVPVFSMKRPSFLHDFAITKKYALF 338
K+D +GE FA Y ++ +P+L YF+F+ +G K DV + +K P+ +HDFAIT+ + +
Sbjct: 301 KVDPVSGELFALSYDVVSKPYLKYFKFNKDGTKSKDVDI-PLKVPTMMHDFAITENFVVV 359
Query: 339 ADIQLEINPLDMIFGGSPVRSDPSKVPRIGILPRYADNESKMKWFDVPGLNIMHLINAWD 398
D Q+ +M+ GGSPV D KV R GIL + A + ++W + P HL NAW+
Sbjct: 360 PDQQVVFKLGEMMRGGSPVVYDKEKVSRFGILDKNATDSDGIRWIEAPECFCFHLWNAWE 419
Query: 399 EEDGKTVTIVAPNILCVEHMMERL-ELIHEMIEKVRINVETGIVTRQPLSAR----NLDL 453
E++ V ++ + + + E + ++ ++R+N++TG TR+ + NL+
Sbjct: 420 EKETDEVVVIGSCMTPADSIFNECDENLKSVLSEIRLNLKTGKSTRRAIIREEEQVNLEA 479
Query: 454 AVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGCRLYGEDCYGGEPFFVARE 513
++N LG+K +F Y A+ P PK SG K+D+ G EV +YG + +GGEP F+ R
Sbjct: 480 GMVNKSKLGRKTQFAYLALAEPWPKVSGFAKVDLSSG-EVKKYMYGGEKFGGEPMFLPRS 538
Query: 514 D-GEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRRVPYGFHGLFVKESD 572
E EDDGY++++VHDEK+ S V++A + E E A ++LP RVPYGFHG F++ D
Sbjct: 539 AWSEREDDGYILAFVHDEKEWRSELQVVNAMTLELE--ATIELPSRVPYGFHGTFIQSKD 596
Query: 573 ITR 575
+ +
Sbjct: 597 LRK 599
>UniRef100_Q9M6E8 9-cis-epoxycarotenoid dioxygenase [Phaseolus vulgaris]
Length = 615
Score = 377 bits (969), Expect = e-103
Identities = 211/572 (36%), Positives = 319/572 (54%), Gaps = 28/572 (4%)
Query: 24 TASVRTKEKPQTSQPKTLRTTPTKTLL--ALLTTTTQKLKNTNKTLQTSSISALILNTLD 81
T S++T P+ QP + TT T T + TTTT + T T N L
Sbjct: 50 TCSLQTLHYPKQYQPTSTSTTTTPTPIKPTTTTTTTTPHRETKPLSDTKQPFPQKWNFLQ 109
Query: 82 DIINTFIDP------------PIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSL 129
T +D P+ + DP+ ++ NFAPV E Q + G +P +
Sbjct: 110 KAAATGLDMVETALVSHESKHPLPKTADPKVQIAGNFAPV-PEHAADQALPVVGKIPKCI 168
Query: 130 NGVYIRNGPNPQFLPRGPYHLFDGDGMLHAVKITNGKATLCSRYVQTYKYKIENEAGYQI 189
+GVY+RNG NP + P +H FDGDGM+HAVK TNG A+ R+ +T + E G +
Sbjct: 169 DGVYVRNGANPLYEPVAGHHFFDGDGMVHAVKFTNGAASYACRFTETQRLAQEKSLGRPV 228
Query: 190 LPNVFSGFNSLIGLAARGSVAVARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPY 249
P + G+A R + AR L + +G+G+ N L F N L A+ E DLPY
Sbjct: 229 FPKAIGELHGHSGIA-RLLLFYARSLFQLVDGSHGMGVANAGLVYFNNHLLAMSEDDLPY 287
Query: 250 EIKVTPNGDIQTIGRYDFNGKLFMNMTAHPKIDSDTGECFAYRYGLI-RPFLTYFRFDSN 308
+++T NGD+ T+GRYDFNG+L M AHPK+D G+ A Y ++ +P+L YFRF ++
Sbjct: 288 HVRITSNGDLTTVGRYDFNGQLNSTMIAHPKLDPVNGDLHALSYDVVQKPYLKYFRFSAD 347
Query: 309 GVKHNDVPVFSMKRPSFLHDFAITKKYALFADIQLEINPLDMIFGGSPVRSDPSKVPRIG 368
GVK DV + +K P+ +HDFAIT+ + + D Q+ +MI GGSPV D +K R G
