

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144730.2 - phase: 0 /pseudo
(866 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
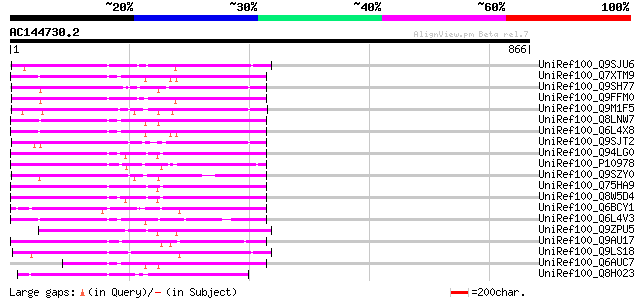
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SJU6 Putative retroelement pol polyprotein [Arabidop... 263 2e-68
UniRef100_Q7XTM9 OSJNBa0033G05.13 protein [Oryza sativa] 256 2e-66
UniRef100_Q9SH77 Putative retroelement pol polyprotein [Arabidop... 253 2e-65
UniRef100_Q9FFM0 Copia-like retrotransposable element [Arabidops... 251 5e-65
UniRef100_Q9M1F5 Copia-like polyprotein [Arabidopsis thaliana] 248 7e-64
UniRef100_Q8LNW7 Putative polyprotein [Oryza sativa] 246 2e-63
UniRef100_Q6L4X8 Putative polyprotein [Oryza sativa] 237 1e-60
UniRef100_Q9SJT2 Putative retroelement pol polyprotein [Arabidop... 235 4e-60
UniRef100_Q94LG0 Putative retroelement pol polyprotein [Oryza sa... 231 5e-59
UniRef100_P10978 Retrovirus-related Pol polyprotein from transpo... 230 2e-58
UniRef100_Q9SZY0 Putative retrotransposon [Arabidopsis thaliana] 229 2e-58
UniRef100_Q75HA9 Putative polyprotein [Oryza sativa] 225 5e-57
UniRef100_Q8W5D4 Putative retrotransposon-related protein [Oryza... 222 3e-56
UniRef100_Q6BCY1 Gag-Pol [Ipomoea batatas] 219 2e-55
UniRef100_Q6L4V3 Putative polyprotein [Oryza sativa] 214 7e-54
UniRef100_Q9ZPU5 Putative retroelement pol polyprotein [Arabidop... 214 9e-54
UniRef100_Q9AU17 Polyprotein-like [Lycopersicon chilense] 214 9e-54
UniRef100_Q9LS18 Retroelement pol polyprotein-like [Arabidopsis ... 209 3e-52
UniRef100_Q6AUC7 Putative polyprotein [Oryza sativa] 207 1e-51
UniRef100_Q8H023 Putative retrovirus-related pol polyprotein [Or... 202 3e-50
>UniRef100_Q9SJU6 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 838
Score = 263 bits (671), Expect = 2e-68
Identities = 168/458 (36%), Positives = 260/458 (56%), Gaps = 30/458 (6%)
Query: 3 DIEKFTGSNDFGLWKVKMRAI---------LIQQKCVEALKGEAQMDVHLTPAEKT---E 50
++EK G D+ LWK K+ A L + + +E + A+ D LT E E
Sbjct: 7 EVEKLDGEGDYVLWKEKLLAHIELLGLLEGLEEDEAIEEEESTAETDSLLTKTEDKVLKE 66
Query: 51 MNDKAVSAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYRMVE 110
KA S +IL LG+ VLR+V +E TA M LD L+M KSL +R LKQ+LY Y+M +
Sbjct: 67 KRGKARSTVILSLGNHVLRKVIKEKTAAGMIRVLDKLFMAKSLPNRIYLKQRLYGYKMSD 126
Query: 111 SKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GTITLE 170
S I E + +F K+I DL N+ V++ DED+A+ L +LP+ F+ KDT+ YGK T+ L+
Sbjct: 127 SMTIEENVNDFFKLISDLENVKVSVPDEDQAIVLLMSLPKQFDQLKDTLKYGK-TTLALD 185
Query: 171 EVQAALRTKELTKFKELK-VEDSGEGLNV-SRERSQNRGKGKGKNSRSKSRSKGDGNKTQ 228
E+ A+R+K L K +++S + L V R RS+ R K +N +S+SRSK K
Sbjct: 186 EITGAIRSKVLELGASGKMLKNSSDALFVQDRGRSEKRDKSSERN-KSQSRSKSREKKV- 243
Query: 229 YKCFICHNPGHFKKDCP--ERKGNGGGNPSVQIASNEEGYES-AGALTV------TSWEP 279
C++C GHFKK C + K G N +SN G + A AL V + E
Sbjct: 244 --CWVCGKEGHFKKQCYVWKEKNKKGNNSEKGESSNVIGQAADAAALAVREESNADNQEV 301
Query: 280 EKGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLL 339
+ W++D+GCS+H+ PR+++F + + G V + N +I+ IG+IR++ D+ LL
Sbjct: 302 DNEWIMDTGCSFHMTPRRDWFVEFDESQTGRVKMANQTYSEIKGIGSIRIQNDDNTTVLL 361
Query: 340 KDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLYILEGSTVI 399
K+VRY+P + +NLIS+ + G + + G +++ G + + KG K+ LY+L+G V
Sbjct: 362 KNVRYVPSMSKNLISMGTLEDQGCWFQSKAGTLKVVKGCMTLLKGKKVGTLYLLQGVVVT 421
Query: 400 ADASVASVDTLDVTKLWHLRLGHVSERGIWLN*LNKGC 437
+A+ A + D +K+WH RL H+S+R I + + KGC
Sbjct: 422 GNAN-AVTSSKDESKIWHSRLCHMSQRNIDVL-IKKGC 457
>UniRef100_Q7XTM9 OSJNBa0033G05.13 protein [Oryza sativa]
Length = 1181
Score = 256 bits (655), Expect = 2e-66
Identities = 157/451 (34%), Positives = 254/451 (55%), Gaps = 30/451 (6%)
Query: 1 KWDIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMDVHLTPAEKTEMNDKAVSAII 60
K+D+ F LW+VKMRA+L QQ+ +AL G + + EK + + KA+S I
Sbjct: 5 KYDLPLLDRDTRFSLWQVKMRAVLAQQELDDALSGFDKRTQDWSNDEK-KRDRKAMSYIH 63
Query: 61 LCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTE 120
L L + +L+EV +E TA + KL+ + MTK L + LKQ+L+ +++ + +M+ L+
Sbjct: 64 LHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLSA 123
Query: 121 FNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GTITLEEVQAALRTKE 180
F +I+ DL +++V +++D AL L C+LP S+ NF+DT+LY + T+TL+EV AL KE
Sbjct: 124 FKEIVADLESMEVKYDEKDLALILLCSLPSSYANFRDTILYSR-DTLTLKEVYDALHAKE 182
Query: 181 LTKFKELKVEDSGEGLNVSRERSQNRGKGKGKNSRSKSRSKGDGN-------KTQYK-CF 232
++K EG N E RG + KN+ +KSR K + + +YK C
Sbjct: 183 -----KMKKMVPSEGSNSQAEGLVVRGSQQEKNTNNKSRDKSSSSYRGRSKSRGRYKSCK 237
Query: 233 ICHNPGHFKKDC--PERKGNGGGNPSVQIASNEEGY------ESAGALTVTSW----EPE 280
C GH C + K G + EEG E + A + ++ +
Sbjct: 238 YCKRDGHDISKCWKLQDKDKRTGKYIPKGKKEEEGKAAVVTDEKSDAELLVAYAGCAQTS 297
