

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144731.8 - phase: 0
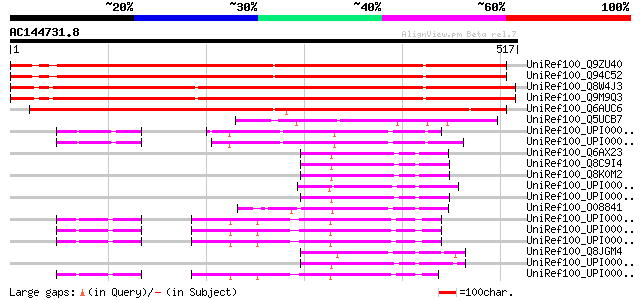
(517 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9ZU40 Expressed protein [Arabidopsis thaliana] 711 0.0
UniRef100_Q94C52 Hypothetical protein At2g01270 [Arabidopsis tha... 708 0.0
UniRef100_Q8W4J3 Hypothetical protein At1g15020; T15D22.8 [Arabi... 693 0.0
UniRef100_Q9M9Q3 T15D22.7 protein [Arabidopsis thaliana] 693 0.0
UniRef100_Q6AUC6 Hypothetical protein OSJNBa0079H23.16 [Oryza sa... 627 e-178
UniRef100_Q5UCB7 Thioredoxin-like protein [Chlamydomonas reinhar... 135 4e-30
UniRef100_UPI0000362968 UPI0000362968 UniRef100 entry 124 7e-27
UniRef100_UPI0000362967 UPI0000362967 UniRef100 entry 124 9e-27
UniRef100_Q6AX23 MGC86371 protein [Xenopus laevis] 118 4e-25
UniRef100_Q8C9I4 Mus musculus 3 days neonate thymus cDNA, RIKEN ... 115 2e-24
UniRef100_Q8K0M2 Quiescin Q6-like 1 [Mus musculus] 115 2e-24
UniRef100_UPI00003AB2AE UPI00003AB2AE UniRef100 entry 115 4e-24
UniRef100_UPI00001CF07C UPI00001CF07C UniRef100 entry 114 9e-24
UniRef100_O08841 GEC-3 [Cavia porcellus] 113 1e-23
UniRef100_UPI00004322F6 UPI00004322F6 UniRef100 entry 112 2e-23
UniRef100_UPI00004322F5 UPI00004322F5 UniRef100 entry 112 2e-23
UniRef100_UPI00004322F3 UPI00004322F3 UniRef100 entry 112 2e-23
UniRef100_Q8JGM4 Quiescin/sulfhydryl oxidase [Gallus gallus] 112 3e-23
UniRef100_UPI0000246FED UPI0000246FED UniRef100 entry 110 8e-23
UniRef100_UPI00004322F4 UPI00004322F4 UniRef100 entry 110 8e-23
>UniRef100_Q9ZU40 Expressed protein [Arabidopsis thaliana]
Length = 495
Score = 711 bits (1834), Expect = 0.0
Identities = 343/507 (67%), Positives = 409/507 (80%), Gaps = 17/507 (3%)
Query: 1 MNLLYFALFLCTISVSSSATSSSSSSFPGRSILREVHDNDQTGTTDHDYAVELNATNFDA 60
M+L++ LF + +SS++ S R ILRE+ D D AVELN TNFD+
Sbjct: 1 MSLVHLLLFAGLVIAASSSSPGS------RLILREISDQK-------DKAVELNTTNFDS 47
Query: 61 VLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLTRVDCASKINI 120
VLKDTPA +AVVEFFA+WCPACRNYKPHYEKVA+LFNGPDA+HPGI+L+TRVDCA K N
Sbjct: 48 VLKDTPAKYAVVEFFAHWCPACRNYKPHYEKVARLFNGPDAIHPGIVLMTRVDCAMKTNT 107
Query: 121 KLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDARTADRLLNWINKQMSSSF 180
KLCDKFSV HYPMLFWGPP KFV GSWEPK++KS+I VIDD RTA+RLLNWINKQ+ SS+
Sbjct: 108 KLCDKFSVSHYPMLFWGPPTKFVSGSWEPKKDKSEILVIDDGRTAERLLNWINKQIGSSY 167
Query: 181 GLDDQKFQNEHLSSNVSDPEQIARAIYDVEEATSLAFDIILENKMIK-PETRASLVKFLQ 239
GLDDQKF+NEH SN++D QI++A+YDVEEAT+ AFDIIL +K IK ET AS ++F+Q
Sbjct: 168 GLDDQKFKNEHALSNLTDYNQISQAVYDVEEATAEAFDIILAHKAIKSSETSASFIRFIQ 227
Query: 240 VLTAHHPSRRCRKGAGDLLVSFADLYPTDFWSSHKQEDDKSSVKNFQICGKEVPRGYWMF 299
+L AHH SRRCRKGA ++LV++ DL P+ S+++ ++ NF ICGK+VPRGY+MF
Sbjct: 228 LLAAHHLSRRCRKGAAEILVNYDDLCPSGN-CSYEKSGGNDTLGNFPICGKDVPRGYYMF 286
Query: 300 CRGSKNETRGFSCGLWVLLHSLSVRIEDGESQFTFNAICDFVHNFFVCEECRQHFYDMCS 359
CRGSKN+TRGFSCGLWVL+HSLSVRIEDGES F F ICDFV+NFF+C+ECR HF DMC
Sbjct: 287 CRGSKNDTRGFSCGLWVLMHSLSVRIEDGESHFAFTTICDFVNNFFMCDECRLHFNDMCL 346
Query: 360 SVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIWPTKQLCPSCYLGHD 419
SV TPF KARDFVLW+WS+HNKVNERL K+EASLGTGDPKFPK IWP K+LCP CYL +
Sbjct: 347 SVKTPFKKARDFVLWVWSTHNKVNERLLKDEASLGTGDPKFPKIIWPPKELCPLCYLSSN 406
Query: 420 QKSNKIEWNQDEVYKALKNYYEKTLVSLYKEKDIAGNDGTKGAALEDLIVGTNAVVVPVG 479
QKS IEW+ + VYK LKNYY LVSLYKEK ++ + +A EDL V TNA+VVP+G
Sbjct: 407 QKS--IEWDHEHVYKFLKNYYGPKLVSLYKEKSVSRSKEETVSATEDLTVATNALVVPIG 464
Query: 480 AALAIAVASCAFGALACYWRSQQKSRK 506
AALAIA+ASCAFGALACYWR+QQK+RK
Sbjct: 465 AALAIAIASCAFGALACYWRTQQKNRK 491
>UniRef100_Q94C52 Hypothetical protein At2g01270 [Arabidopsis thaliana]
Length = 495
Score = 708 bits (1827), Expect = 0.0
Identities = 342/507 (67%), Positives = 408/507 (80%), Gaps = 17/507 (3%)
Query: 1 MNLLYFALFLCTISVSSSATSSSSSSFPGRSILREVHDNDQTGTTDHDYAVELNATNFDA 60
