

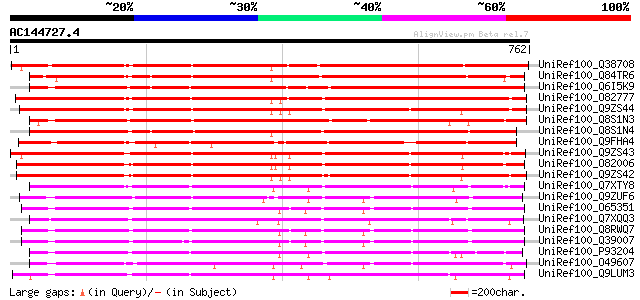
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144727.4 - phase: 0
(762 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q38708 Subtilisin-like protease [Alnus glutinosa] 690 0.0
UniRef100_Q84TR6 Subtilase [Casuarina glauca] 689 0.0
UniRef100_Q6I5K9 Putative subtilisin-like proteinase [Oryza sativa] 678 0.0
UniRef100_O82777 Subtilisin-like protease [Lycopersicon esculentum] 616 e-175
UniRef100_Q9ZS44 SBT4B protein [Lycopersicon esculentum] 607 e-172
UniRef100_Q8S1N3 Putative subtilisin-like protease [Oryza sativa] 603 e-171
UniRef100_Q8S1N4 Putative subtilisin-like protease [Oryza sativa] 603 e-171
UniRef100_Q9FHA4 Subtilisin-type protease-like [Arabidopsis thal... 596 e-169
UniRef100_Q9ZS43 SBT4C protein [Lycopersicon esculentum] 595 e-168
UniRef100_O82006 Subtilisin-like protease [Lycopersicon esculentum] 587 e-166
UniRef100_Q9ZS42 SBT4E protein [Lycopersicon esculentum] 575 e-162
UniRef100_Q7XTY8 OSJNBa0019K04.9 protein [Oryza sativa] 555 e-156
UniRef100_Q9ZUF6 Subtilisin-like serine protease, putative [Arab... 532 e-149
UniRef100_O65351 Cucumisin-like serine protease [Arabidopsis tha... 530 e-149
UniRef100_Q7XQQ3 OSJNBa0084A10.2 protein [Oryza sativa] 528 e-148
UniRef100_Q8RWQ7 AT5g67360/K8K14_8 [Arabidopsis thaliana] 525 e-147
UniRef100_Q39007 Subtilisin-like protease [Arabidopsis thaliana] 519 e-145
UniRef100_P93204 SBT1 protein [Lycopersicon esculentum] 518 e-145
UniRef100_O49607 Subtilisin proteinase-like [Arabidopsis thaliana] 518 e-145
UniRef100_Q9LUM3 Subtilisin proteinase-like protein [Arabidopsis... 516 e-145
>UniRef100_Q38708 Subtilisin-like protease [Alnus glutinosa]
Length = 761
Score = 690 bits (1781), Expect = 0.0
Identities = 371/775 (47%), Positives = 515/775 (65%), Gaps = 35/775 (4%)
Query: 3 SNICLWLWFSYIT----SLHVIFTLALSDNYIIHMNLSDMPKSFSNQHSWYESTLAQVTT 58
+ ICL F + + +LH T YI+HM+ S MPK+F++ H+WY S + +
Sbjct: 5 NGICLPYLFLFASCICLALHASSTSMEKSTYIVHMDKSHMPKAFTSHHNWYSSIVDCL-- 62
Query: 59 TNNNLNNSTSSKIFYTYTNVMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQ 118
N T+S YTY +V++GFSA+LS +E ++L+ GF+S+ D LDTTH+P+
Sbjct: 63 ---NSEKPTTSSFVYTYNHVLHGFSASLSHQELDTLRESPGFVSAYRDRNATLDTTHTPR 119
Query: 119 FLGLNPYRGAWPTSDFGKDIIVGVIDTGVWPESESFRDDGMT-KIPSKWKGQLCQFENSN 177
FL LNP G WP S++G+D+I+GVID+GVWPES+SF+DDGMT ++P++WKG +C E
Sbjct: 120 FLSLNPTGGLWPASNYGEDVIIGVIDSGVWPESDSFKDDGMTAQVPARWKG-ICSREG-- 176
Query: 178 IQSINLSLCNKKLIGARFFNKGFLAKHSNISTTILNSTRDTNGHGTHTSTTAAGSKVDGA 237
N S+CN KLIGAR+FN G +A N +T +NS RDT GHGTHT++TAAG+ V+GA
Sbjct: 177 ---FNSSMCNSKLIGARYFNNGIMAAIPN-ATFSMNSARDTLGHGTHTASTAAGNYVNGA 232
Query: 238 SFFGYANGTARGIASSSRVAIYKTAWGKDGDALSSDIIAAIDAAISDGVDILSISLGSDD 297
S+FGY GTARGIA +RVA+YK W + +SD++A ID AI+DGVD++SISLG D
Sbjct: 233 SYFGYGKGTARGIAPRARVAVYKVTWPEG--RYTSDVLAGIDQAIADGVDVISISLGYDG 290
Query: 298 LLLYKDPVAIATFAAMEKGIFVSTSAGNNGPSFKSIHNGIPWVITVAAGTLDREFLGTVT 357
+ LY+DP+AIA+FAAMEKG+ VSTSAGN GP F ++HNGIPWV+TVAAG +DR F GT+T
Sbjct: 291 VPLYEDPIAIASFAAMEKGVVVSTSAGNAGPFFGNMHNGIPWVLTVAAGNIDRSFAGTLT 350
Query: 358 LGNGVSLTGLSFYLGNFSANNFPIVF---MGMCDNVKELNTVKRKIVVCEGNNETLHEQM 414
LGN ++TG + + + + +V+ + C++ + L+ +V+CE ++ Q+
Sbjct: 351 LGNDQTITGWTMFPASAIIESSQLVYNKTISACNSTELLSDAVYSVVICEAITP-IYAQI 409
Query: 415 FNVYKAKVVGGVFISNILDINDVDN--SFPSIIINPVNGEIVKAYIKSHNSNASSIANMS 472
+ ++ V G + ISN + ++ S P ++I+P + A IK ++ +A +
Sbjct: 410 DAITRSNVAGAILISNHTKLFELGGGVSCPCLVISPKDAA---ALIKYAKTDEFPLAGLK 466
Query: 473 FKKTAFGVKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNFGTEVF- 531
F++T G K P+V +YSSRGPS S P +LKPD+ APG+ +LA+W N + GT V+
Sbjct: 467 FQETITGTKPAPAVAYYSSRGPSPSYPGILKPDVMAPGSLVLASWIPNEATAQIGTNVYL 526
Query: 532 -NNFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNG 590
+++N++ GTSM+CPH +GVAALLK AH WSP++IRSA+MTT++ LDNT I + G
Sbjct: 527 SSHYNMVSGTSMACPHASGVAALLKAAHPEWSPAAIRSAMMTTANPLDNTLNPIHENGKK 586
Query: 591 NRAATPFALGAGHINPNRALDPGLVYDIGVQDYINLLCALNFTQKNISAITRSSFNDCSK 650
A+P A+GAGHI+PNRALDPGLVYD QDYINLLC++N+ + I AI RS CS
Sbjct: 587 FHLASPLAMGAGHIDPNRALDPGLVYDATPQDYINLLCSMNYNKAQILAIVRSDSYTCSN 646
Query: 651 -PSLDLNYPSFIAFSNARNSSRTTNEFHRTVTNVGEKKTTYFASITPIKGFRVTVIPNKL 709
PS DLNYPSFIAF N+ R+ N F RTVTNVG+ TY A++T K RV V P L
Sbjct: 647 DPSSDLNYPSFIAFHNS-TCRRSVNTFQRTVTNVGDGAATYKATVTAPKDSRVIVSPQTL 705
Query: 710 VFKKKNEKISYKLK-IEGPRMTQKNKVAFGYLSW--RDGKHVVRSPIVVTNINFN 761
F K EK SY L I R T++ ++FG L W +GKH+VRSPIVV+ + N
Sbjct: 706 AFGSKYEKQSYNLTIINFTRDTKRKDISFGALVWANENGKHMVRSPIVVSPLRIN 760
>UniRef100_Q84TR6 Subtilase [Casuarina glauca]
Length = 764
Score = 689 bits (1777), Expect = 0.0
Identities = 369/745 (49%), Positives = 511/745 (68%), Gaps = 37/745 (4%)
Query: 29 YIIHMNLSDMPKSFSNQHSWYESTLAQVTTTNNNLNNST-----SSKIFYTYTNVMNGFS 83
YI+HM+ S MPK+F++ HSWY L+ + + N+ ST +S YTY +V++GFS
Sbjct: 35 YIVHMDKSHMPKAFTSHHSWY---LSIIDSLNSERPTSTEELKSASSFLYTYNHVLHGFS 91
Query: 84 ANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPYRGAWPTSDFGKDIIVGVI 143
L E+ ESLK GFIS+ D LDTTH+P+FL L+P G WPTS++G+D+I+GVI
Sbjct: 92 VALCQEDVESLKNTPGFISAYQDRNATLDTTHTPEFLSLSPSWGLWPTSNYGEDVIIGVI 151
Query: 144 DTGVWPESESFRDDGMT-KIPSKWKGQLCQFENSNIQSINLSLCNKKLIGARFFNKGFLA 202
D+GVWPESESF DDGM +P++WKG +CQ + N S CN KLIGAR+FN G LA
Sbjct: 152 DSGVWPESESFNDDGMNASVPARWKG-ICQVG----EQFNSSHCNSKLIGARYFNNGILA 206
Query: 203 KHSNISTTILNSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTA 262
+ NI T +NS RDT GHGTHT++TAAG+ V+ SFFGY GTARGIA +R+A+YK
Sbjct: 207 ANPNI-TFGMNSARDTIGHGTHTASTAAGNYVNDVSFFGYGKGTARGIAPRARLAVYKVN 265
Query: 263 WGKDGDALSSDIIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFAAMEKGIFVSTS 322
W ++G +SD++A ID AI+DGVD++SIS+G D L++DP+AIA+FAAMEKG+ VSTS
Sbjct: 266 W-REG-RYASDVLAGIDQAIADGVDVISISMGFDGAPLHEDPIAIASFAAMEKGVLVSTS 323
Query: 323 AGNNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLSFYLGNFSANNFPIV 382
AGN GP F ++HNGIPWV+TVA GT+DR F GT+TLGN +TG + + + N P+V
Sbjct: 324 AGNEGPFFGNLHNGIPWVLTVAGGTVDRSFAGTLTLGNDQIITGWTLFPASAVIQNLPLV 383
Query: 383 F---MGMCDNVKELNTVKRKIVVCEGNNETLHEQMFNVYKAKVVGGVFISNILDINDVDN 439
+ + C++ + L+ I++CE ++ +Q+ ++ ++ VVG + ISN + +++
Sbjct: 384 YDKNISACNSPELLSEAIYTIIICE-QARSIRDQIDSLARSNVVGAILISNNTNSSELGE 442
Query: 440 -SFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSC 498
+ P ++I+P + E V IK N N + A+M F+KT G K P+V Y+SRGPS S
Sbjct: 443 VTCPCLVISPKDAEAV---IKYANFNEIAFASMKFQKTFLGAKPAPAVASYTSRGPSPSY 499
Query: 499 PYVLKPDITAPGTSILAAWPTNVPVSNFGTEVF--NNFNLIDGTSMSCPHVAGVAALLKG 556
P VLKPD+ APG+ ILAAW + GT V+ +++N++ GTSM+CPH +G+AALLK
Sbjct: 500 PGVLKPDVMAPGSQILAAWVPTDATAQIGTNVYLSSHYNMVSGTSMACPHASGIAALLKA 559
Query: 557 AHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNRALDPGLVY 616
AH WSP++IRSA++TT++ LDNT++ I+D G ++ A+P A+GAG+I+PN AL+PGLVY
Sbjct: 560 AHPEWSPAAIRSAMITTANPLDNTQKPIRDNGLDHQVASPLAMGAGNIDPNCALEPGLVY 619
Query: 617 DIGVQDYINLLCALNFTQKNISAITRSSFNDCSKPSLDLNYPSFIAFSNARNSSRTTNEF 676
D QDYINLLC++NF + I AI R+ +CS PS DLNYPSFIAF N +N + +F
Sbjct: 620 DATPQDYINLLCSMNFDRTQILAIIRTRSYNCSNPSSDLNYPSFIAFHNGKNDT-VVKKF 678
Query: 677 HRTVTNVGEKKTTYFASITPIKGFRVTVIPNKLVFKKKNEKISYKLKIE---GPRMTQKN 733
RTVTNVG+ Y ASI +G RV V P LVFK+K E+ S+ L ++ GP+M
Sbjct: 679 RRTVTNVGDAVAIYNASIAAPRGSRVVVYPQTLVFKEKYEQKSFTLTMKFKRGPKM---- 734
Query: 734 KVAFGYLSW--RDGKHVVRSPIVVT 756
+FG L W +GKH+VRSPIVV+
Sbjct: 735 DTSFGALVWTHENGKHIVRSPIVVS 759
>UniRef100_Q6I5K9 Putative subtilisin-like proteinase [Oryza sativa]
Length = 761
Score = 678 bits (1749), Expect = 0.0