Query: 281 KGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLLK 340
W+LD+ C+YH+CP +++F E+ +GG V +G++ C++ IGT+++KMFD L
Sbjct: 298 DQWILDTACTYHMCPNRDWFATYEVVQGGTVLMGDDTPCEVAGIGTVQIKMFDGCIRTLS 357
Query: 341 DVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGS-KIHGLYILEGSTVI 399
DVR+IP L+R+LIS+ D GY G+++++ G+L++ K S K LY L+G+T++
Sbjct: 358 DVRHIPNLKRSLISLCTLDRKGYKYSGGDGILKVTKGSLVVMKASIKSANLYHLQGTTIL 417
Query: 400 ADASVA--SVDTLDVTKLWHLRLGHVSERGI 428
+ + S+ D T LWH+RLGH+SE G+
Sbjct: 418 GNVATVSDSLSNSDATNLWHMRLGHMSEIGL 448
>UniRef100_Q9SH77 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1356
Score = 253 bits (645), Expect = 2e-65
Identities = 163/457 (35%), Positives = 255/457 (55%), Gaps = 41/457 (8%)
Query: 3 DIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMDVHLTPAEKT------------- 49
++EKF G D+ +WK K+ A + ALK + +++
Sbjct: 7 EVEKFDGRGDYTMWKEKLLAHMDILGLNTALKESESTGEKKSVLDESDEDYEEKLEKFEA 66
Query: 50 --EMNDKAVSAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYR 107
E KA SAI+L + D+VLR++ +ESTA +M LD LYM+K+L +R KQ+LY ++
Sbjct: 67 LEEKKKKARSAIVLSVTDRVLRKIKKESTAAAMLLALDKLYMSKALPNRIYPKQKLYSFK 126
Query: 108 MVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GTI 167
M E+ + + EF +II DL N++V + DED+A+ L ALP++F+ KDT+ Y +I
Sbjct: 127 MSENLSVEGNIDEFLQIITDLENMNVIISDEDQAILLLTALPKAFDQLKDTLKYSSGKSI 186
Query: 168 -TLEEVQAALRTKEL---TKFKELKVEDSGEGLNVSRERSQNRGKGKGKNSRSKSRSKGD 223
TL+EV AA+ +KEL + K +KV+ EGL V +++++N+GKG+ K K + K
Sbjct: 187 LTLDEVAAAIYSKELELGSVKKSIKVQ--AEGLYV-KDKNENKGKGEQKG---KGKGKKG 240
Query: 224 GNKTQYKCFICHNPGHFKKDCPER-----------KG-NGGGNPSVQIASNEEGYESAGA 271
+K + C+ C GHF+ CP + KG + GG ++ A+ GY + A
Sbjct: 241 KSKKKPGCWTCGEEGHFRSSCPNQNKPQFKQSQVVKGESSGGKGNLAEAA---GYYVSEA 297
Query: 272 LTVTSWEPEKGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKM 331
L+ T E W+LD+GCSYH+ ++E+F + GG V +GN +++ +GTIR+K
Sbjct: 298 LSSTEVHLEDEWILDTGCSYHMTYKREWFHEFNEDAGGSVRMGNKTVSRVRGVGTIRVKN 357
Query: 332 FDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLY 391
D +L +VRYIP + RNL+S+ F+ GY E G++RI G ++ G + LY
Sbjct: 358 SDGLTIVLTNVRYIPDMDRNLLSLGTFEKAGYKFESEDGILRIKAGNQVLLTGRRYDTLY 417
Query: 392 ILEGSTVIADASVASVDTLDVTKLWHLRLGHVSERGI 428
+L V A S+A V D T LWH RL H+S++ +
Sbjct: 418 LLNWKPV-ASESLAVVKRADDTVLWHQRLCHMSQKNM 453
>UniRef100_Q9FFM0 Copia-like retrotransposable element [Arabidopsis thaliana]
Length = 1342
Score = 251 bits (642), Expect = 5e-65
Identities = 164/457 (35%), Positives = 248/457 (53%), Gaps = 38/457 (8%)
Query: 3 DIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMDVHLTPAEKT------------- 49
++EKF G D+ LWK K+ A + +E L E + V + E +
Sbjct: 7 EVEKFDGDGDYILWKEKLLAHMEMLGLLEGLGEEEEAVVEDSTTEISDGGNQDPETATSK 66
Query: 50 -------EMNDKAVSAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQ 102
E KA S IIL LG+ VLR+V ++ TA M LD L+M KSL +R LKQ+
Sbjct: 67 LEDKILKEKRGKARSTIILSLGNNVLRKVIKQKTAAGMIKVLDQLFMAKSLPNRIYLKQR 126
Query: 103 LYFYRMVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYG 162
LY Y+M E+ + E + +F K+I DL N+ V + DED+A+ L +LPR F+ K+T+ Y
Sbjct: 127 LYGYKMSENMTMEENVNDFFKLISDLENVKVVVPDEDQAIVLLMSLPRQFDQLKETLKYC 186
Query: 163 K*GTITLEEVQAALRTKELTKFKELK-VEDSGEGLNV-SRERSQNRGKGKGKNSRSKSRS 220
K T+ LEE+ +A+R+K L K ++++ +GL V R RS+ RGKG KN +S+S+S
Sbjct: 187 K-TTLHLEEITSAIRSKILELGASGKLLKNNSDGLFVQDRGRSETRGKGPNKN-KSRSKS 244
Query: 221 KGDGNKTQYKCFICHNPGHFKKDC---PERKGNGGGNPSVQIASNEEGYESAGALTVT-- 275
KG G C+IC GHFKK C ER G + + ++ A AL V+
Sbjct: 245 KGAGK----TCWICGKEGHFKKQCYVWKERNKQGSTSERGEASTVTARVTDAAALVVSRA 300
Query: 276 ----SWEPEKGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKM 331
+ W+LD+GCS+H+ RK++ + G V +GN+ +++ IG +R+K
Sbjct: 301 LLGFAEVTPDTWILDTGCSFHMTCRKDWIIDFKETASGKVRMGNDTYSEVKGIGDVRIKN 360
Query: 332 FDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLY 391
D LL DVRYIP++ +NLIS+ + G ++G++ I L + G K LY
Sbjct: 361 EDGSTILLTDVRYIPEMSKNLISLGTLEDKGCWFESKKGILTIFKNDLTVLTGKKESTLY 420
Query: 392 ILEGSTVIADASVASVDTLDVTKLWHLRLGHVSERGI 428
L+G+T+ +A+V + D T LWH RLGH+ +G+
Sbjct: 421 FLQGTTLAGEANVIDKEK-DETSLWHSRLGHIGAKGL 456
>UniRef100_Q9M1F5 Copia-like polyprotein [Arabidopsis thaliana]
Length = 1363
Score = 248 bits (632), Expect = 7e-64
Identities = 158/466 (33%), Positives = 254/466 (53%), Gaps = 48/466 (10%)
Query: 3 DIEKFTGSNDFGLWKVKM---------RAILIQQKCVEALKGEAQMDVHLTPAEKTEMND 53
++EKF G D+ +WK K+ A+L + + + +++ E+ +M
Sbjct: 7 EVEKFDGRGDYTMWKEKLLAHIDMLGLSAVLRESETPMGKERDSEKSDEDEKEEREKMEA 66
Query: 54 ------KAVSAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYR 107
KA S I+L + D+VLR++ +E++A +M LD LYM+K+L +R LKQ+LY ++