M+L++ LF + +SS++ S R ILRE+ D D AVELN TNFD+
Sbjct: 1 MSLVHLLLFAGLVIAASSSSPGS------RLILREISDQK-------DKAVELNTTNFDS 47
Query: 61 VLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLTRVDCASKINI 120
VLKDTPA +AVVEFFA+WCPACRNYKPHYEKVA+LFN PDA+HPGI+L+TRVDCA K N
Sbjct: 48 VLKDTPAKYAVVEFFAHWCPACRNYKPHYEKVARLFNDPDAIHPGIVLMTRVDCAMKTNT 107
Query: 121 KLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDARTADRLLNWINKQMSSSF 180
KLCDKFSV HYPMLFWGPP KFV GSWEPK++KS+I VIDD RTA+RLLNWINKQ+ SS+
Sbjct: 108 KLCDKFSVSHYPMLFWGPPTKFVSGSWEPKKDKSEILVIDDGRTAERLLNWINKQIGSSY 167
Query: 181 GLDDQKFQNEHLSSNVSDPEQIARAIYDVEEATSLAFDIILENKMIK-PETRASLVKFLQ 239
GLDDQKF+NEH SN++D QI++A+YDVEEAT+ AFDIIL +K IK ET AS ++F+Q
Sbjct: 168 GLDDQKFKNEHALSNLTDYNQISQAVYDVEEATAEAFDIILAHKAIKSSETSASFIRFIQ 227
Query: 240 VLTAHHPSRRCRKGAGDLLVSFADLYPTDFWSSHKQEDDKSSVKNFQICGKEVPRGYWMF 299
+L AHH SRRCRKGA ++LV++ DL P+ S+++ ++ NF ICGK+VPRGY+MF
Sbjct: 228 LLAAHHLSRRCRKGAAEILVNYDDLCPSGN-CSYEKSGGNDTLGNFPICGKDVPRGYYMF 286
Query: 300 CRGSKNETRGFSCGLWVLLHSLSVRIEDGESQFTFNAICDFVHNFFVCEECRQHFYDMCS 359
CRGSKN+TRGFSCGLWVL+HSLSVRIEDGES F F ICDFV+NFF+C+ECR HF DMC
Sbjct: 287 CRGSKNDTRGFSCGLWVLMHSLSVRIEDGESHFAFTTICDFVNNFFMCDECRLHFNDMCL 346
Query: 360 SVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIWPTKQLCPSCYLGHD 419
SV TPF KARDFVLW+WS+HNKVNERL K+EASLGTGDPKFPK IWP K+LCP CYL +
Sbjct: 347 SVKTPFKKARDFVLWVWSTHNKVNERLLKDEASLGTGDPKFPKIIWPPKELCPLCYLSSN 406
Query: 420 QKSNKIEWNQDEVYKALKNYYEKTLVSLYKEKDIAGNDGTKGAALEDLIVGTNAVVVPVG 479
QKS IEW+ + VYK LKNYY LVSLYKEK ++ + +A EDL V TNA+VVP+G
Sbjct: 407 QKS--IEWDHEHVYKFLKNYYGPKLVSLYKEKSVSRSKEETVSATEDLTVATNALVVPIG 464
Query: 480 AALAIAVASCAFGALACYWRSQQKSRK 506
AALAIA+ASCAFGALACYWR+QQK+RK
Sbjct: 465 AALAIAIASCAFGALACYWRTQQKNRK 491
>UniRef100_Q8W4J3 Hypothetical protein At1g15020; T15D22.8 [Arabidopsis thaliana]
Length = 528
Score = 693 bits (1788), Expect = 0.0
Identities = 332/516 (64%), Positives = 414/516 (79%), Gaps = 12/516 (2%)
Query: 1 MNLLYFALFLCTISVSSSATSSSSSSFPGRSILREVHDNDQTGTTDHDYAVELNATNFDA 60
M+L++ L L +S+ ++A+ S S RSILR++ N D A+ELNATNFD+
Sbjct: 1 MSLIHLFLLLGLLSLEAAASFSPGS----RSILRDIGSNV---ADQKDNAIELNATNFDS 53
Query: 61 VLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLTRVDCASKINI 120
V +D+PA +AV+EFFA+WCPACRNYKPHYEKVA+LFNG DAV+PG++L+TRVDCA K+N+
Sbjct: 54 VFQDSPAKYAVLEFFAHWCPACRNYKPHYEKVARLFNGADAVYPGVVLMTRVDCAIKMNV 113
Query: 121 KLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDARTADRLLNWINKQMSSSF 180
KLCDKFS+ HYPMLFW PP +FVGGSW PKQEK++I V+++ RTAD LLNWINKQ+ SS+
Sbjct: 114 KLCDKFSINHYPMLFWAPPKRFVGGSWGPKQEKNEISVVNEWRTADLLLNWINKQIGSSY 173
Query: 181 GLDDQKFQNEHLSSNVSDPEQIARAIYDVEEATSLAFDIILENKMIKP-ETRASLVKFLQ 239
GLDDQK N L SN+SD EQI++AI+D+EEAT AFDIIL +K IK ET AS ++FLQ
Sbjct: 174 GLDDQKLGN--LLSNISDQEQISQAIFDIEEATEEAFDIILAHKAIKSSETSASFIRFLQ 231
Query: 240 VLTAHHPSRRCRKGAGDLLVSFADLYPTDFWSSHKQEDDKSSVKNFQICGKEVPRGYWMF 299
+L AHHPSRRCR G+ ++LV+F D+ P+ S ++ K S++NF ICGK+VPRGY+ F
Sbjct: 232 LLVAHHPSRRCRTGSAEILVNFDDICPSGECSYDQESGAKDSLRNFHICGKDVPRGYYRF 291
Query: 300 CRGSKNETRGFSCGLWVLLHSLSVRIEDGESQFTFNAICDFVHNFFVCEECRQHFYDMCS 359
CRGSKNETRGFSCGLWVL+HSLSVRIEDGESQF F AICDF++NFF+C++CR+HF+DMC
Sbjct: 292 CRGSKNETRGFSCGLWVLMHSLSVRIEDGESQFAFTAICDFINNFFMCDDCRRHFHDMCL 351
Query: 360 SVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIWPTKQLCPSCYLGHD 419
SV TPF KARD LWLWS+HNKVNERL K+E SLGTGDPKFPK IWP KQLCPSCYL
Sbjct: 352 SVKTPFKKARDIALWLWSTHNKVNERLKKDEDSLGTGDPKFPKMIWPPKQLCPSCYLSST 411
Query: 420 QKSNKIEWNQDEVYKALKNYYEKTLVSLYKEKDIAGNDGTKGAALEDLIVGTNAVVVPVG 479
+K+ I+W+ D+VYK LK YY + LVS+YK+ + + AA E++ V TNA+VVPVG
Sbjct: 412 EKN--IDWDHDQVYKFLKKYYGQKLVSVYKKNGESVSKEEVIAAAEEMAVPTNALVVPVG 469
Query: 480 AALAIAVASCAFGALACYWRSQQKSRKYFHHLHSLK 515
AALAIA+ASCAFGALACYWR+QQK+RKY ++ H LK
Sbjct: 470 AALAIALASCAFGALACYWRTQQKNRKYNYNPHYLK 505