Sbjct: 348 GVKSPDVEI-PLKEPTMMHDFAITENFVVVPDQQVVFKLTEMITGGSPVVYDKNKTSRFG 406
Query: 369 ILPRYADNESKMKWFDVPGLNIMHLINAWDEEDGKTVTIVAPNILCVEHMMERL-ELIHE 427
IL + A + + M+W D P HL NAW+E + + ++ + + + E +
Sbjct: 407 ILDKNAKDANAMRWIDAPECFCFHLWNAWEEPETDEIVVIGSCMTPADSIFNECDESLKS 466
Query: 428 MIEKVRINVETGIVTRQPLSAR----NLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVV 483
++ ++R+N+ TG TR+P+ + NL+ ++N LG+K +F Y A+ P PK SG
Sbjct: 467 VLSEIRLNLRTGKSTRRPIISDAEQVNLEAGMVNRNKLGRKTQFAYLALAEPWPKVSGFA 526
Query: 484 KIDVLKGEEVGCRLYGEDCYGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESRFLVMDAK 543
K+D+ G EV +YGE+ +GGEP F+ +GEEE DGY++++VHDEK+ +S +++A+
Sbjct: 527 KVDLFSG-EVQKYMYGEEKFGGEPLFL--PNGEEEGDGYILAFVHDEKEWKSELQIVNAQ 583
Query: 544 SPEFEIVAEVKLPRRVPYGFHGLFVKESDITR 575
+ + E A +KLP RVPYGFHG F+ D+ +
Sbjct: 584 NLKLE--ASIKLPSRVPYGFHGTFIHSKDLRK 613
>UniRef100_Q9C6Z1 9-cis-epoxycarotenoid dioxygenase, putative [Arabidopsis thaliana]
Length = 589
Score = 376 bits (966), Expect = e-103
Identities = 205/490 (41%), Positives = 293/490 (58%), Gaps = 13/490 (2%)
Query: 91 PIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHL 150
P+ +VDPRH +S N+APV ++ + V G +P ++GVY+RNG NP F P +HL
Sbjct: 102 PLPKTVDPRHQISGNYAPVPEQSVKSSLSV-DGKIPDCIDGVYLRNGANPLFEPVSGHHL 160
Query: 151 FDGDGMLHAVKITNGKATLCSRYVQTYKYKIENEAGYQILPNVFSGFNSLIGLAARGSVA 210
FDGDGM+HAVKITNG A+ R+ +T + E + G I P + G+A R +
Sbjct: 161 FDGDGMVHAVKITNGDASYSCRFTETERLVQEKQLGSPIFPKAIGELHGHSGIA-RLMLF 219
Query: 211 VARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQTIGRYDFNGK 270
AR L G N NG G+ N L F +RL A+ E DLPY+++VT NGD++TIGR+DF+G+
Sbjct: 220 YARGLFGLLNHKNGTGVANAGLVYFHDRLLAMSEDDLPYQVRVTDNGDLETIGRFDFDGQ 279
Query: 271 LFMNMTAHPKIDSDTGECFAYRYGLI-RPFLTYFRFDSNGVKHNDVPVFSMKRPSFLHDF 329
L M AHPKID T E FA Y ++ +P+L YF+F G K DV + + P+ +HDF
Sbjct: 280 LSSAMIAHPKIDPVTKELFALSYDVVKKPYLKYFKFSPEGEKSPDVEI-PLASPTMMHDF 338
Query: 330 AITKKYALFADIQLEINPLDMIFGGSPVRSDPSKVPRIGILPRYADNESKMKWFDVPGLN 389
AIT+ + + D Q+ DM G SPV+ D K+ R GILPR A + S+M W + P
Sbjct: 339 AITENFVVIPDQQVVFKLSDMFLGKSPVKYDGEKISRFGILPRNAKDASEMVWVESPETF 398
Query: 390 IMHLINAWDEEDGKTVTIVAPNILCVEHMMERL-ELIHEMIEKVRINVETGIVTRQPL-- 446
HL NAW+ + V ++ + + + E ++ ++ ++R+N++TG TR+ +
Sbjct: 399 CFHLWNAWESPETDEVVVIGSCMTPADSIFNECDEQLNSVLSEIRLNLKTGKSTRRTIIP 458
Query: 447 --SARNLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGCRLYGEDCYG 504