Sbjct: 67 FEEKKRKARSTIVLSVSDRVLRKIKKETSAAAMLEALDRLYMSKALPNRIYLKQKLYSFK 126
Query: 108 MVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GTI 167
M E+ I + EF I+ DL N++V + DED+A+ L +LP+ F+ KDT+ Y T+
Sbjct: 127 MSENLSIEGNIDEFLHIVADLENLNVLVSDEDQAILLLMSLPKPFDQLKDTLKYSSGKTV 186
Query: 168 -TLEEVQAALRTKELTKFKELK--VEDSGEGLNVSRERSQNRG----KGKGKNSRSKSRS 220
+L+EV AA+ ++EL +F +K ++ EGL V +++++NRG K KGK RSKS+S
Sbjct: 187 LSLDEVAAAIYSREL-EFGSVKKSIKGQAEGLYV-KDKAENRGRSEQKDKGKGKRSKSKS 244
Query: 221 KGDGNKTQYKCFICHNPGHFKKDCPER-----------KGNGGGNPSVQIASNEEGYESA 269
K C+IC GH K CP + KG G + + ESA
Sbjct: 245 KRG-------CWICGEDGHLKSTCPNKNKPQFKNQGSNKGESSGGKGNLVEGSVNFVESA 297
Query: 270 G-----ALTVTSWEPEKGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVI 324
G AL+ T E W++D+GC YH+ ++E+ E + E GG V +GN +++ +
Sbjct: 298 GMFVSEALSSTDIHLEDEWIMDTGCIYHMTHKREWLEDFDEEAGGSVRMGNKSISRVKGV 357
Query: 325 GTIRLKMFDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKG 384
GT+R+ + L++VRYIP + RNL+S+ F+ G+ E G++RI G ++ +G
Sbjct: 358 GTVRIVNDNGLTVTLQNVRYIPDMDRNLLSLGTFEKAGHKFESENGMLRIKSGNQVLLEG 417
Query: 385 SKIHGLYILEGSTVIADASVASVDTLDVTKLWHLRLGHVSERGIWL 430
+ LYIL G D S+A D T LWH RL H+S++ + L
Sbjct: 418 RRYDTLYILHGKPA-TDESLAVARANDDTVLWHRRLCHMSQKNMSL 462
>UniRef100_Q8LNW7 Putative polyprotein [Oryza sativa]
Length = 1280
Score = 246 bits (628), Expect = 2e-63
Identities = 150/451 (33%), Positives = 244/451 (53%), Gaps = 30/451 (6%)
Query: 1 KWDIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMDVHLTPAEKTEMNDKAVSAII 60
K+D+ F LW+VKMRA+L QQ +AL G + + EK + + KA+S I
Sbjct: 40 KYDLPLLDRDTRFSLWQVKMRAVLAQQDLDDALSGFDKRTQDWSNDEKKK-DRKAMSYIH 98
Query: 61 LCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTE 120
L L + +L+EV +E TA + KL+ + MTK L + LKQ+L+ +++ + +M+ L+
Sbjct: 99 LHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLST 158
Query: 121 FNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GTITLEEVQAALRTKE 180
F +I+ DL +I+V ++ED L L C+LP S+ NF+DT+LY T+ L+EV AL KE
Sbjct: 159 FKEIVADLESIEVKYDEEDLGLILLCSLPSSYANFRDTILYSH-DTLILKEVYDALHAKE 217
Query: 181 LTKFKELKVEDSGEGLNVSRERSQNRGKGKGKNSRSKSRSKGDG-------NKTQYK-CF 232
++K EG N E RG+ + KN++++SR K ++ +YK C
Sbjct: 218 -----KMKKMVPSEGSNSQAEGLVVRGRQQEKNTKNQSRDKSSSSYRGRSKSRGRYKSCK 272
Query: 233 ICHNPGHFKKDCPE------------RKGNGGGNPSVQIASNEEGYESAGALTVTSWEPE 280
C GH +C + KG + ++E+ +
Sbjct: 273 YCKRDGHDISECWKLQDKDKRTGKYIPKGKKEEEGKAAVVTDEKSDTELLVAYAGCAQTS 332
Query: 281 KGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLLK 340
W+LD+ +YH+CP +++F E +GG V +G++ C++ IGT+++KMFD L
Sbjct: 333 DQWILDTAWTYHMCPNRDWFATYEALQGGTVLMGDDTPCEVAGIGTVQIKMFDGYIRTLS 392
Query: 341 DVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGS-KIHGLYILEGSTVI 399
DVR+IP L+R+LIS+ D GY G+++++ G+L++ K K LY L G+T++
Sbjct: 393 DVRHIPNLKRSLISLCTLDRKGYKYSGGDGILKVTKGSLVVMKADIKSANLYHLRGTTIL 452
Query: 400 ADASVA--SVDTLDVTKLWHLRLGHVSERGI 428
+ + S+ D T LWH+RLGH+SE G+
Sbjct: 453 GNVAAVSDSLSNSDATNLWHMRLGHMSEIGL 483
>UniRef100_Q6L4X8 Putative polyprotein [Oryza sativa]
Length = 1211
Score = 237 bits (604), Expect = 1e-60
Identities = 153/452 (33%), Positives = 247/452 (53%), Gaps = 32/452 (7%)
Query: 1 KWDIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMDVHLTPAEKTEMNDKAVSAII 60
K+D+ F LW+VKMRA+L QQ +AL G + + EK + + K +S I
Sbjct: 5 KYDLPLLDRDTRFSLWQVKMRAVLAQQDLDDALSGFDKRTQDWSNDEK-KRDRKTMSYIH 63
Query: 61 LCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTE 120
L L + +L+EV +E TA + KL+ + MTK L + LKQ+L+ +++ + +M+ L+
Sbjct: 64 LHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDGSVMDHLSA 123
Query: 121 FNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GTITLEEVQAALRTKE 180
F KI+ DL +++V ++ED L L C+LP S+ NF+DT+LY T+TL+EV AL KE
Sbjct: 124 FKKIVADLESMEVKYDEEDLCLILLCSLPSSYANFRDTILYSC-DTLTLKEVYDALHAKE 182
Query: 181 LTKFKELKVEDSGEGLNVSRERSQNRGKGKGKNSRSKSRSKGDG-------NKTQYK-CF 232
++K EG N E RG+ + KN+ SKSR K ++ +YK C
Sbjct: 183 -----KIKKMVPSEGSNSQAEGLVVRGRQQEKNTNSKSRDKSSSSYRGRSKSRGRYKSCK 237
Query: 233 ICHNPGHFKKDC--PERKGNGGGNPSVQIASNEEGY------ESAGALTVTSW----EPE 280
GH +C + K G + EEG E + A + ++ +
Sbjct: 238 YYKRDGHDISECWKLQDKDKRTGKYVPKGKKEEEGKAAVVTDEKSDAELLVAYAGCAQTS 297
Query: 281 KGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLLK 340
W+LD+ C+YH+C +++F E +GG V +G++ C++ + T+++KMFD L
Sbjct: 298 DQWILDTACTYHMCLNRDWFATYEAVQGGTVLMGDDTPCEVAGVETVQIKMFDGCIRTLS 357
Query: 341 DVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGS-KIHGLYILEGSTVI 399
DVR+IP L+R+LIS+ D Y G+++++ G+L++ K K LY + G+T++
Sbjct: 358 DVRHIPNLKRSLISLCTLDRKVYKYSGGDGILKVTKGSLVVMKADIKSANLYHVRGTTIL 417