>UniRef100_Q9M9Q3 T15D22.7 protein [Arabidopsis thaliana]
Length = 536
Score = 693 bits (1788), Expect = 0.0
Identities = 332/516 (64%), Positives = 414/516 (79%), Gaps = 12/516 (2%)
Query: 1 MNLLYFALFLCTISVSSSATSSSSSSFPGRSILREVHDNDQTGTTDHDYAVELNATNFDA 60
M+L++ L L +S+ ++A+ S S RSILR++ N D A+ELNATNFD+
Sbjct: 1 MSLIHLFLLLGLLSLEAAASFSPGS----RSILRDIGSNV---ADQKDNAIELNATNFDS 53
Query: 61 VLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLTRVDCASKINI 120
V +D+PA +AV+EFFA+WCPACRNYKPHYEKVA+LFNG DAV+PG++L+TRVDCA K+N+
Sbjct: 54 VFQDSPAKYAVLEFFAHWCPACRNYKPHYEKVARLFNGADAVYPGVVLMTRVDCAIKMNV 113
Query: 121 KLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDARTADRLLNWINKQMSSSF 180
KLCDKFS+ HYPMLFW PP +FVGGSW PKQEK++I V+++ RTAD LLNWINKQ+ SS+
Sbjct: 114 KLCDKFSINHYPMLFWAPPKRFVGGSWGPKQEKNEISVVNEWRTADLLLNWINKQIGSSY 173
Query: 181 GLDDQKFQNEHLSSNVSDPEQIARAIYDVEEATSLAFDIILENKMIKP-ETRASLVKFLQ 239
GLDDQK N L SN+SD EQI++AI+D+EEAT AFDIIL +K IK ET AS ++FLQ
Sbjct: 174 GLDDQKLGN--LLSNISDQEQISQAIFDIEEATEEAFDIILAHKAIKSSETSASFIRFLQ 231
Query: 240 VLTAHHPSRRCRKGAGDLLVSFADLYPTDFWSSHKQEDDKSSVKNFQICGKEVPRGYWMF 299
+L AHHPSRRCR G+ ++LV+F D+ P+ S ++ K S++NF ICGK+VPRGY+ F
Sbjct: 232 LLVAHHPSRRCRTGSAEILVNFDDICPSGECSYDQESGAKDSLRNFHICGKDVPRGYYRF 291
Query: 300 CRGSKNETRGFSCGLWVLLHSLSVRIEDGESQFTFNAICDFVHNFFVCEECRQHFYDMCS 359
CRGSKNETRGFSCGLWVL+HSLSVRIEDGESQF F AICDF++NFF+C++CR+HF+DMC
Sbjct: 292 CRGSKNETRGFSCGLWVLMHSLSVRIEDGESQFAFTAICDFINNFFMCDDCRRHFHDMCL 351
Query: 360 SVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIWPTKQLCPSCYLGHD 419
SV TPF KARD LWLWS+HNKVNERL K+E SLGTGDPKFPK IWP KQLCPSCYL
Sbjct: 352 SVKTPFKKARDIALWLWSTHNKVNERLKKDEDSLGTGDPKFPKMIWPPKQLCPSCYLSST 411
Query: 420 QKSNKIEWNQDEVYKALKNYYEKTLVSLYKEKDIAGNDGTKGAALEDLIVGTNAVVVPVG 479
+K+ I+W+ D+VYK LK YY + LVS+YK+ + + AA E++ V TNA+VVPVG
Sbjct: 412 EKN--IDWDHDQVYKFLKKYYGQKLVSVYKKNGESVSKEEVIAAAEEMAVPTNALVVPVG 469
Query: 480 AALAIAVASCAFGALACYWRSQQKSRKYFHHLHSLK 515
AALAIA+ASCAFGALACYWR+QQK+RKY ++ H LK
Sbjct: 470 AALAIALASCAFGALACYWRTQQKNRKYNYNPHYLK 505
>UniRef100_Q6AUC6 Hypothetical protein OSJNBa0079H23.16 [Oryza sativa]
Length = 513
Score = 627 bits (1618), Expect = e-178
Identities = 299/492 (60%), Positives = 367/492 (73%), Gaps = 8/492 (1%)
Query: 21 SSSSSSFPGRSILREVHDNDQTGTTDHDYAVELNATNFDAVLKDTPATFAVVEFFANWCP 80
++S ++ P + R + + G D D AV+LNATNFDA LK + +AVVEFFA+WCP
Sbjct: 18 AASLAAAPRGAAARSLGGREGPGEVDADAAVDLNATNFDAFLKASLEPWAVVEFFAHWCP 77
Query: 81 ACRNYKPHYEKVAKLFNGPDAVHPGIILLTRVDCASKINIKLCDKFSVGHYPMLFWGPPP 140
ACRNYKPHYEKVAKLFNG DA HPG+IL+ RVDCASK+NI LC++FSV HYP L WGPP
Sbjct: 78 ACRNYKPHYEKVAKLFNGRDAAHPGLILMARVDCASKVNIDLCNRFSVDHYPFLLWGPPT 137
Query: 141 KFVGGSWEPKQEKSDIHVIDDARTADRLLNWINKQMSSSFGLDDQKFQNEH-LSSNVSDP 199
KF W+PKQE ++I +IDD RTA+RLL WIN QM SSF L+D+K++NE+ L N SDP
Sbjct: 138 KFASAKWDPKQENNEIKLIDDGRTAERLLKWINNQMKSSFSLEDKKYENENMLPKNASDP 197
Query: 200 EQIARAIYDVEEATSLAFDIILENKMIKPETRASLVKFLQVLTAHHPSRRCRKGAGDLLV 259
EQI +AIYDVEEAT+ A IILE K IKP+ R SL++FLQ+L A HPS+RCR+G+ +LL+
Sbjct: 198 EQIVQAIYDVEEATAQALQIILERKTIKPKNRDSLIRFLQILVARHPSKRCRRGSAELLI 257
Query: 260 SFADLYPTDFWSSHKQEDDKS----SVKNFQICGKEVPRGYWMFCRGSKNETRGFSCGLW 315
+F D + ++ S QE K + +N ICGKEVPRGYW+FCRGSK+ETRGFSCGLW
Sbjct: 258 NFDDHWSSNL-SLSSQEGSKLLESVAEENHWICGKEVPRGYWLFCRGSKSETRGFSCGLW 316
Query: 316 VLLHSLSVRIEDGESQFTFNAICDFVHNFFVCEECRQHFYDMCSSVSTPFNKARDFVLWL 375
VL+HSL+VRI DGESQ TF +ICDF+HNFF+CEECR+HFY+MCSSVS PF AR+ LWL
Sbjct: 317 VLMHSLTVRIGDGESQSTFTSICDFIHNFFICEECRKHFYEMCSSVSAPFRTARELSLWL 376
Query: 376 WSSHNKVNERLSKEEASLGTGDPKFPKTIWPTKQLCPSCYLGHDQKSNKIEWNQDEVYKA 435
WS+HNKVN RL KEE +GTGDP FPK WP QLCPSCY ++WN+D VY+
Sbjct: 377 WSTHNKVNMRLMKEEKDMGTGDPLFPKVTWPPNQLCPSCYRSSKVTDGAVDWNEDAVYQF 436
Query: 436 LKNYYEKTLVSLYKEKDIAG-NDGTKGAALEDLIVGTNAVVVPVGAALAIAVASCAFGAL 494