Identities = 362/739 (48%), Positives = 495/739 (65%), Gaps = 33/739 (4%)
Query: 29 YIIHMNLSDMPKSFSNQHSWYESTLAQVTTTNNNLNNSTSSKIFYTYTNVMNGFSANLSP 88
YI+HM+ S MP++F++Q SWYESTLA + + +FY Y N M+GF+A ++
Sbjct: 38 YIVHMDKSAMPRAFASQASWYESTLAAA---------APGADMFYVYDNAMHGFAARVTA 88
Query: 89 EEHESLKTFSGFISSIPD--LPLKLDTTHSPQFLGLNPYRGA-WPTSDFGKDIIVGVIDT 145
+E E L+ GF+S PD ++ DTTH+P+FLG++ G W S++G+D+IVGV+DT
Sbjct: 89 DELEKLRGSRGFVSCYPDDARAVRRDTTHTPEFLGVSASSGGLWEASEYGEDVIVGVVDT 148
Query: 146 GVWPESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSLCNKKLIGARFFNKGFLAKHS 205
GVWPES SFRDDG+ +P++WKG C+ S +CN+KL+GAR FNKG +A +
Sbjct: 149 GVWPESASFRDDGLPPVPARWKGY-CE---SGTAFDAGKVCNRKLVGARKFNKGLVAA-T 203
Query: 206 NISTTILNSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWGK 265
N+ T +NS RDT+GHGTHTS+TAAGS V GASFFGYA GTARG+A +RVA+YK W
Sbjct: 204 NL-TIAVNSPRDTDGHGTHTSSTAAGSPVAGASFFGYAPGTARGMAPRARVAMYKALW-- 260
Query: 266 DGDALSSDIIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFAAMEKGIFVSTSAGN 325
D SDI+AAID AI+DGVD+LS+SLG +D+ Y+DP+AI FAAM++G+FVSTSAGN
Sbjct: 261 DEGTYPSDILAAIDQAIADGVDVLSLSLGLNDVPFYRDPIAIGAFAAMQRGVFVSTSAGN 320
Query: 326 NGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLSFYLGNFSA-NNFPIVFM 384
+GP +HNG PW +TVA+GT DREF G V LG+G ++ G S Y G+ S + VF+
Sbjct: 321 DGPDPGFLHNGTPWTLTVASGTGDREFAGIVRLGDGTTVIGQSMYPGSPSTIASSGFVFL 380
Query: 385 GMCDNVKELNTVKRKIVVCEGNNETLHEQMFNVYKAKVVGGVFISN--ILDINDVDNSFP 442
G CDN L + K+V+C+ ++L +F V AK G+F+SN ++++ +FP
Sbjct: 381 GACDNDTALARNRDKVVLCDAT-DSLSAAIFAVQVAKARAGLFLSNDSFRELSE-HFTFP 438
Query: 443 SIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYVL 502
+I++P + + YIK + +SI F T G K P V YSSRGPS SCP VL
Sbjct: 439 GVILSPQDAPALLQYIKRSRAPRASI---KFGVTILGTKPAPVVATYSSRGPSASCPTVL 495
Query: 503 KPDITAPGTSILAAWPTNVPVSNFGTE-VFNNFNLIDGTSMSCPHVAGVAALLKGAHNGW 561
KPD+ APG+ ILA+WP NV VS G++ +++ FN+I GTSMSCPH +GVAAL+K H W
Sbjct: 496 KPDVLAPGSLILASWPENVSVSTVGSQQLYSRFNVISGTSMSCPHASGVAALIKAVHPEW 555
Query: 562 SPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNRALDPGLVYDIGVQ 621
SP+++RSA+MTT+ +DNT IKD+G NR ATP A+G+GHI+PNRA+DPGLVYD G
Sbjct: 556 SPAAVRSAMMTTASAVDNTNAPIKDMGRANRGATPLAMGSGHIDPNRAVDPGLVYDAGAD 615
Query: 622 DYINLLCALNFTQKNISAITR--SSFNDCSKPSLDLNYPSFIAFSNARNSSRTTNEFHRT 679
DY+ L+CA+N+T I + + SS DC+ +LDLNYPSFIAF + ++ F R
Sbjct: 616 DYVKLMCAMNYTAAQIKTVAQSPSSAVDCAGATLDLNYPSFIAFFDPGATAPAARTFTRA 675
Query: 680 VTNVGEKKTTYFASITPIKGFRVTVIPNKLVFKKKNEKISYKLKIEGPRMTQKNKVAFGY 739
VTNVG+ +Y A + + G V+V P +LVF +K+E Y + I G + ++V G
Sbjct: 676 VTNVGDAPASYSAKVKGLGGLTVSVSPERLVFGRKHETQKYTVVIRGQMKNKTDEVLHGS 735
Query: 740 LSWRD--GKHVVRSPIVVT 756
L+W D GK+ VRSPIV T
Sbjct: 736 LTWVDDAGKYTVRSPIVAT 754
>UniRef100_O82777 Subtilisin-like protease [Lycopersicon esculentum]
Length = 761
Score = 616 bits (1589), Expect = e-175
Identities = 343/767 (44%), Positives = 488/767 (62%), Gaps = 33/767 (4%)
Query: 9 LWFSYITSLHVIFTLALSDNYIIHMNLSDMPKSFSNQHSWYESTLAQV-TTTNNNLNNST 67
L FS+ S H+ LA YI+H++ S MP F++ H W+ ST+ + + ++++
Sbjct: 7 LLFSWALSAHLFLALAQRSTYIVHLDKSLMPNVFTDHHHWHSSTIDSIKASVPSSVDRFH 66
Query: 68 SS-KIFYTYTNVMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPYR 126
S+ K+ Y+Y NV++GFSA LS +E +LK GFIS+ D ++ TTH+ FL LNP
Sbjct: 67 SAPKLVYSYDNVLHGFSAVLSKDELAALKKLPGFISAYKDRTVEPHTTHTSDFLKLNPSS 126
Query: 127 GAWPTSDFGKDIIVGVIDTGVWPESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSLC 186
G WP S G+D+IV V+D+G+WPES SF+DDGM +IP +WKG +C+ N S+C
Sbjct: 127 GLWPASGLGQDVIVAVLDSGIWPESASFQDDGMPEIPKRWKG-ICKPGT----QFNASMC 181
Query: 187 NKKLIGARFFNKGFLAKHSNISTTILNSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGT 246
N+KLIGA +FNKG LA ++ T +NS RDT+GHGTH ++ AG+ G S FGYA GT
Sbjct: 182 NRKLIGANYFNKGILANDPTVNIT-MNSARDTDGHGTHCASITAGNFAKGVSHFGYAPGT 240
Query: 247 ARGIASSSRVAIYKTAWGKDGDALSSDIIAAIDAAISDGVDILSISLGSDDLLLYKDPVA 306
ARG+A +R+A+YK ++ + +SD+IAA+D A++DGVD++SIS G + LY+D ++
Sbjct: 241 ARGVAPRARLAVYKFSFNE--GTFTSDLIAAMDQAVADGVDMISISYGYRFIPLYEDAIS 298
Query: 307 IATFAAMEKGIFVSTSAGNNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTG 366
IA+F AM KG+ VS SAGN GP S++NG PW++ VA+G DR F GT+TLGNG+ + G
Sbjct: 299 IASFGAMMKGVLVSASAGNRGPGIGSLNNGSPWILCVASGHTDRTFAGTLTLGNGLKIRG 358
Query: 367 LSFYLGNFSANNFPIVF---MGMCDNVKELNTV---KRKIVVCEGNNETLHEQMFNVYKA 420
S + + P+++ + C + + L+ V + IV+C+ N + +QM + +A
Sbjct: 359 WSLFPARAFVRDSPVIYNKTLSDCSSEELLSQVENPENTIVICDDNGD-FSDQMRIITRA 417
Query: 421 KVVGGVFISNILDI-NDVDNSFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFG 479
++ +FIS + P +++N G+ V Y+K ++ + A ++F++T
Sbjct: 418 RLKAAIFISEDPGVFRSATFPNPGVVVNKKEGKQVINYVK---NSVTPTATITFQETYLD 474
Query: 480 VKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNFGTEVF--NNFNLI 537
K P V S+RGPS S + KPDI APG ILAA+P NV ++ GT + ++ L
Sbjct: 475 TKPAPVVAASSARGPSRSYLGISKPDILAPGVLILAAYPPNVFATSIGTNILLSTDYILE 534
Query: 538 DGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPF 597
GTSM+ PH AG+AA+LK AH WSPS+IRSA+MTT+D LDNT++ IKD N N+AATP
Sbjct: 535 SGTSMAAPHAAGIAAMLKAAHPEWSPSAIRSAMMTTADPLDNTRKPIKDSDN-NKAATPL 593
Query: 598 ALGAGHINPNRALDPGLVYDIGVQDYINLLCALNFTQKNISAITRSS-FNDCSKPSLDLN 656
+GAGH++PNRALDPGLVYD QDY+NLLC+LNFT++ I RSS ++CS PS DLN
Sbjct: 594 DMGAGHVDPNRALDPGLVYDATPQDYVNLLCSLNFTEEQFKTIARSSASHNCSNPSADLN 653
Query: 657 YPSFIA-FSNARNSSRTTNEFHRTVTNVGEKKTTYFASITPIKGFRVTVIPNKLVFKKKN 715
YPSFIA +S N + +F RTVTNVG+ TY A + K ++V P LVFK KN
Sbjct: 654 YPSFIALYSIEGNFTLLEQKFKRTVTNVGKGAATYKAKLKAPKNSTISVSPQILVFKNKN 713
Query: 716 EKISYKLKIE--GPRMTQKNKVAFGYLSW--RDGKHVVRSPIVVTNI 758
EK SY L I G +N G ++W ++G H VRSPIV + I
Sbjct: 714 EKQSYTLTIRYIGDEGQSRN---VGSITWVEQNGNHSVRSPIVTSPI 757
>UniRef100_Q9ZS44 SBT4B protein [Lycopersicon esculentum]
Length = 777
Score = 607 bits (1566), Expect = e-172
Identities = 342/768 (44%), Positives = 487/768 (62%), Gaps = 40/768 (5%)
Query: 15 TSLHVIFTLALSDNYIIHMNLSDMPKSFSNQHSWYESTLAQV-TTTNNNLNNSTSS-KIF 72
T + T+A YI+H++ S MP F++ H W+ ST+ + + ++++ S+ K+
Sbjct: 18 THMFCFLTIAQRSTYIVHLDKSLMPNVFTDHHHWHSSTIDSIKASVPSSVDRFHSAPKLV 77
Query: 73 YTYTNVMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPYRGAWPTS 132
Y+Y NV +GFSA LS E +LK GF+S+ D ++ TTH+ FL LNP G WP S
Sbjct: 78 YSYDNVFHGFSAVLSQNELAALKKLPGFVSAYEDRTVEPHTTHTSDFLKLNPSSGLWPAS 137
Query: 133 DFGKDIIVGVIDTGVWPESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSLCNKKLIG 192
G+D+I+ V+D G+WPES SF+DDGM +IP +WKG +C+ N S+CN+KLIG
Sbjct: 138 GLGQDVIIAVLDGGIWPESASFQDDGMPEIPKRWKG-ICRPGTQ----FNTSMCNRKLIG 192
Query: 193 ARFFNKGFLAKHSNISTTILNSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIAS 252
A +FNKG LA ++ + +NS RDTNGHGTH ++ AAG+ AS FGYA G ARG+A
Sbjct: 193 ANYFNKGILADDPTVNIS-MNSARDTNGHGTHCASIAAGNFAKDASHFGYAPGIARGVAP 251
Query: 253 SSRVAIYKTAWGKDGDALSSDIIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFAA 312
+R+A+YK ++ + +SD+IAA+D A++DGVD++SIS G + LY+D ++IA+F A
Sbjct: 252 RARIAVYKFSFSEG--TFTSDLIAAMDQAVADGVDMISISFGYRFIPLYEDAISIASFGA 309
Query: 313 MEKGIFVSTSAGNNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLSFYLG 372
M KG+ VS SAGN GPS S+ NG PW++ VAAG DR F GT+TLGNG+ + G S +
Sbjct: 310 MMKGVLVSASAGNRGPSVGSLGNGSPWILCVAAGHTDRRFAGTLTLGNGLKIRGWSLFPA 369
Query: 373 NFSANNFPIVF---MGMCDNVKELNTV---KRKIVVCEGNNET----LHEQMFNVYKAKV 422
+ +++ + CD+V+ L+ V +R IV+C+ N + Q+FN+ +A+V
Sbjct: 370 RAYVRDSLVIYNKTLATCDSVELLSQVPDAERTIVICDYNADEDGFGFASQIFNINQARV 429
Query: 423 VGGVFISNILDI-NDVDNSFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAF-GV 480
G+FIS + S+P ++IN G+ V Y+K ++AS A ++F++T G