NL+ ++N LG+K R+ Y AI P PK SG K+D+ G EV YG YG
Sbjct: 459 GSVQMNLEAGMVNRNLLGRKTRYAYLAIAEPWPKVSGFAKVDLSTG-EVKNHFYGGKKYG 517
Query: 505 GEPFFVARE-DGEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRRVPYGF 563
GEPFF+ R + + EDDGY++S+VHDE+ ES +++A + E E A VKLP RVPYGF
Sbjct: 518 GEPFFLPRGLESDGEDDGYIMSFVHDEESWESELHIVNAVTLELE--ATVKLPSRVPYGF 575
Query: 564 HGLFVKESDI 573
HG FV +D+
Sbjct: 576 HGTFVNSADM 585
>UniRef100_Q5URR0 9-cis-epoxycarotenoid dioxygenase [Brassica rapa subsp. pekinensis]
Length = 562
Score = 370 bits (949), Expect = e-101
Identities = 205/549 (37%), Positives = 311/549 (56%), Gaps = 19/549 (3%)
Query: 33 PQTSQPKTLRTTPTKTLLALLTTTTQKLKNTNKTLQTSSISALILNTLDD-IINTFIDPP 91
P PK T+P A++ K +T + +A L+ + +++ P
Sbjct: 19 PALHFPKQSSTSP-----AIIVNPKTKESDTKQMSLFQRAAAAALDAAEGFLVSHERQHP 73
Query: 92 IKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLF 151
+ + DP ++ NFAPV +E P + + G +P S+ GVY+RNG NP P +H F
Sbjct: 74 LPKTADPSVQIAGNFAPV-NEQPLRRNLPVVGKIPDSIKGVYVRNGANPLHEPVTGHHFF 132
Query: 152 DGDGMLHAVKITNGKATLCSRYVQTYKYKIENEAGYQILPNVFSGFNSLIGLAARGSVAV 211
DGDGM+HAV +G A+ R+ T ++ E + G + P + G+A R +
Sbjct: 133 DGDGMVHAVTFEDGSASYACRFTLTNRFIQERQLGRPVFPKAIGELHGHTGIA-RLMLFY 191
Query: 212 ARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQTIGRYDFNGKL 271
AR G +P +G G+ N L F NRL A+ E DLPY++++TP GD++T+GRYDF+G+L
Sbjct: 192 ARAAAGLVDPAHGTGVANAGLVYFNNRLLAMSEDDLPYQVRITPGGDLKTVGRYDFDGQL 251
Query: 272 FMNMTAHPKIDSDTGECFAYRYGLI-RPFLTYFRFDSNGVKHNDVPVFSMKRPSFLHDFA 330
M AHPK+D ++GE FA Y ++ +P+L YFRF +G K DV + + +P+ +HD
Sbjct: 252 ESTMIAHPKVDPESGELFALSYDVVSKPYLKYFRFSPDGEKSPDVEI-QLDQPTIMHDLP 310
Query: 331 ITKKYALFADIQLEINPLDMIFGGSPVRSDPSKVPRIGILPRYADNESKMKWFDVPGLNI 390
IT+ + + D Q+ +MI GGSPV D KV R GIL +YA + S ++W + P
Sbjct: 311 ITENFVVLPDQQVVFKLQEMIRGGSPVIYDKEKVARFGILDKYAADSSGIRWIEAPDCFC 370
Query: 391 MHLINAWDEEDGKTVTIVAPNILCVEHMM-ERLELIHEMIEKVRINVETGIVTRQPLSAR 449
HL NAW+E + + V ++ + + + E E + ++ ++R+N+ TG TR+P+ +
Sbjct: 371 FHLWNAWEEPETEEVVVIGSCMTPPDSIFNESDENLKSVLSEIRLNLRTGESTRRPIISD 430
Query: 450 -----NLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGCRLYGEDCYG 504
NL+ ++N LG+K +F Y A+ P PK SG K+D+ G EV LYG+D YG
Sbjct: 431 GDQQVNLEAGMVNRNMLGRKTKFAYLALAEPWPKVSGFAKVDLATG-EVKKHLYGDDRYG 489
Query: 505 GEPFFVAREDGEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRRVPYGFH 564
GEP F+ E EDDG+++ +VHDEK +S V+DAK+ E VA V+LP RVPYGFH