L NYY K LVS YKE + K ED + +NA VP+GAAL +A+ASC FGAL
Sbjct: 437 LVNYYGKKLVSSYKETYMESLQQQEKKIVSEDSSI-SNAASVPIGAALGVAIASCTFGAL 495
Query: 495 ACYWRSQQKSRK 506
AC+WR+QQK+RK
Sbjct: 496 ACFWRAQQKNRK 507
>UniRef100_Q5UCB7 Thioredoxin-like protein [Chlamydomonas reinhardtii]
Length = 438
Score = 135 bits (339), Expect = 4e-30
Identities = 90/288 (31%), Positives = 132/288 (45%), Gaps = 29/288 (10%)
Query: 231 RASLVKFLQVLTAHHPSRRCRKGAGDLLVSFADLYPTDFWSSHKQEDDKSSVKNFQICGK 290
R +L L HPS C+ G+ LL + +P ED + +CG
Sbjct: 19 REALQLLLAAWAEAHPSPSCKAGSARLLEGYEAAWPA------AAEDAPPGLTRAALCGP 72
Query: 291 E---VPRGYWMFCRGSKNETRGFSCGLWVLLHSLSVRIEDGESQFTFNAICDFVHNFFVC 347
RG W C GS+ ++RGFSCGLW L+H+L+ R+ S + + F +FF+C
Sbjct: 73 PGGLAWRGLWSACAGSQPDSRGFSCGLWFLIHTLAARMPSPTSVLEY--LRAFNTHFFLC 130
Query: 348 EECRQHFYDMCSSVSTPFNKA--RDFVLWLWSSHNKVNERLSKEEASLG---TGDPKFPK 402
E C++HF + +S A RD VLWLW +HN+VNERL E G TGDP++PK
Sbjct: 131 EPCQKHFGRILASPEAAAATASRRDLVLWLWRTHNEVNERLRGIETRYGHSTTGDPEWPK 190
Query: 403 TIWPTKQLCPSCYLGHDQKSNK--------IEWNQDEVYKALKNYYEKTLV----SLYKE 450
+WP + CP+C + + S W+++ VY+ L Y + KE
Sbjct: 191 EVWPAPEACPACRVQYKPGSGSGSGSGALPGSWDEEAVYQYLTAAYGPGSAPGADGIVKE 250
Query: 451 KDIAGNDGTKGAALEDLIVGTNAVVVPVGAALA-IAVASCAFGALACY 497
+ G G G L + + + V P + + VA AL Y
Sbjct: 251 AAVGGGGGGGGVLLSEKGLYSRRVPPPTSTLYSMLPVAGVLAAALMLY 298
>UniRef100_UPI0000362968 UPI0000362968 UniRef100 entry
Length = 456
Score = 124 bits (311), Expect = 7e-27
Identities = 79/247 (31%), Positives = 123/247 (48%), Gaps = 16/247 (6%)
Query: 201 QIARAIYDVEEATSLAFDIILE---NKMIKPETRASLVKFLQVLTAHHPSRRCRKGAGDL 257
Q++R +Y + ++L + + +E + + SL K++ VL + P R
Sbjct: 213 QVSR-VYMADLESTLDYSLRVELAAHSVFSGHALVSLKKYISVLVKYFPGRPMVMNLLKS 271
Query: 258 LVSFADLYPTDFWSSHKQEDDKSSVKNFQICGKEVPRGY-WMFCRGSKNETRGFSCGLWV 316
L S+ P D S E K Q +P+G W+ C+GS+ RG+ CG+W
Sbjct: 272 LNSWLQDQPGDEISYEALE--KIIDNRAQSPNTTLPQGARWVGCQGSQPHYRGYPCGVWT 329
Query: 317 LLHSLSVRIEDGE---SQFTFNAICDFVHNFFVCEECRQHFYDMCSSVSTPFNKARDFVL 373
L H LSV+ + E ++ + + +VH+FF C C +HF +M + N VL
Sbjct: 330 LFHVLSVQAKKDEGTDAKEVLSTMRGYVHHFFGCRLCAKHFEEMAQKSLSEVNTLSAAVL 389
Query: 374 WLWSSHNKVNERLSKEEASLGTGDPKFPKTIWPTKQLCPSCYLGHDQKSNKIEWNQDEVY 433
WLW HN+VN RL A + DPKFPK WP+ ++CPSC+ + + ++ WNQD V
Sbjct: 390 WLWKRHNQVNNRL----AGALSEDPKFPKIQWPSPEMCPSCHSVMENREHR--WNQDRVL 443
Query: 434 KALKNYY 440
L +YY
Sbjct: 444 SFLLSYY 450
Score = 48.5 bits (114), Expect = 5e-04
Identities = 30/88 (34%), Positives = 45/88 (51%), Gaps = 7/88 (7%)
Query: 48 DYAVELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAK-LFNGPDAVHPGI 106
D + LNA N + VL ++ A V EF+A+WC C + P Y+ +A+ + AV
Sbjct: 2 DQIISLNAENVETVLVNSTAAI-VAEFYASWCGHCVAFSPVYKSLARDIKEWKPAVD--- 57
Query: 107 ILLTRVDCASKINIKLCDKFSVGHYPML 134
L VDCA+ +LC + + YP L
Sbjct: 58 --LAAVDCAATETRQLCFDYGIKGYPTL 83
>UniRef100_UPI0000362967 UPI0000362967 UniRef100 entry
Length = 767
Score = 124 bits (310), Expect = 9e-27
Identities = 82/266 (30%), Positives = 128/266 (47%), Gaps = 17/266 (6%)
Query: 206 IYDVEEATSLAFDIILE---NKMIKPETRASLVKFLQVLTAHHPSRRCRKGAGDLLVSFA 262
+Y + ++L + + +E + + SL K++ VL + P R L S+
Sbjct: 313 VYMADLESTLDYSLRVELAAHSVFSGHALVSLKKYISVLVKYFPGRPMVMNLLKSLNSWL 372
Query: 263 DLYPTDFWSSHKQEDDKSSVKNFQICGKEVPRGY-WMFCRGSKNETRGFSCGLWVLLHSL 321
P D S E K Q +P+G W+ C+GS+ RG+ CG+W L H L
Sbjct: 373 QDQPGDEISYEALE--KIIDNRAQSPNTTLPQGARWVGCQGSQPHYRGYPCGVWTLFHVL 430
Query: 322 SVRIEDGE---SQFTFNAICDFVHNFFVCEECRQHFYDMCSSVSTPFNKARDFVLWLWSS 378
SV+ + E ++ + + +VH+FF C C +HF +M + N VLWLW
Sbjct: 431 SVQAKKDEGTDAKEVLSTMRGYVHHFFGCRLCAKHFEEMAQKSLSEVNTLSAAVLWLWKR 490
Query: 379 HNKVNERLSKEEASLGTGDPKFPKTIWPTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKN 438
HN+VN RL A + DPKFPK WP+ ++CPSC+ + + ++ WNQD V L +
Sbjct: 491 HNQVNNRL----AGALSEDPKFPKIQWPSPEMCPSCHSVMENREHR--WNQDRVLSFLLS 544
Query: 439 YY--EKTLVSLYKEKDIAGNDGTKGA 462