Sbjct: 430 KAGIFISEDPTVFTSSSFSYPGVVINKKEGKQVINYVK---NSASPTATITFQETYMDGE 486
Query: 481 KSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNV-PVSNFGTEVFNNFNLIDG 539
+ P + +S+RGPS S + KPDI APG ILAA+P N+ S E+ +++ L G
Sbjct: 487 RPAPILARFSARGPSRSYLGIPKPDIMAPGVLILAAFPPNIFSESIQNIELSSDYELKSG 546
Query: 540 TSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPFAL 599
TSM+ PH AG+AA+LKGAH WSPS+IRSA+MTT++ LD+T++ I++ + N ATP +
Sbjct: 547 TSMAAPHAAGIAAMLKGAHPEWSPSAIRSAMMTTANHLDSTQKPIRE--DDNMIATPLDM 604
Query: 600 GAGHINPNRALDPGLVYDIGVQDYINLLCALNFTQKNISAITRSS--FNDCSKPSLDLNY 657
GAGH++PNRALDPGLVYD QDYINL+C++NFT++ RSS +N+CS PS DLNY
Sbjct: 605 GAGHVDPNRALDPGLVYDATPQDYINLICSMNFTEEQFKTFARSSANYNNCSNPSADLNY 664
Query: 658 PSFIA---FSNARNSSRTTNEFHRTVTNVGEKKTTYFASITPIKGFRVTVIPNKLVFKKK 714
PSFIA FS N + +F RT+TNVG+ TY I K V+V P LVFK K
Sbjct: 665 PSFIALYPFSLEGNFTWLEQKFRRTLTNVGKGGATYKVKIETPKNSTVSVSPRTLVFKGK 724
Query: 715 NEKISYKLKIE--GPRMTQKNKVAFGYLSW--RDGKHVVRSPIVVTNI 758
N+K SY L I G KN FG ++W +G H VRSPIV + I
Sbjct: 725 NDKQSYNLTIRYIGDSDQSKN---FGSITWVEENGNHTVRSPIVTSTI 769
>UniRef100_Q8S1N3 Putative subtilisin-like protease [Oryza sativa]
Length = 760
Score = 603 bits (1555), Expect = e-171
Identities = 341/752 (45%), Positives = 470/752 (62%), Gaps = 43/752 (5%)
Query: 29 YIIHMNLSDMPKS------FSNQHSWYESTLAQVTTTNNNLNNSTSSKIFYTYTNVMNGF 82
YI+HM+ S MP ++ SWY +TL + +++ Y Y N M+GF
Sbjct: 27 YIVHMDKSAMPSGGGGGNGSTSLESWYAATLRAA---------APGARMIYVYRNAMSGF 77
Query: 83 SANLSPEEHESLKTFSGFISSIPDLPL-KLDTTHSPQFLGLNPYRGAWPTSDFGKDIIVG 141
+A LS E+H L GF+SS D P+ + DTTH+P+FLG++ G W T+ +G +IVG
Sbjct: 78 AARLSAEQHARLSRSPGFLSSYLDAPVTRRDTTHTPEFLGVSGAGGLWETASYGDGVIVG 137
Query: 142 VIDTGVWPESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSLCNKKLIGARFFNKGFL 201
V+DTGVWPES S+RDDG+ +P++WKG C+ S + CN+KLIGAR F+ G
Sbjct: 138 VVDTGVWPESGSYRDDGLPPVPARWKGY-CE---SGTRFDGAKACNRKLIGARKFSAGLA 193
Query: 202 AKHSNISTTI-LNSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYK 260
A + TI +NS RDT+GHGTHTS+TAAGS V GAS+FGYA G ARG+A +RVA+YK
Sbjct: 194 AALGRRNITIAVNSPRDTDGHGTHTSSTAAGSPVPGASYFGYAPGVARGMAPRARVAVYK 253
Query: 261 TAWGKDGDALSSDIIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFAAMEKGIFVS 320
+ + G ++DI+AAID AI+DGVD+LSISLG ++ L+ DPVAI +FAAM+ GIFVS
Sbjct: 254 VLFDEGG--YTTDIVAAIDQAIADGVDVLSISLGLNNRPLHTDPVAIGSFAAMQHGIFVS 311
Query: 321 TSAGNNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLSFYLGNFS-ANNF 379
TSAGN+GP +HNG PW +TVAAGT+DREF G V LG+G ++ G S Y G+ +
Sbjct: 312 TSAGNDGPGLSVLHNGAPWALTVAAGTVDREFSGIVELGDGTTVIGESLYAGSPPITQST 371
Query: 380 PIVFMGMCDNVKELNTVKRKIVVC--EGNNETLHEQMFNVYKAKVVGGVFISN-ILDIND 436
P+V++ CDN + + KIV+C + ++ L + V A GG+F++N +
Sbjct: 372 PLVYLDSCDNFTAIRRNRDKIVLCDAQASSFALQVAVQFVQDANAAGGLFLTNDPFRLLF 431
Query: 437 VDNSFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSN 496
+FP +++P +G + YI+ + + IA F+ T K P YSSRGP+
Sbjct: 432 EQFTFPGALLSPHDGPAILRYIQRSGAPTAKIA---FRATLLNTKPAPEAAAYSSRGPAV 488
Query: 497 SCPYVLKPDITAPGTSILAAWPTNVPVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLKG 556
SCP VLKPDI APG+ +LA+W +V V + + FN+I GTSM+ PH AGVAALL+
Sbjct: 489 SCPTVLKPDIMAPGSLVLASWAESVAVVG---NMTSPFNIISGTSMATPHAAGVAALLRA 545
Query: 557 AHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNRALDPGLVY 616
H WSP++IRSA+MTT+ LDNT I D+ AATP A+G+GHI+PNRA DPGLVY
Sbjct: 546 VHPEWSPAAIRSAMMTTAATLDNTGRSINDMARAGHAATPLAMGSGHIDPNRAADPGLVY 605
Query: 617 DIGVQDYINLLCALNFTQKNISAITRSS---FNDCSKPSLDLNYPSFIAFSNARNSSRT- 672
D DY+ L+CA+ + +I A+T+ S N S DLNYPSFIA+ + R+++
Sbjct: 606 DAVPGDYVELMCAMGYNLSDIRAVTQWSTYAVNCSGASSPDLNYPSFIAYFDRRSAAAAA 665
Query: 673 --TNEFHRTVTNVGEKKTTYFASIT-PIKGFRVTVIPNKLVFKKKNEKISYKLKIEGPRM 729
T F R VTNVG +Y A + + G V+V P++LVF KK E Y L + G ++
Sbjct: 666 AETKTFVRVVTNVGAGAASYRAKVKGNLGGLAVSVTPSRLVFGKKGETQKYTLVLRG-KI 724
Query: 730 TQKNKVAFGYLSWRD--GKHVVRSPIVVTNIN 759
+KV G L+W D GK+ VRSPIV T ++
Sbjct: 725 KGADKVLHGSLTWVDDAGKYTVRSPIVATTLS 756
>UniRef100_Q8S1N4 Putative subtilisin-like protease [Oryza sativa]
Length = 753
Score = 603 bits (1554), Expect = e-171
Identities = 335/725 (46%), Positives = 459/725 (63%), Gaps = 24/725 (3%)
Query: 29 YIIHMNLSDMPKSFSNQHSWYESTLAQVTTTNNNLNNSTSSKIFYTYTNVMNGFSANLSP 88
YI+HM+ S MP S+ WY +T+A +T +I YTY ++GF+A LS
Sbjct: 34 YIVHMDKSAMPAHHSDHREWYSATVATLTPGAPR-GGRGGPRIVYTYDEALHGFAATLSA 92
Query: 89 EEHESLKTFSGFISSIPDLPLKL--DTTHSPQFLGLNPYRGAWPTSDFGKDIIVGVIDTG 146
E +L+ GF+S+ PD + DTTHS +FL L+P+ G WP + FG+ +I+GVIDTG
Sbjct: 93 SELGALRLAPGFVSAYPDRRADVLHDTTHSTEFLRLSPFGGLWPAARFGEGVIIGVIDTG 152
Query: 147 VWPESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSLCNKKLIGARFFNKGFLAKHSN 206
VWPES SF D GM +PS+W+G+ C+ Q L +CN+KLIGAR+FN+G +A +
Sbjct: 153 VWPESASFDDGGMPPVPSRWRGE-CEAG----QDFTLDMCNRKLIGARYFNRGLVAANPT 207
Query: 207 ISTTILNSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWGKD 266
+ T +NSTRDT GHGTHTS+TA GS ASFFGY GTA G+A + VA+YK W +
Sbjct: 208 V-TVSMNSTRDTLGHGTHTSSTAGGSPAPCASFFGYGRGTASGVAPRAHVAMYKAMWPEG 266
Query: 267 GDALSSDIIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFAAMEKGIFVSTSAGNN 326
A SD++AA+DAAI+DGVD++SIS G D + LY+DPVAIA FAA+E+GI VS SAGN+
Sbjct: 267 RYA--SDVLAAMDAAIADGVDVISISSGFDGVPLYEDPVAIAAFAAIERGILVSASAGND 324
Query: 327 GPSFKSIHNGIPWVITVAAGTLDRE-FLGTVTLGNGV--SLTGLSFYLGNFSANNFPIVF 383
GP ++HNGIPW++TVAAG +DR+ F G++ LG+ ++TG++ Y N + +V+
Sbjct: 325 GPRLGTLHNGIPWLLTVAAGMVDRQMFAGSIYLGDDTRSTITGITRYPENAWIKDMNLVY 384
Query: 384 ---MGMCDNVKELNTVKRKIVVCEGNNETLHEQMFNVYKAKVVGGVFISNILDINDVDNS 440
+ C++ L T+ + IVVC L +QM +A V +FISN I + +
Sbjct: 385 NDTISACNSSTSLATLAQSIVVCYDTGILL-DQMRTAAEAGVSAAIFISNTTLITQSEMT 443
Query: 441 FPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPY 500
FP+I++NP + + +YI NS+A A + F++T G + P V YSSRGPS S
Sbjct: 444 FPAIVVNPSDAASLLSYI---NSSARPTATIKFQQTIIGTRPAPVVAAYSSRGPSRSYEG 500
Query: 501 VLKPDITAPGTSILAAWPTNVPVSNFG-TEVFNNFNLIDGTSMSCPHVAGVAALLKGAHN 559
VLKPDI APG SILAAW P++ G T + ++F + GTSM+CPH AGVAALL+ AH
Sbjct: 501 VLKPDIMAPGDSILAAWAPVAPLAQVGSTALGSDFAVESGTSMACPHAAGVAALLRAAHP 560
Query: 560 GWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNRALDPGLVYDIG 619
WSP+ I+SA+MTT+ +DNT I D G+G+ AA+P A+GAG ++PN A+DPGLVYD G
Sbjct: 561 DWSPAMIKSAMMTTATAVDNTFRPIGDAGHGDAAASPLAIGAGQVDPNAAMDPGLVYDAG 620
Query: 620 VQDYINLLCALNFTQKNISAITRSSFNDCSKPSLDLNYPSFIAFSNARNSSRTTNEFHRT 679
+D++ LLC+ NFT I AITRS +CS + D+NYPSFIA A ++S F RT
Sbjct: 621 PEDFVELLCSTNFTAAQIMAITRSKAYNCSFSTNDMNYPSFIAVFGANDTSGDM-RFSRT 679
Query: 680 VTNVGEKKTTYFASITPIKGFRVTVIPNKLVFKKKNEKISYKLKIEGPRMTQKNKVAFGY 739
VTNVG TY A VTV P LVF + + S+ + + T + AFG
Sbjct: 680 VTNVGAGAATYRAFSVSPSNVEVTVSPETLVFTEVGQTASFLVDLNLTAPT-GGEPAFGA 738
Query: 740 LSWRD 744
+ W D
Sbjct: 739 VIWAD 743
>UniRef100_Q9FHA4 Subtilisin-type protease-like [Arabidopsis thaliana]
Length = 736
Score = 596 bits (1537), Expect = e-169
Identities = 335/749 (44%), Positives = 460/749 (60%), Gaps = 61/749 (8%)
Query: 14 ITSLHVIFTLALSDNYIIHMNLSDMPKSFSNQHSWYESTLAQVTTTNNNLNNSTSSKIFY 73
+ S V A + YIIHM+LS P FS+ SW+ +TL V T KI Y
Sbjct: 10 VFSFFVAIVTAETSPYIIHMDLSAKPLPFSDHRSWFSTTLTSVITNRK-------PKIIY 62
Query: 74 TYTNVMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPYRGAWPTSD 133
YT+ ++GFSA L+ E + LK G++S DLP+KL TT SP+F+GLN G WP S+
Sbjct: 63 AYTDSVHGFSAVLTNSELQRLKHKPGYVSFTKDLPVKLHTTFSPKFIGLNSTSGTWPVSN 122
Query: 134 FGKDIIVGVIDTGVWPESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSLCNKKLIGA 193
+G I++G+IDTG+WP+S SF DDG+ +PSKWKG C+F +S SLCNKKLIGA