Sbjct: 490 GEPLFLPGEGA--EDDGHILCFVHDEKTWKSCVNVIDAKTMSAEPVAVVELPHRVPYGFH 547
Query: 565 GLFVKESDI 573
FV E +
Sbjct: 548 AFFVTEEQL 556
>UniRef100_Q9AXZ4 9-cis-epoxycarotenoid dioxygenase [Persea americana]
Length = 625
Score = 368 bits (945), Expect = e-100
Identities = 200/494 (40%), Positives = 288/494 (57%), Gaps = 16/494 (3%)
Query: 91 PIKPSVDPRHVLSQNFAPVLDELPPTQCKV-IKGTLPPSLNGVYIRNGPNPQFLPRGPYH 149
P+ + DP ++ NFAPV + P Q + + G +P L+GVY+RNG NP F P +H
Sbjct: 137 PLPKTADPEVQIAGNFAPVAEH--PVQHGIPVAGRIPRCLDGVYVRNGANPLFEPIAGHH 194
Query: 150 LFDGDGMLHAVKITNGKATLCSRYVQTYKYKIENEAGYQILPNVFSGFNSLIGLAARGSV 209
FDGDGM+HAV+ NG A+ RY +T + E + I P + G+A R +
Sbjct: 195 FFDGDGMIHAVRFRNGSASYSCRYTETRRLVQERQLSRPIFPKAIGELHGHSGIA-RLLL 253
Query: 210 AVARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQTIGRYDFNG 269
R L G N G+G+ N L F RL A+ E DLPY +++TP+GD++T+GR+DF+
Sbjct: 254 FYTRGLFGLVNADEGMGVANAGLVYFNRRLLAMSEDDLPYHVRITPSGDLKTVGRHDFDN 313
Query: 270 KLFMNMTAHPKIDSDTGECFAYRYGLIR-PFLTYFRFDSNGVKHNDVPVFSMKRPSFLHD 328
+L +M AHPK+D ++GE F+ Y + R P+L YF F +G K DV + + RP+ +HD
Sbjct: 314 QLRSSMIAHPKLDPESGELFSLSYDVARKPYLKYFHFAPDGWKSPDVEI-PLDRPTMIHD 372
Query: 329 FAITKKYALFADIQLEINPLDMIFGGSPVRSDPSKVPRIGILPRYADNESKMKWFDVPGL 388
FAIT+ + + D Q+ +MI GGSPV D +K R GILP+YA + S+M W D P
Sbjct: 373 FAITENFVVIPDQQVVFKLEEMIRGGSPVVYDKNKTSRFGILPKYAPDASEMIWVDAPDC 432
Query: 389 NIMHLINAWDEEDGKTVTIVAPNILCVEHMM-ERLELIHEMIEKVRINVETGIVTRQ--- 444
HL NAW+E + V +V + + + E E + ++ ++R+N TG TR+
Sbjct: 433 FCFHLWNAWEEPESGEVVVVGSCMTPPDSIFNENEESLKSILTEIRLNTRTGESTRRTII 492
Query: 445 -PLSARNLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGCRLYGEDCY 503
P NL+ ++N LG+K RF Y AI P PK SG+ K+D+ G EV +YGE +
Sbjct: 493 DPQKPLNLEAGMVNRNRLGRKTRFAYLAIAEPWPKVSGIAKVDLGTG-EVNRFVYGERQF 551
Query: 504 GGEPFFVAREDGE--EEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRRVPY 561
GGEP+F+ RE EDDGY+VS++HDEK S L+++A + E A V LP RVPY
Sbjct: 552 GGEPYFIPREPSTSGREDDGYVVSFMHDEKTSRSELLILNAMNMRLE--ASVMLPSRVPY 609
Query: 562 GFHGLFVKESDITR 575
GFHG F+ D+ +
Sbjct: 610 GFHGTFISSRDLAK 623
>UniRef100_Q9M9F5 F3F9.10 [Arabidopsis thaliana]
Length = 657
Score = 366 bits (939), Expect = 1e-99
Identities = 211/567 (37%), Positives = 317/567 (55%), Gaps = 19/567 (3%)
Query: 19 IPQFNTASVRTKEKPQTS-QPKTLRTTPTKTLLALLTTTTQKLKNTNKTLQTSSISALIL 77
+P + ++SV K +S Q T + K L +T K N N L +A ++
Sbjct: 96 VPCYVSSSVNKKSSVSSSLQSPTFKPPSWKKLCNDVTNLIPKTTNQNPKLNPVQRTAAMV 155