YY + L + + G G +GA
Sbjct: 545 YYSSQDILTAHRALRRGGGGGGRRGA 570
Score = 48.5 bits (114), Expect = 5e-04
Identities = 30/88 (34%), Positives = 45/88 (51%), Gaps = 7/88 (7%)
Query: 48 DYAVELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAK-LFNGPDAVHPGI 106
D + LNA N + VL ++ A V EF+A+WC C + P Y+ +A+ + AV
Sbjct: 52 DQIISLNAENVETVLVNSTAAI-VAEFYASWCGHCVAFSPVYKSLARDIKEWKPAVD--- 107
Query: 107 ILLTRVDCASKINIKLCDKFSVGHYPML 134
L VDCA+ +LC + + YP L
Sbjct: 108 --LAAVDCAATETRQLCFDYGIKGYPTL 133
>UniRef100_Q6AX23 MGC86371 protein [Xenopus laevis]
Length = 661
Score = 118 bits (296), Expect = 4e-25
Identities = 61/162 (37%), Positives = 77/162 (46%), Gaps = 18/162 (11%)
Query: 297 WMFCRGSKNETRGFSCGLWVLLHSLSVRIE-----------DGESQFTFNAICDFVHNFF 345
W+ CRGSK+ RG+ C LW L HSL+V+ + E + + ++ FF
Sbjct: 394 WVGCRGSKSNLRGYPCSLWKLFHSLTVQAAVKPDALANTAFEAEPRAVLQTMRRYIREFF 453
Query: 346 VCEECRQHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIW 405
C EC +HF M +LWLW HN VN RLS + DPKFPK W
Sbjct: 454 GCRECAKHFEAMAKETVDSVKTPDQAILWLWRKHNVVNNRLSGAPSE----DPKFPKVQW 509
Query: 406 PTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYYEKTLVSL 447
PT LC +C H Q WN+DEV LK YY +SL
Sbjct: 510 PTSDLCSAC---HSQTGGVHSWNEDEVLAFLKRYYGNQEISL 548
Score = 40.0 bits (92), Expect = 0.16
Identities = 20/63 (31%), Positives = 35/63 (54%), Gaps = 6/63 (9%)
Query: 71 VVEFFANWCPACRNYKPHYEKVAK-LFNGPDAVHPGIILLTRVDCASKINIKLCDKFSVG 129
++EF+++WC C NY P ++ +A+ + + A+ G++ DCA + N C F V
Sbjct: 59 LLEFYSSWCGHCINYAPTWKALARDVRDWEPAIKIGVL-----DCAEEDNYGACKDFGVT 113
Query: 130 HYP 132
YP
Sbjct: 114 LYP 116
>UniRef100_Q8C9I4 Mus musculus 3 days neonate thymus cDNA, RIKEN full-length enriched
library, clone:A630053N20 product:hypothetical protein,
full insert sequence [Mus musculus]
Length = 527
Score = 115 bits (289), Expect = 2e-24
Identities = 60/164 (36%), Positives = 82/164 (49%), Gaps = 19/164 (11%)
Query: 297 WMFCRGSKNETRGFSCGLWVLLHSLSVRIE-----------DGESQFTFNAICDFVHNFF 345
W+ C+GS+ E RG+ C LW L H+L+V+ +G Q AI ++ FF
Sbjct: 244 WVGCQGSRLELRGYPCSLWKLFHTLTVQASTHPEALAGTGFEGHPQAVLQAIRRYIRTFF 303
Query: 346 VCEECRQHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIW 405
C+EC +HF +M VLWLW HN VN RL+ + DPKFPK W
Sbjct: 304 GCKECGEHFEEMAKESMDSVKTPDQAVLWLWRKHNMVNSRLAGHLSE----DPKFPKVPW 359
Query: 406 PTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYYEK-TLVSLY 448
PT LCP+C H++ WN+ +V LK +Y + LV Y
Sbjct: 360 PTPDLCPAC---HEEIKGLDSWNEGQVLLFLKQHYSRDNLVDAY 400
>UniRef100_Q8K0M2 Quiescin Q6-like 1 [Mus musculus]
Length = 639
Score = 115 bits (289), Expect = 2e-24
Identities = 60/164 (36%), Positives = 82/164 (49%), Gaps = 19/164 (11%)
Query: 297 WMFCRGSKNETRGFSCGLWVLLHSLSVRIE-----------DGESQFTFNAICDFVHNFF 345
W+ C+GS+ E RG+ C LW L H+L+V+ +G Q AI ++ FF
Sbjct: 409 WVGCQGSRLELRGYPCSLWKLFHTLTVQASTHPEALAGTGFEGHPQAVLQAIRRYIRTFF 468
Query: 346 VCEECRQHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIW 405
C+EC +HF +M VLWLW HN VN RL+ + DPKFPK W
Sbjct: 469 GCKECGEHFEEMAKESMDSVKTPDQAVLWLWRKHNMVNSRLAGHLSE----DPKFPKVPW 524
Query: 406 PTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYYEK-TLVSLY 448
PT LCP+C H++ WN+ +V LK +Y + LV Y
Sbjct: 525 PTPDLCPAC---HEEIKGLDSWNEGQVLLFLKQHYSRDNLVDAY 565
>UniRef100_UPI00003AB2AE UPI00003AB2AE UniRef100 entry
Length = 656
Score = 115 bits (287), Expect = 4e-24
Identities = 60/177 (33%), Positives = 91/177 (50%), Gaps = 21/177 (11%)
Query: 294 RGYWMFCRGSKNETRGFSCGLWVLLHSLSVR------------IEDGESQFTFNAICDFV 341
R W+ C+GS+ E RG++C LW L H+L+V+ +ED + + ++
Sbjct: 369 RTEWVGCQGSRPELRGYTCSLWKLFHTLTVQAALRPTALISTGLEDNP-RIVLEVMRRYI 427
Query: 342 HNFFVCEECRQHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFP 401
H+FF C+ C QHF +M VLWLW HN VN RL+ + T DPKFP
Sbjct: 428 HHFFGCKACAQHFEEMAKESMDSVQTLDKAVLWLWEKHNVVNNRLAGDL----TEDPKFP 483