Sbjct: 123 YGAGIVIGIIDTGIWPDSPSFHDDGVGSVPSKWKG-ACEFNSS-------SLCNKKLIGA 174
Query: 194 RFFNKGFLAKHSNISTTIL---NSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGI 250
+ FNKG A + ++ T + +S DT GHGTH + AAG+ V AS+F YA GTA GI
Sbjct: 175 KVFNKGLFANNPDLRETKIGQYSSPYDTIGHGTHVAAIAAGNHVKNASYFSYAQGTASGI 234
Query: 251 ASSSRVAIYKTAWGKDGDALSSDIIAAIDAAISDGVDILSISLG--------SDDLLLYK 302
A + +AIYK AW + SSD+IAAID AI DGV ++S+SLG +D L
Sbjct: 235 APHAHLAIYKAAW--EEGIYSSDVIAAIDQAIRDGVHVISLSLGLSFEDDDDNDGFGLEN 292
Query: 303 DPVAIATFAAMEKGIFVSTSAGNNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGV 362
DP+A+A+FAA++KG+FV TS GN+GP + S+ NG PW++TV AGT+ R+F GT+T GN V
Sbjct: 293 DPIAVASFAAIQKGVFVVTSGGNDGPYYWSLINGAPWIMTVGAGTIGRQFQGTLTFGNRV 352
Query: 363 SLTGLSFYLGNFSANNFPIVFM--GMCDNVKELNTVKRKIVVCEGNNETLHEQMFNVYKA 420
S + S + G F + FP+ ++ G +N T+ +IVVC N + ++ +
Sbjct: 353 SFSFPSLFPGEFPSVQFPVTYIESGSVEN----KTLANRIVVC-NENINIGSKLHQIRST 407
Query: 421 KVVGGVFISNIL--DINDVDNSFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAF 478
V I++ L + + + FP I + E +++Y S+ +NA+ A + F+KT
Sbjct: 408 GAAAVVLITDKLLEEQDTIKFQFPVAFIGSKHRETIESYASSNKNNAT--AKLEFRKTVI 465
Query: 479 GVKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNF-GTEVFNNFNLI 537
G K P V YSSRGP S P +LKPDI APGT IL+AWP+ ++ +F+ FNL+
Sbjct: 466 GTKPAPEVGTYSSRGPFTSFPQILKPDILAPGTLILSAWPSVEQITGTRALPLFSGFNLL 525
Query: 538 DGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPF 597
GTSM+ PHVAGVAAL+K H WSPS+I+SAIMTT+ LDN P
Sbjct: 526 TGTSMAAPHVAGVAALIKQVHPNWSPSAIKSAIMTTALTLDN----------------PL 569
Query: 598 ALGAGHINPNRALDPGLVYDIGVQDYINLLC-ALNFTQKNISAITRSSFND-CSKPSLDL 655
A+GAGH++ N+ L+PGL+YD QD+IN LC ++K I+ ITRS+ +D C KPS L
Sbjct: 570 AVGAGHVSTNKVLNPGLIYDTTPQDFINFLCHEAKQSRKLINIITRSNISDACKKPSPYL 629
Query: 656 NYPSFIAFSNARNSSRTTNEFHRTVTNVGEKKTTYFASITPIKGFRVTVIPNKLVFKKKN 715
NYPS IA+ + SS F RT+TNVGE K +Y + +KG V V P KL+F +KN
Sbjct: 630 NYPSIIAYFTSDQSS--PKIFKRTLTNVGEAKRSYIVRVRGLKGLNVVVEPKKLMFSEKN 687
Query: 716 EKISYKLKIEGPRMTQKNKVAFGYLSWRD 744
EK+SY +++E PR Q+N V +G +SW D
Sbjct: 688 EKLSYTVRLESPRGLQEN-VVYGLVSWVD 715
>UniRef100_Q9ZS43 SBT4C protein [Lycopersicon esculentum]
Length = 779
Score = 595 bits (1533), Expect = e-168
Identities = 334/785 (42%), Positives = 494/785 (62%), Gaps = 43/785 (5%)
Query: 1 MASNICLWLWFSYIT------SLHVIFTLALSDNYIIHMNLSDMPKSFSNQHSWYESTLA 54
M + I LWL+ Y+ S H+ +A YI+H++ S MP F + H W+ ST+
Sbjct: 1 METRILLWLYSPYLVLLSWALSAHLFLAIAQRSTYIVHLDKSLMPNVFLDDHHWHSSTIE 60
Query: 55 QVTTTNNNLNNSTSS--KIFYTYTNVMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLD 112
+ + + S K+ Y+Y +V +GFSA LS +E +LK GFIS+ D ++ D
Sbjct: 61 SIKAAVPSSADRFHSAPKLVYSYDHVFHGFSAVLSKDELAALKKSPGFISAYKDRTVEPD 120
Query: 113 TTHSPQFLGLNPYRGAWPTSDFGKDIIVGVIDTGVWPESESFRDDGMTKIPSKWKGQLCQ 172
TT++ +L LNP G WP S G+D+I+GV+D G+WPES SF+DDG+ +IP +WKG +C
Sbjct: 121 TTYTSDYLKLNPSSGLWPASGLGQDVIIGVLDGGIWPESASFQDDGIPEIPKRWKG-IC- 178
Query: 173 FENSNIQSINLSLCNKKLIGARFFNKGFLAKHSNISTTILNSTRDTNGHGTHTSTTAAGS 232
+ N S+CN+KL+GA +FNKG LA ++ + +NS RDTNGHGTH ++ AAG+
Sbjct: 179 ---TPGTQFNTSMCNRKLVGANYFNKGLLADDPTLNIS-MNSARDTNGHGTHCASIAAGN 234
Query: 233 KVDGASFFGYANGTARGIASSSRVAIYKTAWGKDGDALSSDIIAAIDAAISDGVDILSIS 292
G S FGYA GTARG+A +R+A+YK ++ ++G +L+SD+IAA+D A++DGVD++SIS
Sbjct: 235 FAKGVSHFGYAQGTARGVAPQARIAVYKFSF-REG-SLTSDLIAAMDQAVADGVDMISIS 292
Query: 293 LGSDDLLLYKDPVAIATFAAMEKGIFVSTSAGNNGPSFKSIHNGIPWVITVAAGTLDREF 352
+ + LY+D ++IA+F AM KG+ VS SAGN GPS+ ++ NG PW++ VAAG DR F
Sbjct: 293 FSNRFIPLYEDAISIASFGAMMKGVLVSASAGNRGPSWGTLGNGSPWILCVAAGFTDRTF 352
Query: 353 LGTVTLGNGVSLTGLSFYLGNFSANNFPIVF---MGMCDN---VKELNTVKRKIVVCEGN 406
GT+TLGNG+ + G S + +FP+++ + C + + + + I++C+ N
Sbjct: 353 AGTLTLGNGLKIRGWSLFPARAFVRDFPVIYNKTLSDCSSDELLSQFPDPQNTIIICDYN 412
Query: 407 NET----LHEQMFNVYKAKVVGGVFISNILDINDVDN-SFPSIIINPVNGEIVKAYIKSH 461
Q+F+V +A+ + G+FIS + V + + P ++I+ G+ V Y+K
Sbjct: 413 KLEDGFGFDSQIFHVTQARFIAGIFISEDPAVFRVASFTHPGVVIDEKEGKQVINYVK-- 470
Query: 462 NSNASSIANMSFKKTAFG-VKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTN 520
++ + A ++F++T + +P + YSSRGPS S + KPDI APG ILAA P N
Sbjct: 471 -NSVAPTATITFQETYVDRERPSPFLLGYSSRGPSRSYAGIAKPDIMAPGALILAAVPPN 529
Query: 521 V-PVSNFGTEVFNNFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDN 579
+ VS ++ ++ L GTSM+ PH AG+AA+LKGAH WSPS+IRSA+MTT++ L++
Sbjct: 530 ISSVSIENLQLTTDYELKSGTSMAAPHAAGIAAMLKGAHPDWSPSAIRSAMMTTANHLNS 589
Query: 580 TKEHIKDIGNGNRAATPFALGAGHINPNRALDPGLVYDIGVQDYINLLCALNFTQKNISA 639
+E I + + + A+P +G+GH++PNRALDPGLVYD QDYINL+C+LNFT++
Sbjct: 590 AQEPITE--DDDMVASPLGIGSGHVDPNRALDPGLVYDATPQDYINLICSLNFTEEQFKT 647
Query: 640 ITRSS--FNDCSKPSLDLNYPSFIAF---SNARNSSRTTNEFHRTVTNVGEKKTTYFASI 694
RSS +++CS PS DLNYPSFIAF S A N +F RT+TNVG+ TY I
Sbjct: 648 FARSSANYHNCSNPSADLNYPSFIAFYSYSQAGNYPWLEQKFRRTLTNVGKDGATYEVKI 707
Query: 695 TPIKGFRVTVIPNKLVFKKKNEKISYKLKIEGPRMTQKNKVAFGYLSW--RDGKHVVRSP 752
K ++V P LVFK KNEK SY L I R +K G ++W ++G H VRSP
Sbjct: 708 ESPKNSTISVSPQTLVFKNKNEKQSYTLTIR-YRGDEKGG-QDGSITWVEKNGNHSVRSP 765
Query: 753 IVVTN 757
+V+T+
Sbjct: 766 MVITS 770
>UniRef100_O82006 Subtilisin-like protease [Lycopersicon esculentum]
Length = 779
Score = 587 bits (1512), Expect = e-166
Identities = 331/768 (43%), Positives = 491/768 (63%), Gaps = 37/768 (4%)
Query: 11 FSYITSLHVIFTLALSDNYIIHMNLSDMPKSFSNQHSWYESTLAQV-TTTNNNLNNSTSS 69
FS+ S H+ +A YI+H++ S MP F++ H W+ ST+ + + ++++ S+
Sbjct: 17 FSWALSAHLYLAIAQRSTYIVHLDKSLMPNVFTDHHHWHSSTIDSIKASVPSSVDRFHSA 76
Query: 70 -KIFYTYTNVMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPYRGA 128
K+ Y+Y +V +GFSA LS +E +LK GFIS+ D ++ DTT++ +L LNP G
Sbjct: 77 PKLVYSYDHVFHGFSAVLSKDELAALKKSPGFISAYKDRTVEPDTTYTFGYLKLNPSYGL 136
Query: 129 WPTSDFGKDIIVGVIDTGVWPESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSLCNK 188
WP S G+D+I+GV+D+G+WPES SF+DDG+ +IP +WKG +C N Q N S+CN+
Sbjct: 137 WPASGLGQDMIIGVLDSGIWPESASFQDDGIPEIPKRWKG-IC---NPGTQ-FNTSMCNR 191
Query: 189 KLIGARFFNKGFLAKHSNISTTILNSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGTAR 248
KLIGA +FNKG LA+ N++ + +NS RDTNGHGTH+++ AAG+ G S FGYA GTAR
Sbjct: 192 KLIGANYFNKGLLAEDPNLNIS-MNSARDTNGHGTHSASIAAGNFAKGVSHFGYAQGTAR 250
Query: 249 GIASSSRVAIYKTAWGKDGDALSSDIIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIA 308
G+A +R+A+YK ++ ++G +L+SD+IAA+D A++DGVD++SIS + + LY+D ++IA
Sbjct: 251 GVAPQARIAVYKFSF-REG-SLTSDLIAAMDQAVADGVDMISISFSNRFIPLYEDAISIA 308
Query: 309 TFAAMEKGIFVSTSAGNNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLS 368
+F AM KG+ VS SAGN G S+ ++ NG PW++ VAAG DR F GT+TLGNG+ + G S
Sbjct: 309 SFGAMMKGVLVSASAGNRGHSWGTVGNGSPWILCVAAGFTDRTFAGTLTLGNGLKIRGWS 368
Query: 369 FYLGNFSANNFPIVF---MGMCDN---VKELNTVKRKIVVCEGNNET----LHEQMFNVY 418
+ +FP+++ + C + + + + I++C+ N Q+F+V
Sbjct: 369 LFPARAFVRDFPVIYNKTLSDCSSDALLSQFPDPQNTIIICDYNKLEDGFGFDSQIFHVT 428
Query: 419 KAKVVGGVFISNILDINDVDN-SFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTA 477
+A+ G+FIS + V + + ++I+ G+ V Y+K ++ S A ++F++T
Sbjct: 429 QARFKAGIFISEDPAVFRVASFTHLGVVIDKKEGKQVINYVK---NSVSPTATITFQETY 485
Query: 478 FG-VKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNVP-VSNFGTEVFNNFN 535
+ +P + YSSRGPS S + KPDI APG ILAA P N+P VS ++ ++
Sbjct: 486 VDRERPSPFLLGYSSRGPSRSYAGIAKPDIMAPGALILAAVPPNIPSVSIENLQLTTDYE 545
Query: 536 LIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAAT 595
L GTSM+ PH AG+AA+LKGAH WSPS+IRSA+MTT++ L++ ++ I + + + A+
Sbjct: 546 LKSGTSMAAPHAAGIAAMLKGAHPDWSPSAIRSAMMTTANHLNSAQDPITE--DDDMVAS 603
Query: 596 PFALGAGHINPNRALDPGLVYDIGVQDYINLLCALNFTQKNISAITRSS--FNDCSKPSL 653