Query: 400 ADASVASVDTL---DVTKLWHLRLGHVSERGI 428
+ + S D+L D T LWH+RLGH+SE G+
Sbjct: 418 GNIAAVS-DSLYNSDATNLWHMRLGHMSEIGL 448
>UniRef100_Q9SJT2 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1333
Score = 235 bits (600), Expect = 4e-60
Identities = 158/446 (35%), Positives = 247/446 (54%), Gaps = 41/446 (9%)
Query: 3 DIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQM-----DVHLTPAEKTE------- 50
++EKF G D+ +WK K+ A L ALK E + ++ LT E+ E
Sbjct: 7 EVEKFDGRGDYTMWKEKLMAHLDILGLSVALKEEDDLVEKVAEMQLTEEEEKEEVLRREL 66
Query: 51 ---MNDKAVSAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYR 107
KA SAI+L + D+VLR++ +E +A +M LD LYM+K+L +R KQ+LY ++
Sbjct: 67 LEEKRRKARSAIVLSVTDRVLRKIKKEQSAAAMLGVLDKLYMSKALPNRIYQKQKLYSFK 126
Query: 108 MVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*G-T 166
M E+ I + EF +II DL N +V + DED+A+ L +LP+ F+ +DT+ YG T
Sbjct: 127 MSENLSIEGNIDEFLRIIADLENTNVLVSDEDQAILLLMSLPKPFDQLRDTLKYGLGRVT 186
Query: 167 ITLEEVQAALRTKELTKFKELK-VEDSGEGLNVSRERSQNRGKGK---GKNSRSKSRSKG 222
++L+EV AA+ +KEL K ++ EGL V +E+++ RG+ + N+ KSRSK
Sbjct: 187 LSLDEVVAAIYSKELELGSNKKSIKGQAEGLFV-KEKTETRGRTEQRGNNNNNKKSRSK- 244
Query: 223 DGNKTQYKCFICHNPGHFKKDCPERKGNGGGNPSVQIASNEEGYESAGALTVTSWEPEKG 282
++++ C+IC NG N S G + AL+ T E
Sbjct: 245 --SRSKKGCWIC-----------GESSNGSSN-----YSEANGLYVSEALSSTDIHLEDE 286
Query: 283 WVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLLKDV 342
WV+D+GCSYH+ ++E+FE L + GG V +GN K++ IGTIR+K L +V
Sbjct: 287 WVMDTGCSYHMTYKREWFEDLNEDAGGSVRMGNKTVSKVRGIGTIRVKNEAGMVVRLTNV 346
Query: 343 RYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLYILEGSTVIADA 402
RYIP++ RNL+S+ F+ GY ++E G + I G ++ + + LY+L+ V +
Sbjct: 347 RYIPEMDRNLLSLGTFEKSGYSFKLENGTLSIIAGDSVLLTVRRCYTLYLLQWRPV-TEE 405
Query: 403 SVASVDTLDVTKLWHLRLGHVSERGI 428
S++ V D T LWH RLGH+S++ +
Sbjct: 406 SLSVVKRQDDTILWHRRLGHMSQKNM 431
>UniRef100_Q94LG0 Putative retroelement pol polyprotein [Oryza sativa]
Length = 1326
Score = 231 bits (590), Expect = 5e-59
Identities = 156/448 (34%), Positives = 245/448 (53%), Gaps = 31/448 (6%)
Query: 1 KWDIEKFTGSNDFGLWKVKMRAILIQQKCV-EALKGEAQMDVHLTPAEKTEMNDKAVSAI 59
K+D+ F LW+VKMRAIL Q + EAL+ + AE+ + KA+ I
Sbjct: 5 KYDLPLLDYKTRFSLWQVKMRAILAQTSDLDEALESFGKKKSTEWTAEEKRKDRKALLLI 64
Query: 60 ILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLT 119
L L + +L+EV +E TA + KL+S+ M+K L + +K +L+ +++ ES ++ ++
Sbjct: 65 QLHLSNDILQEVLQEKTAAELWLKLESICMSKDLTSKMHIKMKLFSHKLQESGSVLNHIS 124
Query: 120 EFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GTITLEEVQAALRTK 179
F +I+ DL +I+V +DED L L C+LP S+ NF+DT+L + +TL EV AL+ +
Sbjct: 125 VFKEIVVDLVSIEVQFDDEDLGLLLLCSLPSSYANFRDTILLSR-DELTLAEVYEALQNR 183
Query: 180 ELTKFKELKVEDS----GEGLNV---SRERSQNRGKGKGKNSRSKSRSKGDGNKTQYKCF 232
E K K + D+ GE L V S +R+ N + K S+S+ RSK G K C
Sbjct: 184 E--KMKGMVQSDASSSKGEALQVRGRSEQRTYNDSSDRDK-SQSRGRSKSRGKKF---CK 237
Query: 233 ICHNPGHFKKDC------PERKGNGGGNPSVQIASNEEGYESAGALTVTSW--EPEKGWV 284
C HF ++C +RK +G + ++ E +S L V + W+
Sbjct: 238 YCKKKNHFIEECWKLQNKEKRKSDG----KASVVTSAENSDSGDCLVVFAGCVASHDEWI 293
Query: 285 LDSGCSYHICPRKEYFEMLE-LEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLLKDVR 343
LD+ CS+HIC +++F + ++ G VV +G++ +I IG++++K D LKDVR
Sbjct: 294 LDTACSFHICINRDWFSSYKSVQNGDVVRMGDDNPREIVGIGSVQIKTHDGMTRTLKDVR 353
Query: 344 YIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGS-KIHGLYILEGSTVIADA 402
+IP + RNLIS+S D GY GV+++S G+L+ G LY+L GST+
Sbjct: 354 HIPGMARNLISLSTLDAEGYKYSSSGGVVKVSKGSLVYMIGDMNSANLYVLRGSTLHGSV 413
Query: 403 SVASV--DTLDVTKLWHLRLGHVSERGI 428
+ A+V D T LWH+RLGH+SE G+
Sbjct: 414 TAAAVSKDEPIKTNLWHMRLGHMSELGM 441
>UniRef100_P10978 Retrovirus-related Pol polyprotein from transposon TNT 1-94
[Contains: Protease (EC 3.4.23.-); Reverse transcriptase
(EC 2.7.7.49); Endonuclease] [Nicotiana tabacum]
Length = 1328
Score = 230 bits (586), Expect = 2e-58
Identities = 159/449 (35%), Positives = 234/449 (51%), Gaps = 37/449 (8%)
Query: 1 KWDIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMDVHLTPAEKTEMNDKAVSAII 60
K+++ KF G N F W+ +MR +LIQQ + L +++ + + +++++A SAI
Sbjct: 5 KYEVAKFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAEDWADLDERAASAIR 64
Query: 61 LCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTE 120
L L D V+ + E TA + +L+SLYM+K+L ++ LK+QLY M E + L
Sbjct: 65 LHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKKQLYALHMSEGTNFLSHLNV 124
Query: 121 FNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GTITLEEVQAALRTKE 180
FN +I LAN+ V +E+EDKA+ L +LP S++N T+L+GK TI L++V +AL E
Sbjct: 125 FNGLITQLANLGVKIEEEDKAILLLNSLPSSYDNLATTILHGK-TTIELKDVTSALLLNE 183