Query: 402 KTIWPTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYY-EKTLVSLYKEKDIAGND 457
K WPT LCP+C H++ WN+ +V + +K++Y + ++ Y E +D
Sbjct: 484 KVQWPTPDLCPAC---HEEIKGLHSWNEAQVLQFMKHHYNSENILYRYTESQTDSSD 537
>UniRef100_UPI00001CF07C UPI00001CF07C UniRef100 entry
Length = 725
Score = 114 bits (284), Expect = 9e-24
Identities = 58/164 (35%), Positives = 83/164 (50%), Gaps = 19/164 (11%)
Query: 297 WMFCRGSKNETRGFSCGLWVLLHSLSVRIE-----------DGESQFTFNAICDFVHNFF 345
W+ C+GS+ E RG+ C LW L H+L+V+ +G+ Q + ++ FF
Sbjct: 409 WVGCQGSRLELRGYPCSLWKLFHTLTVQASTHPEALVGTGFEGDPQAVLQTMRRYIRTFF 468
Query: 346 VCEECRQHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIW 405
C+EC +HF +M A VLWLW HN VN RL+ + DPKFPK W
Sbjct: 469 GCKECGEHFEEMAKESMDSVKTADQAVLWLWRKHNMVNSRLAGHLSE----DPKFPKVPW 524
Query: 406 PTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYYEK-TLVSLY 448
PT LCP C H++ W++++V LK +Y + LV Y
Sbjct: 525 PTPDLCPVC---HEEIKGLDSWDEEQVLVFLKQHYSRDNLVDTY 565
>UniRef100_O08841 GEC-3 [Cavia porcellus]
Length = 613
Score = 113 bits (283), Expect = 1e-23
Identities = 75/233 (32%), Positives = 111/233 (47%), Gaps = 37/233 (15%)
Query: 233 SLVKFLQVLTAHHPSRRCRKGAGDLLVSFADLYPTDFW--SSHKQEDDKSSVKNFQ---- 286
+L KF+ VLT + P + L+ +F L T+ W HK++ S K
Sbjct: 330 ALKKFVTVLTKYFPGQ-------PLVRNF--LQSTNEWLKRQHKKKMPYSFFKTAMDSRN 380
Query: 287 ---ICGKEVPRGYWMFCRGSKNETRGFSCGLWVLLHSLSVRIED---------GESQFTF 334
+ KEV W+ C+GS++ RGF C LW+L H L+V+ Q
Sbjct: 381 EEAVITKEVN---WVGCQGSESHFRGFPCSLWILFHFLTVQASQKNAESSQKPANGQEVL 437
Query: 335 NAICDFVHNFFVCEECRQHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLG 394
AI ++V FF C +C HF M + D VLWLW+SHN+VN RL A
Sbjct: 438 QAIRNYVRFFFGCRDCANHFEQMAAGSMHRVKSPNDAVLWLWTSHNRVNARL----AGAP 493
Query: 395 TGDPKFPKTIWPTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYYEKTLVSL 447
+ DP+FPK WP +LC +C H++ S + W+ D + LK ++ + + L
Sbjct: 494 SEDPQFPKVQWPPPELCSAC---HNELSGEPVWDVDATLRFLKTHFSPSNIVL 543
Score = 39.3 bits (90), Expect = 0.28
Identities = 23/81 (28%), Positives = 45/81 (55%), Gaps = 7/81 (8%)
Query: 53 LNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAK-LFNGPDAVHPGIILLTR 111
L A + + ++P+ +AV EFFA+WC C + P ++ +AK + + A++ L
Sbjct: 46 LQADTVRSTVLNSPSAWAV-EFFASWCGHCIAFAPTWKALAKDIKDWRPALN-----LAA 99
Query: 112 VDCASKINIKLCDKFSVGHYP 132
++CA + N +C F++ +P
Sbjct: 100 LNCADETNNAVCRDFNIAGFP 120
>UniRef100_UPI00004322F6 UPI00004322F6 UniRef100 entry
Length = 504
Score = 112 bits (281), Expect = 2e-23
Identities = 79/279 (28%), Positives = 128/279 (45%), Gaps = 42/279 (15%)
Query: 186 KFQNEHLSSNVSDPEQIARAIYDVEEATSLAFDIILEN-------------KMIKPETRA 232
K +N LS + + QI ++E+ + + LEN KMIK +
Sbjct: 244 KNENHKLSISTTKQLQIIEQQKNIEKNEDYLYQLDLENTLKYSISHEIPLHKMIKDKKMD 303
Query: 233 SLVKFLQVLTAHHPSRRCR---KGAGDLLVSFADLYPTDFWSSHKQ-EDDKSSVKNFQIC 288
+L K+L VL + P + + D+++ +++ +F K E++ S + +
Sbjct: 304 ALKKYLNVLAEYFPLKYGNIFLETIRDIILKRSNISGEEFSQIVKSIEEEMSPIYSGP-- 361
Query: 289 GKEVPRGYWMFCRGSKNETRGFSCGLWVLLHSLSVRI------EDGESQFTFNAICDFVH 342
W+ C+GSK E RG+ CGLW + H L+V + E + A+ ++
Sbjct: 362 ------SKWIGCKGSKEEYRGYPCGLWTMFHMLTVNFAILNKDAEHEPRKILEAMYGYIQ 415
Query: 343 NFFVCEECRQHFYDMCS-SVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFP 401
FF C +C QHF M S + + D +LWLWS+HN+VN RLS + T DP++
Sbjct: 416 YFFGCADCSQHFVQMASKNKMFEVSNINDSILWLWSAHNEVNARLSGD----NTEDPEYK 471
Query: 402 KTIWPTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYY 440
K +P K CP+C + WN++ V LK Y
Sbjct: 472 KIQYPAKIYCPNC------RYENSTWNEENVLHYLKTKY 504
Score = 51.2 bits (121), Expect = 7e-05
Identities = 30/89 (33%), Positives = 49/89 (54%), Gaps = 9/89 (10%)
Query: 48 DYAVELNATNF-DAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAK-LFNGPDAVHPG 105
D V LN TNF +V +DT + +VEF+ +WC C + P ++ A ++ D
Sbjct: 6 DDVVILNVTNFKSSVYEDTKSW--LVEFYNSWCGYCLRFAPIWKDFANDIYAWRD----- 58