P +G+GH++PNRALDPGLVYD QDYINL+C+LNFT++ RSS +++CS PS
Sbjct: 604 PLGIGSGHVDPNRALDPGLVYDATPQDYINLICSLNFTEEQFKTFARSSANYHNCSNPSA 663
Query: 654 DLNYPSFIAF---SNARNSSRTTNEFHRTVTNVGEKKTTYFASITPIKGFRVTVIPNKLV 710
DLNYPSFIAF S N +F RT+TNVG+ TY I K ++V P LV
Sbjct: 664 DLNYPSFIAFYSYSQEGNYPWLEQKFRRTLTNVGKGGATYKVKIESPKNSTISVSPQTLV 723
Query: 711 FKKKNEKISYKLKIEGPRMTQKNKVAFGYLSW--RDGKHVVRSPIVVT 756
FK KNEK SY L I N G ++W ++G VRSPIV+T
Sbjct: 724 FKNKNEKQSYTLTIR--YRGDFNSGQTGSITWVEKNGNRSVRSPIVLT 769
>UniRef100_Q9ZS42 SBT4E protein [Lycopersicon esculentum]
Length = 777
Score = 575 bits (1481), Expect = e-162
Identities = 332/773 (42%), Positives = 491/773 (62%), Gaps = 38/773 (4%)
Query: 10 WFSYITSLHVIFTLALSDN--YIIHMNLSDMPKSFSNQHSWYESTLAQV-TTTNNNLNNS 66
+ S+ S H++ LA++ YI+H++ S MP F++ H W+ ST+ + + ++LN
Sbjct: 11 FLSWFLSAHLLCYLAIAQRSTYIVHLDKSLMPNVFTDHHHWHSSTIDSIKASVPSSLNRF 70
Query: 67 TS-SKIFYTYTNVMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPY 125
S K+ Y+Y +V +GFSA LS +E ++LK GFIS+ D ++ DTT++ +L LNP
Sbjct: 71 HSVPKLVYSYDHVFHGFSAVLSKDELKALKKSPGFISAYKDRTVEPDTTYTSDYLKLNPS 130
Query: 126 RGAWPTSDFGKDIIVGVIDTGVWPESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSL 185
G WP S G+D+I+GV+D G+WPES SFRDDG+ +IP +W G +C N Q N S+
Sbjct: 131 SGLWPASGLGQDVIIGVLDGGIWPESASFRDDGIPEIPKRWTG-IC---NPGTQ-FNTSM 185
Query: 186 CNKKLIGARFFNKGFLAKHSNISTTILNSTRDTNGHGTHTSTTAAGSKVDGASFFGYANG 245
CN+KLIGA +FNKG LA ++ + +NS RDTNGHGTH ++ AAG+ G S FGYA G
Sbjct: 186 CNRKLIGANYFNKGLLADDPTLNIS-MNSARDTNGHGTHCASIAAGNFAKGVSHFGYAQG 244
Query: 246 TARGIASSSRVAIYKTAWGKDGDALSSDIIAAIDAAISDGVDILSISLGSDDLLLYKDPV 305
TARG+A +R+A+YK ++ ++G +L+SD+IAA+D A++DGVD++SIS + LY+D +
Sbjct: 245 TARGVAPRARIAVYKFSF-REG-SLTSDLIAAMDQAVADGVDMISISFSYRFIPLYEDAI 302
Query: 306 AIATFAAMEKGIFVSTSAGNNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLT 365
+IA+F AM KG+ VS SAGN GPS+ S+ NG PW++ VA+G DR F GT+ LGNG+ +
Sbjct: 303 SIASFGAMMKGVLVSASAGNRGPSWGSLGNGSPWILCVASGYTDRTFAGTLNLGNGLKIR 362
Query: 366 GLSFYLGNFSANNFPIVF---MGMCDNVKELNTV---KRKIVVCEGNNET----LHEQMF 415
G S + + +++ + C + + L+ V + I++C+ N + Q+
Sbjct: 363 GWSLFPARAFVRDSLVIYSKTLATCMSDELLSQVPDPESTIIICDYNADEDGFGFSSQIS 422
Query: 416 NVYKAKVVGGVFISNILDI-NDVDNSFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFK 474
+V +A+ G+FIS + D S P ++I+ G+ V Y+K NS A ++ ++F+
Sbjct: 423 HVEEARFKAGIFISEDPGVFRDASFSHPGVVIDKKEGKKVINYVK--NSVAPTV-TITFQ 479
Query: 475 KT-AFGVKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNV-PVSNFGTEVFN 532
+T G + P + SSRGPS S + KPDI APG ILAA P N+ S +
Sbjct: 480 ETYVDGERPAPVLAGSSSRGPSRSYLGIAKPDIMAPGVLILAAVPPNLFSQSIQNIALAT 539
Query: 533 NFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNR 592
++ L GTSM+ PH AG+AA+LKGAH WSPS+IRSA+MTT++ L++ ++ I++ + N
Sbjct: 540 DYELKSGTSMAAPHAAGIAAMLKGAHPEWSPSAIRSAMMTTANHLNSAQKPIRE--DDNF 597
Query: 593 AATPFALGAGHINPNRALDPGLVYDIGVQDYINLLCALNFTQKNISAITRS--SFNDCSK 650
ATP +GAGH++PNRALDPGLVYD QD+INL+C++NFT++ RS S+++CS
Sbjct: 598 VATPLDMGAGHVDPNRALDPGLVYDATPQDHINLICSMNFTEEQFKTFARSSASYDNCSN 657
Query: 651 PSLDLNYPSFIA---FSNARNSSRTTNEFHRTVTNVGEKKTTYFASITPIKGFRVTVIPN 707
PS DLNYPSFIA FS N + +F RT+TNVG+ TY K V+V P
Sbjct: 658 PSADLNYPSFIALYPFSLEENFTWLEQKFRRTLTNVGKGGATYKVQTETPKNSIVSVSPR 717
Query: 708 KLVFKKKNEKISYKLKIEGPRMTQKNKVAFGYLSW--RDGKHVVRSPIVVTNI 758
LVFK+KN+K SY L I + +++ G ++W +G H VRSPIV++ I
Sbjct: 718 TLVFKEKNDKQSYTLSIRSIGDSDQSR-NVGSITWVEENGNHSVRSPIVISRI 769
>UniRef100_Q7XTY8 OSJNBa0019K04.9 protein [Oryza sativa]
Length = 776
Score = 555 bits (1430), Expect = e-156
Identities = 319/754 (42%), Positives = 446/754 (58%), Gaps = 42/754 (5%)
Query: 29 YIIHMNLSDMPKSFSNQHSWYESTLAQVTTTN-NNLNNSTSSKIFYTYTNVMNGFSANLS 87
YI+ M S+MP SF H WY ST+ V+++ + + S++I Y Y +GF+A L
Sbjct: 34 YIVQMAASEMPSSFDFYHEWYASTVKSVSSSQLEDEEDDASTRIIYNYETAFHGFAAQLD 93
Query: 88 PEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPY--RGAWPTSDFGKDIIVGVIDT 145
EE E + G ++ IP+ L+L TT SP FLG+ P W S D++VGV+DT
Sbjct: 94 EEEAELMAEADGVLAVIPETVLQLHTTRSPDFLGIGPEVSNRIWSDSLADHDVVVGVLDT 153
Query: 146 GVWPESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSLCNKKLIGARFFNKGFLAKHS 205
G+WPES SF D G+ +P+KWKG LCQ + + CN+K++GAR F G+ A
Sbjct: 154 GIWPESPSFSDKGLGPVPAKWKG-LCQTG----RGFTTANCNRKIVGARIFYNGYEASSG 208
Query: 206 NIS-TTILNSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWG 264
I+ TT L S RD +GHGTHT+ TAAGS V A+ +GYA G ARG+A +RVA YK W
Sbjct: 209 PINETTELKSPRDQDGHGTHTAATAAGSPVQDANLYGYAGGVARGMAPRARVAAYKVCWA 268
Query: 265 KDGDALSSDIIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFAAMEKGIFVSTSAG 324
G SSDI+AA+D A+SDGVD+LSISLG Y D ++IA+F AM+ G+FV+ SAG
Sbjct: 269 --GGCFSSDILAAVDRAVSDGVDVLSISLGGGASRYYLDSLSIASFGAMQMGVFVACSAG 326
Query: 325 NNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLSFYLG--NFS-ANNFPI 381
N GP S+ N PW+ TV A T+DR+F TVTLGNG ++TG+S Y G N S +P+
Sbjct: 327 NAGPDPISLTNLSPWITTVGASTMDRDFPATVTLGNGANITGVSLYKGLRNLSPQEQYPV 386
Query: 382 VFMG----------MC-DNVKELNTVKRKIVVCEGNNETLHEQMFNVYKAKVVGGVFISN 430
V++G +C + + + V KIV+C+ ++ V +A +G + +
Sbjct: 387 VYLGGNSSMPDPRSLCLEGTLQPHDVSGKIVICDRGISPRVQKGQVVKEAGGIGMILANT 446
Query: 431 ILDINDV---DNSFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVD 487
+ ++ + P++ + G K+Y K S A +SF T G++ +P V
Sbjct: 447 AANGEELVADSHLLPAVAVGEAEGIAAKSYSK---SAPKPTATLSFGGTKLGIRPSPVVA 503
Query: 488 FYSSRGPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNFGTEVFN-NFNLIDGTSMSCPH 546
+SSRGP+ +LKPD+ APG +ILAAW + S+ ++ FN++ GTSMSCPH
Sbjct: 504 AFSSRGPNILTLEILKPDVVAPGVNILAAWSGDASPSSLSSDSRRVGFNILSGTSMSCPH 563
Query: 547 VAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINP 606
VAGVAAL+K +H WSP+ I+SA+MTT+ + DNT +KD G +A+TPF GAGHI+P
Sbjct: 564 VAGVAALIKASHPDWSPAQIKSALMTTAYVHDNTYRPMKDAATG-KASTPFEHGAGHIHP 622
Query: 607 NRALDPGLVYDIGVQDYINLLCALNFTQKNISAITRSSFNDCS---KPSLDLNYPSF-IA 662
RAL PGLVYDIG DY+ LC + T + T++S C + DLNYP+ +
Sbjct: 623 VRALTPGLVYDIGQADYLEFLCTQHMTPMQLRTFTKNSNMTCRHTFSSASDLNYPAISVV 682
Query: 663 FSNARNSSRTTNEFHRTVTNVGEKKTTYFASITPIKGFRVTVIPNKLVFKKKNEKISYKL 722
F++ + + T RTVTNVG +TY +T KG V V PN L F N+K+SYK+
Sbjct: 683 FADQPSKALTV---RRTVTNVGPPSSTYHVKVTKFKGADVIVEPNTLHFVSTNQKLSYKV 739
Query: 723 KIEGPRMTQKNKVAFGYLSWRDGKHVVRSPIVVT 756
+ + QK FG LSW DG H+VRSP+V+T
Sbjct: 740 TVT-TKAAQK-APEFGALSWSDGVHIVRSPVVLT 771
>UniRef100_Q9ZUF6 Subtilisin-like serine protease, putative [Arabidopsis thaliana]
Length = 754
Score = 532 bits (1370), Expect = e-149
Identities = 322/769 (41%), Positives = 440/769 (56%), Gaps = 67/769 (8%)
Query: 15 TSLHVIFTLALSDNYIIHMNLSDMPKSFSNQHSWYESTLAQVTTTNNNLNNSTSSKIFYT 74
T L ++ YII +N SD P+SF H WY S L ++ S + YT
Sbjct: 16 TFLFLLLHTTAKKTYIIRVNHSDKPESFLTHHDWYTSQL------------NSESSLLYT 63
Query: 75 YTNVMNGFSANLSPEEHESLKTFSGFISSIPDLPL-KLDTTHSPQFLGLNPYRGAWPTSD 133
YT +GFSA L E +SL + S I I + PL L TT +P+FLGLN G
Sbjct: 64 YTTSFHGFSAYLDSTEADSLLSSSNSILDIFEDPLYTLHTTRTPEFLGLNSEFGVHDLGS 123
Query: 134 FGKDIIVGVIDTGVWPESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSLCNKKLIGA 193
+I+GV+DTGVWPES SF D M +IPSKWKG+ C+ S+ S LCNKKLIGA
Sbjct: 124 SSNGVIIGVLDTGVWPESRSFDDTDMPEIPSKWKGE-CE-SGSDFDS---KLCNKKLIGA 178
Query: 194 RFFNKGF-LAKHSNISTTILN-STRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIA 251
R F+KGF +A S+ + S RD +GHGTHTSTTAAGS V ASF GYA GTARG+A
Sbjct: 179 RSFSKGFQMASGGGFSSKRESVSPRDVDGHGTHTSTTAAGSAVRNASFLGYAAGTARGMA 238
Query: 252 SSSRVAIYKTAWGKDGDALSSDIIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFA 311
+ +RVA YK W SDI+AA+D AI DGVD+LS+SLG Y+D +AI F+