Query: 181 LTKFKELKVEDSG-----EGLNVSRERSQNRGKGKGKNSRSKSRSKGDGNKTQYKCFICH 235
+ K E+ G EG S +RS N G +SK+RSK C+ C+
Sbjct: 184 KMR---KKPENQGQALITEGRGRSYQRSSNNYGRSGARGKSKNRSK----SRVRNCYNCN 236
Query: 236 NPGHFKKDCPE-RKGNG---------------GGNPSVQIASNEEGYESAGALTVTSWEP 279
PGHFK+DCP RKG G N +V + NEE E L+ P
Sbjct: 237 QPGHFKRDCPNPRKGKGETSGQKNDDNTAAMVQNNDNVVLFINEE--EECMHLS----GP 290
Query: 280 EKGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLL 339
E WV+D+ S+H P ++ F + G V +GN KI IG I +K +L
Sbjct: 291 ESEWVVDTAASHHATPVRDLFCRYVAGDFGTVKMGNTSYSKIAGIGDICIKTNVGCTLVL 350
Query: 340 KDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLYILEGSTVI 399
KDVR++P LR NLIS D GY + R++ G+L+IAKG LY
Sbjct: 351 KDVRHVPDLRMNLISGIALDRDGYESYFANQKWRLTKGSLVIAKGVARGTLYRTNAEICQ 410
Query: 400 ADASVASVDTLDVTKLWHLRLGHVSERGI 428
+ + A D + V LWH R+GH+SE+G+
Sbjct: 411 GELNAAQ-DEISV-DLWHKRMGHMSEKGL 437
>UniRef100_Q9SZY0 Putative retrotransposon [Arabidopsis thaliana]
Length = 1230
Score = 229 bits (585), Expect = 2e-58
Identities = 155/457 (33%), Positives = 232/457 (49%), Gaps = 57/457 (12%)
Query: 3 DIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMDVHLTPAEK-------------T 49
++EKF G D+ LWK K+ A + AL+ + L E+
Sbjct: 7 EMEKFDGHGDYTLWKEKLMAHMDLLGLTVALRETQSVSDPLESEEEGKESEKGDKEALME 66
Query: 50 EMNDKAVSAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYRMV 109
E KA S I+L + D+VLR+ +E TA SM LD LYM+K+L +R LKQ+LY Y+M
Sbjct: 67 EKRQKARSTIVLSVSDQVLRKSKKEKTAPSMLEALDKLYMSKALPNRIYLKQKLYSYKMQ 126
Query: 110 ESKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK-*GTIT 168
E+ + + EF ++I DL N +V + DED+A+ L +LP+ F+ KDT+ YG T++
Sbjct: 127 ENLSVEGNIDEFLRLIADLENTNVLVSDEDQAILLLMSLPKQFDQLKDTLKYGSGRTTLS 186
Query: 169 LEEVQAALRTKELTKFKELK-VEDSGEGLNVSRERSQNRG----KGKGKNSRSKSRSKGD 223
++EV AA+ +KEL K + EGL V +++ + RG K KG RS+SRSKG
Sbjct: 187 VDEVVAAIYSKELELGSNKKSIRGQAEGLYV-KDKPETRGMSEQKEKGNKGRSRSRSKG- 244
Query: 224 GNKTQYKCFICHNPGHFKKDCPER---------KGNGGGNPSVQIASNE---EGYESAGA 271
C+IC GHFK CP + + +G + I N GY + A
Sbjct: 245 ----WKGCWICGEEGHFKTSCPNKGKQQNKGKDQASGSKGEAATIKGNTSEGSGYYVSEA 300
Query: 272 LTVTSWEPEKGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKM 331
L T WV+D+GC+YH+ +KE+FE L + GG V +GN K +
Sbjct: 301 LHSTDVNLGNEWVMDTGCNYHMTHKKEWFEELSEDAGGTVRMGNKSTSKFR--------- 351
Query: 332 FDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLY 391
V+YIP + RNL+S+ + GY + GV+ + G + GS+ LY
Sbjct: 352 ----------VKYIPDMDRNLLSMGTLEEHGYSFESKNGVLVVKEGTRTLLIGSRHEKLY 401
Query: 392 ILEGSTVIADASVASVDTLDVTKLWHLRLGHVSERGI 428
+L+G ++ S+ D T LWH RLGH+S++ +
Sbjct: 402 LLQGKPEVSH-SMTVERRNDDTVLWHRRLGHISQKNM 437
>UniRef100_Q75HA9 Putative polyprotein [Oryza sativa]
Length = 1322
Score = 225 bits (573), Expect = 5e-57
Identities = 150/446 (33%), Positives = 240/446 (53%), Gaps = 27/446 (6%)
Query: 1 KWDIEKFTGSNDFGLWKVKMRAILIQQKCV-EALKGEAQMDVHLTPAEKTEMNDKAVSAI 59
K+D+ F LW+VKMRA+L Q + EAL+ + AE+ + KA+S I
Sbjct: 5 KYDLPLLDYKTRFSLWQVKMRAVLAQTSDLDEALESFGKKKTTEWTAEEKRKDRKALSLI 64
Query: 60 ILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLT 119
L L + +L+EV ++ TA + KL+S+ M+K L + +K +L+ +++ ES ++ ++
Sbjct: 65 QLHLSNDILQEVLQKKTAAELWLKLESICMSKDLTSKMHIKMKLFSHKLHESGSVLNHIS 124
Query: 120 EFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GTITLEEVQAALRTK 179
F +I+ DL +++V +DED L L C+LP S+ NF+ T+L + +TL EV AL+ +
Sbjct: 125 VFKEIVADLVSMEVQFDDEDLGLLLLCSLPSSYANFRHTILLSR-DELTLAEVYEALQNR 183
Query: 180 ELTK--FKELKVEDSGEGLNV---SRERSQNRGKGKGKNSRSKSRSKGDGNKTQYKCFIC 234
E K + GE L V S +R+ N K S+S+ RSK G K C C
Sbjct: 184 EKMKGMVQSYASSSKGEALQVRGRSEQRTYNDSNDHDK-SQSRGRSKSRGKKF---CKYC 239
Query: 235 HNPGHFKKDC------PERKGNGGGNPSVQIASNEEGYESAGALTVTSW--EPEKGWVLD 286
HF ++C +RK +G + ++ E +S L V + W+LD
Sbjct: 240 KKKNHFIEECWKLQNKEKRKSDG----KASVVTSAENSDSGDCLVVFAGYVASHDEWILD 295
Query: 287 SGCSYHICPRKEYFEMLE-LEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLLKDVRYI 345
+ CS+HIC +++F + ++ VV +G++ +I IG++++K D LKDVR+I
Sbjct: 296 TACSFHICINRDWFSSYKSVQNEDVVRMGDDNPREIVGIGSVQIKTHDGMTRTLKDVRHI 355
Query: 346 PKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGS-KIHGLYILEGSTVIADASV 404
P + RNLIS+S D GY GV+++S G+L+ G LY+L GST+ +
Sbjct: 356 PGMARNLISLSTLDAEGYKYSGSGGVVKVSKGSLVYMIGDMNSANLYVLRGSTLHGSVTA 415
Query: 405 ASV--DTLDVTKLWHLRLGHVSERGI 428
A+V D T LWH+RLGH+SE G+
Sbjct: 416 AAVTKDEPSKTNLWHMRLGHMSELGM 441
>UniRef100_Q8W5D4 Putative retrotransposon-related protein [Oryza sativa]
Length = 1229
Score = 222 bits (566), Expect = 3e-56
Identities = 152/448 (33%), Positives = 242/448 (53%), Gaps = 31/448 (6%)
Query: 1 KWDIEKFTGSNDFGLWKVKMRAILIQQKCV-EALKGEAQMDVHLTPAEKTEMNDKAVSAI 59