Query: 106 IILLTRVDCASKINIKLCDKFSVGHYPML 134
I+++ +DCA N +C ++ + HYPML
Sbjct: 59 IVVVAAIDCADDDNNPICREYEIMHYPML 87
>UniRef100_UPI00004322F5 UPI00004322F5 UniRef100 entry
Length = 469
Score = 112 bits (281), Expect = 2e-23
Identities = 79/279 (28%), Positives = 128/279 (45%), Gaps = 42/279 (15%)
Query: 186 KFQNEHLSSNVSDPEQIARAIYDVEEATSLAFDIILEN-------------KMIKPETRA 232
K +N LS + + QI ++E+ + + LEN KMIK +
Sbjct: 209 KNENHKLSISTTKQLQIIEQQKNIEKNEDYLYQLDLENTLKYSISHEIPLHKMIKDKKMD 268
Query: 233 SLVKFLQVLTAHHPSRRCR---KGAGDLLVSFADLYPTDFWSSHKQ-EDDKSSVKNFQIC 288
+L K+L VL + P + + D+++ +++ +F K E++ S + +
Sbjct: 269 ALKKYLNVLAEYFPLKYGNIFLETIRDIILKRSNISGEEFSQIVKSIEEEMSPIYSGP-- 326
Query: 289 GKEVPRGYWMFCRGSKNETRGFSCGLWVLLHSLSVRI------EDGESQFTFNAICDFVH 342
W+ C+GSK E RG+ CGLW + H L+V + E + A+ ++
Sbjct: 327 ------SKWIGCKGSKEEYRGYPCGLWTMFHMLTVNFAILNKDAEHEPRKILEAMYGYIQ 380
Query: 343 NFFVCEECRQHFYDMCS-SVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFP 401
FF C +C QHF M S + + D +LWLWS+HN+VN RLS + T DP++
Sbjct: 381 YFFGCADCSQHFVQMASKNKMFEVSNINDSILWLWSAHNEVNARLSGD----NTEDPEYK 436
Query: 402 KTIWPTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYY 440
K +P K CP+C + WN++ V LK Y
Sbjct: 437 KIQYPAKIYCPNC------RYENSTWNEENVLHYLKTKY 469
Score = 51.2 bits (121), Expect = 7e-05
Identities = 30/89 (33%), Positives = 49/89 (54%), Gaps = 9/89 (10%)
Query: 48 DYAVELNATNF-DAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAK-LFNGPDAVHPG 105
D V LN TNF +V +DT + +VEF+ +WC C + P ++ A ++ D
Sbjct: 8 DDVVILNVTNFKSSVYEDTKSW--LVEFYNSWCGYCLRFAPIWKDFANDIYAWRD----- 60
Query: 106 IILLTRVDCASKINIKLCDKFSVGHYPML 134
I+++ +DCA N +C ++ + HYPML
Sbjct: 61 IVVVAAIDCADDDNNPICREYEIMHYPML 89
>UniRef100_UPI00004322F3 UPI00004322F3 UniRef100 entry
Length = 514
Score = 112 bits (281), Expect = 2e-23
Identities = 79/279 (28%), Positives = 128/279 (45%), Gaps = 42/279 (15%)
Query: 186 KFQNEHLSSNVSDPEQIARAIYDVEEATSLAFDIILEN-------------KMIKPETRA 232
K +N LS + + QI ++E+ + + LEN KMIK +
Sbjct: 254 KNENHKLSISTTKQLQIIEQQKNIEKNEDYLYQLDLENTLKYSISHEIPLHKMIKDKKMD 313
Query: 233 SLVKFLQVLTAHHPSRRCR---KGAGDLLVSFADLYPTDFWSSHKQ-EDDKSSVKNFQIC 288
+L K+L VL + P + + D+++ +++ +F K E++ S + +
Sbjct: 314 ALKKYLNVLAEYFPLKYGNIFLETIRDIILKRSNISGEEFSQIVKSIEEEMSPIYSGP-- 371
Query: 289 GKEVPRGYWMFCRGSKNETRGFSCGLWVLLHSLSVRI------EDGESQFTFNAICDFVH 342
W+ C+GSK E RG+ CGLW + H L+V + E + A+ ++
Sbjct: 372 ------SKWIGCKGSKEEYRGYPCGLWTMFHMLTVNFAILNKDAEHEPRKILEAMYGYIQ 425
Query: 343 NFFVCEECRQHFYDMCS-SVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFP 401
FF C +C QHF M S + + D +LWLWS+HN+VN RLS + T DP++
Sbjct: 426 YFFGCADCSQHFVQMASKNKMFEVSNINDSILWLWSAHNEVNARLSGD----NTEDPEYK 481
Query: 402 KTIWPTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYY 440
K +P K CP+C + WN++ V LK Y
Sbjct: 482 KIQYPAKIYCPNC------RYENSTWNEENVLHYLKTKY 514
Score = 51.2 bits (121), Expect = 7e-05
Identities = 30/89 (33%), Positives = 49/89 (54%), Gaps = 9/89 (10%)
Query: 48 DYAVELNATNF-DAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAK-LFNGPDAVHPG 105
D V LN TNF +V +DT + +VEF+ +WC C + P ++ A ++ D
Sbjct: 16 DDVVILNVTNFKSSVYEDTKSW--LVEFYNSWCGYCLRFAPIWKDFANDIYAWRD----- 68
Query: 106 IILLTRVDCASKINIKLCDKFSVGHYPML 134
I+++ +DCA N +C ++ + HYPML
Sbjct: 69 IVVVAAIDCADDDNNPICREYEIMHYPML 97
>UniRef100_Q8JGM4 Quiescin/sulfhydryl oxidase [Gallus gallus]
Length = 743
Score = 112 bits (280), Expect = 3e-23
Identities = 64/177 (36%), Positives = 89/177 (50%), Gaps = 18/177 (10%)
Query: 297 WMFCRGSKNETRGFSCGLWVLLHSLSVRIEDGESQF-----TFNAICDFVHNFFVCEECR 351
W+ CRGS+ RG+ CGLW + H L+V+ G N + +V +FF C+EC
Sbjct: 404 WVGCRGSEPHFRGYPCGLWTIFHLLTVQAAQGGPDEELPLEVLNTMRCYVKHFFGCQECA 463
Query: 352 QHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIWPTKQLC 411
QHF M + R+ VLWLWS HN+VN RL A T DP+FPK WP +C