Sbjct: 239 TRARVATYKVCWSTG--CFGSDILAAMDRAILDGVDVLSLSLGGGSAPYYRDTIAIGAFS 296
Query: 312 AMEKGIFVSTSAGNNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLSFY- 370
AME+G+FVS SAGN+GP+ S+ N PWV+TV AGTLDR+F LGNG LTG+S Y
Sbjct: 297 AMERGVFVSCSAGNSGPTRASVANVAPWVMTVGAGTLDRDFPAFANLGNGKRLTGVSLYS 356
Query: 371 --------------LGNFSANNFPIVFMGMCDNVKELNTVKRKIVVCEGNNETLHEQMFN 416
GN S++N + G D+ + V+ KIVVC+ E+
Sbjct: 357 GVGMGTKPLELVYNKGNSSSSN--LCLPGSLDS----SIVRGKIVVCDRGVNARVEKGAV 410
Query: 417 VYKAKVVGGVFISNILDINDV---DNSFPSIIINPVNGEIVKAYIKSHNSNASSIANMSF 473
V A +G + + ++ + P+I + G++++ Y+K S++ A + F
Sbjct: 411 VRDAGGLGMIMANTAASGEELVADSHLLPAIAVGKKTGDLLREYVK---SDSKPTALLVF 467
Query: 474 KKTAFGVKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAW-----PTNVPVSNFGT 528
K T VK +P V +SSRGP+ P +LKPD+ PG +ILA W PT + + T
Sbjct: 468 KGTVLDVKPSPVVAAFSSRGPNTVTPEILKPDVIGPGVNILAGWSDAIGPTGLDKDSRRT 527
Query: 529 EVFNNFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIG 588
+ FN++ GTSMSCPH++G+A LLK AH WSPS+I+SA+MTT+ +LDNT + D
Sbjct: 528 Q----FNIMSGTSMSCPHISGLAGLLKAAHPEWSPSAIKSALMTTAYVLDNTNAPLHDAA 583
Query: 589 NGNRAATPFALGAGHINPNRALDPGLVYDIGVQDYINLLCALNFTQKNISAITRSSFNDC 648
+ N + P+A G+GH++P +AL PGLVYDI ++YI LC+L++T +I AI + +C
Sbjct: 584 D-NSLSNPYAHGSGHVDPQKALSPGLVYDISTEEYIRFLCSLDYTVDHIVAIVKRPSVNC 642
Query: 649 SKPSLD---LNYPSFIAFSNARNSSRTTNEFHRTVTNVGEKKTTYFASITPIKGFRVTVI 705
SK D LNYPSF + R T E VTNVG + Y ++ ++V
Sbjct: 643 SKKFSDPGQLNYPSFSVLFGGKRVVRYTRE----VTNVGAASSVYKVTVNGAPSVGISVK 698
Query: 706 PNKLVFKKKNEKISYKLKIEGPR-MTQKNKVAFGYLSWRDGKHVVRSPI 753
P+KL FK EK Y + + ++ NK FG ++W + +H VRSP+
Sbjct: 699 PSKLSFKSVGEKKRYTVTFVSKKGVSMTNKAEFGSITWSNPQHEVRSPV 747
>UniRef100_O65351 Cucumisin-like serine protease [Arabidopsis thaliana]
Length = 757
Score = 530 bits (1366), Expect = e-149
Identities = 320/765 (41%), Positives = 448/765 (57%), Gaps = 56/765 (7%)
Query: 18 HVIFTLALSDNYIIHMNLSDMPKSFSNQHSWYESTLAQVTTTNNNLNNSTSSKIFYTYTN 77
HV + + YI+HM S MP SF +WY+S+L + S S+++ YTY N
Sbjct: 21 HVSSSSSDQGTYIVHMAKSQMPSSFDLHSNWYDSSLRSI---------SDSAELLYTYEN 71
Query: 78 VMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPYRG-AWPTSDFGK 136
++GFS L+ EE +SL T G IS +P+ +L TT +P FLGL+ + +P +
Sbjct: 72 AIHGFSTRLTQEEADSLMTQPGVISVLPEHRYELHTTRTPLFLGLDEHTADLFPEAGSYS 131
Query: 137 DIIVGVIDTGVWPESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSLCNKKLIGARFF 196
D++VGV+DTGVWPES+S+ D+G IPS WKG C+ + SLCN+KLIGARFF
Sbjct: 132 DVVVGVLDTGVWPESKSYSDEGFGPIPSSWKGG-CEAGTN----FTASLCNRKLIGARFF 186
Query: 197 NKGFLAKHSNISTTILN-STRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSR 255
+G+ + I + + S RD +GHGTHTS+TAAGS V+GAS GYA+GTARG+A +R
Sbjct: 187 ARGYESTMGPIDESKESRSPRDDDGHGTHTSSTAAGSVVEGASLLGYASGTARGMAPRAR 246
Query: 256 VAIYKTAWGKDGDALSSDIIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFAAMEK 315
VA+YK W G SSDI+AAID AI+D V++LS+SLG Y+D VAI FAAME+
Sbjct: 247 VAVYKVCWL--GGCFSSDILAAIDKAIADNVNVLSMSLGGGMSDYYRDGVAIGAFAAMER 304
Query: 316 GIFVSTSAGNNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLSFYLGNFS 375
GI VS SAGN GPS S+ N PW+ TV AGTLDR+F LGNG + TG+S + G
Sbjct: 305 GILVSCSAGNAGPSSSSLSNVAPWITTVGAGTLDRDFPALAILGNGKNFTGVSLFKGEAL 364
Query: 376 ANN-FPIVFMGMCDNVKELN----------TVKRKIVVCE-GNNETLHEQMFNVYKAKVV 423
+ P ++ G N N VK KIV+C+ G N + Q +V KA
Sbjct: 365 PDKLLPFIYAGNASNATNGNLCMTGTLIPEKVKGKIVMCDRGINARV--QKGDVVKAAGG 422
Query: 424 GGVFISNIL----DINDVDNSFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFG 479
G+ ++N ++ + P+ + G+I++ Y+ ++ + A++S T G
Sbjct: 423 VGMILANTAANGEELVADAHLLPATTVGEKAGDIIRHYV---TTDPNPTASISILGTVVG 479
Query: 480 VKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAW-----PTNVPVSNFGTEVFNNF 534
VK +P V +SSRGP++ P +LKPD+ APG +ILAAW PT + + E F
Sbjct: 480 VKPSPVVAAFSSRGPNSITPNILKPDLIAPGVNILAAWTGAAGPTGLASDSRRVE----F 535
Query: 535 NLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAA 594
N+I GTSMSCPHV+G+AALLK H WSP++IRSA+MTT+ + + DI G + +
Sbjct: 536 NIISGTSMSCPHVSGLAALLKSVHPEWSPAAIRSALMTTAYKTYKDGKPLLDIATG-KPS 594
Query: 595 TPFALGAGHINPNRALDPGLVYDIGVQDYINLLCALNFTQKNISAITRSSFN-DCSK--P 651
TPF GAGH++P A +PGL+YD+ +DY+ LCALN+T I +++R ++ D SK
Sbjct: 595 TPFDHGAGHVSPTTATNPGLIYDLTTEDYLGFLCALNYTSPQIRSVSRRNYTCDPSKSYS 654
Query: 652 SLDLNYPSFIAFSNARNSSRTTNEFHRTVTNVGEKKTTYFASITPIKGFRVTVIPNKLVF 711
DLNYPSF + + + T RTVT+VG T + G +++V P L F
Sbjct: 655 VADLNYPSFAVNVDGVGAYKYT----RTVTSVGGAGTYSVKVTSETTGVKISVEPAVLNF 710
Query: 712 KKKNEKISYKLKIEGPRMTQKNKVAFGYLSWRDGKHVVRSPIVVT 756
K+ NEK SY + +FG + W DGKHVV SP+ ++
Sbjct: 711 KEANEKKSYTVTFTVDSSKPSGSNSFGSIEWSDGKHVVGSPVAIS 755
>UniRef100_Q7XQQ3 OSJNBa0084A10.2 protein [Oryza sativa]
Length = 776
Score = 528 bits (1361), Expect = e-148
Identities = 306/757 (40%), Positives = 439/757 (57%), Gaps = 45/757 (5%)
Query: 29 YIIHMNLSDMPKSFSNQHSWYESTLAQVTTTNNNLNNSTSSKIFYTYTNVMNGFSANLSP 88
Y++ M++S MP F+ WY S L+ + + + +++ YTY++ MNGFSA L+
Sbjct: 29 YVVRMDVSAMPAPFATHDGWYRSVLSSASA--RDAAAAPAAEHLYTYSHAMNGFSAVLTA 86
Query: 89 EEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPYRGAWPTSDFGKDIIVGVIDTGVW 148
+ E ++ G ++ P+ +L TT +P FLGL+ GAWP S +G D++VG++DTGVW
Sbjct: 87 RQVEEIRRADGHVAVFPETYARLHTTRTPAFLGLSAGAGAWPASRYGADVVVGIVDTGVW 146
Query: 149 PESESFRDDGMTK-IPSKWKGQLCQFENSNIQSINLSLCNKKLIGARFFNKGFLAKHSNI 207
PES SF D G+ +P++WKG C+ S S+CN+KL+GAR F+KG + NI
Sbjct: 147 PESASFSDAGVAAPVPARWKGA-CEAG----ASFRPSMCNRKLVGARSFSKGLRQRGLNI 201
Query: 208 STTILNSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWGKDG 267
S +S RD GHG+HTS+TAAG+ V GAS+FGYANGTA G+A +RVA+YK + D
Sbjct: 202 SDDDYDSPRDYYGHGSHTSSTAAGAAVPGASYFGYANGTATGVAPMARVAMYKAVFSADT 261
Query: 268 -DALSSDIIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFAAMEKGIFVSTSAGNN 326
++ S+D++AA+D AI+DGVD++S+SLG + + VAI FAA+ +GI V+ SAGN+
Sbjct: 262 LESASTDVLAAMDQAIADGVDVMSLSLGFPESPYDTNVVAIGAFAAVRRGILVTCSAGND 321
Query: 327 GPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGV----SLTGLSFYLGNFSANNFPIV 382
G ++ NG PW+ TV A T+DR F TVTLG G S+ G S Y G A +
Sbjct: 322 GSDSYTVLNGAPWITTVGASTIDRAFTATVTLGAGAGGARSIVGRSVYPGRVPA-GAAAL 380
Query: 383 FMGMCDNVKE--------LNTVKRKIVVCEGNNETLHEQMFNVYKAKVVGGVFISNILDI 434
+ G + KE V+ K V C +HEQM+ V G + SN+ +I
Sbjct: 381 YYGRGNRTKERCESGSLSRKDVRGKYVFCNAGEGGIHEQMYEVQSNGGRGVIAASNMKEI 440
Query: 435 ND-VDNSFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYSSRG 493
D D P +++ P +G ++ Y + A+ A++ F T GVK P+V ++SSRG
Sbjct: 441 MDPSDYVTPVVLVTPSDGAAIQRYA---TAAAAPRASVRFAGTELGVKPAPAVAYFSSRG 497
Query: 494 PSNSCPYVLKPDITAPGTSILAAWPTNVPVSNFG---TEVFNNFNLIDGTSMSCPHVAGV 550
PS P +LKPD+ APG ILAAW N V T+++ N+ L+ GTSM+ PHVAGV
Sbjct: 498 PSPVSPAILKPDVVAPGVDILAAWVPNKEVMELDGGETKLYTNYMLVSGTSMASPHVAGV 557
Query: 551 AALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNRAL 610
AALL+ AH WSP+++RSA+MTT+ + DN + G TP G+GH++PN+A
Sbjct: 558 AALLRSAHPDWSPAAVRSAMMTTAYVKDNADDADLVSMPGGSPGTPLDYGSGHVSPNQAT 617
Query: 611 DPGLVYDIGVQDYINLLCA-LNFTQKNISAITRSSFNDC-----SKPSLDLNYPSFIAFS 664
DPGLVYDI DY+ LC L +T + ++AI C + DLNYPSF+
Sbjct: 618 DPGLVYDITADDYVAFLCGELRYTSRQVAAIAGHRAG-CPAGAGAASHRDLNYPSFMVIL 676
Query: 665 NARNSSRTTNEFHRTVTNVGEKKTTYFASITPIKGFRVTVIPNKLVFKKKNEKISYKLKI 724
N NS+ T F RT+TNV Y S+T G V V P L F K + + +
Sbjct: 677 NKTNSA--TRTFTRTLTNVAGSPAKYAVSVTAPAGMAVKVTPATLSFAGKGSTQGFSVTV 734
Query: 725 EGPRMTQK----NKVA-FGYLSWRD--GKHVVRSPIV 754
+ ++ + N + +G+LSW + G+HVVRSPIV
Sbjct: 735 QVSQVKRSRDGDNYIGNYGFLSWNEVGGQHVVRSPIV 771
>UniRef100_Q8RWQ7 AT5g67360/K8K14_8 [Arabidopsis thaliana]