K+D+ F LW+VKMRA+L Q + EAL+ + AE+ + KA+S I
Sbjct: 2 KYDLPLQDYKTRFSLWQVKMRAVLAQTSDLDEALESFGKKKTTEWTAEEKRKDRKALSLI 61
Query: 60 ILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLT 119
L L + +L++V +E TA + KL+S+ M+K L + +K +L+ +++ ES ++ ++
Sbjct: 62 QLHLSNDILQKVLQEKTAAELWFKLESICMSKDLTSKMHIKMKLFSHKLQESGSVLNHIS 121
Query: 120 EFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GTITLEEVQAALRTK 179
F +II DL +++V +DED L L C+LP + NF+DT+L + +TL EV AL+ +
Sbjct: 122 VFKEIIADLVSMEVQFDDEDLGLLLLCSLPSLYANFRDTILLSR-DELTLAEVYEALQNR 180
Query: 180 ELTKFKELKVEDS----GEGLNV---SRERSQNRGKGKGKNSRSKSRSKGDGNKTQYKCF 232
E K K + D+ G+ L V S +R+ N + K S+S+ RSK G K C
Sbjct: 181 E--KMKGMVQSDASSSKGKALQVRGRSEQRTYNDSNDRDK-SQSRGRSKSRGKKF---CK 234
Query: 233 ICHNPGHFKKDC------PERKGNGGGNPSVQIASNEEGYESAGALTVTSW--EPEKGWV 284
C HF ++C +RK +G + ++ E +SA L + W+
Sbjct: 235 YCKKKNHFIEECWKLQNKEKRKSDG----KASVVTSAENSDSADCLVFFAGCVASHDEWI 290
Query: 285 LDSGCSYHICPRKEYFEM-LELEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLLKDVR 343
LD+ C + IC +++F ++ G VV +G+N +I IG++++K D LKDVR
Sbjct: 291 LDTACLFLICINRDWFSSHKSVQNGDVVRMGDNNPREIMGIGSVQIKTHDGMTRTLKDVR 350
Query: 344 YIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGS-KIHGLYILEGSTVIADA 402
+IP + RNLIS+S D GY GV+++S G+L+ G LY+L GST+
Sbjct: 351 HIPGMARNLISLSTLDAEGYKYSGSGGVVKVSKGSLVYMIGDMNSANLYVLRGSTLHGSL 410
Query: 403 SVASV--DTLDVTKLWHLRLGHVSERGI 428
+ A+V D T LWH+RLGH+SE G+
Sbjct: 411 TAAAVSKDEPSKTNLWHMRLGHMSELGM 438
>UniRef100_Q6BCY1 Gag-Pol [Ipomoea batatas]
Length = 1298
Score = 219 bits (559), Expect = 2e-55
Identities = 155/442 (35%), Positives = 242/442 (54%), Gaps = 31/442 (7%)
Query: 1 KWDIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMDVHLTPAEK-TEMNDKAVSAI 59
K++IEKF G N F LWK+K++AIL + C+ A+ ++ V T +K +EMN+ A++ +
Sbjct: 4 KFEIEKFNGKN-FSLWKLKVKAILRKDNCLAAI---SERPVDFTDDKKWSEMNEDAMADL 59
Query: 60 ILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLT 119
L + D VL + + TA + + L+ LY KSL ++ LK++LY RM ES + E L
Sbjct: 60 YLSIADGVLSSIEEKKTANEIWDHLNRLYEAKSLHNKIFLKRKLYTLRMSESTSVTEHLN 119
Query: 120 EFNKIIDDLANIDVNLEDEDKALHLPCALPRSFE----NFKDTMLYGK*GTITLEEVQAA 175
N + L ++ +E +++A L +LP S++ N + +L + ++V AA
Sbjct: 120 TLNTLFSQLTSLSCKIEPQERAELLLQSLPDSYDQLIINLTNNILTDY---LVFDDVAAA 176
Query: 176 LRTKELT-KFKELKVED--SGEGLNVSRERSQNRGKGKGKNSRSKSRSKGDGNKTQYKCF 232
+ +E K KE + + E L V R RS RG+ G+ RSKS +K C+
Sbjct: 177 VLEEESRRKNKEDRQVNLQQAEALTVMRGRSTERGQSSGRG-RSKS------SKKNLTCY 229
Query: 233 ICHNPGHFKKDCPERKGNGGGNPSVQIASNEEGYESAGALTVTSWEPEKG----WVLDSG 288
C GH KKDC N NP +AS + + + E K W++DSG
Sbjct: 230 NCGKKGHLKKDCWNLAQNS--NPQGNVASTSDDGSALCCEASIAREGRKRFADIWLIDSG 287
Query: 289 CSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLLKDVRYIPKL 348
+YH+ RKE+F E GG V ++ A +I IGTI+LKM+D ++DVR++ L
Sbjct: 288 ATYHMTSRKEWFHHYEPISGGSVYSCDDHALEIIGIGTIKLKMYDGTVQTVQDVRHVKGL 347
Query: 349 RRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKI-HGLYILEGSTV-IADASVAS 406
++NL+S + D ++GVM+I GAL++ KG KI LY+L+G T+ A+ASVA+
Sbjct: 348 KKNLLSYGILDNSATQIETQKGVMKIFQGALVVMKGEKIAANLYMLKGETLQEAEASVAA 407
Query: 407 VDTLDVTKLWHLRLGHVSERGI 428
D T LWH +LGH+S++G+
Sbjct: 408 CSP-DSTLLWHQKLGHMSDQGM 428
>UniRef100_Q6L4V3 Putative polyprotein [Oryza sativa]
Length = 1243
Score = 214 bits (546), Expect = 7e-54
Identities = 139/439 (31%), Positives = 238/439 (53%), Gaps = 34/439 (7%)
Query: 1 KWDIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMDVHLTPAEKTEMNDKAVSAII 60
K+D+ F LW+VKMRA+L QQ +AL G + + EK + + KA+S I
Sbjct: 5 KYDLPLLDRDTRFSLWQVKMRAVLAQQDLDDALSGFDKRTQDWSNDEK-KRDRKAISYIH 63
Query: 61 LCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTE 120
L L + +L+EV +E TA + KL+ + MTK L + LKQ+L+ +++ + + +M+ L+
Sbjct: 64 LHLSNNILQEVLKEETAAGLWLKLEQICMTKDLTSKMHLKQKLFLHKLQDDESVMDHLSA 123
Query: 121 FNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GTITLEEVQAALRTKE 180
F +I+ DL +++V +++D L L C+LP S+ NF+ T+LY + T+TL+EV A KE
Sbjct: 124 FKEIVADLESMEVKYDEDDLGLILLCSLPSSYANFRGTILYSR-DTLTLKEVYDAFHAKE 182
Query: 181 LTKFKELKVEDSGEGLNVSRERSQNRGKGKGKNSRSKSRSKGDG-------NKTQYK-CF 232
++K + EG N E RG+ + KN++++SR K ++ +YK C
Sbjct: 183 -----KMKKMVTSEGSNSQAEGLVVRGRQQKKNTKNQSRDKSSSSYRGRTKSRGRYKSCK 237
Query: 233 ICHNPGHFKKDCPERKGNGGGNPSVQIASNEEGYESAGALTVTSWEPEKGWVLDSGCSYH 292
C GH +C + + + I ++ E A+ + V +GC+
Sbjct: 238 YCKRDGHDISECWKLQ-DKDKRTGKYIPKGKKEEEGKAAVVTDEKSDAELLVAYAGCAQ- 295
Query: 293 ICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLLKDVRYIPKLRRNL 352
+++F E +GG V +G++ C++ IGT+++KMFD L DV++IP L+R+L
Sbjct: 296 -TSDQDWFATYEALQGGTVLMGDDTPCEVAGIGTVQIKMFDGCIRTLSDVQHIPNLKRSL 354