Sbjct: 464 QHFEAMAAKSMDQVKSRREAVLWLWSHHNEVNARL----AGGDTEDPQFPKLQWPPPDMC 519
Query: 412 PSCYLGHDQKSNKIEWNQDEVYKALKNYYEKTLVSLYKEKDI----AGNDGTKGAAL 464
P C H ++ W++ V LK ++ +L +LY + I AG + A L
Sbjct: 520 PQC---HREERGVHTWDEAAVLSFLKEHF--SLGNLYLDHAIPIPMAGEEAAASARL 571
Score = 41.6 bits (96), Expect = 0.056
Identities = 24/82 (29%), Positives = 43/82 (52%), Gaps = 5/82 (6%)
Query: 53 LNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLTRV 112
L A + L +P+ +AV EFFA+WC C ++ P + +A+ P +++ +
Sbjct: 55 LGADTAERRLLGSPSAWAV-EFFASWCGHCIHFAPTWRALAE---DVREWRPA-VMIAAL 109
Query: 113 DCASKINIKLCDKFSVGHYPML 134
DCA + N ++C F + +P L
Sbjct: 110 DCADEANQQVCADFGITGFPTL 131
>UniRef100_UPI0000246FED UPI0000246FED UniRef100 entry
Length = 589
Score = 110 bits (276), Expect = 8e-23
Identities = 58/180 (32%), Positives = 86/180 (47%), Gaps = 22/180 (12%)
Query: 297 WMFCRGSKNETRGFSCGLWVLLHSLSVRIE-----------DGESQFTFNAICDFVHNFF 345
W+ C+GS++E RG+ C LW L H+L+V + + Q + +VH FF
Sbjct: 306 WVGCQGSRSELRGYPCSLWKLFHTLTVEASTHPDALVGTGFEDDPQAVLQTMRRYVHTFF 365
Query: 346 VCEECRQHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIW 405
C+EC +HF +M +LWLW HN VN RL+ + DP+FPK W
Sbjct: 366 GCKECGEHFEEMAKESMDSVKTPDQAILWLWKKHNMVNGRLAGHLSE----DPRFPKLQW 421
Query: 406 PTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYYEK-TLVSLYKEKDIAGNDGTKGAAL 464
PT LCP+C H++ W++ V LK +Y + L+ Y D ++G L
Sbjct: 422 PTPDLCPAC---HEEIKGLASWDEGHVLTFLKQHYGRDNLLDTYSADQ---GDSSEGGTL 475
>UniRef100_UPI00004322F4 UPI00004322F4 UniRef100 entry
Length = 512
Score = 110 bits (276), Expect = 8e-23
Identities = 78/276 (28%), Positives = 127/276 (45%), Gaps = 42/276 (15%)
Query: 186 KFQNEHLSSNVSDPEQIARAIYDVEEATSLAFDIILEN-------------KMIKPETRA 232
K +N LS + + QI ++E+ + + LEN KMIK +
Sbjct: 255 KNENHKLSISTTKQLQIIEQQKNIEKNEDYLYQLDLENTLKYSISHEIPLHKMIKDKKMD 314
Query: 233 SLVKFLQVLTAHHPSRRCR---KGAGDLLVSFADLYPTDFWSSHKQ-EDDKSSVKNFQIC 288
+L K+L VL + P + + D+++ +++ +F K E++ S + +
Sbjct: 315 ALKKYLNVLAEYFPLKYGNIFLETIRDIILKRSNISGEEFSQIVKSIEEEMSPIYSGP-- 372
Query: 289 GKEVPRGYWMFCRGSKNETRGFSCGLWVLLHSLSVRI------EDGESQFTFNAICDFVH 342
W+ C+GSK E RG+ CGLW + H L+V + E + A+ ++
Sbjct: 373 ------SKWIGCKGSKEEYRGYPCGLWTMFHMLTVNFAILNKDAEHEPRKILEAMYGYIQ 426
Query: 343 NFFVCEECRQHFYDMCS-SVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFP 401
FF C +C QHF M S + + D +LWLWS+HN+VN RLS + T DP++
Sbjct: 427 YFFGCADCSQHFVQMASKNKMFEVSNINDSILWLWSAHNEVNARLSGD----NTEDPEYK 482
Query: 402 KTIWPTKQLCPSCYLGHDQKSNKIEWNQDEVYKALK 437
K +P K CP+C + WN++ V LK
Sbjct: 483 KIQYPAKIYCPNC------RYENSTWNEENVLHYLK 512
Score = 51.2 bits (121), Expect = 7e-05
Identities = 30/89 (33%), Positives = 49/89 (54%), Gaps = 9/89 (10%)
Query: 48 DYAVELNATNF-DAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAK-LFNGPDAVHPG 105
D V LN TNF +V +DT + +VEF+ +WC C + P ++ A ++ D
Sbjct: 10 DDVVILNVTNFKSSVYEDTKSW--LVEFYNSWCGYCLRFAPIWKDFANDIYAWRD----- 62
Query: 106 IILLTRVDCASKINIKLCDKFSVGHYPML 134
I+++ +DCA N +C ++ + HYPML
Sbjct: 63 IVVVAAIDCADDDNNPICREYEIMHYPML 91
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.135 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 920,453,861
Number of Sequences: 2790947
Number of extensions: 39432868
Number of successful extensions: 105251
Number of sequences better than 10.0: 678
Number of HSP's better than 10.0 without gapping: 283
Number of HSP's successfully gapped in prelim test: 395
Number of HSP's that attempted gapping in prelim test: 104248
Number of HSP's gapped (non-prelim): 1084
length of query: 517
length of database: 848,049,833
effective HSP length: 132
effective length of query: 385
effective length of database: 479,644,829
effective search space: 184663259165
effective search space used: 184663259165
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC144731.8