Length = 757
Score = 525 bits (1353), Expect = e-147
Identities = 319/765 (41%), Positives = 447/765 (57%), Gaps = 56/765 (7%)
Query: 18 HVIFTLALSDNYIIHMNLSDMPKSFSNQHSWYESTLAQVTTTNNNLNNSTSSKIFYTYTN 77
HV + + YI+HM S MP SF +WY+S+L + S S+++ YTY N
Sbjct: 21 HVSSSSSDQGTYIVHMAKSQMPSSFDLHSNWYDSSLRSI---------SDSAELLYTYEN 71
Query: 78 VMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPYRG-AWPTSDFGK 136
++GFS L+ EE +SL T G IS +P+ +L TT +P FLGL+ + +P +
Sbjct: 72 AIHGFSTRLTQEEADSLMTQPGVISVLPEHRYELHTTRTPLFLGLDEHTADLFPEAGSYS 131
Query: 137 DIIVGVIDTGVWPESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSLCNKKLIGARFF 196
D++VGV+DTGVWPES+S+ D+G IPS WKG C+ + SLCN+KLIGARFF
Sbjct: 132 DVVVGVLDTGVWPESKSYSDEGFGPIPSSWKGG-CEAGTN----FTASLCNRKLIGARFF 186
Query: 197 NKGFLAKHSNISTTILN-STRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSR 255
+G+ + I + + S RD +GHGTHTS+TAAGS V+GAS GYA+GTARG+A +R
Sbjct: 187 ARGYESTMGPIDESKESRSPRDDDGHGTHTSSTAAGSVVEGASLLGYASGTARGMAPRAR 246
Query: 256 VAIYKTAWGKDGDALSSDIIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFAAMEK 315
VA+YK W G SSDI+AAID AI+D V++LS+SLG Y+D VAI FAAME+
Sbjct: 247 VAVYKVCWL--GGCFSSDILAAIDKAIADNVNVLSMSLGGGMSDYYRDGVAIGAFAAMER 304
Query: 316 GIFVSTSAGNNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLSFYLGNFS 375
GI VS SAGN GPS S+ N PW+ TV AGTLDR+F LGNG + TG+S + G
Sbjct: 305 GILVSCSAGNAGPSSSSLSNVAPWITTVGAGTLDRDFPALAILGNGKNFTGVSLFKGEAL 364
Query: 376 ANN-FPIVFMGMCDNVKELN----------TVKRKIVVCE-GNNETLHEQMFNVYKAKVV 423
+ P ++ G N N VK KIV+C+ G N + Q +V KA
Sbjct: 365 PDKLLPFIYAGNASNATNGNLCMTGTLIPEKVKGKIVMCDRGINARV--QKGDVVKAAGG 422
Query: 424 GGVFISNIL----DINDVDNSFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFG 479
G+ ++N ++ + P+ + G+I++ Y+ ++ + A++S T G
Sbjct: 423 VGMILANTAANGEELVADAHLLPATTVGEKAGDIIRHYV---TTDPNPTASISILGTVVG 479
Query: 480 VKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAW-----PTNVPVSNFGTEVFNNF 534
VK +P V +SSRGP++ P +LKPD+ APG +ILAAW PT + + E F
Sbjct: 480 VKPSPVVAAFSSRGPNSITPNILKPDLIAPGVNILAAWTGAAGPTGLASDSRRVE----F 535
Query: 535 NLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAA 594
N+I GTSMSCPHV+G+AALLK H SP++IRSA+MTT+ + + DI G + +
Sbjct: 536 NIISGTSMSCPHVSGLAALLKSVHPECSPAAIRSALMTTAYKTYKDGKPLLDIATG-KPS 594
Query: 595 TPFALGAGHINPNRALDPGLVYDIGVQDYINLLCALNFTQKNISAITRSSFN-DCSK--P 651
TPF GAGH++P A +PGL+YD+ +DY+ LCALN+T I +++R ++ D SK
Sbjct: 595 TPFDHGAGHVSPTTATNPGLIYDLTTEDYLGFLCALNYTSPQIRSVSRRNYTCDPSKSYS 654
Query: 652 SLDLNYPSFIAFSNARNSSRTTNEFHRTVTNVGEKKTTYFASITPIKGFRVTVIPNKLVF 711
DLNYPSF + + + T RTVT+VG T + G +++V P L F
Sbjct: 655 VADLNYPSFAVNVDGVGAYKYT----RTVTSVGGAGTYSVKVTSETTGVKISVEPAVLNF 710
Query: 712 KKKNEKISYKLKIEGPRMTQKNKVAFGYLSWRDGKHVVRSPIVVT 756
K+ NEK SY + +FG + W DGKHVV SP+ ++
Sbjct: 711 KEANEKKSYTVTFTVDSSKPSGSNSFGSIEWSDGKHVVGSPVAIS 755
>UniRef100_Q39007 Subtilisin-like protease [Arabidopsis thaliana]
Length = 746
Score = 519 bits (1337), Expect = e-145
Identities = 316/765 (41%), Positives = 445/765 (57%), Gaps = 58/765 (7%)
Query: 18 HVIFTLALSDNYIIHMNLSDMPKSFSNQHSWYESTLAQVTTTNNNLNNSTSSKIFYTYTN 77
HV + + YI+HM S P SF +WY+S+L + S S+++ YTY N
Sbjct: 12 HVSSSSSDQGTYIVHMAKSQTPSSFDLHSNWYDSSLRSI---------SDSAELLYTYEN 62
Query: 78 VMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPYRG-AWPTSDFGK 136
++GFS L+ EE +SL T G IS +P+ +L TT +P FLGL+ + +P +
Sbjct: 63 AIHGFSTRLTQEEADSLMTQPGVISVLPEHRYELHTTRTPLFLGLDEHTADLFPEAGSYS 122
Query: 137 DIIVGVIDTGVWPESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSLCNKKLIGARFF 196
D++VGV+DTGVWPES+S+ D+G IPS WKG C+ + SLCN+KLIGARFF
Sbjct: 123 DVVVGVLDTGVWPESKSYSDEGFGPIPSSWKGG-CEAGTN----FTASLCNRKLIGARFF 177
Query: 197 NKGFLAKHSNISTTILN-STRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSR 255
+G+ + I + + S RD +GHGTHTS+TAAGS V+GAS GYA+GTARG+ +
Sbjct: 178 ARGYESTMGPIDESKESRSPRDDDGHGTHTSSTAAGSVVEGASLLGYASGTARGMLHA-- 235
Query: 256 VAIYKTAWGKDGDALSSDIIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFAAMEK 315
+A+YK W G SSDI+AAID AI+D V++LS+SLG Y+D VAI FAAME+
Sbjct: 236 LAVYKVCWL--GGCFSSDILAAIDKAIADNVNVLSMSLGGGMSDYYRDGVAIGAFAAMER 293
Query: 316 GIFVSTSAGNNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLSFYLGNFS 375
GI VS SAGN GPS S+ N PW+ TV AGTLDR+F LGNG + TG+S + G
Sbjct: 294 GILVSCSAGNAGPSSSSLSNVAPWITTVGAGTLDRDFPALAILGNGKNFTGVSLFKGEAL 353
Query: 376 ANN-FPIVFMGMCDNVKELN----------TVKRKIVVCE-GNNETLHEQMFNVYKAKVV 423
+ P ++ G N N VK KIV+C+ G N + Q +V KA
Sbjct: 354 PDKLLPFIYAGNASNATNGNLCMTGTLIPEKVKGKIVMCDRGINARV--QKGDVVKAAGG 411
Query: 424 GGVFISNIL----DINDVDNSFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFG 479
G+ ++N ++ + P+ + G+I++ Y+ ++ + A++S T G
Sbjct: 412 VGMILANTAANGEELVADAHLLPATTVGEKAGDIIRHYV---TTDPNPTASISILGTVVG 468
Query: 480 VKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAW-----PTNVPVSNFGTEVFNNF 534
VK +P V +SSRGP++ P +LKPD+ APG +ILAAW PT + + E F
Sbjct: 469 VKPSPVVAAFSSRGPNSITPNILKPDLIAPGVNILAAWTGAAGPTGLASDSRRVE----F 524
Query: 535 NLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAA 594
N+I GTSMSCPHV+G+AALLK H WSP++IRSA+MTT+ + + DI G + +
Sbjct: 525 NIISGTSMSCPHVSGLAALLKSVHPEWSPAAIRSALMTTAYKTYKDGKPLLDIATG-KPS 583
Query: 595 TPFALGAGHINPNRALDPGLVYDIGVQDYINLLCALNFTQKNISAITRSSFN-DCSK--P 651
TPF GAGH++P A +PGL+YD+ +DY+ LCALN+T I +++R ++ D SK
Sbjct: 584 TPFDHGAGHVSPTTATNPGLIYDLTTEDYLGFLCALNYTSPQIRSVSRRNYTCDPSKSYS 643
Query: 652 SLDLNYPSFIAFSNARNSSRTTNEFHRTVTNVGEKKTTYFASITPIKGFRVTVIPNKLVF 711
DLNYPSF + + + T RTVT+VG T + G +++V P L F
Sbjct: 644 VADLNYPSFAVNVDGAGAYKYT----RTVTSVGGAGTYSVKVTSETTGVKISVEPAVLNF 699
Query: 712 KKKNEKISYKLKIEGPRMTQKNKVAFGYLSWRDGKHVVRSPIVVT 756
K+ NEK SY + +FG + W DGKHVV SP+ ++
Sbjct: 700 KEANEKKSYTVTFTVDSSKPSGSNSFGSIEWSDGKHVVGSPVAIS 744
>UniRef100_P93204 SBT1 protein [Lycopersicon esculentum]
Length = 766
Score = 518 bits (1335), Expect = e-145
Identities = 313/755 (41%), Positives = 436/755 (57%), Gaps = 54/755 (7%)
Query: 29 YIIHMNLSDMPKSFSNQHSWYESTLAQVTTTNNNLNNSTSSKIFYTYTNVMNGFSANLSP 88
YIIHM+ +MP F + WY+S+L V S S+ + YTY +V++G+S L+
Sbjct: 31 YIIHMDKFNMPADFDDHTQWYDSSLKSV---------SKSANMLYTYNSVIHGYSTQLTA 81
Query: 89 EEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLN--PYRGAWPTSDFGKDIIVGVIDTG 146
+E ++L G + ++ +L TT SP FLGL R +P ++ ++I+GV+DTG
Sbjct: 82 DEAKALAQQPGILLVHEEVIYELHTTRSPTFLGLEGRESRSFFPQTEARSEVIIGVLDTG 141
Query: 147 VWPESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSLCNKKLIGARFFNKGFLAKHSN 206
VWPES+SF D G+ ++P+ WKG+ CQ ++ + S CN+KLIGARFF++G+ A
Sbjct: 142 VWPESKSFDDTGLGQVPASWKGK-CQTG----KNFDASSCNRKLIGARFFSQGYEAAFGA 196
Query: 207 ISTTILN-STRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWGK 265
I TI + S RD GHGTHT+TTAAGS V GAS GYA GTARG+AS +RVA YK W
Sbjct: 197 IDETIESKSPRDDEGHGTHTATTAAGSVVTGASLLGYATGTARGMASHARVAAYKVCW-- 254
Query: 266 DGDALSSDIIAAIDAAISDGVDILSISLGSDDLLLYKDPVAIATFAAMEKGIFVSTSAGN 325
G SSDI+A +D A+ DGV++LS+SLG ++D VAI F+A +GIFVS SAGN
Sbjct: 255 TGGCFSSDILAGMDQAVIDGVNVLSLSLGGTISDYHRDIVAIGAFSAASQGIFVSCSAGN 314
Query: 326 NGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLSFYLGN-FSANNFPIVFM 384
GPS ++ N PW+ TV AGT+DREF + +GNG L G+S Y G ++ P+V+
Sbjct: 315 GGPSSGTLSNVAPWITTVGAGTMDREFPAYIGIGNGKKLNGVSLYSGKALPSSVMPLVYA 374
Query: 385 GMCDNVKELN----------TVKRKIVVCEGNNETLHEQMFNVYKAKVVGGVFISNILDI 434
G N V KIVVC+ ++ V A + G+ ++N
Sbjct: 375 GNVSQSSNGNLCTSGSLIPEKVAGKIVVCDRGMNARAQKGLVVKDAGGI-GMILANTDTY 433
Query: 435 NDV----DNSFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYS 490
D + P+ + G ++K YI SN++ A ++F T GV+ +P V +S