Query: 353 ISISMFDGLGYCTRIERGVMRISHGALIIAK-GSKIHGLYILEGSTVIADASVA--SVDT 409
IS+ G+++++ G+L++ K K LY L G+T++ + + S+
Sbjct: 355 ISL-------------YGILKVTKGSLVVMKVDIKSANLYHLRGTTILGNVAAVFDSLSN 401
Query: 410 LDVTKLWHLRLGHVSERGI 428
D T LWH+RLGH+SE G+
Sbjct: 402 SDATNLWHMRLGHMSEIGL 420
>UniRef100_Q9ZPU5 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1335
Score = 214 bits (545), Expect = 9e-54
Identities = 135/404 (33%), Positives = 213/404 (52%), Gaps = 22/404 (5%)
Query: 48 KTEMNDKAVSAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYR 107
+ E DKA + I L + DKVLR++ TA LD L+M +SL HR + Y ++
Sbjct: 42 RLERCDKAKNVIFLNVADKVLRKIELCKTAAEAWETLDRLFMIRSLPHRVYTQLSFYTFK 101
Query: 108 MVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GT- 166
M E+K I E + +F KI+ DL ++ +++ DE +A+ L +LP ++ +TM Y
Sbjct: 102 MQENKKIDENIDDFLKIVADLNHLQIDVTDEVQAILLLSSLPARYDGLVETMKYSNSREK 161
Query: 167 ITLEEVQAALRTKELTKFKELKVEDSGEGLNVSRERSQNRGKGKGKNSRSKSRSKG-DGN 225
+ L++V A R KE + + G + +R R + +G +++SRSK DG
Sbjct: 162 LRLDDVMVAARDKERELSQNNRPVVEG---HFARGRPDGKNNNQGNKGKNRSRSKSADGK 218
Query: 226 KTQYKCFICHNPGHFKKDC------PERKGNGGGNPSVQIASNEEGYESAGALTVTSW-- 277
+ C+IC GHFKK C + K G N +A + E + A L T
Sbjct: 219 RV---CWICGKEGHFKKQCYKWIERNKSKQQGSDNGESSLAKSTEAFNPAMVLLATDETL 275
Query: 278 ----EPEKGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFD 333
WVLD+GCS+H+ PRK++F+ + G V +GN+ ++ IG+I+++ D
Sbjct: 276 VVTDSIANEWVLDTGCSFHMTPRKDWFKDFKELSSGYVKMGNDTYSPVKGIGSIKIRNSD 335
Query: 334 DRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLYIL 393
+L DVRY+P + RNLIS+ + G + + G+++I G I KG K LYIL
Sbjct: 336 GSQVILTDVRYMPNMTRNLISLGTLEDRGCWFKSQDGILKIVKGCSTILKGQKRDTLYIL 395
Query: 394 EGSTVIADASVASVDTLDVTKLWHLRLGHVSERGIWLN*LNKGC 437
+G T + S +S + D T LWH RLGH+S++G+ + + KGC
Sbjct: 396 DGVTEEGE-SHSSAEVKDETALWHSRLGHMSQKGMEIL-VKKGC 437
>UniRef100_Q9AU17 Polyprotein-like [Lycopersicon chilense]
Length = 1328
Score = 214 bits (545), Expect = 9e-54
Identities = 146/442 (33%), Positives = 224/442 (50%), Gaps = 22/442 (4%)
Query: 1 KWDIEKFTGSND-FGLWKVKMRAILIQQKCVEALKGEAQMDVHLTPAEKTEMNDKAVSAI 59
K+++ KF G F +W+ +M+ +LIQQ +AL G+++ + + E+++KA SAI
Sbjct: 5 KYEVAKFNGDKPVFSMWQRRMKDLLIQQGLHKALGGKSKKPESMKLEDWEELDEKAASAI 64
Query: 60 ILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLT 119
L L D V+ + E +A + KL++LYM+K+L ++ LK+QLY M E + L
Sbjct: 65 RLHLTDDVVNNIVDEESACGIWTKLENLYMSKTLTNKLYLKKQLYTLHMDEGTNFLSHLN 124
Query: 120 EFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GTITLEEVQAALRTK 179
N +I LAN+ V +E+EDK + L +LP S++ T+L+GK +I L++V +AL
Sbjct: 125 VLNGLITQLANLGVKIEEEDKRIVLLNSLPSSYDTLSTTILHGK-DSIQLKDVTSALLLN 183
Query: 180 ELTKFKELKVEDSGE-GLNVSRERSQNRGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPG 238
E + K E+ G+ + SR RS R S ++ +SK C+ C PG
Sbjct: 184 EKMR---KKPENHGQVFITESRGRSYQRSSSNYGRSGARGKSKVRSKSKARNCYNCDQPG 240
Query: 239 HFKKDCPERKGNGG--------GNPSVQIASNEEGY----ESAGALTVTSWEPEKGWVLD 286
HFK+DCP K G N + + +N++ E + + E E WV+D
Sbjct: 241 HFKRDCPNPKRGKGESSGQKNDDNTAAMVQNNDDVVLLINEEEECMHLAGTESE--WVVD 298
Query: 287 SGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLLKDVRYIP 346
+ SYH P ++ F + G V +GN KI IG I K +LKDVR++P
Sbjct: 299 TAASYHATPVRDLFCRYVAGDYGNVKMGNTSYSKIAGIGDICFKTNVGCTLVLKDVRHVP 358
Query: 347 KLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLYILEGSTVIADASVAS 406
LR NLIS D GY R++ GAL+IAKG LY + I + +
Sbjct: 359 DLRMNLISGIALDQDGYENYFANQKWRLTKGALVIAKGVARGTLY--RTNAEICQGELNA 416
Query: 407 VDTLDVTKLWHLRLGHVSERGI 428
+ LWH R+GH SE+G+
Sbjct: 417 AHEENSADLWHKRMGHTSEKGL 438
>UniRef100_Q9LS18 Retroelement pol polyprotein-like [Arabidopsis thaliana]
Length = 1338
Score = 209 bits (532), Expect = 3e-52
Identities = 136/462 (29%), Positives = 235/462 (50%), Gaps = 35/462 (7%)
Query: 5 EKFTGSNDFGLWKVKMRAILIQQKCVEALK--------------------GEAQMDVHLT 44
E+F G D+ LWK K+ A L +ALK E + + H
Sbjct: 18 ERFDGRGDYTLWKRKLLAQLEVMGISDALKEKEEKKEAVETERVKVVSSSSERRREEHKK 77
Query: 45 PAEKTEMNDKAVSAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLY 104
+ E +KA S IIL + D +LR + E TA M + LD LY++ L+ R LK++L+
Sbjct: 78 DHSREEKENKARSVIILSVADNILRRIRTEETAAGMISVLDKLYLSDPLSSRISLKRKLF 137
Query: 105 FYRMVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK* 164
++M E+K + E + +F +I++DL +DV + DEDKA L +LPR E K ++ Y +
Sbjct: 138 EFKMSENKAVEENIEDFFRIVEDLEKLDVYVSDEDKAFMLLLSLPRKLEQLKYSLDYCE- 196
Query: 165 GTITLEEVQAALRTKELTKFK-ELKVEDSGEGLNVSRERSQNRGKGKGKNSRSKSRSKGD 223
+TL V A+ KEL + E + E+ + L++ R + R + + ++ K + + +
Sbjct: 197 EPLTLGRVMTAIYKKELEVAQIERQTEEEEKRLSL---RERERSDYREEQAKGKEKVRSE 253