Query: 78 NTLDDIINTFIDP-----PIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGV 132
LD + N I P + DP ++ NF PV E P + GT+P + GV
Sbjct: 156 --LDAVENAMISHERRRHPHPKTADPAVQIAGNFFPV-PEKPVVHNLPVTGTVPECIQGV 212
Query: 133 YIRNGPNPQFLPRGPYHLFDGDGMLHAVKITNGKATLCSRYVQTYKYKIENEAGYQILPN 192
Y+RNG NP P +HLFDGDGM+HAV+ NG + R+ +T + E E G + P
Sbjct: 213 YVRNGANPLHKPVSGHHLFDGDGMVHAVRFDNGSVSYACRFTETNRLVQERECGRPVFPK 272
Query: 193 VFSGFNSLIGLAARGSVAVARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIK 252
+ +G+A + + R L G +P G+G+ N L F L A+ E DLPY +K
Sbjct: 273 AIGELHGHLGIA-KLMLFNTRGLFGLVDPTGGLGVANAGLVYFNGHLLAMSEDDLPYHVK 331
Query: 253 VTPNGDIQTIGRYDFNGKLFMNMTAHPKIDSDTGECFAYRYGLI-RPFLTYFRFDSNGVK 311
VT GD++T GRYDF+G+L M AHPKID +T E FA Y ++ +P+L YFRF S+G K
Sbjct: 332 VTQTGDLETSGRYDFDGQLKSTMIAHPKIDPETRELFALSYDVVSKPYLKYFRFTSDGEK 391
Query: 312 HNDVPVFSMKRPSFLHDFAITKKYALFADIQLEINPLDMIFGGSPVRSDPSKVPRIGILP 371
DV + + +P+ +HDFAIT+ + + D Q+ +MI GGSPV D K R GIL
Sbjct: 392 SPDVEI-PLDQPTMIHDFAITENFVVIPDQQVVFRLPEMIRGGSPVVYDEKKKSRFGILN 450
Query: 372 RYADNESKMKWFDVPGLNIMHLINAWDEEDGKTVTIVAPNILCVEHMM-ERLELIHEMIE 430
+ A + S ++W +VP HL N+W+E + V ++ + + + E E + ++
Sbjct: 451 KNAKDASSIQWIEVPDCFCFHLWNSWEEPETDEVVVIGSCMTPPDSIFNEHDETLQSVLS 510
Query: 431 KVRINVETGIVTRQPLSAR--NLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVL 488
++R+N++TG TR+P+ + NL+ ++N LG+K R+ Y A+ P PK SG K+D+
Sbjct: 511 EIRLNLKTGESTRRPVISEQVNLEAGMVNRNLLGRKTRYAYLALTEPWPKVSGFAKVDLS 570
Query: 489 KGEEVGCRLYGEDCYGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFE 548
G E+ +YGE YGGEP F+ DG EED GY++ +VHDE+K +S +++A + + E
Sbjct: 571 TG-EIRKYIYGEGKYGGEPLFLPSGDG-EEDGGYIMVFVHDEEKVKSELQLINAVNMKLE 628
Query: 549 IVAEVKLPRRVPYGFHGLFVKESDITR 575
A V LP RVPYGFHG F+ + D+++
Sbjct: 629 --ATVTLPSRVPYGFHGTFISKEDLSK 653
>UniRef100_O49505 Neoxanthin cleavage enzyme - like protein [Arabidopsis thaliana]
Length = 583
Score = 365 bits (937), Expect = 2e-99
Identities = 209/560 (37%), Positives = 317/560 (56%), Gaps = 24/560 (4%)
Query: 32 KPQTSQPKTLRTT---------PTKTLLALLTTTTQKLKNTNKTLQTSSISALILNTLDD 82
+P QPK ++ T + L TT T N + +A+ ++ +
Sbjct: 26 RPIKRQPKVIKCTVQIDVTELTKKRQLFTPRTTATPPQHNPLRLNIFQKAAAIAIDAAER 85
Query: 83 -IINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQ 141
+I+ D P+ + DPR ++ N++PV E + ++GT+P ++GVYIRNG NP
Sbjct: 86 ALISHEQDSPLPKTADPRVQIAGNYSPV-PESSVRRNLTVEGTIPDCIDGVYIRNGANPM 144