Sbjct: 434 GDELVADAHLIPTAAVGQTAGNLIKQYIA---SNSNPTATIAFGGTKLGVQPSPVVAAFS 490
Query: 491 SRGPSNSCPYVLKPDITAPGTSILAAWPTNVPVSNFGTEVFN-NFNLIDGTSMSCPHVAG 549
SRGP+ P VLKPD+ APG +ILA W V + + N FN+I GTSMSCPHV+G
Sbjct: 491 SRGPNPITPDVLKPDLIAPGVNILAGWTGKVGPTGLQEDTRNVGFNIISGTSMSCPHVSG 550
Query: 550 VAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPFALGAGHINPNRA 609
+AALLK AH WSP++IRSA+MTTS + I+D+ G ++TPF GAGH+NP A
Sbjct: 551 LAALLKAAHPEWSPAAIRSALMTTSYSTYKNGKTIEDVATG-MSSTPFDYGAGHVNPTAA 609
Query: 610 LDPGLVYDIGVQDYINLLCALNFTQKNISAITRSSFNDCSKPS----LDLNYPSFIA--- 662
+ PGLVYD+ V DYIN LCAL+++ I I + + C + DLNYPSF
Sbjct: 610 VSPGLVYDLTVDDYINFLCALDYSPSMIKVIAKRDIS-CDENKEYRVADLNYPSFSIPME 668
Query: 663 ---FSNARNSSRTTNEFHRTVTNVGEKKTTYFASI-TPIKGFRVTVIPNKLVFKKKNEKI 718
+A +S+ T + RT+TNVG TY AS+ + + ++ V P L F +KNEK
Sbjct: 669 TAWGEHADSSTPTVTRYTRTLTNVG-NPATYKASVSSETQDVKILVEPQTLTFSRKNEKK 727
Query: 719 SYKLKIEGPRMTQKNKVAFGYLSWRDGKHVVRSPI 753
+Y + +F L W DG+HVV SPI
Sbjct: 728 TYTVTFTA-TSKPSGTTSFARLEWSDGQHVVASPI 761
>UniRef100_O49607 Subtilisin proteinase-like [Arabidopsis thaliana]
Length = 764
Score = 518 bits (1333), Expect = e-145
Identities = 311/764 (40%), Positives = 435/764 (56%), Gaps = 62/764 (8%)
Query: 29 YIIHMNLSDMPKSFSNQHSWYESTLAQVTTTNNNLNNSTSSKIFYTYTNVMNGFSANLSP 88
+I ++ MP F + WY + A+ S+I + Y V +GFSA ++P
Sbjct: 28 FIFRIDGGSMPSIFPTHYHWYSTEFAE------------ESRIVHVYHTVFHGFSAVVTP 75
Query: 89 EEHESLKTFSGFISSIPDLPLKLDTTHSPQFLGLNPYRGAWPTSDFGKDIIVGVIDTGVW 148
+E ++L+ ++ D +L TT SPQFLGL +G W SD+G D+I+GV DTG+W
Sbjct: 76 DEADNLRNHPAVLAVFEDRRRELHTTRSPQFLGLQNQKGLWSESDYGSDVIIGVFDTGIW 135
Query: 149 PESESFRDDGMTKIPSKWKGQLCQFENSNIQSINLSLCNKKLIGARFFNKG-FLAKHSNI 207
PE SF D + IP +W+G +C+ + N CN+K+IGARFF KG A I
Sbjct: 136 PERRSFSDLNLGPIPKRWRG-VCE-SGARFSPRN---CNRKIIGARFFAKGQQAAVIGGI 190
Query: 208 STTI-LNSTRDTNGHGTHTSTTAAGSKVDGASFFGYANGTARGIASSSRVAIYKTAWGKD 266
+ T+ S RD +GHGTHTS+TAAG AS GYA+G A+G+A +R+A YK W KD
Sbjct: 191 NKTVEFLSPRDADGHGTHTSSTAAGRHAFKASMSGYASGVAKGVAPKARIAAYKVCW-KD 249
Query: 267 GDALSSDIIAAIDAAISDGVDILSISLGSDDLL---LYKDPVAIATFAAMEKGIFVSTSA 323
L SDI+AA DAA+ DGVD++SIS+G D + Y DP+AI ++ A KGIFVS+SA
Sbjct: 250 SGCLDSDILAAFDAAVRDGVDVISISIGGGDGITSPYYLDPIAIGSYGAASKGIFVSSSA 309
Query: 324 GNNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTLGNGVSLTGLSFYLG-NFSANNFPIV 382
GN GP+ S+ N PWV TV A T+DR F LG+G L G+S Y G + FP+V
Sbjct: 310 GNEGPNGMSVTNLAPWVTTVGASTIDRNFPADAILGDGHRLRGVSLYAGVPLNGRMFPVV 369
Query: 383 FMG--------MC-DNVKELNTVKRKIVVCEGNNETLHEQMFNVYKAKVVGGVF---ISN 430
+ G +C +N + V+ KIV+C+ + + V KA VG + SN
Sbjct: 370 YPGKSGMSSASLCMENTLDPKQVRGKIVICDRGSSPRVAKGLVVKKAGGVGMILANGASN 429
Query: 431 ILDINDVDNSFPSIIINPVNGEIVKAYIKSHNSNASSIANMSFKKTAFGVKSTPSVDFYS 490
+ + P+ + G+ +KAY SH + IA++ F+ T G+K P + +S
Sbjct: 430 GEGLVGDAHLIPACAVGSNEGDRIKAYASSH---PNPIASIDFRGTIVGIKPAPVIASFS 486
Query: 491 SRGPSNSCPYVLKPDITAPGTSILAAW-----PTNVPVSNFGTEVFNNFNLIDGTSMSCP 545
RGP+ P +LKPD+ APG +ILAAW PT +P TE FN++ GTSM+CP
Sbjct: 487 GRGPNGLSPEILKPDLIAPGVNILAAWTDAVGPTGLPSDPRKTE----FNILSGTSMACP 542
Query: 546 HVAGVAALLKGAHNGWSPSSIRSAIMTTSDILDNTKEHIKDIGNGNRAATPFALGAGHIN 605
HV+G AALLK AH WSP+ IRSA+MTT++++DN+ + D G ++ATP+ G+GH+N
Sbjct: 543 HVSGAAALLKSAHPDWSPAVIRSAMMTTTNLVDNSNRSLIDESTG-KSATPYDYGSGHLN 601
Query: 606 PNRALDPGLVYDIGVQDYINLLCALNFTQKNISAITRSSFN--DCSKPSL-DLNYPSFIA 662
RA++PGLVYDI DYI LC++ + K I ITR+ KPS +LNYPS A
Sbjct: 602 LGRAMNPGLVYDITNDDYITFLCSIGYGPKTIQVITRTPVRCPTTRKPSPGNLNYPSITA 661
Query: 663 FSNARNSSRTTNEFHRTVTNVGEKKTTYFASITPIKGFRVTVIPNKLVFKKKNEKISYKL 722
+ RT TNVG+ + Y A I +G VTV P +LVF ++ SY +
Sbjct: 662 VFPTNRRGLVSKTVIRTATNVGQAEAVYRARIESPRGVTVTVKPPRLVFTSAVKRRSYAV 721
Query: 723 KIEGPRMTQKNKV------AFGYLSWRD-GKHVVRSPIVVTNIN 759
+ + +N V FG ++W D GKHVVRSPIVVT ++
Sbjct: 722 TV---TVNTRNVVLGETGAVFGSVTWFDGGKHVVRSPIVVTQMD 762
>UniRef100_Q9LUM3 Subtilisin proteinase-like protein [Arabidopsis thaliana]
Length = 775
Score = 516 bits (1329), Expect = e-145
Identities = 306/787 (38%), Positives = 443/787 (55%), Gaps = 52/787 (6%)
Query: 5 ICLWLWFSYITSLHVIFTLALSDN---YIIHMNLSDMPKSFSNQHSWYESTLAQVTTTNN 61
+ + +F ++ +L + A S N YI+H++ P F WY S+LA +T
Sbjct: 1 MAFFFYFFFLLTLSSPSSSASSSNSLTYIVHVDHEAKPSIFPTHFHWYTSSLASLT---- 56
Query: 62 NLNNSTSSKIFYTYTNVMNGFSANLSPEEHESLKTFSGFISSIPDLPLKLDTTHSPQFLG 121
S+ I +TY V +GFSA L+ ++ L IS IP+ L TT SP+FLG
Sbjct: 57 ----SSPPSIIHTYDTVFHGFSARLTSQDASQLLDHPHVISVIPEQVRHLHTTRSPEFLG 112
Query: 122 LNPY--RGAWPTSDFGKDIIVGVIDTGVWPESESFRDDGMTKIPSKWKGQLCQFENSNIQ 179
L G SDFG D+++GVIDTGVWPE SF D G+ +P KWKGQ Q
Sbjct: 113 LRSTDKAGLLEESDFGSDLVIGVIDTGVWPERPSFDDRGLGPVPIKWKGQCIAS-----Q 167
Query: 180 SINLSLCNKKLIGARFFNKGFLAKHSNIS-TTILNSTRDTNGHGTHTSTTAAGSKVDGAS 238
S CN+KL+GARFF G+ A + ++ TT S RD++GHGTHT++ +AG V AS
Sbjct: 168 DFPESACNRKLVGARFFCGGYEATNGKMNETTEFRSPRDSDGHGTHTASISAGRYVFPAS 227
Query: 239 FFGYANGTARGIASSSRVAIYKTAWGKDGDALSSDIIAAIDAAISDGVDILSISLGSDDL 298
GYA+G A G+A +R+A YK W + SDI+AA D A++DGVD++S+S+G +
Sbjct: 228 TLGYAHGVAAGMAPKARLAAYKVCW--NSGCYDSDILAAFDTAVADGVDVISLSVGGVVV 285
Query: 299 LLYKDPVAIATFAAMEKGIFVSTSAGNNGPSFKSIHNGIPWVITVAAGTLDREFLGTVTL 358
Y D +AI F A+++GIFVS SAGN GP ++ N PW+ TV AGT+DR+F V L
Sbjct: 286 PYYLDAIAIGAFGAIDRGIFVSASAGNGGPGALTVTNVAPWMTTVGAGTIDRDFPANVKL 345
Query: 359 GNGVSLTGLSFY--LGNFSANNFPIVFMG-----------MC-DNVKELNTVKRKIVVCE 404
GNG ++G+S Y G +P+V+ G +C + + N VK KIV+C+
Sbjct: 346 GNGKMISGVSVYGGPGLDPGRMYPLVYGGSLLGGDGYSSSLCLEGSLDPNLVKGKIVLCD 405
Query: 405 GNNETLHEQMFNVYKAKVVGGVFISNILDINDV---DNSFPSIIINPVNGEIVKAYIKSH 461
+ + V K +G + + + D + + P+ + G+ ++ YI
Sbjct: 406 RGINSRATKGEIVRKNGGLGMIIANGVFDGEGLVADCHVLPATSVGASGGDEIRRYISES 465
Query: 462 NSNASS---IANMSFKKTAFGVKSTPSVDFYSSRGPSNSCPYVLKPDITAPGTSILAAWP 518
+ + SS A + FK T G++ P V +S+RGP+ P +LKPD+ APG +ILAAWP
Sbjct: 466 SKSRSSKHPTATIVFKGTRLGIRPAPVVASFSARGPNPETPEILKPDVIAPGLNILAAWP 525
Query: 519 TNVPVSNFGTE-VFNNFNLIDGTSMSCPHVAGVAALLKGAHNGWSPSSIRSAIMTTSDIL 577
+ S ++ FN++ GTSM+CPHV+G+AALLK AH WSP++IRSA++TT+ +
Sbjct: 526 DRIGPSGVTSDNRRTEFNILSGTSMACPHVSGLAALLKAAHPDWSPAAIRSALITTAYTV 585
Query: 578 DNTKEHIKDIGNGNRAATPFALGAGHINPNRALDPGLVYDIGVQDYINLLCALNFTQKNI 637
DN+ E + D GN ++ G+GH++P +A+DPGLVYDI DYIN LC N+T+ NI
Sbjct: 586 DNSGEPMMDESTGNTSSV-MDYGSGHVHPTKAMDPGLVYDITSYDYINFLCNSNYTRTNI 644
Query: 638 SAITRSSFNDCSKPSL-----DLNYPSFIAFSNARNSSRTTNEFHRTVTNVGEKKTTYFA 692
ITR DC +LNYPSF S+ + F RTVTNVG+ + Y
Sbjct: 645 VTITRRQ-ADCDGARRAGHVGNLNYPSFSVVFQQYGESKMSTHFIRTVTNVGDSDSVYEI 703
Query: 693 SITPIKGFRVTVIPNKLVFKKKNEKISYKLKIEGPRMTQK---NKVAFGYLSWRDGKHVV 749
I P +G VTV P KL F++ +K+S+ ++++ + V G++ W DGK V
Sbjct: 704 KIRPPRGTTVTVEPEKLSFRRVGQKLSFVVRVKTTEVKLSPGATNVETGHIVWSDGKRNV 763
Query: 750 RSPIVVT 756
SP+VVT
Sbjct: 764 TSPLVVT 770
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,307,103,438
Number of Sequences: 2790947
Number of extensions: 57966228
Number of successful extensions: 164788
Number of sequences better than 10.0: 1005
Number of HSP's better than 10.0 without gapping: 423
Number of HSP's successfully gapped in prelim test: 582
Number of HSP's that attempted gapping in prelim test: 161049
Number of HSP's gapped (non-prelim): 1984
length of query: 762
length of database: 848,049,833
effective HSP length: 135
effective length of query: 627
effective length of database: 471,271,988
effective search space: 295487536476
effective search space used: 295487536476
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 79 (35.0 bits)
Medicago: description of AC144727.4