Query: 224 GNKTQYKCFICHNPGHFKKDCPERKGNGGGNPSVQI-ASNEEGYESAGALTVTSWEPEKG 282
+ + C+ C GH K +C + K N SV+ S+ + S G++ + S +
Sbjct: 254 AREKKGPCWRCGQKGHVKTECFQEKKNKSRKKSVRYEESSAQSIVSGGSVFMVSEAAARA 313
Query: 283 -------WVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFDDR 335
W+ D+GC+ H+ RKE+FE L E G V + N+ +++ IG++R+ D
Sbjct: 314 SKGSSEEWICDTGCTSHMSSRKEWFEDLVFSESGNVSMANDTTLQVKGIGSVRILNDDGT 373
Query: 336 DFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLYILEG 395
LL +V YIP + +NLIS+ + G + + G++++ G + + K K+ LY+L+G
Sbjct: 374 TVLLTNVMYIPGMSKNLISLGTLENKGCWFKSKNGILKVIKGCITLMKAEKVGTLYMLKG 433
Query: 396 STVIADASVASVDTLDVTKLWHLRLGHVSERGIWLN*LNKGC 437
V A A + TK+ H++ H+S+ + + + KGC
Sbjct: 434 KAVTARRR-AVQGPKEETKMEHIKPAHMSQTSLEIP-VKKGC 473
>UniRef100_Q6AUC7 Putative polyprotein [Oryza sativa]
Length = 1241
Score = 207 bits (527), Expect = 1e-51
Identities = 120/363 (33%), Positives = 197/363 (54%), Gaps = 29/363 (7%)
Query: 89 MTKSLAHRQCLKQQLYFYRMVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCAL 148
MTK L + LKQ+L+ +++ + +M+ L+ F +I+ DL +++V ++ED L L C+L
Sbjct: 1 MTKDLTSKMHLKQKLFLHKLQDDGSVMDHLSAFKEIVADLESMEVKYDEEDLGLILLCSL 60
Query: 149 PRSFENFKDTMLYGK*GTITLEEVQAALRTKELTKFKELKVEDSGEGLNVSRERSQNRGK 208
P S+ NF+DT+LY + T+TL+EV AL KE ++K EG N E RG+
Sbjct: 61 PSSYANFRDTILYSR-DTLTLKEVYDALHAKE-----KMKKMVPSEGSNSQAEGLVVRGR 114
Query: 209 GKGKNSRSKSRSKGDG-------NKTQYK-CFICHNPGHFKKDCPER------------K 248
+ KN+ +KSR K ++ +YK C C GH +C + K
Sbjct: 115 QQEKNTNNKSRDKSSSIYRGRSKSRGRYKSCKYCKRDGHDISECWKLQDKDKRTRKYIPK 174
Query: 249 GNGGGNPSVQIASNEEGYESAGALTVTSWEPEKGWVLDSGCSYHICPRKEYFEMLELEEG 308
G + ++E+ + W+LD+ C+YH+CP +++F E +G
Sbjct: 175 GKKEEEGKAAVVTDEKSDAELLVAYAGCAQTSDQWILDTACTYHMCPNRDWFATYEAVQG 234
Query: 309 GVVCLGNNKACKIQVIGTIRLKMFDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIE 368
G V +G++ C++ IGT+++KMFD L DVR+IP L+R+LIS+ D GY
Sbjct: 235 GTVLMGDDTPCEVAGIGTVQIKMFDGCIRTLLDVRHIPNLKRSLISLCTLDRKGYKYSGG 294
Query: 369 RGVMRISHGALIIAKGS-KIHGLYILEGSTVIADASVA--SVDTLDVTKLWHLRLGHVSE 425
G+++++ G+L++ K K LY L G+T++ + + S+ D T LWH+RLGH+SE
Sbjct: 295 DGILKVTKGSLVVMKADIKYANLYHLRGTTILGNVAAVSDSLSNSDATNLWHMRLGHMSE 354
Query: 426 RGI 428
G+
Sbjct: 355 IGL 357
>UniRef100_Q8H023 Putative retrovirus-related pol polyprotein [Oryza sativa]
Length = 556
Score = 202 bits (515), Expect = 3e-50
Identities = 127/388 (32%), Positives = 213/388 (54%), Gaps = 17/388 (4%)
Query: 14 GLWKVKMRAILIQQKCVEALKGEAQMDVHLTPAEKTEMNDKAVSAIILCLGDKVLREVSR 73
G +++++ +L QQ +AL E M + + EM +A + I L L D V+ +V
Sbjct: 152 GSSQMRLKDLLAQQGISKAL--EETMPEKMDVGKWVEMKAQATAIIRLSLSDFVMYQVMD 209
Query: 74 ESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTEFNKIIDDLANIDV 133
E T + +KL SLYM+KSL + LKQQLY +M E + + + FN+++ DL+ +DV
Sbjct: 210 EKTPKEIWDKLASLYMSKSLTSKLYLKQQLYGLQMQEESDLRKHVDVFNQLVVDLSKLDV 269
Query: 134 NLEDEDKALHLPCALPRSFENFKDTMLYGK*GTITLEEVQAALRTKELTKFKELKVEDSG 193
L+DEDKA+ L C+LP SFE+ T+ +GK T+ EE+ ++L ++L + K+ + ++
Sbjct: 270 KLDDEDKAIILLCSLPPSFEHVVTTLTHGK-DTVKTEEIISSLLARDLRRSKKNEAMEAS 328
Query: 194 EGLNVSRERSQNRGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCPERKGNGGG 253
+ ++ + + G SKS+ KG +C+ CH GH +++CP K GG
Sbjct: 329 QAESLLVKAKHDHEAGV-----SKSKEKGA------RCYKCHEFGHIRRNCPLLKKRKGG 377
Query: 254 NPSVQIASNEEGYESAGALTVTSWEPEKGWVLDSGCSYHICPRKEYFEMLELEEGGVVCL 313
S+ + S LTV++ + + W+LDS SYH+ + E+F + + GVV L
Sbjct: 378 IASLAARGDNSDSSSHEILTVSNEKSGEAWMLDSASSYHVTSKWEWFSSYKSGDFGVVYL 437
Query: 314 GNNKACKIQVIGTIRLKMFDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRI--ERGV 371
GN+ + ++ +G I+ KM+D + LL DVR++P LR++LIS+ G+ ++ +R
Sbjct: 438 GNDTSYRVIGVGDIKFKMYDGNEVLLSDVRHVPGLRKSLISLGSLHETGWLYQVDSDRKT 497
Query: 372 MRISHGALIIAKGSKIHG-LYILEGSTV 398
M I + G + LY L+GS V
Sbjct: 498 MNIMKDGKTVMTGERTSSCLYKLQGSAV 525
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.344 0.151 0.505
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,263,392,363
Number of Sequences: 2790947
Number of extensions: 48701793
Number of successful extensions: 227211
Number of sequences better than 10.0: 811
Number of HSP's better than 10.0 without gapping: 393
Number of HSP's successfully gapped in prelim test: 421
Number of HSP's that attempted gapping in prelim test: 224969
Number of HSP's gapped (non-prelim): 1432
length of query: 866
length of database: 848,049,833
effective HSP length: 136
effective length of query: 730
effective length of database: 468,481,041
effective search space: 341991159930
effective search space used: 341991159930
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 79 (35.0 bits)
Medicago: description of AC144730.2