Query: 142 FLPRGPYHLFDGDGMLHAVKITNGKATLCSRYVQTYKYKIENEAGYQILPNVFSGFNSLI 201
F P +HLFDGDGM+HAVKITNG A+ R+ +T + E G + P +
Sbjct: 145 FEPTAGHHLFDGDGMVHAVKITNGSASYACRFTKTERLVQEKRLGRPVFPKAIGELHGHS 204
Query: 202 GLAARGSVAVARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQT 261
G+A R + AR L G N NG+G+ N L F NRL A+ E DLPY++K+T GD+QT
Sbjct: 205 GIA-RLMLFYARGLCGLINNQNGVGVANAGLVYFNNRLLAMSEDDLPYQLKITQTGDLQT 263
Query: 262 IGRYDFNGKLFMNMTAHPKIDSDTGECFAYRYGLI-RPFLTYFRFDSNGVKHNDVPVFSM 320
+GRYDF+G+L M AHPK+D T E A Y ++ +P+L YFRF +GVK ++ + +
Sbjct: 264 VGRYDFDGQLKSAMIAHPKLDPVTKELHALSYDVVKKPYLKYFRFSPDGVKSPELEI-PL 322
Query: 321 KRPSFLHDFAITKKYALFADIQLEINPLDMIFGGSPVRSDPSKVPRIGILPRYADNESKM 380
+ P+ +HDFAIT+ + + D Q+ +MI G SPV D KV R+GI+P+ A S++
Sbjct: 323 ETPTMIHDFAITENFVVIPDQQVVFKLGEMISGKSPVVFDGEKVSRLGIMPKDATEASQI 382
Query: 381 KWFDVPGLNIMHLINAWDEEDGKTVTIVAPNILCVEHMM-ERLELIHEMIEKVRINVETG 439
W + P HL NAW+ + + + ++ + + + ER E + ++ ++RIN+ T
Sbjct: 383 IWVNSPETFCFHLWNAWESPETEEIVVIGSCMSPADSIFNERDESLRSVLSEIRINLRTR 442
Query: 440 IVTRQPLSAR---NLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGCR 496
TR+ L NL++ ++N LG+K RF + AI P PK SG K+D+ G E+
Sbjct: 443 KTTRRSLLVNEDVNLEIGMVNRNRLGRKTRFAFLAIAYPWPKVSGFAKVDLCTG-EMKKY 501
Query: 497 LYGEDCYGGEPFFVAREDG---EEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEV 553
+YG + YGGEPFF+ G E EDDGY+ +VHDE+ S +++A + + E A +
Sbjct: 502 IYGGEKYGGEPFFLPGNSGNGEENEDDGYIFCHVHDEETKTSELQIINAVNLKLE--ATI 559
Query: 554 KLPRRVPYGFHGLFVKESDI 573
KLP RVPYGFHG FV +++
Sbjct: 560 KLPSRVPYGFHGTFVDSNEL 579
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.140 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,043,205,301
Number of Sequences: 2790947
Number of extensions: 48735364
Number of successful extensions: 113668
Number of sequences better than 10.0: 378
Number of HSP's better than 10.0 without gapping: 241
Number of HSP's successfully gapped in prelim test: 137
Number of HSP's that attempted gapping in prelim test: 111590
Number of HSP's gapped (non-prelim): 957
length of query: 579
length of database: 848,049,833
effective HSP length: 133
effective length of query: 446
effective length of database: 476,853,882
effective search space: 212676831372
effective search space used: 212676831372
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 78 (34.7 bits)
Medicago: description of AC144759.6


